Mutations in human LMNA cause Emery-Dreifuss muscular dystrophy; however, a mechanistic link between the effect of mutations on lamin filament assembly and disease phenotype has not been established. Here we show that changes in lamin filament structure translate into disease phenotypes in Caenorhabditis elegans by altering the character of the nuclear lamina.
Abstract
Mutations in the human LMNA gene underlie many laminopathic diseases, including Emery-Dreifuss muscular dystrophy (EDMD); however, a mechanistic link between the effect of mutations on lamin filament assembly and disease phenotypes has not been established. We studied the ΔK46 Caenorhabditis elegans lamin mutant, corresponding to EDMD-linked ΔK32 in human lamins A and C. Cryo-electron tomography of lamin ΔK46 filaments in vitro revealed alterations in the lateral assembly of dimeric head-to-tail polymers, which causes abnormal organization of tetrameric protofilaments. Green fluorescent protein (GFP):ΔK46 lamin expressed in C. elegans was found in nuclear aggregates in postembryonic stages along with LEM-2. GFP:ΔK46 also caused mislocalization of emerin away from the nuclear periphery, consistent with a decreased ability of purified emerin to associate with lamin ΔK46 filaments in vitro. GFP:ΔK46 animals had motility defects and muscle structure abnormalities. These results show that changes in lamin filament structure can translate into disease-like phenotypes via altering the localization of nuclear lamina proteins, and suggest a model for how the ΔK32 lamin mutation may cause EDMD in humans.
INTRODUCTION
The nuclear lamina is a meshwork of proteins adjacent to the nucleoplasmic face of the inner nuclear membrane and serves as both a main nuclear structural element as well as an interface between the nucleoplasm and the nuclear membrane (Fawcett, 1966; Aebi et al., 1986). The evolutionarily conserved protein lamin is the major component of the nuclear lamina and is found as two types (A-type and B-type) in humans (Gruenbaum et al., 2003). Mutations in the human A-type lamin gene (LMNA) cause at least 14 diseases, collectively termed laminopathies (Broers et al., 2006; Mattout et al., 2006; Worman and Bonne, 2007). Among the autosomal-dominant laminopathies are the muscle diseases Emery-Dreifuss muscular dystrophy (EDMD), LMNA-related congenital muscular dystrophy (L-CMD), limb-girdle muscular dystrophy, and dilated cardiomyopathy; the aging and skin disorder restrictive dermopathy; the neuropathic Charcot-Marie-Tooth disorder; the lipodystrophies mandibuloacral dysplasia and Dunnigan-type familial partial lipodystrophy; and the systemic premature aging syndromes Hutchison-Gilford progeria syndrome and atypical Werner syndrome (Vlcek and Foisner, 2007; Quijano-Roy et al., 2008). Intriguingly, the wide range of tissue types affected and severity of the disease phenotypes cannot be directly correlated with the position of the mutation in the LMNA gene (Worman and Bonne, 2007).
There are three major models to explain how lamin mutations may lead to tissue-specific phenotypes. The first model gives lamin a role as the main structural element of the nucleus; disrupting this foundation may cause tissues under higher stress (such as the mechanical load on muscle cells) to be more prone to lesions leading to specific tissue degeneration (Ostlund et al., 2001). The second model suggests that lamin is a key regulator of signaling pathways affecting DNA replication, transcription, and chromatin organization, mediated through interactions between lamin, lamin-binding proteins, and chromatin (Cohen et al., 2001; Gruenbaum et al., 2005; Vlcek and Foisner, 2007; Dechat et al., 2008). The third model suggests that lamins A and C are involved in regulating cell type–specific gene expression during adult stem cell differentiation (Gotzmann and Foisner, 2006). These three models are not mutually exclusive; therefore, studying mutant lamin structure in isolation from the physiological effects of the mutation may not fully explain how a mutation can cause a specific laminopathy.
Based on its sequence and predicted structure, lamin is classified as a type-V intermediate filament (IF) protein (Steinert and Roop, 1988). It has a short, unstructured amino-terminal head domain, an α-helical coiled-coil rod domain made of four segments of heptad repeats, and a carboxyl-terminal tail domain containing a nuclear localization signal and an immunoglobulin (Ig)-fold (Strelkov et al., 2004; Ben-Harush et al., 2009). Lamins assemble into a network of 10-nm filaments beneath the inner nuclear membrane, as first described in the Xenopus germinal vesicle (Aebi et al., 1986), but their structure in somatic cells is still unknown. Although the general morphology of the lamin network has been studied in vivo, further elucidation of the higher order assembly pattern and dynamics of filament formation has required analysis of lamin structure in vitro (Moir et al., 1991; Geisler et al., 1998; Stuurman et al., 1998; Karabinos et al., 2003). Intriguingly, under the tested experimental conditions, lamins from most species, including human lamins, form paracrystalline arrays instead of 10-nm filaments in vitro (Moir et al., 1991; Stuurman et al., 1998; Melcer et al., 2007; Taimen et al., 2009). Although these structures have been recapitulated by a vast overexpression of Drosophila and Xenopus lamins in insect cells, their physiological relevance is not clear (Klapper et al., 1997). Importantly, depending on the specific assembly conditions, lamin from Caenorhabditis elegans can form either paracrystalline arrays or 10-nm IF-like filaments (Karabinos et al., 2003; Foeger et al., 2006); both structures have a similar arrangement of tetrameric protofilaments (i.e., consisting of four lamin polypeptides), which are in turn composed of two antiparallel head-to-tail polymers of lamin dimers (Ben-Harush et al., 2009). Therefore, C. elegans is currently the only system in which lamin functions can be studied from the level of in vitro filament assembly to the physiology of the organism.
C. elegans lamin (Ce-lamin) is encoded by a single gene (lmn-1) and shows many functional similarities to both human lamin A and lamin B proteins (Riemer et al., 1993; Liu et al., 2000; Melcer et al., 2007). Because of this conservation, we are able to mutate residues in Ce-lamin homologous to LMNA residues mutated in laminopathies; these mutations have varying effects on in vitro assembly of paracrystalline fibers and filaments (Wiesel et al., 2008; Ben-Harush et al., 2009). Furthermore, many of these mutations were expressed in C. elegans and showed distinct degrees of mislocalization within the nucleus and alterations in protein dynamics (Wiesel et al., 2008). These observations begin to mirror the fact that different lamin mutations cause different diseases in humans. In the current study, we study a new EDMD-linked Ce-lamin mutation in a conserved region of lamins (Figure 1A) to link filament assembly defects with disease-related phenotypes. An in-frame deletion of three base pairs in exon 1 of LMNA (c.94_96delAAG), resulting in the deletion of Lys-32 in the head region of human lamin A (p.Lys32del), leads to EDMD and L-CMD (Vytopil et al., 2002; D'Amico et al., 2005; Quijano-Roy et al., 2008). Here we created the identical deletion of the conserved lysine in C. elegans (K46). We show that ΔK46 filaments undergo abnormal lateral assembly into tetrameric protofilaments, which probably causes the nuclear aggregates seen in C. elegans nuclei expressing green fluorescent protein (GFP):ΔK46. These aggregates also contain LEM-2. We also show that the expression of GFP:ΔK46 caused mislocalization of Ce-emerin away from the nuclear periphery in vivo and a decreased association of purified emerin with ΔK46 filaments in vitro. GFP:ΔK46 animals show motility defects and muscle structure abnormalities, suggesting that changes in lamin filament structure translate into disease-like phenotypes via altering the organization of the nuclear lamina.
FIGURE 1:
ΔK46 lamin forms abnormal filaments. (A) ClustalW (Larkin et al., 2007) alignment of coil 1A of the first rod domain of Ce-lamin (Ce_Lmn-1) with human lamins A and C (Hs_LMNA/C), human lamin B (Hs_LMNB), Xenopus lamin A (Xl_lam-A), and Drosophila lamin (Dm_lam). Conserved residues in all lamins are in bold; lysine 46 of Ce-lamin and homologous residues are shaded. (B) Electron micrographs of negative-stained wt and ΔK46 lamin filaments reveal thicker filaments of ΔK46 lamin (arrows). Scale bar = 100 nm. (C) Thick (30 nm) sections of tomograms of ΔK46 filaments show the altered structure of the IF-like filaments. The periodic arrangement of globular tail domains along protofilaments can be clearly seen as pairs (arrowheads) separated by an average distance of 14 nm. Scale bar = 50 nm. (D) Sections (10 nm) through tomograms of Ce-lamin filaments reveal a 7-nm shift of the globular tail domains in the ΔK46 filaments (left panel) compared with wt filaments (right panel). The magnification is similar in both sections. (E) 1-wt (left) and ΔK46 (right) lamin dimers assemble into a polar head-to-tail polymer of dimers. 2-Two antiparallel head-to tail polymers form a protofilament. The distances measured by the tomographic analysis in (D) are superimposed on a schematic 2D view of the head-to-tail polymer comprising the protofilaments. Both wt and ΔK46 Ce-lamin dimers associate into two head-to-tail polymers with 55 nm between the N-terminal head (yellow) and C-terminal tail (pink) domains. 3-The lateral assembly of these polymers into tetrameric protofilaments is disrupted in ΔK46 filaments.
RESULTS
An EDMD-linked mutation affects the in vitro assembly of Ce-lamin filaments
The deletion of Lys-32 in human lamins A and C (ΔK32) causes EDMD and L-CMD (Vytopil et al., 2002; D'Amico et al., 2005; Quijano-Roy et al., 2008). This lysine residue is in the highly conserved coil 1A region at the N terminus of the first lamin rod domain and corresponds to K46 in Ce-lamin (Figure 1A). To study the effect of this mutation on lamin filament assembly, wild-type (wt) and ΔK46 Ce-lamin proteins were bacterially expressed, purified to near homogeneity, and assembled into IF-like filaments. They were first visualized by negative staining electron microscopy (Figure 1B), as previously described (Wiesel et al., 2008). Whereas wild-type Ce-lamin assembled into homogeneous 10-nm-thick, IF-like filaments, the ΔK46 mutant lamin assembled into filaments with a mean width of 14 nm. More than 40% of ΔK46 filaments were larger than wt (n = 350; arrows in Figure 1B). To view these filaments without staining and at a resolution that would allow more detailed structural analysis, we moved to three-dimensional (3D) analysis using cryo-electron tomography (cryo-ET), a technique we previously used to analyze supramolecular organization of Ce-lamin filaments (Ben-Harush et al., 2009). Figure 1C shows a 30-nm-thick section through a reconstructed volume of ΔK46 IF-like lamin filaments. Repeating patterns of globular tail domains (arrowheads) imply relatively normal association of ΔK46 lamin dimers into polymers (see model, Figure 1E). The average spacing between tail domains in the ΔK46 mutant lamin, however, alternated between 14 and 34 nm, in contrast to 21-27 nm in wt lamin (Figure 1D). The 7-nm shift in the position of the paired globular tail domains along the tetrameric protofilaments suggests that the ΔK46 mutation alters the lateral assembly of head-to-tail polymers of lamin dimers into protofilaments. By imposing these distances onto a two-dimensional (2D) model (Figure 1E), it becomes apparent how the normal ΔK46 polymers can misassemble into abnormally spaced protofilaments, likely affecting both the structural integrity of the nuclear lamina and the ability of lamin binding partners to associate with the nuclear lamina.
In vivo localization of GFP:ΔK46 lamin
To assess the effect of the ΔK46 mutation on phenotypes in a whole organism, we created a mutant C. elegans strain expressing a GFP-tagged copy of wt lamin (GFP:wt) or a GFP-tagged copy of lamin lacking residue K46 (GFP:ΔK46) under the control of the baf-1 promoter, which has a spatiotemporal expression pattern identical to that of lmn-1 (Margalit et al., 2007; Wiesel et al., 2008). Western blot analysis shows that the strain has a low level of GFP:ΔK46 overexpression of 10–15% of the endogenous lamin (arrow, Figure 2A). The localization of GFP:ΔK46 protein within the mutant strains was visualized by fluorescence microscopy of live animals (Figure 2). GFP:ΔK46 expression could be detected in all cell types in embryonic, larval, and adult-stage worms (Figure 2, B–D), similar to the endogenous lamin (unpublished data). In embryonic nuclei, GFP:ΔK46 expression was similar to that of GFP:wt, with essentially uniform peripheral staining (Figure 2B). In L1 larval stage animals, GFP:ΔK46 was seen in large aggregates in 59% of scored nuclei as compared with 16% mostly smaller aggregates for GFP:wt (Figure 2, C and E). In individual adult worms, GFP:ΔK46 expression was low or missing in some nuclei and had an overall higher expression in the head and tail regions, which was also the case in GFP:wt worms (Figure 2D). Additionally, GFP:ΔK46 was found in large aggregates at the nuclear periphery in 62% of scored nuclei (Figure 2, C–E). In contrast, GFP:wt localization was found in a relatively smooth pattern around the nuclear periphery and not in aggregates (17% of nuclei contained smaller aggregates; Figure 2E). Immunostaining with anti–Ce-lamin antibodies, which recognize both endogenous and GFP:ΔK46 lamin, revealed that the presence of GFP:ΔK46 did not affect the proper localization of endogenous lamin at the nuclear periphery (unpublished data). Because the GFP:ΔK46 lamin is expressed at only 10% of endogenous levels, the mutant aggregations stained with anti-lamin antibodies are less apparent than when the GFP is visualized natively (Figure 2) or with anti-GFP antibodies (Figures 3 and 4). Finally, the mobility of GFP:ΔK46 lamin was monitored by fluorescence recovery after photobleaching (FRAP). As we have previously published, wt lamin is stable and immobile within the nucleus, and several disease-linked mutations drastically perturb this stability (Wiesel et al., 2008). GFP:ΔK46 lamin, however, remained immobile in nuclei with or without GFP aggregations, similar to GFP:wt (unpublished data).
FIGURE 2:
GFP:ΔK46 lamin shows altered localization. (A) Representative Western blot analysis of GFP:lamin (arrow), lamin, and emerin protein levels in worm extracts from GFP:wt and GFP:ΔK46 animals. (B–D) Deconvoluted images of GFP-expressing worms. (B) GFP:ΔK46 can be seen in all cells in embryos with relatively normal nuclear localization, similar to GFP:wt. (C) GFP:ΔK46 is expressed in all cell types in L1 larval stage animals, with obvious nuclear aggregations in many nuclei (white arrows and inset). (D) Both GFP:wt and GFP:ΔK46 expression is strongest in the head (left panels) and tail (right panels) of early adult animals and does not appear in 100% of cells. Examples of GFP:ΔK46 aggregates are indicated with white arrows. Representative nuclei are enlarged in the inset (red outline) and show uniform shape even in the presence of GFP:ΔK46 aggregates. All scale bars are 10 μm in main images and 1 μm in insets. (E) Quantification of GFP aggregation confirms the visual data in (C) and (D).
FIGURE 3:
LEM-2 localizes to GFP aggregates in nuclei expressing GFP:ΔK46. (A) L4 larval stage wt (GFP:wt, left panels) and mutant (GFP:ΔK46, right panels) animals were costained with DAPI (blue), anti-GFP (green), and anti-LEM-2 (red) antibodies. A representative slice from 3D deconvoluted images reveals that LEM-2 associates with the GFP-tagged lamin (yellow) in both GFP:wt and GFP:ΔK46 animals (arrowheads) and is found in the GFP aggregates in GFP:ΔK46 nuclei (arrows). Scale bars = 10 μm for all panels. (B) Quantification of GFP and LEM-2 aggregation confirms the visual data in A.
FIGURE 4:
Emerin is mislocalized in nuclei expressing GFP:ΔK46. (A) L4 larval stage wt (GFP:wt, left panels) and mutant (GFP:ΔK46, right panels) animals were costained with anti-GFP (green) and anti-Ce-emerin (red) antibodies. A representative slice from 3D deconvoluted images showing the region posterior to the pharynx reveals that the localization of GFP:wt and emerin do not always overlap. Emerin staining is seen in most GFP-positive cells in wt animals (arrowheads); however, emerin staining is defective or absent (arrows) in many GFP-positive cells in mutant animals. Scale bars = 10 μm for all panels. (B) Quantification of Ce-emerin localization in GFP-positive cells reveals a significant difference in the colocalization of GFP and emerin in GFP:ΔK46 (n = 136) and GFP:wt (n = 86) nuclei (****p < 0.0001; Student's two-tailed t test).
LEM-2 colocalizes with GFP aggregates in GFP:ΔK46 nuclei
Mammalian lamin A is required for the peripheral localization of LEM-domain proteins, including emerin and LEM-2 (Wilson and Foisner, 2010). Likewise, Ce-lamin is required for the peripheral localization of emerin and LEM-2 (Liu et al., 2003). We therefore tested how expression of ΔK46 affects emerin and LEM-2 localization by coimmunostaining L4 larval-stage animals for each of these proteins. Staining of GFP:wt animals revealed uniform nuclear envelope localization of both GFP:wt lamin and LEM-2 (Figure 3, left panels). LEM-2 appeared to remain associated with GFP:ΔK46 lamin, including in the GFP aggregates characteristic of these nuclei (Figure 3A, right panels). Quantification confirmed that 62.5% of GFP:ΔK46 nuclei had LEM-2 aggregates, as opposed to only 18.5% in GFP:wt nuclei (Figure 3B).
Emerin is lost from the nuclear lamina in GFP:ΔK46 nuclei
We next investigated the effect of GFP:ΔK46 on emerin localization by costaining L4 larval-stage animals with anti–Ce-emerin and anti–GFP antibodies (Figure 4, A and B). Distinct patterns of emerin staining (red) were observed in GFP:wt and GFP:ΔK46 L4 animals: either normal, peripheral emerin staining colocalizing with GFP:lamin (arrowheads in Figure 4A) or “emerin defective,” with either diffuse emerin staining and/or incomplete colocalization with GFP:lamin or a complete lack of emerin staining above background levels (arrows in Figure 4A). Whereas emerin staining was almost ubiquitous in GFP:wt nuclei, many GFP:ΔK46 nuclei had an absence of emerin signal. GFP-positive nuclei were scored for emerin localization in deconvoluted z-slices, revealing a significant delocalization of emerin in GFP:ΔK46-expressing nuclei: Nearly 78% of GFP-positive nuclei (n = 136) had defective Ce-emerin localization in GFP:ΔK46 animals, compared with 27.5% in GFP:wt (n = 86) (Figure 4B). Interestingly, the abnormal emerin localization was not accompanied by a drastic overall decline in emerin protein levels, as measured by Western blot analysis (Figure 2A). Therefore, GFP:ΔK46 does not affect the expression of emerin, but rather it causes a reduced ability of emerin to properly associate with the nuclear lamina.
Ce-emerin has reduced association with ΔK46 filaments in vitro
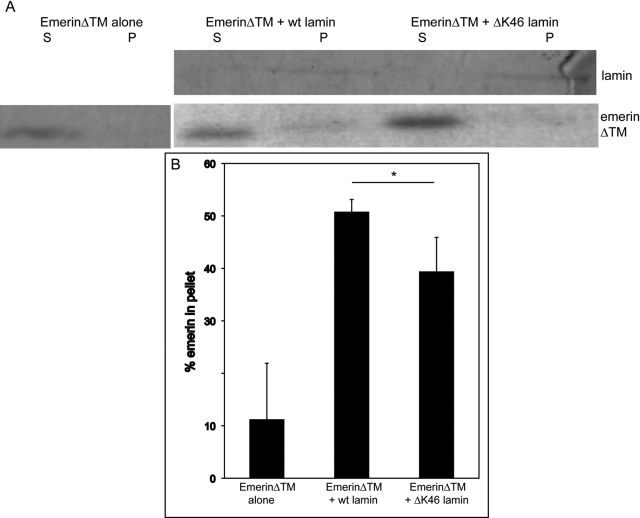
Due to the mislocalization of emerin in GFP:ΔK46-expressing adult nuclei, we sought to study the potential mechanism by which this could occur. We performed an in vitro cosedimentation experiment to determine the ability of emerin to associate with assembled lamin filaments. Heterologously expressed Ce-emerin was incubated alone or in the presence of wt or ΔK46 mutant lamin filaments. After sedimentation, the majority of emerin was found in the supernatant fraction (Figure 5). In the presence of wt lamin, a fraction of emerin is found in the pellet, along with lamin. The fraction of emerin found in the pellet was significantly reduced when incubated with ΔK46 lamin filaments (p = 0.04) (Figure 5), suggesting that the ΔK46 mutation directly affects the ability of lamin to bind emerin.
FIGURE 5:
Decreased association of emerin with ΔK46 lamin filaments in vitro. Heterologously expressed emerinΔTM was incubated alone or with wt lamin filaments or with ΔK46 lamin filaments. The samples were sedimented, and supernatant and pellet fractions were separated. (A) Representative Coomassie blue–stained SDS–PAGE gel. (B) Quantification of three individual experiments shows a significant reduction in the amount of emerin in the pellet when it is incubated with DK46 lamin filaments from when it is incubated with wt filaments (*p < 0.05; Student's two-tailed t test). Error bars represent SD.
GFP:ΔK46 expression causes motility and muscle phenotypes in C. elegans
There was a slight decrease in the brood size of GFP:ΔK46 animals: 152.2 ± 16 compared with the wt value of 198.2 ± 12.8 (p = 0.029) (Supplemental Figure S1A) and no difference in the viability of laid embryos (unpublished data). GFP localization (Figure 2B) and emerin staining of C. elegans embryos did not reveal any drastic defects in nuclear size and shape or in lamina structure. DAPI staining of DNA did not reveal gross chromosomal aberrations such as chromatin bridges or aggregations (Supplemental Figure S1B and unpublished data), which have previously been visualized in various C. elegans mutant strains (Margalit et al., 2005). The normal staining pattern of emerin in embryos was surprising given the mislocalization seen in L4 larval stage animals (Figure 4). The Ce-emerin antibody was detected with a Cy3-conjugated secondary antibody, and there was no detectable signal in the Cy2/FITC channel to produce bleedthrough signal into the Cy3 channel. Therefore, both GFP:ΔK46 lamin and emerin localization is normal in embryos and is disrupted only after early embryonic development.
We next sought to determine whether motility and muscle phenotypes, which are associated with the EDMD disease in humans, could be observed in the GFP:ΔK46 strain. Most cases of EDMD in humans are caused by either loss of emerin or by dominant mutations in lamin A, including ΔK32. The most drastic EDMD phenotypes in humans are those related to muscle function; indeed, the ΔK32 mutation is associated with a variety of clinical symptoms affecting muscles, including contractures, progressive limb and muscle weakness, and cardiomyopathy associated with cardiac conduction defects (Vytopil et al., 2002; D'Amico et al., 2005; Quijano-Roy et al., 2008). To determine whether the C. elegans GFP:ΔK46 animals have muscle abnormalities, motility assays were performed on L4 larval stage worms. To control for the effect of the GFP tag on lamin function, the strain overexpressing wt lamin tagged with GFP (GFP:wt) was also included in the experiments. Animals were placed on clean agar plates, and the number of times they moved their head to propagate forward movement was scored. This crawling motility was not altered in GFP:wt or GFP:ΔK46 animals (Figure 6A). The ability of worms to move in liquid has been proposed as a more sensitive measure of C. elegans motility (Pierce-Shimomura et al., 2008). Therefore we performed a swimming motility assay in which wt (N2), GFP:wt, and GFP:ΔK46 animals were placed in a drop of water and allowed to swim for 15 min, after which the number of thrashes was scored by the number of times a worm moved its head to one side (Pierce-Shimomura et al., 2008). The GFP:ΔK46 animals were significantly impaired in their ability to move: Whereas GFP:wt worms had an average thrash-rate of 94.8 ± 3.5 head turns/min (not significantly different from N2), GFP:ΔK46 worms moved at a rate of 70.1 ± 4.9 turns/min (p = 0.00011) (Figure 6B). To ensure that this motility defect was caused specifically by the ΔK46 mutation and was not a result of the position of gene integration or another acquired mutation(s), the GFP:ΔK46 gene was specifically targeted for RNAi-mediated knockdown by feeding worms with bacteria carrying a dsRNA vector complementary to the GFP coding region. As a negative control, worms were fed with bacteria carrying an empty vector (L4440). Figure 6C shows that RNAi down-regulation of GFP:ΔK46 significantly rescued the swimming motility (p = 0.001).
FIGURE 6:
Reduced swimming motility of GFP:ΔK46 animals. (A) Crawling motility of L4 larval stage worms was measured as the number of head turns per minute after placement on an agar plate. GFP:ΔK46 and wt (N2) animals showed similar motility. (B) Swimming motility of L4 larval stage worms was measured as the number of head turns to one side per minute after 15 min in a drop of water. GFP:ΔK46 animals were significantly impaired in their ability to move (****p = 0.0001). (C) Swimming motility was measured after feeding with an RNAi construct targeting GFP:ΔK46 (GFP) or an empty vector (L4440). Depletion of GFP:ΔK46 significantly rescued the swimming defect of GFP:ΔK46 L4 larval stage animals (**p = 0.005; ***p = 0.001; Student's two-tailed t test). Error bars represent SEM.
On the basis of this motility phenotype, we hypothesized that we should be able to detect structural lesions in muscle of GFP:ΔK46 animals. Visualizing worms with differential interference contrast (DIC) microscopy revealed striking lesions along the cuticle of early adult animals, in 100% of worms examined (Figure 7). These lesions were apparent in L1 larvae and did not coincide with molting, and were never seen in GFP:wt worms or in GFP:ΔK46 worms fed with RNAi against GFP (unpublished data). We did not observe cell corpses or any other indicators that programmed cell death was occurring in these regions (Conradt and Xue, 2005). Instead, these lesions resemble the muscle attachment (Mua) phenotype, which is an abnormal muscle attachment to the hypodermis and cuticle seen in worm strains lacking genes important in maintaining muscle integrity (Zaidel-Bar et al., 2010).
FIGURE 7:
DIC analysis of early adult animals reveals structural abnormalities in GFP:ΔK46 animals. wt (GFP:wt) and GFP:ΔK46 animals were immobilized and photographed 1 d after reaching the L4 larval stage. Arrowheads indicate points of lesions, where muscles are detached from the cuticle, seen exclusively in mutant strains, in all worms examined. Scale bar = 10 μm for all panels.
To assess the ultrastructure of muscle fibers, sarcomeres, and attachment sites, we prepared worms for transmission electron microscopy (TEM). In both lateral- and cross-thin sections of GFP:wt L4 worms, muscle cells were uniformly arranged, with sarcomeres containing even, parallel filaments (black arrows) clearly defined between dense bodies (white arrows) and cell-cell attachments, similar to wt animals (Moerman and Williams, 2006) (Figure 8, A and D). In GFP:ΔK46 animals, this organized pattern was severely disrupted (Figure 8, B, C, E, and F). The assembly of filaments, dense bodies, and attachment sites was impaired so that sarcomeres were grossly misshapen. In many instances, dense bodies were absent; in other cases, they were misplaced (white arrows). There were many aberrant membrane-bound structures – some contiguous with cell membranes and others as vesicular structures within the sarcomeres (black arrowheads). Filament organization was also disrupted (black arrows). Lesions at the edges of muscle cells were evident, perhaps responsible for the disrupted cuticle attachment seen in the DIC images in Figure 7 (black arrowheads). Finally, major disorganization is evident in the subcuticle region between the cuticle and the muscle fiber (white arrowheads, especially in Figure 8, B and C), again consistent with the cuticle attachment phenotype. Taken together, our data show that the ΔK46 mutation in Ce-lamin causes motility phenotypes that are coupled with muscle disorganization, which resemble the effects of the disease-causing ΔK32 mutation in human lamins A and C.
FIGURE 8:
TEM reveals muscle abnormalities in GFP:ΔK46 animals. wt longitudinal (long) (A) and transverse (trans) (D) sections show normal fiber (black arrows), dense body/attachment (white arrows), and subcuticle organization (white arrowhead with bracket), with few membrane-bound vesicles (black arrowheads). GFP:ΔK46 expression disrupts muscle organization, as evident in both longitudinal (B and C) and transverse (E and F) sections: Fibers are less uniformly arranged (black arrows); dense bodies and attachments are absent or mislocalized, causing sarcomere disorganization (white arrows); more aberrant membrane-bound structures are seen within sarcomeres and at cell edges (black arrowheads); and gross abnormalities between the sarcomere and the cuticle edge occur (white arrowheads). Sizes of all scale bars are indicated in the respective panels.
DISCUSSION
Although autosomal-dominant EDMD was described more than 40 yr ago (Emery and Dreifuss, 1966), and the disease-causing locus mapped to human LMNA more than a decade ago (Bonne et al., 1999), the molecular mechanisms linking lamin mutations to disease phenotypes remain a mystery. In vitro and ex vivo examination of lamin filaments have begun to reveal that lamin mutations can cause different diseases by having various effects on the assembly of paracrystalline fibers and protofilaments (Wiesel et al., 2008; Ben-Harush et al., 2009; Taimen et al., 2009). Correlating abnormalities in in vitro lamin filament assembly with disease-like phenotypes in model organisms could provide a basis for understanding how different lamin mutations cause diseases through changes in the structural integrity of the nucleus and/or protein-protein or protein-DNA complexes. To date, no lamin mutation has been comprehensively studied from filament assembly to organism physiology. We focused on a single lamin mutation, ΔK32 in human lamins A and C, corresponding to the conserved ΔK46 mutation in the Ce-lamin, to present a model for how this lamin mutation causes EDMD.
The ΔK46 lamin mutation leads to defective lamin filament assembly, as seen both by negative-stain TEM and cryo-ET (Figure 1). Cryo-ET enabled us to resolve and thus measure the distances between paired tail domains, revealing that the initial association of lamin dimers to form polymers was normal, but the lateral assembly of these head-to-tail polymers was shifted. This phenotype is new, as we have previously described lamin mutants that have different effects on overall filament organization (Wiesel et al., 2008; Ben-Harush et al., 2009). These data also suggest that the loss of the K46 residue shifted the interaction site between one head-to-tail polymer and the rod domain of the other head-to-tail polymer. Mapping the interaction sites between the two head-to-tail polymers that make the protofilament is a major goal for future studies. We hypothesize that the ΔK32 mutation in human lamin A will also show abnormal lateral assembly of the lamin dimer polymers because it is positioned in a conserved sequence. Because the conditions for assembling human lamin A filaments in vitro have not been identified, testing this hypothesis is currently not possible.
To associate protofilament assembly phenotypes with disease-related phenotypes, we expressed a GFP-tagged version of ΔK46 in C. elegans. The expression of GFP:ΔK46 in these worms was analyzed during early life stages. GFP expression was detected in all cell types in all stages (Figure 2), similar to wt lamin (Liu et al., 2000). In adult animals, however, GFP:ΔK46 expression was missing in some cells, which varied among individual animals. A difference was also seen at the level of GFP:ΔK46 localization: Some nuclei were wt in appearance with smooth peripheral GFP localization, whereas other nuclei contained GFP aggregates (Figure 2). This variation in expression and localization correlates with the variability in clinical symptoms in striated muscle laminopathies, including EDMD (Vytopil et al., 2002).
Immunofluorescence analysis showed that the endogenous Ce-lamin localization was not affected by the expression of GFP:ΔK46, similar to what has been described for other lamin mutations (Wiesel et al., 2008). This finding is in contrast to what is seen in fibroblasts cultured from patients expressing ΔK32 lamin, in which the localization of endogenous lamin A and lamin B is disrupted (Muchir et al., 2004). What is similar, however, is that the shape of the nucleus is not affected, expression levels of endogenous lamin and emerin do not change, and, importantly, a portion of peripheral emerin is mislocalized in both patient fibroblasts (Capanni et al., 2003) and in GFP:ΔK46 L4 larval stage worms (Figure 4). The mislocalization of emerin is especially interesting, as mutations in human emerin are associated with the X-linked form of EDMD (Bione et al., 1994); furthermore, this mislocalization occurs following early embryogenesis because embryonic emerin showed normal localization (Supplemental Figure S1B). The mislocalization of emerin seems to be a direct consequence of an impaired ability to associate with lamin filaments, which is not sufficient for emerin displacement in embryonic cells (Figure 5). LEM-2 appears to remain associated with the ΔK46 mutant lamin, but is mislocalized into GFP:ΔK46 lamin aggregates at the nuclear envelope (Figure 3). Together, these data suggest that altered ΔK46 lamin filaments affect the ability of lamin binding partners to associate with the lamina and to maintain proper localization at the nuclear envelope. Indeed, loss of emerin and LEM-2 in muscle cells causes muscle differentiation and maintenance phenotypes both in mice (Huber et al., 2009) and in C. elegans (R.B. and Y.G., unpublished observations). Although we have provided a basic link between the in vitro and in vivo phenotypes, much future research remains to be done to establish the precise mechanisms by which the altered character of the lamina caused by the ΔK46 mutation affects signaling pathways, gene expression, and/or structural integrity.
In this study, we also correlate the altered assembly of ΔK46 filaments and the resulting affects on lamina structure with disease-like phenotypes in the GFP:ΔK46 strain. The fertility of mutant animals was only slightly lower than that of GFP:wt worms, and we could not detect any apparent embryonic phenotype (Supplemental Figure S1). Given the role of the nuclear lamina in meiosis (reviewed in Penkner et al., 2009), it is intriguing that this mutation does not drastically affect fertility, even though it is expressed in many cell types (Figure 2). We next studied phenotypes related to the hallmark muscle weakening and wasting seen in EDMD patients. Worms expressing GFP:ΔK46 were impaired in their ability to move in water, but not to crawl on agar plates (Figure 6). These two distinct movements differ both in their kinetics and in the patterns of muscle and neuromuscular activation, suggesting specificity in the role of lamin in these processes (Ghosh and Emmons, 2008; Pierce-Shimomura et al., 2008; Mejat et al., 2009).
Because of the motility phenotype, we hypothesized that worms expressing GFP:ΔK46 would have muscle lesions. We analyzed structural elements of worm muscle at two levels: light microscopy (Figure 7) and TEM (Figure 8). At the level of light microscopy, lesions could be observed resembling those seen in animals lacking either MUA-3 (Bercher et al., 2001) or RNF-5 (Zaidel-Bar et al., 2010). Although it is unknown whether the ΔK46 lamin mutation acts through these particular proteins, comparing the ΔK46 phenotype with these published descriptions of the Mua phenotype reveals that this mutant lamin disrupts attachment of the muscle or other underlying tissues to the cuticle, thus implicating lamin in muscle integrity and function. If GFP:ΔK46 affects the attachment of the muscle or hypodermis to the cuticle, proper contractile force to generate movement may not be obtained, leading to motility defects. Analyzing muscle cell structure with TEM revealed disorganization of sarcomeres, filaments, dense bodies, and attachment sites. The ability of worms to crawl but not swim may indicate that these lesions are insufficient to cause a motility phenotype unless the worms are under higher mechanical stress, as the rate of movement in swimming is up to four times that for crawling (Ghosh and Emmons, 2008; Pierce-Shimomura et al., 2008).
Together, these data establish the GFP:ΔK46 C. elegans strain as a potential model for studying human EDMD caused by the ΔK32 lamin mutation and provide a basis for future studies of other laminopathic mutations. C. elegans recapitulates the autosomal-dominant human disease state by having mutant lamin expressed in the background of endogenous wt lamin (Wang et al., 2006). Although the 10–15% level of overexpression does not duplicate the human disease state of 50% mutant/50% wt lamin, it is enough to provide striking dominant phenotypes. In this way, we are able to observe phenotypes based on the interaction between mutant and endogenous lamin instead of the absence or replacement of the wt lamin. Future research with this model strain will reveal the molecular mechanisms linking lamin filament organization with the EDMD-like muscle phenotypes.
MATERIALS AND METHODS
Constructs and bacterial expression of Ce-lamin and Ce-emerin proteins
Ce-lamin cDNA in pET24d (Foeger et al., 2006) was mutagenized by site-directed mutagenesis using the Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA) and the following primers: Fwd 5′-cttcacgtcttcaagaggatcatttgacttcactc-3′; Rev 5′-gagtgaagtcaaatgatcctcttgaagacgtgaag-3′; constructs were verified by DNA sequencing. Ce-emerin 1-325 (missing the C-terminal transmembrane domain to create a soluble protein) in pET15b was a gift from K. Wilson, Johns Hopkins University. Plasmids were used to transform Escherichia coli BL21(DE3)-(codon plus-RIL) (Stratagene, La Jolla, CA), expressed by isopropyl-β-d-thio-galactoside (IPTG) induction, and the proteins were purified with nickel-nitrilotriacetic acid (Ni-NTA; Qiagen, Valencia, CA) beads as described (Foeger et al., 2006).
Worm strains were generated as described (Wiesel et al., 2008). Briefly, wt and ΔK46 lmn-1 were cloned into the pAD010 vector, which includes the baf-1 promoter (Margalit et al., 2007), the unc-119 gene, and a gfp gene with C. elegans introns. Constructs were introduced into worms by microparticle bombardment as described (Praitis et al., 2001); resulting strains were out-crossed to wt (N2) worms three times.
Assembly of Ce-lamin filaments
Bacterially expressed and purified Ce-lamin (0.1–0.2 mg/ml) in urea-containing buffer was dialyzed at room temperature against buffer containing 2 mM Tris–HCl, pH 9.0, 1 mM dithiothreitol for 4 h, followed by dialysis for 16 h against a buffer containing 15 mM Tris–HCl pH 7.4, 1 mM dithiothreitol, as described (Foeger et al., 2006).
Co-sedimentation
Heterologously expressed Ce-emerinΔTM was incubated overnight alone or with equal concentrations of Ce-lamin wt or mutant filaments in a total volume of up to 500 μl. Samples were centrifuged in an L8-70 Beckman ultracentrifuge in an LP42-TI rotor at 30,000 rpm, 23°C for 25 min. The supernatant and pellet fractions were separated, the pellet was resuspended in elution buffer containing 8 M urea, and equal volumes of wt and mutant pellet and supernatant samples were run on a 12% SDS–PAGE gel and visualized by staining with Coomassie blue. Average band intensities were calculated with ImageJ software.
Electron microscopy
L4 larval stage worms were prepared using a two-step fixation in formaldehyde/glutaraldehyde followed by osmium tetroxide as described (Cohen et al., 2002; Haithcock et al., 2005). Filaments were prepared for electron microscopy as described (Foeger et al., 2006). The samples were visualized with a transmission electron microscope (Technai 12; Philips, Eindhoven, The Netherlands) equipped with a MegaView II CCD camera (SIS, Münster, Germany). Measurements of filament width were taken at 50 places in each micrograph from at least five independent experiments.
Cryo-ET
Lamin filaments were prepared and images were processed as described (Ben-Harush et al., 2009). Briefly, lamins were applied to a glow-discharged, 200-mesh, carbon-coated copper grid (Quantifoil, Jena, Germany) to which colloidal gold was added, followed immediately by vitrification (Dubochet et al., 1988). Data were collected using a 300 kV FEI (Eindhoven, Netherlands) Polara transmission electron microscope equipped with a field-emission gun, and a Gatan (Pleasanton, CA) postcolumn GIF 2002 energy filter. Tilt series images were collected over an angular range of 60° to –60°, with a 2° increment, with a defocus value of –10 μm. The resulting pixel size at the specimen level was 4.25 Å. Thus, the theoretical resolution limit, as set by the contrast transfer function, was ∼44 Å. The projection images (2048 × 2048) were aligned to a common frame using fiducial gold markers, and reconstructed by weighted back-projection, as implemented by the TOM toolbox software package (Nickell et al., 2005).
Antibodies, Western blots, and immunofluorescence
Ce-emerin 2570, Ce-lamin 3932, and LEM-2 3597 antibodies were previously described (Lee et al., 2000; Tzur et al., 2002); anti-GFP was obtained from Roche (Basel, Switzerland); horseradish peroxidase–, Cy2-, and Cy3-conjugated secondary antibodies were obtained from The Jackson Laboratory (Bar Harbor, ME). For Western blotting, the protein amount from mixed-stage worm extracts was determined using the Bradford assay. Equal amounts were loaded on a 10% acrylamide gel, followed by transfer to polyvinylidene difluoride membranes. Primary antibodies were used at dilutions of 1:500 (vol/vol) (Ce-emerin) and 1:5000 (vol/vol) (Ce-lamin); secondary horseradish peroxidase at 1:10,000 (vol/vol). Quantification was performed by measuring the average intensity of the bands and normalizing to Ce-lamin with ImageJ software.
For immunostaining, worms were fixed with methanol/acetone essentially as described (Duerr, 2006). Briefly, worms were freeze-cracked on poly-l-lysine slides with liquid nitrogen, followed by methanol/acetone incubation at –20°C. Slides were blocked in 1% Tween-20 in phosphate-buffered saline (PBS-T) followed by 10% milk in PBS. Primary antibodies were diluted 1:100 (vol/vol) (Ce-emerin), 1:200 (vol/vol) (LEM-2), 1:400 (vol/vol) (Ce-lamin), or 1:600 (vol/vol) (GFP) in 10% milk, and 10 μl was pipetted to slides, which were covered with a plastic coverslip and incubated in a humidity chamber for 3 h at room temperature. After several PBS/PBS-T washes, Cy2- or Cy3-conjugated secondary antibodies were diluted 1:200 (vol/vol) in 10% milk, and 10 μl was pipetted to slides. Slides were then covered and incubated in a humidity chamber overnight at 4°C. PBS/PBS-T washes were repeated, and 400 μl of 1 μg/ml DAPI (where applicable) was added for 10 min followed by a final PBS wash. Slides were mounted with n-propylgallate and sealed for visualization.
Microscopy
Images were taken with an Axiocam CCD camera mounted on a Zeiss (Thornwood, NY) Axioplan II microscope equipped for fluorescence and DIC. Deconvolution (3D) was performed using AutoQuant X2.2 (Media Cybernetics, Bethesda, MD). For live imaging, worms were paralyzed with 1 mM levamisole before mounting on agar pad slides for visualization.
Fertility experiments
Healthy L4 larval stage worms with GFP expression were picked individually to plates and transferred to a fresh plate each subsequent day during their gravid life span. Progeny from each day were scored after 24 h to ensure they had time to hatch. Four independent experiments were performed, each containing 10 GFP:wt and 10 GFP:ΔK46 worms.
Motility experiments and RNAi feeding
For crawling motility, healthy L4 animals with GFP expression were transferred to an NGM plate without food. After a few seconds, the number of head turns in 1 min was scored. Six independent experiments were performed, each containing 10 GFP:wt and 10 GFP:ΔK46 worms.
For swimming motility, healthy L4 animals with GFP expression were transferred to wells containing 200 μl of water. After 15 min, the number of head turns to one side per minute was scored. Four independent experiments were performed, each containing 20 GFP:wt and 20 GFP:ΔK46 worms.
For RNAi feeding experiments, the empty control vector L4440 or the L4417 GFP feeding vector (gift of A. Fire, Stanford University) were used to transform E. coli HT115(DE3) cells as described (Liu et al., 2000). Transformed bacteria were plated on NGM plates containing 50 μg/ml ampicillin and 120 μg/ml IPTG to induce expression of the RNAi constructs. Five healthy L4 larval GFP:wt or GFP:ΔK46 animals were picked to a L4440 or L4417 plate and removed after ∼24 h when progeny appeared. Progeny were selected based on GFP expression at the L4 larval stage, and the swimming motility assay was repeated as described earlier in text, with 20 GFP:wt or GFP:ΔK46 worms fed with each construct used in six independent experiments.
Supplementary Material
Acknowledgments
We thank Roi Babaoff for helping to generate the GFP:ΔK46 animals. We also thank Gisèle Bonne for excellent comments. This study was funded by grants from the Muscular Dystrophy Association (MDA), Israel Science Foundation (ISF), USA-Israel Binational Science Foundation (BSF), the European Union Sixth Framework Program (Euro-laminopathies #018690), the Legacy (Morasha) fund of the Israeli Science Foundation (to Y.G.), and the German-Israel Foundation (GIF) and Fritz-Thyssen Foundation (to O.M. and Y.G.).
Abbreviations used:
- 2D
two-dimensional
- 3D
three-dimensional
- Ce-lamin
Caenorhabditis elegans lamin
- DIC
differential interference contrast
- EDMD
Emery-Dreifuss muscular dystrophy
- ET
electron tomography
- GFP
green fluorescent protein
- IF
intermediate filament
- IPTG
isopropyl-β-d-thio-galactoside
- L-CMD
LMNA-related congenital muscular dystrophy
- TEM
transmission electron microscopy
- wt
wild-type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-01-0064) on June 8, 2011.
REFERENCES
- Aebi U, Cohn J, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- Ben-Harush K, Wiesel N, Frenkiel-Krispin D, Moeller D, Soreq E, Aebi U, Herrmann H, Gruenbaum Y, Medalia O. The supramolecular organization of the C. elegans nuclear lamin filament. J Mol Biol. 2009;386:1392–1402. doi: 10.1016/j.jmb.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Bercher M, Wahl J, Vogel BE, Lu C, Hedgecock EM, Hall DH, Plenefisch JD. mua-3, a gene required for mechanical tissue integrity in Caenorhabditis elegans, encodes a novel transmembrane protein of epithelial attachment complexes. J Cell Biol. 2001;154:415–426. doi: 10.1083/jcb.200103035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, Toniolo D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet. 1994;8:323–327. doi: 10.1038/ng1294-323. [DOI] [PubMed] [Google Scholar]
- Bonne G, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- Capanni C, et al. Failure of lamin A/C to functionally assemble in R482L mutated familial partial lipodystrophy fibroblasts: altered intermolecular interaction with emerin and implications for gene transcription. Exp Cell Res. 2003;291:122–134. doi: 10.1016/s0014-4827(03)00395-1. [DOI] [PubMed] [Google Scholar]
- Cohen M, Lee KK, Wilson KL, Gruenbaum Y. Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem Sci. 2001;26:41–47. doi: 10.1016/s0968-0004(00)01727-8. [DOI] [PubMed] [Google Scholar]
- Cohen M, Tzur YB, Neufeld E, Feinstein N, Delannoy MR, Wilson KL, Gruenbaum Y. Transmission electron microscope studies of the nuclear envelope in Caenorhabditis elegans embryos. J Struct Biol. 2002;140:232–240. doi: 10.1016/s1047-8477(02)00516-6. [DOI] [PubMed] [Google Scholar]
- Conradt B, Xue D. WormBook. October 6, 2005. Programmed cell death. Available at: http://www.wormbook.org/chapters/www_programcelldeath/programcelldeath.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico A, et al. Two patients with “Dropped head syndrome” due to mutations in LMNA or SEPN1 genes. Neuromuscul Disord. 2005;15:521–524. doi: 10.1016/j.nmd.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Duerr JS. WormBook. June 19, 2006. Immunohistochemistry. Available at: http://www.wormbook.org/chapters/www_immunohistochemistry/immunohistochemistry.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery AE, Dreifuss FE. Unusual type of benign x-linked muscular dystrophy. J Neurol Neurosurg Psychiatry. 1966;29:338–342. doi: 10.1136/jnnp.29.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DW. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am J Anat. 1966;119:129–145. doi: 10.1002/aja.1001190108. [DOI] [PubMed] [Google Scholar]
- Foeger N, Wiesel N, Lotsch D, Mucke N, Kreplak L, Aebi U, Gruenbaum Y, Herrmann H. Solubility properties and specific assembly pathways of the B-type lamin from Caenorhabditis elegans. J Struct Biol. 2006;155:340–350. doi: 10.1016/j.jsb.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Geisler N, Schunemann J, Weber K, Haner M, Aebi U. Assembly and architecture of invertebrate cytoplasmic intermediate filaments reconcile features of vertebrate cytoplasmic and nuclear lamin-type intermediate filaments. J Mol Biol. 1998;282:601–617. doi: 10.1006/jmbi.1998.1995. [DOI] [PubMed] [Google Scholar]
- Ghosh R, Emmons SW. Episodic swimming behavior in the nematode C. elegans. J Exp Biol. 2008;211:3703–3711. doi: 10.1242/jeb.023606. [DOI] [PubMed] [Google Scholar]
- Gotzmann J, Foisner R. A-type lamin complexes and regenerative potential: a step towards understanding laminopathic diseases? Histochem Cell Biol. 2006;125:33–41. doi: 10.1007/s00418-005-0050-8. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Goldman RD, Meyuhas R, Mills E, Margalit A, Fridkin A, Dayani Y, Prokocimer M, Enosh A. The nuclear lamina and its functions in the nucleus. Int Rev Cytol. 2003;226:1–62. doi: 10.1016/s0074-7696(03)01001-5. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, Gruenbaum Y, Liu J. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber MD, Guan T, Gerace L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol Cell Biol. 2009;29:5718–5728. doi: 10.1128/MCB.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabinos A, Schunemann J, Meyer M, Aebi U, Weber K. The single nuclear lamin of Caenorhabditis elegans forms in vitro stable intermediate filaments and paracrystals with a reduced axial periodicity. J Mol Biol. 2003;325:241–247. doi: 10.1016/s0022-2836(02)01240-8. [DOI] [PubMed] [Google Scholar]
- Klapper M, Exner K, Kempf A, Gehrig C, Stuurman N, Fisher PA, Krohne G. Assembly of A- and B-type lamins studied in vivo with the baculovirus system. J Cell Sci. 1997;110(Pt 20):2519–2532. doi: 10.1242/jcs.110.20.2519. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee KK, Gruenbaum Y, Spann P, Liu J, Wilson KL. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell. 2000;11:3089–3099. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lee KK, Segura-Totten M, Neufeld E, Wilson KL, Gruenbaum Y. MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2003;100:4598–4603. doi: 10.1073/pnas.0730821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rolef Ben-Shahar T, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit A, Liu J, Fridkin A, Wilson KL, Gruenbaum Y. A lamin-dependent pathway that regulates nuclear organization, cell cycle progression and germ cell development. Novartis Found Symp. 2005;264:231–240. [PubMed] [Google Scholar]
- Margalit A, Neufeld E, Feinstein N, Wilson KL, Podbilewicz B, Gruenbaum Y. Barrier to autointegration factor blocks premature cell fusion and maintains adult muscle integrity in C. elegans. J Cell Biol. 2007;178:661–673. doi: 10.1083/jcb.200704049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattout A, Dechat T, Adam SA, Goldman RD, Gruenbaum Y. Nuclear lamins, diseases and aging. Curr Opin Cell Biol. 2006;18:335–341. doi: 10.1016/j.ceb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Mejat A, et al. Lamin A/C-mediated neuromuscular junction defects in Emery-Dreifuss muscular dystrophy. J Cell Biol. 2009;184:31–44. doi: 10.1083/jcb.200811035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcer S, Gruenbaum Y, Krohne G. Invertebrate lamins. Exp Cell Res. 2007;313:2157–2166. doi: 10.1016/j.yexcr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Moerman DG, Williams BD. WormBook. January 16, 2006. Sarcomere assembly in C. elegans muscle. Available at: http://www.wormbook.org/chapters/www_SarcomereAssembCelegansmusc/SarcomereAssembCelegansmusc.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Donaldson AD, Stewart M. Expression in Escherichia coli of human lamins A and C: influence of head and tail domains on assembly properties and paracrystal formation. J Cell Sci. 1991;99(2):363–372. doi: 10.1242/jcs.99.2.363. [DOI] [PubMed] [Google Scholar]
- Muchir A, et al. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve. 2004;30:444–450. doi: 10.1002/mus.20122. [DOI] [PubMed] [Google Scholar]
- Nickell S, Forster F, Linaroudis A, Net WD, Beck F, Hegerl R, Baumeister W, Plitzko JM. TOM software toolbox: acquisition and analysis for electron tomography. J Struct Biol. 2005;149:227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Ostlund C, Bonne G, Schwartz K, Worman HJ. Properties of lamin A mutants found in Emery-Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J Cell Sci. 2001;114:4435–4445. doi: 10.1242/jcs.114.24.4435. [DOI] [PubMed] [Google Scholar]
- Penkner AM, et al. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 2009;139:920–933. doi: 10.1016/j.cell.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Chen BL, Mun JJ, Ho R, Sarkis R, McIntire SL. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc Natl Acad Sci USA. 2008;105:20982–20987. doi: 10.1073/pnas.0810359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijano-Roy S, et al. De novo LMNA mutations cause a new form of congenital muscular dystrophy. Ann Neurol. 2008;64:177–186. doi: 10.1002/ana.21417. [DOI] [PubMed] [Google Scholar]
- Riemer D, Dodemont H, Weber K. A nuclear lamin of the nematode Caenorhabditis elegans with unusual structural features; cDNA cloning and gene organization. Eur J Cell Biol. 1993;62:214–223. [PubMed] [Google Scholar]
- Steinert PM, Roop DR. Molecular and cellular biology of intermediate filaments. Annu Rev Biochem. 1988;57:593–625. doi: 10.1146/annurev.bi.57.070188.003113. [DOI] [PubMed] [Google Scholar]
- Strelkov SV, Schumacher J, Burkhard P, Aebi U, Herrmann H. Crystal structure of the human lamin A coil 2B dimer: implications for the head-to-tail association of nuclear lamins. J Mol Biol. 2004;343:1067–1080. doi: 10.1016/j.jmb.2004.08.093. [DOI] [PubMed] [Google Scholar]
- Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- Taimen P, et al. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc Natl Acad Sci USA. 2009;106:20788–20793. doi: 10.1073/pnas.0911895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur YB, Hersh BM, Horvitz HR, Gruenbaum Y. Fate of the nuclear lamina during Caenorhabditis elegans apoptosis. J Struct Biol. 2002;137:146–153. doi: 10.1006/jsbi.2002.4452. [DOI] [PubMed] [Google Scholar]
- Vlcek S, Foisner R. Lamins and lamin-associated proteins in aging and disease. Curr Opin Cell Biol. 2007;19:298–304. doi: 10.1016/j.ceb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Vytopil M, et al. Frequent low penetrance mutations in the Lamin A/C gene, causing Emery Dreifuss muscular dystrophy. Neuromuscul Disord. 2002;12:958–963. doi: 10.1016/s0960-8966(02)00178-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Herron AJ, Worman HJ. Pathology and nuclear abnormalities in hearts of transgenic mice expressing M371K lamin A encoded by an LMNA mutation causing Emery-Dreifuss muscular dystrophy. Hum Mol Genet. 2006;15:2479–2489. doi: 10.1093/hmg/ddl170. [DOI] [PubMed] [Google Scholar]
- Wiesel N, Mattout A, Melcer S, Melamed-Book N, Herrmann H, Medalia O, Aebi U, Gruenbaum Y. Laminopathic mutations interfere with the assembly, localization, and dynamics of nuclear lamins. Proc Natl Acad Sci USA. 2008;105:180–185. doi: 10.1073/pnas.0708974105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL, Foisner R. Lamin-binding proteins. Cold Spring Harb Perspect Biol. 2010;2:a000554. doi: 10.1101/cshperspect.a000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Miller S, Kaminsky R, Broday L. Molting-specific down-regulation of C. elegans body-wall muscle attachment sites: the role of RNF-5 E3 ligase. Biochem Biophys Res Commun. 2010;395:509–514. doi: 10.1016/j.bbrc.2010.04.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.