The Ssy1-Ptr3-Ssy5 (SPS) sensor of extracellular amino acids coordinates the sequential activity of general signaling factors and the 26S proteasome in a novel proteolytic activation cascade to activate the intracellular signaling protease Ssy5, which endoproteolytically activates two latent transcription factors.
Abstract
Regulated proteolysis serves as a mechanism to control cellular processes. The SPS (Ssy1-Ptr3-Ssy5) sensor in yeast responds to extracellular amino acids by endoproteolytically activating transcription factors Stp1 and Stp2 (Stp1/2). The processing endoprotease Ssy5 is regulated via proteasomal degradation of its noncovalently associated N-terminal prodomain. We find that degradation of the prodomain requires a conserved phosphodegron comprising phosphoacceptor sites and ubiquitin-accepting lysine residues. Upon amino acid induction, the phosphodegron is modified in a series of linked events by a set of general regulatory factors involved in diverse signaling pathways. First, an amino acid–induced conformational change triggers phosphodegron phosphorylation by the constitutively active plasma membrane–localized casein kinase I (Yck1/2). Next the prodomain becomes a substrate for polyubiquitylation by the Skp1/Cullin/Grr1 E3 ubiquitin ligase complex (SCFGrr1). Finally, the modified prodomain is concomitantly degraded by the 26S proteasome. These integrated events are requisite for unfettering the Ssy5 endoprotease, and thus Stp1/2 processing. The Ssy5 phosphoacceptor motif resembles the Yck1/2- and Grr1-dependent degrons of regulators in the Snf3/Rgt2 glucose-sensing pathway. Our work defines a novel proteolytic activation cascade that regulates an intracellular signaling protease and illustrates how general signaling components are recruited to distinct pathways that achieve conditional and specific signaling outputs.
INTRODUCTION
Regulated proteolysis serves as an important mechanism used to control many cellular processes, including the separation of chromosomes during cell division and modulation of gene expression in response to environmentally induced signals (Brivanlou and Darnell, 2002; Nasmyth, 2005). Proteolysis can result in the complete degradation of proteins, and site-specific endoproteolytic processing events can generate proteolytic fragments with altered biological activities. Paradoxically, protein degradation can have activating functions. For example, degradation of an autoinhibitory domain or regulatory inhibitory subunit within a protein complex can liberate an intrinsic catalytic activity. Understanding the mechanisms governing regulated proteolysis is an important task in cell biology.
The SPS (Ssy1-Ptr3-Ssy5) signaling pathway enables yeast cells to respond to the presence of extracellular amino acids and induce their uptake (Forsberg and Ljungdahl, 2001). By studying this signal transduction pathway, we and others have unraveled a proteolytic mechanism responsible for the activation of the two homologous transcription factors Stp1 and Stp2 by receptor-activated proteolysis (RAP) (Andréasson and Ljungdahl, 2002; Andréasson et al., 2006; Pfirrmann et al., 2010). According to our current understanding of RAP and the signaling events carried out by the SPS sensor, extracellular amino acids are detected at the plasma membrane by the receptor Ssy1. Amino acid binding stabilizes a conformation of Ssy1 that, together with the peripheral membrane protein Ptr3, activates the Ssy5 endoprotease (Klasson et al., 1999; Abdel-Sater et al., 2004; Andréasson et al., 2006; Wu et al., 2006). On activation, Ssy5 processes transcription factors Stp1 and Stp2, resulting in the removal of N-terminal sequences required for cytoplasmic retention. Consequently, processed Stp1 and Stp2 can enter the nucleus, bind promoters of genes encoding a set of broad specificity amino acid permeases, and activate transcription (Andréasson and Ljungdahl, 2002, 2004; Wielemans et al., 2010; Tumusiime et al., 2011). The molecular mechanisms underlying RAP, and therefore the activation of the Ssy5 endoprotease activation, are under intense study.
The RAP-activated endoprotease Ssy5 exhibits homology to chymotrypsin-like serine proteases and is expressed as an inactive precursor in the form of a zymogen. Directly after translation and folding, Ssy5 cleaves itself into an N-terminal prodomain and a C-terminal catalytic (Cat)-domain. Strikingly, the N-terminal prodomain remains associated with the Cat-domain forming a primed but inactive protease subcomplex of the SPS sensor that is associated with Stp1 and Stp2 (Andréasson et al., 2006; Poulsen et al., 2006). On amino acid induction, the prodomain is degraded by the 26S proteasome and the Cat-domain is able to process Stp1 and Stp2. Removal of the prodomain by synthetically inducing its proteasomal degradation activates the protease in a manner that bypasses the requirement of the other SPS sensor components. These findings unambiguously demonstrate that the prodomain functions as an inhibitory subunit of the SPS sensor (Pfirrmann et al., 2010).
Consistent with an inhibitory function of the prodomain, a tight inverse correlation exists between prodomain levels and Stp1 processing. We have previously proposed a model in which the prodomain attains two alternative conformations, a stable inhibitory conformation and an unstable noninhibitory conformation. According to this model, extracellular amino acids trigger events leading to a conformational switch from the inhibitory to the noninhibitory conformation. The first 130 amino acids of the N-terminal prodomain are essential for proper regulation of this conformational switch. Several mutations in this region destabilize the prodomain independent of Ssy1 and constitutively activate Stp1 processing. Conversely, even though the intrinsic activity of the Cat-domain remains intact, mutations that interfere with prodomain degradation prevent Stp1 processing. Together these observations suggest that the crucial event of the RAP mechanism is the proteasomal degradation of the prodomain (Pfirrmann et al., 2010).
Canonical 26S proteasomal degradation relies on polyubiquitylation of target lysine residues in the substrate that labels the protein for proteolysis. In this process a set of enzymes is required to generally activate (E1), conjugate (E2), and specifically ligate (E3) the ubiquitin modifier to the substrate or its growing polyubiquitin chain (Wolf, 2004). Hence, proteasomal degradation of the Ssy5 prodomain likely involves a specific E3 ubiquitin ligase that mediates polyubiquitylation of the prodomain in response to SPS signaling. A candidate is the general Skp1/Cdc53/F-box protein (SCF) complex, an E3 ubiquitin ligase that is required for Ssy5 activation (Iraqui et al., 1999; Bernard and André, 2001; Abdel-Sater et al., 2004, 2011; Andréasson et al., 2006). SCF complexes achieve substrate specificity through association with exchangeable F-box proteins that enable specific binding and subsequent polyubiquitylation of target proteins (Jonkers and Rep, 2009). Grr1 is one of the best-characterized F-box proteins in yeast, and Grr1 is known to recognize and bind phosphorylated substrates. Notably, several proteins have been found to be inducibly phosphorylated by casein kinase I, Yck1, or Yck2, leading to their SCFGrr1-dependent proteasomal degradation (Moriya and Johnston, 2004; Spielewoy et al., 2010). Both Yck1/2 and SCFGrr1 are required for SPS sensor signaling (Abdel-Sater et al., 2004, 2011; Spielewoy et al., 2004), and importantly, Grr1 is required for prodomain down-regulation (Andréasson et al., 2006; Abdel-Sater et al., 2011).
Recently, Abdel-Sater et al. proposed that Yck1/2- and SCFGrr1-dependent polyubiquitylation of the Ssy5 prodomain induces a conformational change that activates Ssy5. According to their model, the degradation of the Ssy5 prodomain is not the activating step, but a secondary event (Abdel-Sater et al., 2011). Discrepancies between this hypothesis and our RAP model, in which proteasomal degradation of the prodomain is crucial, required that we independently evaluate the role of the general signaling components Yck1/2 and SCFGrr1 and their function in facilitating signaling. In contrast to the Abdel-Sater hypothesis, our results delineate a novel proteolytic activation cascade that ultimately transduces amino acid–induced signals through the proteasomal degradation of the inhibitory prodomain. At the heart of this mechanism is the activation of an intrinsic phosphodegron, which strikingly resembles the Yck1/2- and Grr1-dependent degrons required for the degradation of transcriptional regulators in the Snf3/Rgt2 glucose-sensing pathway (Moriya and Johnston, 2004). Our work provides a paradigm illustration of how general signaling factors are recruited to distinct pathways that achieve conditional and specific signaling outputs.
RESULTS
Casein kinase I activity is required for amino acid–induced degradation of the Ssy5 prodomain
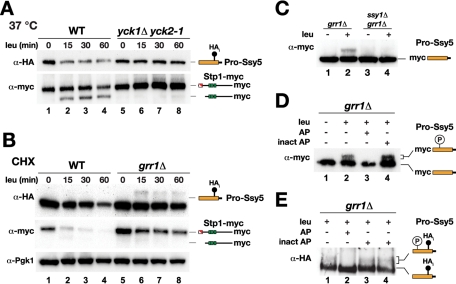
Casein kinase I (Yck1/2) function is required for Ssy5 activation and prodomain phosphorylation (Abdel-Sater et al., 2011). According to our RAP model, amino acid–induced protease activation is triggered by prodomain degradation (Andréasson et al., 2006; Pfirrmann et al., 2010). Consequently, we set out to test if Yck1/2 activity is required for degradation of the Ssy5 prodomain. We compared prodomain levels in amino acid (leucine)-induced wild-type (WT) cells and in yck1Δ yck2-1 cells with conditional temperature-sensitive casein kinase I activity. Consistent with our previous findings, in WT cells, leucine induced the degradation of the well-characterized and fully functional HA-tagged prodomain (Ssy5-HAi) (Pfirrmann et al., 2010). A clear reduction in prodomain levels was observed already 15 min after induction, which strictly correlated in time with Stp1 processing (Figure 1A, lanes 1–4). In contrast, in yck1Δ yck2-1 cells, induction with leucine did not lead to reduced prodomain levels or Stp1 processing (Figure 1A, lanes 5–8). Our results indicate that Yck1/2 act upstream of Ssy5 in the SPS signaling pathway, possibly by directly phosphorylating the prodomain for the induction of its degradation.
FIGURE 1:
Casein kinase I and Grr1 are required for amino acid–induced degradation of the Ssy5 prodomain. (A) Immunoblot analysis of cell extracts from HKY77 (ssy5Δ YCK1 YCK2) (WT) and CAY320 (yck1Δ yck2-1) carrying pSH120 (HAi-SSY5-GST) and pCA204 (STP1-MYC). Cells were pregrown in SD medium at room temperature and incubated at 37°C for 30 min, then samples were taken at the indicated time points after leucine addition. (B) Immunoblot analysis of cell extracts from HKY77 (ssy5Δ) and CAY276 (ssy5Δ grr1Δ) carrying plasmids as in (A); extracts were prepared from cells grown in SD medium after the addition of CHX and leucine at the indicated time points. (C) Immunoblot analysis of extracts from CAY86 (grr1Δ) and CAY267 (ssy1Δ grr1Δ) carrying pHK048 (MYC-SSY5) with (+) or without (–) leucine induction. (D) Immunoblot analysis of immunoprecipitated prodomain from extracts from cultures of CAY276 (ssy5Δ grr1Δ) carrying pHK048 (MYC-SSY5) and pRS317 induced with leucine as indicated. Immunoprecipitated material was incubated with active (AP) or heat-inactivated (inact AP) alkaline phosphatase as indicated. (E) Immunoblot analysis of immunoprecipitated prodomain from extracts obtained from CAY276 (ssy5Δ grr1Δ) carrying pSH120 (HAi-SSY5-GST) and pCA204 (STP1-MYC). Immunoprecipitated material was treated with active (AP), heat (lane 3), or EDTA (lane 4) inactivated alkaline phosphatase (inact AP). Immunoreactive forms of phosphorylated and dephosphorylated Ssy5 prodomain and Stp1 are schematically represented at their corresponding positions of migration.
SCFGrr1 mediates the degradation of phosphorylated prodomain
On SPS sensor induction, Ssy5 prodomain accumulates as a phosphorylated form in grr1Δ cells (Abdel-Sater et al., 2011). To test whether this posttranslational modification is linked to prodomain degradation, we examined prodomain levels after leucine addition in a time-course experiment in the presence of the translation inhibitor cycloheximide (CHX). In contrast to WT cells, the prodomain levels were stable in grr1Δ cells, and no Stp1 processing was evident (Figure 1B). In extracts prepared from grr1Δ cells, a higher-molecular-weight species of the prodomain was detected. The modified prodomain was not detected in extracts from WT cells, indicating that it is rapidly degraded. Formation of the modified prodomain was dependent on SPS sensor signaling because it did not appear in ssy1Δ grr1Δ cells, which lack the amino acid receptor component of the SPS sensor (Figure 1C). We tested whether the modified prodomain was phosphorylated. Functional HAi- and myc-epitope–tagged Ssy5 were immunoprecipitated from extracts prepared from leucine-induced grr1Δ cells. Incubation with protein phosphatase resulted in the disappearance of the slower migrating species, indicating that the aberrant migration induced by leucine was indeed due to phosphorylation (Figure 1, D and E).
Importantly, under no circumstances did we detect phosphorylated prodomain in extracts prepared from WT cells. Thus the accumulation of phosphorylated prodomain appears to be restricted to grr1Δ cells, suggesting that SCFGrr1 is involved in degradation of the phosphorylated prodomain upon signaling. This experimental evidence, and the fact that Yck1/2 are required for prodomain degradation, led us to predict two coupled and amino acid–induced events: first, Yck1/2 directly phosphorylate the prodomain, and second, Grr1 specifically and physically interacts with the phosphorylated prodomain to catalyze its ubiquitylation, creating a signal for prodomain degradation.
Spatial proximity of Yck1 to Ssy5 constitutively activates the protease
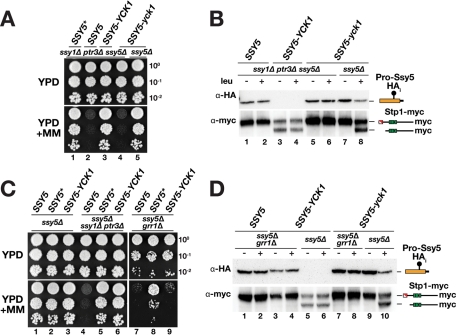
We set out to test our first prediction that Yck1/2 directly phosphorylate the prodomain. We reasoned that the spatial proximity of the kinase to its potential substrate Ssy5 might positively modulate pathway activation, and we investigated the effect of fusing Yck1 to the C terminus of Ssy5. A chimeric Ssy5-Yck1 protein lacking the two C-terminal cysteine residues of Yck1 was constructed; these cysteines normally serve as sites for palmitoylation that anchors the kinase to the plasma membrane (Vancura et al., 1994). To test whether Ssy5-Yck1 bypasses the requirement of upstream signaling components, and thus exhibits constitutive signaling characteristics, the chimera was expressed in an ssy1Δ ptr3Δ ssy5Δ strain. This strain lacks a functional SPS pathway and therefore has a severe defect in amino acid uptake and cannot grow on yeast extract–peptone–dextrose (YPD) medium containing MM (2-{[({[(4-methoxy-6-methyl)-1,3,5-triazin-2-yl]-amino}carbonyl)amino-]-sulfonyl}-benzoic acid), an inhibitor of branched-chain amino acid synthesis (Jørgensen et al., 1998). Whereas cells expressing WT SSY5 did not grow, we found that cells expressing SSY5-YCK1 grew robustly and similar to cells expressing SSY5*, encoding a well-characterized constitutively active form of Ssy5 (Figure 2A, compare lanes 1, 2, and 3) (Andréasson et al., 2006; Pfirrmann et al., 2010). The constitutive activity of Ssy5-Yck1 was dependent on the kinase activity of the chimeric protein, because a kinase-inactivating K98R mutation (SSY5-yck1) in the ATP binding domain of Yck1 (Wang et al., 1992; Moriya and Johnston, 2004) abolished growth on YPD+MM (Figure 2A, lane 4). Importantly, in the context of an intact SPS sensor, the Ssy5 protease moiety encoded by the kinase-defective SSY5-yck1 allele was fully capable of mediating SPS pathway signaling (Figure 2A, lane 5), which indicates that the K98R mutation does not generally impair Ssy5 function.
FIGURE 2:
Fusion of Yck1 to the C terminus of Ssy5 constitutively activates the protease. (A) Plasmids pCA195 (SSY5*), pSH120 (SSY5), pDO100 (SSY5-YCK1), and pDO099 (SSY5-yck1) were introduced into CAY285 (ssy1Δ ptr3Δ ssy5Δ) or HKY77 (ssy5Δ) together with pCA204 (STP1-MYC). Dilutions of cultures were spotted onto YPD and YPD+MM, then plates were incubated at 30°C for 2–3 d and photographed. (B) Immunoblot analysis of cell extracts from strains in (A) induced with leucine as indicated. (C) Plasmids pSH120 (SSY5), pCA195 (SSY5*), and pDO100 (SSY5-YCK1) were introduced into HKY77 (ssy5Δ), CAY285 (ssy1Δ ptr3Δ ssy5Δ), and CAY276 (ssy5Δ grr1Δ) together with pCA204 (STP1-MYC). Dilutions of cultures were spotted onto YPD and YPD+MM. (D) Immunoblot analysis of cell extracts from strains in (C) and strains HKY77 (ssy5Δ) and CAY276 (ssy5Δ grr1Δ) carrying pDO099 (SSY5-yck1) and pCA204 (STP1-MYC) induced with leucine as indicated. The immunoreactive forms of the Ssy5 prodomain and Stp1 are schematically depicted at their corresponding positions of migration.
To elucidate the mechanism underlying the constitutive activity of Ssy5-Yck1, we monitored Stp1 processing and prodomain levels in ssy1Δ ptr3Δ ssy5Δ cells expressing Ssy5-Yck1. As expected, the constitutive activity of Ssy5-Yck1 correlated with Stp1 processing. Remarkably, the prodomain levels in these cells were so low that the signal was below the detection limit (Figure 2B, lanes 3 and 4). The absence of detectable prodomain was dependent on the kinase activity of the fusion protein; readily detectable levels of prodomain were obtained in extracts from cells expressing the SSY5-yck1 allele (Figure 2B, lanes 5 and 6).
These results show that the mere placement of Yck1 kinase activity in physical proximity to Ssy5 triggers efficient degradation of the prodomain, resulting in constitutive Stp1 processing. This finding suggests that, on SPS sensor induction, Yck1/2 directly phosphorylate the Ssy5 prodomain, triggering its degradation and thus protease activation.
Constitutively phosphorylated Ssy5 requires Grr1 for its activity
The fact that phosphorylated prodomain of Ssy5-Yck1 was efficiently degraded led us to examine whether the constitutive activity of this chimera was dependent on SCFGrr1 ubiquitylation. If so, we expected that the constitutive activity of the Ssy5-Yck1 chimera would require GRR1. This notion was tested by monitoring SPS sensor–dependent growth of grr1Δ ssy5Δ cells expressing the SSY5, SSY5*, or SSY5-YCK1 allele. In contrast to cells expressing the fully constitutive and GRR1-independent Ssy5*, cells expressing Ssy5-Yck1 did not grow on YPD+MM (Figure 2C, dilution series 8 and 9). Subsequent immunoblot analysis revealed that the prodomain was stable and readily detected in grr1Δ cells (Figure 2D, lanes 3 and 4). Consistent with an inhibitory function of the prodomain, Stp1 was not processed in these cells (Figure 2D, lanes 3 and 4). These findings indicate that SCFGrr1 is required for activation of Ssy5-Yck1, presumably by targeting the prodomain for rapid degradation by a mechanism that relies on ubiquitylation of the phosphorylated prodomain. Our results clearly place Grr1 downstream of Ssy1-, Ptr3-, and Yck1/2-dependent phosphorylation of Ssy5 in the SPS signaling pathway.
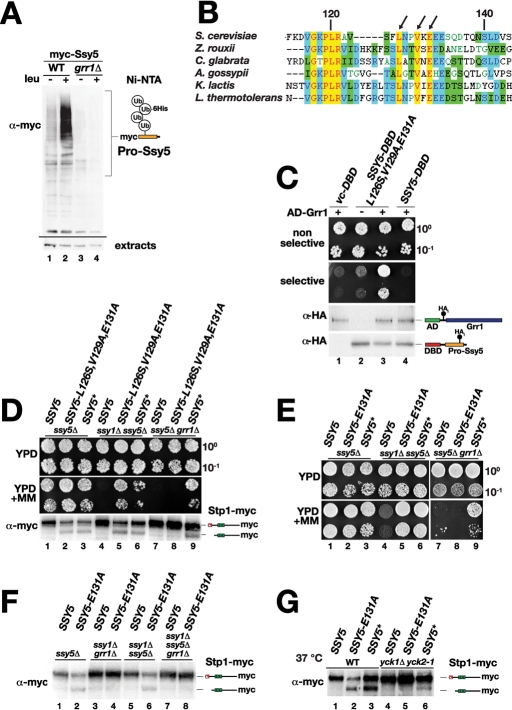
Grr1 physically interacts with induced Ssy5 and catalyzes prodomain polyubiquitylation
Grr1 is required for amino acid–induced prodomain degradation (Andréasson et al., 2006), and it has recently been shown that the prodomain undergoes GRR1-dependent polyubiquitylation upon SPS sensor induction (Abdel-Sater et al., 2011). Based on this knowledge, we posited and tested the possibility that Grr1 directly mediates SCFGrr1-complex–dependent polyubiquitylation and degradation of the phosphorylated prodomain. First, using our strains and characterized epitope-tagged Ssy5 proteins, we confirmed that the prodomain was polyubiquitylated in an amino acid–induced and GRR1-dependent manner (Figure 3A). Next we sought evidence for the requisite physical interaction between Ssy5 and Grr1. We used a directed yeast two-hybrid approach to overcome limitations associated with coimmunoprecipitation approaches (i.e., difficulties to detect low abundance Grr1 and Ssy5) and the anticipated transient nature of their interaction. We examined the ability of WT Ssy5 and Ssy5-L126S V129A E131A, a mutant protein that exhibits Ssy1-independent constitutive signaling properties (Pfirrmann et al., 2010), to interact with Grr1. Although Grr1 did not interact with WT Ssy5, we found that constitutively active Ssy5-L126S V129A E131A interacted efficiently with Grr1, as scored by the robust growth on medium selective for expression of the two-hybrid interaction reporters (Figure 3C). These findings suggest that Grr1 binds directly to an induced form of Ssy5, mediating its polyubiquitylation.
FIGURE 3:
Grr1 physically interacts with Ssy5 and catalyzes its polyubiquitylation. (A) Immunoblot analysis of polyubiquitylated prodomain present in extracts from strains TPY116 (WT) and TPY117 (grr1Δ) carrying plasmids pHK048 (MYC-SSY5) and pTP156 (PCUP1-6His-UBI) induced with leucine as indicated. The immunoreactive forms of polyubiquitylated prodomain are schematically represented at their corresponding position of migration. The levels of a nonspecific immunoreactive band serves as an input control (extracts). (B) Amino acid residues 113–144 of the Ssy5 prodomain (S. cerevisiae) and fungal orthologues were aligned using Clustal X; conserved (yellow) and similar (blue, green) residues are highlighted, and the conserved residues, L126, V129, and E131, are indicated with arrows. (C) Directed two-hybrid analysis of Ssy5 and Grr1 interactions. Plasmids pCA146 (vc-DBD), pTP133 (DBD-SSY5-L126S,V129A,E131A), or pTP134 (DBD-SSY5) were introduced together with pTP122 (AD-GRR1, +) or pJG4-5 (–) into strain CAY235. Growth of transformants was assessed on SC (-His, -Trp) (nonselective) or SC-Gal (-His, -Trp, -Leu, -Lys) (selective), and expression of DNA-binding domain (DBD) and activation domain (AD) constructs were analyzed by immunoblotting. (D) Growth of HKY77 (ssy5Δ), HKY84 (ssy5Δ ssy1Δ), and CAY276 (ssy5Δ grr1Δ) carrying pCA204 (STP1-MYC) and pSH120 (SSY5), pTP112 (SSY5-L126S, V129S, E131A), or pCA195 (SSY5*) on YPD and YPD+MM. Immunoblot analysis of protein extracts from strains grown in SD medium. (E) Growth of HKY77 (ssy5Δ), HKY84 (ssy5Δ ssy1Δ), and CAY276 (ssy5Δ grr1Δ) carrying pCA204 (STP1-MYC) and pSH120 (SSY5), pTP112 (SSY5-E131A), or pCA195 (SSY5*) on YPD and YPD+MM. (F) Immunoblot analysis of extracts from HKY77 (ssy5Δ), CAY276 (ssy5Δ grr1Δ), HKY84 (ssy5Δ ssy1Δ), and TPY120 (ssy5Δ ssy1Δ grr1Δ) carrying pCA204 (STP1-MYC) and pSH120 (SSY5) or pTP115 (SSY5-E131A) grown on SMD. (G) Immunoblot analysis of extracts from HKY77 (ssy5Δ YCK1 YCK2) (WT) and CAY320 (yck1Δ yck2-1) carrying pCA204 (STP1-MYC) and pSH120 (SSY5), pTP115 (SSY5-E131A), or pCA195 (SSY5*). Cells were pregrown in SD at room temperature, and extracts were prepared 30 min after cultures were shifted to 37°C. Immunoreactive forms of Stp1 are schematically depicted at their corresponding positions of migration.
SPS sensor induction triggers the prodomain to switch to a signaling conformation that undergoes phosphorylation-dependent proteasomal degradation
Our ability to detect a direct physical interaction between Ssy5-L126S V129A E131A and Grr1, and the fact that this mutant protein exhibits constitutively down-regulated levels of its prodomain (Pfirrmann et al., 2010), led us to hypothesize that this constitutive Ssy5 mutant adopts a prodomain conformation that attracts Yck1/2 phosphorylation and subsequent SCFGrr1-dependent ubiquitylation. We examined whether the constitutive activity of Ssy5-L126S V129A E131A was SCF Grr1-dependent by monitoring growth on YPD+MM of grr1Δ ssy5Δ cells expressing this mutant protein. We found a strict requirement for Grr1 activity for growth and Stp1 processing (Figure 3D, compare dilution series/lanes 5 and 8). In contrast, control cells expressing the GRR1-independent and constitutive SSY5* allele exhibited robust growth in the presence of MM, and, accordingly, Stp1 processing was observed.
The requirement for an intact SCFGrr1 ubiquitin ligase complex raised the possibility that the activity of constitutive Ssy5 mutant proteins require Yck1/2 function in an event that would obligatorily precede ubiquitylation. We have previously described the isolation of several constitutive mutant alleles affecting the prodomain of SSY5, all carrying mutations in a sequence stretch coding for a short conserved motif around amino acid residue 130 (Figure 3B) (Pfirrmann et al., 2010). The constitutive single point mutant SSY5-E131A allele that exhibits GRR1-dependent growth in the presence of MM (Figure 3E) was used to examine Yck1/2 involvement. The constitutive Stp1 processing activity of the Ssy5-E131A mutant was dependent on Yck1/2 activity (Figure 3G). Taken together, our findings indicate that a single point mutation in a conserved residue of the prodomain activates the Ssy5 protease independently of amino acid–induced signals and upstream SPS sensor component Ssy1, but still retains a requirement for phosphorylation and ubiquitylation for its activity. The data support a model that amino acids induce a conformational change of the Ssy5 prodomain that facilitates subsequent phosphorylation and ubiquitylation-dependent degradation. The switch of the prodomain to the signaling conformation can be mimicked by substitutions close to amino acid residue 130.
A conserved phosphoacceptor motif defines a phosphodegron in the prodomain
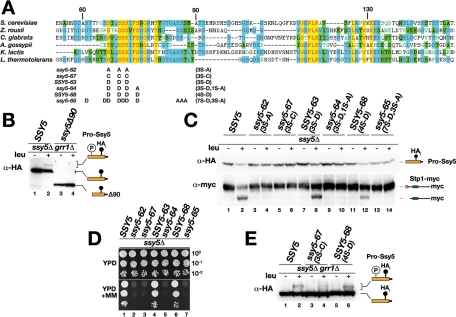
Having established that SPS signaling functions via induced phosphorylation of the prodomain as a trigger for its Grr1-dependent proteasomal degradation, we sought to identify the phosphorylation and ubiquitylation acceptor residues within the prodomain. Alignment of the prodomain sequences of Ssy5 orthologues revealed a conserved sequence motif rich in serine residues potentially serving as phosphoacceptor sites (Figure 4A). We have previously found that the region containing this motif, amino acids 60–90 of the prodomain, is required for signal-induced degradation of the prodomain (Pfirrmann et al., 2010). Consistently, mutation of seven potential phosphoacceptor sites in this motif abolishes phosphorylation and protease activation (Abdel-Sater et al., 2011). We tested whether the first 90 amino acids of the prodomain are important for SPS sensor–induced phosphorylation and found that the prodomain of ssy5Δ90 does not undergo phosphorylation in grr1Δ cells upon leucine induction (Figure 4B, lane 4).
FIGURE 4:
A conserved phosphoacceptor motif in the prodomain is required for its degradation and Stp1 processing. (A) Amino acid residues 53–153 of the Ssy5 prodomain (S. cerevisiae) and fungal orthologues were aligned by using Clustal X; conserved (yellow) and similar (blue, green) residues are highlighted. The serine substitution alleles ssy5-62, -64, -65, -67 and SSY5-63 and -68 are shown. (B) Immunoblot analysis of extracts from CAY276 (ssy5Δ grr1Δ) carrying pCA204 (STP1-MYC) and pSH120 (SSY5) or pSH119 (ssy5Δ90) induced with leucine as indicated. (C) Immunoblot analysis of extracts from strain HKY77 (ssy5Δ) carrying pCA204 (STP1-MYC) and pSH120 (SSY5), pDO067 (ssy5-62), pDO082 (ssy5-67), pDO068 (SSY5-63), pDO069 (ssy5-64), pDO083 (SSY5-68), or pDO072 (ssy5-65) induced with leucine as indicated. (D) Strains in (C) were spotted onto YPD and YPD+MM medium; plates were incubated at 30°C for 2–3 d and photographed. (E) Immunoblot analysis of extracts from strain CAY276 (ssy5Δ grr1Δ) carrying pCA204 (STP1-MYC) and pSH120 (SSY5), pDO082 (SSY5-67), or pDO083 (SSY5-68) induced with leucine as indicated. Immunoreactive forms of Stp1 and Ssy5 prodomain are schematically depicted at their corresponding positions of migration.
This finding prompted us to further investigate the role of the conserved serine residues between amino acids 60 and 90 as potential acceptor sites for phosphorylation and prodomain degradation. We focused our efforts on three highly conserved serine residues (residues 67, 70, and 72) that we found to meet the criteria for Yck1/2 consensus sites (Flotow et al., 1990; Knippschild et al., 2005) or were predicted to be phosphorylated (Ingrell et al., 2007). We replaced these serine residues with either alanine residues (ssy5‑62, 3S-A) or with more structurally related cysteine residues (ssy5-67, 3S-C) and assessed the impact of the mutations on prodomain stability and Stp1-processing activity. Both mutant proteins displayed normal autoprocessing, indicating that the mutations did not induce gross protein folding defects affecting the catalytic potential of the Ssy5 protease (Figure 4C, top panel, lanes 3–6). Importantly, the mutant prodomains were stable and were not degraded in response to SPS-sensor induction (Figure 4C, compare lanes 1 and 2 with lanes 3–6). Consistently, Stp1 processing activity was not detected (Figure 4C, lanes 4 and 6), nor did the mutant proteins support growth in the presence of MM (Figure 4D, dilutions 2 and 3). Taken together, our data indicate that Ser-67, -70, and -72 are required for induced prodomain degradation, likely due to their function as acceptor sites for phosphorylation.
Next we asked how the introduction of negatively charged amino acids at these positions would influence Ssy5 activation. The SSY5-63 (3S-D) allele encodes a mutant protein in which the three serine residues are replaced with aspartate residues. This allele fully complemented the ssy5Δ mutation; similar to WT Ssy5, Ssy5-63 exhibited leucine-induced prodomain degradation and Stp1-processing activity, and supported growth on YPD+MM (Figure 4C, lanes 7 and 8 and 4D, lane 4). This finding raised the possibility that residues with negative charges at putative phosphorylation sites are more compatible with Ssy5 activation than are neutral ones. We tested this notion by creating alleles with different charge profiles; ssy5-64 (3S-D 1S-A) carries an additional serine-to-alanine substitution, and SSY5-68 (4S-D) carries aspartate residues at all four positions. Functional tests showed that ssy5-64 expressed an inactive protein, whereas SSY5-68 expressed a functional one (Figure 4C, compare lanes 9–12, and 4D, dilution series 5 and 6). Apparently, the introduction of negative charges at the putative phosphorylation sites is indeed more compatible with Ssy5 activation.
To examine whether the introduction of a greater number of negatively charged aspartate residues could activate the protease, perhaps independently of phosphorylation, we constructed the ssy5-65 (7S-D 3S-A) allele encoding a protein with seven serine residues replaced with aspartate and three additional serine residues replaced with alanine, eliminating all possible serine phosphoacceptor sites. Although the ssy5-65 allele expressed a protein that was correctly folded, as indicated by proper autolytic processing into Pro- and Cat-domain (Figure 4C, lanes 13 and 14), it did not complement the ssy5Δ mutation (Figure 4D, dilution series 7). This result suggests that negative charges at potential phosphorylation sites cannot substitute for the required phosphorylation in this region of the prodomain as the trigger for protease activation.
We directly assessed the phosphorylation status of the prodomains carrying cysteine (ssy5-67, 3S-C) or aspartate (SSY5-68, 4S-D) substitutions in grr1Δ cells. Compared to WT Ssy5, we reproducibly observed lower levels of induced phosphorylation of the inactive Ssy5-67 prodomain (Figure 4E, compare lanes 4 and 2). The remaining phosphorylation indicates that this mutant still harbors residual functional phosphorylation sites. Apparently, the residual phosphorylation is not sufficient to trigger SCFGrr1 recognition and protease activation. In contrast, the functional Ssy5-68 mutant prodomain displayed induced phosphorylation to the same extent as WT Ssy5. The phosphorylated forms of Ssy5-68 prodomain migrated faster than the corresponding modified WT prodomain, suggesting that fewer phospho-groups were attached to the remaining serine residues (Figure 4E, compare lanes 6 and 2). In context of the negative charges contributed by the aspartate side chains, this residual phosphorylation apparently suffices for downstream signaling events that activate Ssy5.
Taken together, the mutational analysis demonstrates that multisite phosphorylation, and in part the overall negative charge, in the region of amino acid residues 60–90 triggers the degradation of the prodomain. We therefore propose that the prodomain contains a phosphodegron comprising this region, which functions as a recognition module for SCFGrr1, and is therefore required for Ssy5 activation in response to extracellular amino acids.
Ubiquitin acceptor sites in the prodomain are required for activation of Ssy5
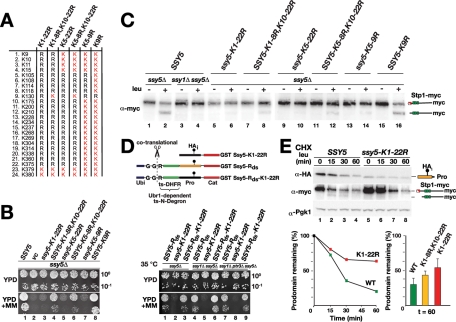
To define the complete phosphodegron in the prodomain, we sought to identify the SCFGrr1 ubiquitylation acceptor lysine residues required for proteasomal degradation. First, we examined whether the lysine residues in the prodomain are required for activation of the protease (Figure 5). We replaced all potential ubiquitin acceptor lysine residues in the prodomain with arginine residues, with the exception of residues K379 and K380 (Figure 5A), which are directly adjacent to the autolytic cleavage site (381/382), to avoid interfering with protease maturation (Poulsen et al., 2006). We found that, although autolytic processing occurred, albeit at reduced levels, the Ssy5-K1-22R mutant protein was nonfunctional (Figure 5B, dilution series 1–3). Accordingly, we could not detect Stp1 processing (Figure 5C, lanes 5 and 6). To rigorously test the catalytic competency of Ssy5-K1-22R, we artificially induced the degradation of its prodomain by fusing a temperature-regulated N degron to the N terminus (Ssy5-Rds-K1-22R) (Figure 5D). We have previously established that this degron can artificially activate WT Ssy5 even in the absence of other SPS sensor components (Pfirrmann et al., 2010). We found that, at 35°C, a temperature that triggers N degron–mediated degradation of the prodomain, Ssy5-K1-22R was activated, and its activity bypassed the requirement of SSY1 or PTR3 (Figure 5D, dilution series 3, 6, and 9). Thus ssy5K1-22R encodes a protease that is fully capable of processing Stp1 but cannot be activated by amino acid–induced signals.
FIGURE 5:
Lysine residues are required for induced prodomain degradation. (A) List of lysine residues (K), and the K-to-R substitutions created in the Ssy5 prodomain. (B) Growth of HKY77 (ssy5Δ) carrying pCA204 (STP1-MYC) and pSH120 (SSY5), pRS316 (vc), pTP144 (ssy5-K1-22), pTP142 (SSY5-K1-8R,K10-22R), pTP143 (ssy5-K5-22R), pTP140 (SSY5-K5-8R,K10-22R), pTP145 (ssy5-K5-9R), or pTP135 (SSY5-K9R) on YPD and YPD+MM medium. (C) Immunoblot analysis of extracts from strains in (B) and strain HKY84 (ssy1Δ ssy5Δ) carrying pCA204 (STP1-MYC) and pSH120 (SSY5) induced with leucine as indicated. (D) Ssy5 N degron constructs are schematically represented and consist of an N-terminal ubiquitin moiety (Ubi), a destabilizing arginine residue (R), and a temperature-sensitive dihydrofolate reductase (ts-DHFR) linker fused at the N terminus of Ssy5 with an internal prodomain HA epitope (HAi) and a C-terminal GST tag. The ubiquitin is cotranslationally cleaved (scissors) following the di-glycine (G-G) residues resulting in proteins with an N-terminal arginine. Growth of HKY77 (ssy5Δ), HKY84 (ssy1Δ ssy5Δ), and CAY285 (ssy1Δ ptr3Δ ssy5Δ) carrying pCA204 (STP1-MYC) and pTP110 (SSY5-Rds), pTP144 (ssy5-K1-22), or pTP149 (SSY5-Rds-K1-22) was assessed; cells were pregrown in SD medium, dilutions were prepared and spotted onto YPD and YPD+MM medium, and the plates were incubated at 35°C for 3 d. (E) Immunoblot analysis of extracts from HKY77 (ssy5Δ) carrying pCA204 (STP1-MYC) and pSH120 (SSY5) or pTP144 (ssy5-K1-22); extracts were prepared from cells grown in SD medium after the addition of CHX and leucine at the indicated time points. The signal intensities of the HA-tagged prodomain were quantified, normalized to a Pgk1 loading control, and plotted relative to the signal intensities before induction (line plot, left panel). Similarly, the signal intensities of HA-tagged prodomain in HKY77 (ssy5Δ) carrying pCA204 (STP1-MYC) and pSH120 (SSY5), pTP142 (SSY5-K1-8R, K10-22R), or pTP144 (ssy5-K1-22) were quantified; the mean relative levels remaining after 60 min are plotted (bar graph, right panel). Error bars show standard deviations based on data from three independent transformants.
We investigated the underlying cause of the blocked activation of Ssy5-K1-22R, considering that an impaired polyubiquitylation of the prodomain is predicted to exert a stabilizing effect during SPS sensor–inducing conditions. We tested this prediction by monitoring prodomain levels in a CHX chase upon SPS sensor induction, and found that the prodomain of Ssy5-K1-22R was stabilized upon amino acid stimulation and no Stp1 processing occurred (Figure 5E, lanes 5–8). Hence, the presence of lysine residues in the prodomain is a requirement for Ssy5 activation, which is consistent with a mechanism involving SCFGrr1 ubiquitylation and degradation by the 26S proteasome.
Lysine residues adjacent to the phosphoacceptor motif serve as ubiquitylation sites required for induced prodomain degradation
To identify the lysine residues that serve as ubiquitylation acceptor sites upon signaling, we systematically reintroduced lysine residues into the prodomain of the nonfunctional Ssy5-K1-22R. First, a single lysine at position K130 was reintroduced. K130 is localized close to the phosphoacceptor motif and in a cluster of residues that we have previously found can be mutated to constitutively activate the protease (Figure 3) (Pfirrmann et al., 2010). The resulting allele SSY5-K1-8R, K10-22R encodes a functional Ssy5 as judged by the ability to support growth on YPD+MM and leucine-induced Stp1 processing (Figure 5B, dilution series 4, and 5C, lanes 7 and 8). This finding prompted us to examine whether K130 is the only ubiquitylation site used during Ssy5 activation. We replaced K130 with arginine in the context of the WT protein creating SSY5-K9R and found that the encoded protein was fully functional (Figure 5B, lane 8, and 5C, lanes 15 and 16). This result indicates that alternative ubiquitylation sites can be used to target the prodomain for degradation.
We considered it likely that, in addition to K130, the four lysine residues that lie between the phosphoacceptor sequence (amino acids 60–90) and K130 could serve as ubiquitylation acceptors. To test this notion, we replaced these five lysine residues (K105–K130) with arginine in an otherwise WT Ssy5 context creating the mutant allele ssy5-K5-9R. Strikingly, this allele did not support growth in the presence of MM (Figure 5B, dilution series 7). This finding indicates that the lysine residues in the region 105–130 are important for Ssy5 protease activation and is consistent with the notion that the ubiquitylation machinery is rather promiscuous. Clearly, one lysine residue in the critical region is sufficient for activation, whereas multiple sites convey a stronger output, which is reflected in more robust growth on YPD+MM (Figure 5B, compare dilution series 4 and 6–8) and a more efficient cleavage of Stp1 (Figure 5C, compare lanes 8 and 12–16). Consistently, after leucine induction, the active Ssy5 mutant with a single lysine (SSY5-K1-8R, K10-22R) exhibited an intermediate level of stability in comparison to WT (SSY5) and the inactive and more stable lysine-less prodomain mutant (ssy5-K1-22R) (Figure 5E, right bar graph).
We suggest that ubiquitylation in this specific region of the prodomain is a prerequisite for efficient targeting of the prodomain for proteasomal destruction. Importantly, the spatial proximity of the requisite ubiquitylation sites to the conserved phosphoacceptor motif defines amino acid residues 60–130 of the prodomain as an intrinsic phosphodegron that is activated upon SPS sensor induction.
DISCUSSION
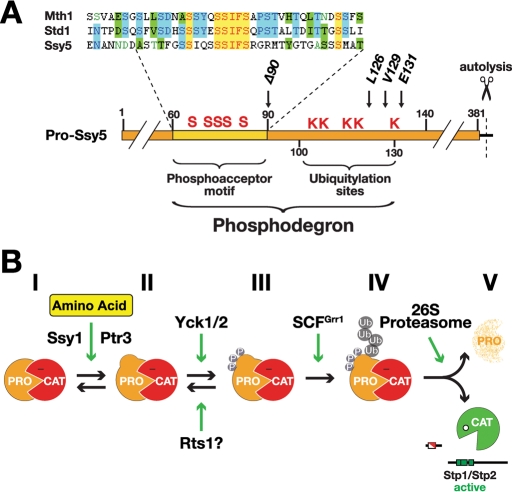
According to our present understanding of RAP, the key event in the activation of Ssy5 is the induced degradation of its prodomain (Andréasson et al., 2006; Pfirrmann et al., 2010). The data presented in this study reveal that prodomain degradation relies on both intrinsic sequence determinants and a set of regulatory factors controlled by the SPS sensor. Specifically, the intrinsic sequence determinants reside in the N terminus of the inhibitory prodomain (amino acid residues 60–130) and function as an inducible phosphodegron (Figure 6A). The phosphodegron consists of a conserved, positively acting sequence motif comprising several phosphoacceptor residues and a cluster of five lysine residues that likely serve as sites for ubiquitylation. On signaling, the phosphodegron becomes activated, resulting in its phosphorylation converting it into a substrate for ubiquitylation that targets the entire prodomain for degradation by the proteasome. The outcome of this chain of rapid and linked events is the liberation of active Cat-domain that processes the downstream transcription factors Stp1 and Stp2.
FIGURE 6:
The phosphodegron in the prodomain controls the activation of Ssy5. (A) Schematic diagram of the Ssy5 prodomain (residues 1–381; scissors indicate the autolytic cleavage site); the phosphodegron (residues 60–130) contains the phosphoacceptor motif (conserved serine residues, S, in the region amino acids 60–90) and the ubiquitylation sites (K). The positions of the conserved L126, V129, and E131 residues and the ssy5Δ90 allele are indicated. The similarity of the phosphoacceptor motif and sequences in Mth1 (amino acids 111–149) and Std1 (amino acids 122–160) are displayed in expanded form; identical (yellow) and similar (blue, green) residues are highlighted. (B) Diagram of the five (I–V) experimentally determined Ssy5 prodomain states leading to SPS sensor–induced Stp1/2-processing activity of the Ssy5 Cat-domain.
The integrated chain of events that activates the phosphodegron on SPS sensor induction can be summarized in a simple model with five prodomain states (Figure 6B). Accordingly, the catalytic activity of the Ssy5 protease is inhibited by the noncovalently associated prodomain in its inhibitory state (I). On amino acid induction, the prodomain receives signals from upstream SPS sensor components Ssy1 and Ptr3, and switches to a signaling conformation (II). Adoption of the signaling conformation exposes the phosphodegron leading to Yck1/2-mediated phosphorylation on multiple sites within the phosphoacceptor motif (III). Phosphorylation is presumably counteracted by phosphatase PP2A with its regulatory subunit Rts1 (Eckert-Boulet et al., 2006; Liu et al., 2008), which, under nonsignaling conditions, may maintain the prodomain in the hypophosphorylated state (II). The shift in the balance between phosphorylation and dephosphorylation rates serves as a switchlike trigger for recognition by the ubiquitin E3 ligase complex SCFGrr1, which recognizes the hyperphosphorylated phosphodegron and directly ubiquitylates at least one of the lysine residues (IV). Concomitantly, the polyubiquitylated prodomain (V) is degraded by the 26S proteasome, unfettering the catalytic activity of the Ssy5 Cat-domain.
The biological significance of the proposed model is based on several lines of key experimental evidence. We have physical evidence for four of five prodomain states (I, III–V) and have genetic evidence to infer the existence of the state II conformation. First, the inhibitory Ssy5 prodomain state I can be detected in extracts from noninduced cells (Abdel-Sater et al., 2004; Andréasson et al., 2006; Poulsen et al., 2006). The stable phosphorylated and ubiquitylated prodomain states, III and IV, can be purified from induced cells carrying downstream blocking mutations (i.e., grr1Δ cells and proteasome inhibited cells, respectively) (Figure 1 and Figure 3; Abdel-Sater et al., 2011). Similarly, upon amino acid induction, we can observe rapid prodomain degradation (V) and stable Cat-domain levels (Andréasson et al., 2006; Pfirrmann et al., 2010). Second, the existence of an unmodified signaling conformation of the prodomain (II) is based on finding that specific mutations in the prodomain constitutively activate the protease (Pfirrmann et al., 2010). Importantly, these mutations bypass the upstream component Ssy1, but not the downstream activity of Yck1/2 and Grr1 (Figure 3); hence, the mutant prodomains must adopt an intermediate signaling conformation (II) that is distinct from state I. Third, we have evidence for direct physical interactions between the required signaling factors, i.e.,Yck1 and Grr1, and the Ssy5 prodomain (Figures 2 and 3). Finally, mutations affecting the phosphoacceptor sites or the adjacent lysine residues within the phosphodegron impair prodomain degradation, and consequently prevent amino acid–induced and SPS sensor–mediated gene expression (Figures 4 and 5).
According to our model, the phosphodegron of Ssy5 is the recognition determinant for the general signaling factors Yck1/2 and SCFGrr1, raising the possibility that other inducible proteasomal substrates carry similar modules. Strikingly, we find that the phosphoacceptor motif in the phosphodegron of Ssy5 exhibits sequence similarity to the regulatory degron sequences found in the two transcriptional repressors Mth1 and Std1 (Figure 6A, sequence alignment). Mth1 and Std1 are the effector components of the yeast Snf3/Rgt2 glucose-sensing pathway. On glucose induction, Std1 and Mth1 undergo SCFGrr1-dependent degradation, resulting in derepression of glucose transporter gene expression (Flick et al., 2003; Lakshmanan et al., 2003). Although functionally and mechanistically distinct from the SPS signaling pathway, the Snf3/Rgt2 glucose-sensing pathway shares certain characteristics, including primary transporter-like sensors and a requirement for Yck1/2 and SCFGrr1 (Johnston and Kim, 2005). The sequence similarity to the regulatory motifs comprising Yck1/2 consensus phosphorylation acceptor serines in Ssy5, Mth1, and Std1 (Moriya and Johnston, 2004) provides an explanation for how these discrete pathways interact with the same signaling factors. Accordingly, casein kinase I and SCFGrr1, which are known to participate as constitutively active signaling components in a variety of cellular processes (Gross and Anderson, 1998; Willems et al., 2004; Knippschild et al., 2005; Jonkers and Rep, 2009), are recruited to distinct signaling pathways by their recognition of inducible and regulated phosphodegrons. This concept raises the possibility that we have identified a phosphodegron that, in a context-dependent manner, can be controlled by several different pathways that generate highly distinct signaling outputs.
All our data are consistent with the model that proteasomal degradation of the prodomain activates the Ssy5 protease. However, the direct involvement of the 26S proteasome in the activation of Ssy5 has recently been questioned. In a recent study, Abdel-Sater et al. propose that prodomain ubiquitylation (state IV) rather than its degradation (state V) is the Ssy5-activating step (Abdel-Sater et al., 2011). This alternative model is based on two observations: Stp1 processing appears to precede prodomain degradation, and Stp1 processing occurs in cells with inhibited proteasome function. We note, however, that neither of these two observations exclude the notion of a critical involvement of the proteasome in Ssy5 activation. First, Stp1 processing activity of the Cat-domain is catalytic (i.e., a small amount of prodomain degradation can generate a substantial amount of processed Stp1). Accordingly, the level of processed Stp1 is predicted to increase before substantial down-regulation of the prodomain. Second, residual proteolytic activity is known to persist in all experimental setups used to decrease proteasome function (i.e., mutational inactivation and MG132 inhibition; Ghislain et al., 1993; Hoppe et al., 2000; Andréasson and Ljungdahl, 2002). As discussed in a previous report, the residual proteasome activity can readily account for the observed Stp1-processing activity (Pfirrmann et al., 2010). Moreover, both unprocessed and processed forms of Stp1 are proteasomal substrates (Pfirrmann et al., 2010; Tumusiime et al., 2011). Consequently, under conditions where the proteasome is inhibited, both forms of Stp1 accumulate. Thus the fact that processed Stp1 accumulates can misleadingly provide the basis for overestimating Ssy5 activity. In summary, although our data do not formally exclude the possibility that polyubiquitylation of the prodomain can suffice to initiate downstream signaling, all available experimental evidence support a model of Ssy5 activation requiring proteasomal degradation of the prodomain.
Pathways involving multiple proteases are frequently organized in cascades (i.e., upstream proteases endoproteolytically activate downstream proteases in a sequential manner). Prominent examples are the blood-clotting cascade (Page et al., 2005), caspases in apoptosis (Boatright and Salvesen, 2003), and proteases in digestion (Stroud et al., 1977). In this study we have defined a novel proteolytic activation cascade with the ubiquitin proteasome system as a central component. To our knowledge, the finding that phosphorylation-dependent proteolysis by the proteasome activates an intracellular signaling protease is novel. A predicating example of the ubiquitin proteasome system activating an intracellular protease, however, is the protease separase (Esp1). Separase is inhibited by its binding to securin (Pds1). Ubiquitylation of securin by the anaphase-promoting complex APC-Cdc20 triggers its subsequent degradation by the proteasome. The degradation of securin liberates separase activity, leading to the degradation of cohesin that links sister chromatids during cell division (Ciosk et al., 1998; Uhlmann et al., 1999; Nasmyth, 2005). Aberrant regulation of separase activity is connected to missegregation and increased loss of chromosomes, a phenomenon frequently found in cancer cells (Nasmyth, 2002). The crucial involvement of proteases in fundamental regulatory events in eukaryotes underlines the importance of studying the general principles of their activation mechanisms.
MATERIALS AND METHODS
Strains and plasmids
The Saccharomyces cerevisiae strains and plasmids used in this work are listed in Table 1 and Table 2, respectively. The yeast strains are isogenic descendants of the S288C-derived strain AA255/PLY115 (Antebi and Fink, 1992), with the exception of the two-hybrid strain CAY235. The sequences of mutagenic oligonucleotides and PCR primers for homologous recombination are available upon request.
TABLE 1:
Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| CAY86 | MATa ura3-52 grr1Δ50::hphMX4 | Andréasson and Ljungdahl, 2002 |
| CAY235 | MATα his3 trp1-100 ura3-52 leu2Δ::6×lexAOp-LEU2 lys2Δ::URA3-p8×lexAOp-LYS2-GAL1 GAL2 | This work |
| CAY267 | MATa ura3-52 ssy1Δ13::hisG grr1Δ50::hphMX4 | This work |
| CAY276 | MATa lys2Δ201 ura3-52 ssy5Δ2::hisG grr1Δ50::hphMX4 | This work |
| CAY285 | MATa lys2Δ201 ura3-52 ptr3Δ15::hisG ssy1Δ13::hisG ssy5Δ2::hisG | Andréasson et al., 2006 |
| CAY320 | MATa lys2Δ201 ura3-52 yck1Δ::hphMX4 yck2-1 | This work |
| HKY77 | MATa lys2Δ201 ura3-52 ssy5Δ2::hisG | Forsberg and Ljungdahl, 2001 |
| HKY84 | MATa lys2Δ201 ura3-52 ssy1Δ13::hisG ssy5Δ2::hisG | Forsberg and Ljungdahl, 2001 |
| HKY85 | MATa lys2Δ201 ura3-52 ptr3Δ15::hisG ssy5Δ2::hisG | Forsberg and Ljungdahl, 2001 |
| TPY116 | MATa lys2Δ201 ura3-52 ssy5Δ2::hisG pdr5Δ3 | This work |
| TPY117 | MATa lys2Δ201 ura3-52 ssy5Δ2::hisG grr1Δ pdr5Δ3 | This work |
| TPY120 | MATa lys2Δ201 ura3-52 ssy1Δ13::hisG ssy5Δ2::hisG grr1::hphMX4 | This work |
TABLE 2:
Plasmids used in this study.
| Plasmid | Description | Reference |
|---|---|---|
| pCA146 | 2μm HIS3 KmR G418R containing PADH-LexA | This work |
| pCA195 | pRS316 (URA3) containing HA-SSY5 | Andréasson et al., 2006 |
| pCA204 | pRS317 (LYS2) containing STP1-MYC-kanMX | Andréasson et al., 2006 |
| pDO067 | pRS316 (URA3) containing HAi-ssy5-62-GST | This work |
| pDO068 | pRS316 (URA3) containing HAi-SSY5-63-GST | This work |
| pDO069 | pRS316 (URA3) containing HAi-ssy5-64-GST | This work |
| pDO072 | pRS316 (URA3) containing HAi-ssy5-65-GST | This work |
| pDO082 | pRS316 (URA3) containing HAi-ssy5-67-GST | This work |
| pDO083 | pRS316 (URA3) containing HAi-SSY5-68-GST | This work |
| pDO099 | pRS316 (URA3) containing HAi-SSY5-yck12-527K98R | This work |
| pDO100 | pRS316 (URA3) containing HAi-SSY5-YCK12-527 | This work |
| pHK048 | pRS316 (URA3) containing MYC-SSY5 | Forsberg and Ljungdahl, 2001 |
| pJG4-5 | yeast two hybrid prey vector (TRP1) | Gyuris et al., 1993 |
| pSH119 | pRS316 (URA3) containing HAi-SSY591-699-GST | Pfirrmann et al., 2010 |
| pSH120 | pRS316 (URA3) containing HAi-SSY5-GST | Pfirrmann et al., 2010 |
| pTP110 | pRS316 (URA3) containing Ubi-Rds-tsDHFR-HAi-SSY5-GST | Pfirrmann et al., 2010 |
| pTP112 | pRS316 (URA3) containing HAi-SSY5-L126S,V129A,E131A-GST | Pfirrmann et al., 2010 |
| pTP115 | pRS316 (URA3) containing HAi-SSY5-E131A-GST | Pfirrmann et al., 2010 |
| pTP122 | pJG4-5 (TRP1) containing PGAL1-AD-HA-GRR12-1152 | This work |
| pTP133 | 2μm HIS3 KmR G418R containing PADH–LexA-SSY5-L126S,V129A,E131A | This work |
| pTP134 | 2μm HIS3 KmR G418R containing PADH-LexA-SSY5 | This work |
| pTP135 | pRS316 (URA3) containing HAi-SSY5-K9R-GST | This work |
| pTP140 | pRS316 (URA3) containing HAi-SSY5-K5-8R,K10-22R-GST | This work |
| pTP142 | pRS316 (URA3) containing HAi-SSY5-K1-8R,K10-22R-GST | This work |
| pTP143 | pRS316 (URA3) containing HAi-ssy5-K5-22R-GST | This work |
| pTP144 | pRS316 (URA3) containing HAi-ssy5-K1-22R- GST | This work |
| pTP145 | pRS316 (URA3) containing HAi-ssy5-K5-9R-GST | This work |
| pTP149 | pRS316 (URA3) containing Ubi-Rds-tsDHFR-HAi-SSY5-K1-22R-GST | This work |
| pTP156 | YEp195 based (URA3 replaced with LYS2) containing PCUP1-6HIS-UBI | This work |
Media
Standard media, including YPD medium; ammonia-based synthetic minimal dextrose (SD) medium, supplemented as required to enable growth of auxotrophic strains; and ammonia-based synthetic complete dextrose (SC) were prepared as described (Andréasson and Ljungdahl, 2002). When needed, l-leucine was added at a concentration of 1.3 mM for 30 min to induce SPS sensor signaling. Sensitivity to MM (100 μg/ml) was monitored on complex medium as described (Andréasson and Ljungdahl, 2004).
Immunoblot analysis
Whole-cell extracts were prepared under denaturing conditions using NaOH and trichloroacetic acid (TCA) as described previously (Silve et al., 1991). Primary antibodies were diluted (vol:vol) as follows: 3F10 anti-HA-horseradish peroxidase (HRP) (Roche Applied Science, Basel, Switzerland), 1:5000; anti–myc-HRP 9E10 mAb (Roche Applied Science), 1:1000; anti-Pgk1 (Molecular Probes, Eugene, OR), 1:5000; anti Penta-His-HRP, 1:1000 (Qiagen, Chatsworth, CA). Immunoreactive bands were visualized by chemiluminescence detection (SuperSignal West Dura Extended-Duration Substrate; Pierce, Rockford, IL) and quantified using an LAS1000 system (Fuji Photo Film Co., Tokyo, Japan).
Dephosphorylation assay
Proteins were immunoprecipitated (Anti-c-Myc-Agarose Affinity Gel; Sigma, St. Louis, MO) from extracts obtained using glass bead lysis in IP-1 buffer (50 mM Tris-HCl, pH 7.5; 250 mM NaCl) containing PhosSTOP (Roche) and Complete Mini EDTA free (Roche). Beads were washed twice with IP-1 buffer and once with dephosphorylation buffer (10 mM Tris-HCl pH 8; 5 mM MgCl2; 0.1 M KCl; 0.02% Triton X-100). Alternatively, cells were lysed in 5% TCA and neutralized with Tris (1 M), and precipitated material was resolubilized in four volumes of IP-2 buffer (50 mM Tris, pH 7.5; 100 mM NaCl; 2 mM EDTA; 1.2% Triton X-100; 1 mM iodoacetic acid) containing PhosSTOP and Complete Mini EDTA free. Immunopurification of HA-tagged Ssy5 was carried out using HA Affinity Matrix (Roche), beads were washed twice with 10 ml of wash buffer (20 mM Tris, pH 7.5; 1 M NaCl), and bound material was eluted by incubating beads two times at 37°C for 15 min in the presence of 250 μl of elution buffer (20 mM Tris, pH 7.5; 0.1 M NaCl) containing 1 mg/ml HA peptide (Roche). Dephosphorylation reactions were performed in 100 μl of dephosphorylation buffer without bovine serum albumin with 10 μl of phosphatase (FastAP, Fermentas Sweden, Helsingborg, Sweden).
CHX chase
Cells were grown to log phase, and CHX was added to the culture at a concentration of 100 μg/ml. Where indicated, leucine was added to a concentration of 1.3 mM (+ leu). Extracts were prepared from samples taken at the time points indicated and were analyzed by immunoblotting.
Directed yeast two-hybrid assay
Two-hybrid interactions between LexA fused to full length Ssy5 variants (pTP133 and pTP134) and an activation domain (AD) fused to full-length Grr1 (pTP122) were tested in strain CAY235. Interactions were assayed for growth on SCD plates lacking tryptophan, histidine, leucine, and lysine, and containing 2% galactose and 1% raffinose as carbon sources.
In vivo polyubiquitylation assay
Ubiquitylated prodomain was affinity purified and detected as described previously (Iglesias et al., 2010). Briefly, 80 OD600 of cells per sample were incubated (30 min) in the presence of 100 μM MG132 (Sigma-Aldrich, St. Louis, MO). Cells were lysed in 10% TCA using glass beads. Precipitated protein was washed with 1 M Tris, resuspended in 1 ml of Buffer A (6 M guanidium HCl; 20 mM Tris, pH 8; 100 mM K2HPO4, 20 mM imidazole, 100 mM NaCl, 0.1% Triton X-100), resolubilized for 1 h (room temperature), then ubiquitylated proteins were purified using 100 μl of NiNTA beads (Qiagen). Beads were washed three times with Buffer A, three times with wash buffer 1 (20 mM Tris, pH 8; 100 mM K2HPO4, 20 mM imidazole, 100 mM NaCl, 0.1% Triton X-100), and three times with wash buffer 2 (20 mM Tris, pH 8; 100 mM K2HPO4, 10 mM imidazole, 1 M NaCl, 0.1% Triton X-100). Bound material was eluted three times with 200 μl of elution buffer (20 mM Tris, pH 8; 100 mM K2HPO4, 500 mM imidazole, 100 mM NaCl) for 10 min.
Acknowledgments
We thank the members of the Ljungdahl laboratory for constructive comments throughout the course of this work and D.H. Wolf (University of Stuttgart, Germany) for plasmids. Nicole Meseberg is gratefully acknowledged for experimental assistance. This research was supported by the Center for Biomembrane Research, Stockholm University (to C.A. and P.O.L.) and by funding from the Swedish Research Council (to P.O.L.).
Abbreviations used:
- CHX
cycloheximide
- HA
hemagglutinin
- HRP
horseradish peroxidase
- MM
(2-{[({[(4-methoxy-6-methyl)-1,3,5-triazin-2-yl]-amino}carbonyl)amino-]-sulfonyl}-benzoic acid
- RAP
receptor-activated proteolysis
- SC
synthetic complete dextrose
- SCF
Skp1/Cdc53/F-box protein
- SD
synthetic minimal dextrose
- SPS
Ssy1-Ptr3-Ssy5
- TCA
trichloroacetic acid
- WT
wild type
- YPD
yeast extract–peptone–dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0282) on June 8, 2011.
REFERENCES
- Abdel-Sater F, El Bakkoury M, Urrestarazu A, Vissers S, André B. Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol Cell Biol. 2004;24:9771–9785. doi: 10.1128/MCB.24.22.9771-9785.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Sater F, Jean C, Merhi A, Vissers S, André B. Amino-acid signalling in yeast: activation of the Ssy5 protease is associated with its phosphorylation-induced ubiquitylation. J Biol Chem. 2011;286:12006–12015. doi: 10.1074/jbc.M110.200592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Heessen S, Ljungdahl PO. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 2006;20:1563–1568. doi: 10.1101/gad.374206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Ljungdahl PO. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 2002;16:3158–3172. doi: 10.1101/gad.239202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson C, Ljungdahl PO. The N-terminal regulatory domain of Stp1p is modular and, fused to an artificial transcription factor, confers full Ssy1p-Ptr3p-Ssy5p sensor control. Mol Cell Biol. 2004;24:7503–7513. doi: 10.1128/MCB.24.17.7503-7513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A, Fink GR. The yeast Ca2+-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol Biol Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard F, André B. Ubiquitin and the SCFGrr1 ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 2001;496:81–85. doi: 10.1016/s0014-5793(01)02412-7. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Brivanlou AH, Darnell JE., Jr Signal transduction and the control of gene expression. Science. 2002;295:813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Eckert-Boulet N, Larsson K, Wu B, Poulsen P, Regenberg B, Nielsen J, Kielland-Brandt MC. Deletion of RTS1, encoding a regulatory subunit of protein phosphatase 2A, results in constitutive amino acid signaling via increased Stp1p processing. Eukaryot Cell. 2006;5:174–179. doi: 10.1128/EC.5.1.174-179.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick KM, Spielewoy N, Kalashnikova TI, Guaderrama M, Zhu Q, Chang HC, Wittenberg C. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol Biol Cell. 2003;14:3230–3241. doi: 10.1091/mbc.E03-03-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ. Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem. 1990;265:14264–14269. [PubMed] [Google Scholar]
- Forsberg H, Ljungdahl PO. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol Cell Biol. 2001;21:814–826. doi: 10.1128/MCB.21.3.814-826.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, Dargemont C, Stutz F. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010;24:1927–1938. doi: 10.1101/gad.583310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingrell CR, Miller ML, Jensen ON, Blom N. NetPhosYeast: prediction of protein phosphorylation sites in yeast. Bioinformatics. 2007;23:895–897. doi: 10.1093/bioinformatics/btm020. [DOI] [PubMed] [Google Scholar]
- Iraqui I, Vissers S, Bernard F, de Craene JO, Boles E, Urrestarazu A, André B. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol. 1999;19:989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Kim JH. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem Soc Trans. 2005;33:247–252. doi: 10.1042/BST0330247. [DOI] [PubMed] [Google Scholar]
- Jonkers W, Rep M. Lessons from fungal F-box proteins. Eukaryot Cell. 2009;8:677–695. doi: 10.1128/EC.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen MU, Bruun MB, Didion T, Kielland-Brandt MC. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast. 1998;14:103–114. doi: 10.1002/(SICI)1097-0061(19980130)14:2<103::AID-YEA203>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Klasson H, Fink GR, Ljungdahl PO. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol Cell Biol. 1999;19:5405–5416. doi: 10.1128/mcb.19.8.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Lakshmanan J, Mosley AL, Özcan S. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr Genet. 2003;44:19–25. doi: 10.1007/s00294-003-0423-2. [DOI] [PubMed] [Google Scholar]
- Liu Z, Thornton J, Spirek M, Butow RA. Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol Cell Biol. 2008;28:551–563. doi: 10.1128/MCB.00929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H, Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci USA. 2004;101:1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. How do so few control so many? Cell. 2005;120:739–746. doi: 10.1016/j.cell.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Page MJ, Macgillivray RT, Di Cera E. Determinants of specificity in coagulation proteases. J Thromb Haemost. 2005;3:2401–2408. doi: 10.1111/j.1538-7836.2005.01456.x. [DOI] [PubMed] [Google Scholar]
- Pfirrmann T, Heessen S, Omnus DJ, Andréasson C, Ljungdahl PO. The prodomain of Ssy5 protease controls receptor-activated proteolysis of transcription factor Stp1. Mol Cell Biol. 2010;30:3299–3309. doi: 10.1128/MCB.00323-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen P, Lo Leggio L, Kielland-Brandt MC. Mapping of an internal protease cleavage site in the Ssy5p component of the amino acid sensor of Saccharomyces cerevisiae and functional characterization of the resulting pro- and protease domains by gain-of-function genetics. Eukaryot Cell. 2006;5:601–608. doi: 10.1128/EC.5.3.601-608.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silve S, Volland C, Garnier C, Jund R, Chevallier MR, Haguenauer-Tsapis R. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol Cell Biol. 1991;11:1114–1124. doi: 10.1128/mcb.11.2.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielewoy N, Flick K, Kalashnikova TI, Walker JR, Wittenberg C. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol Cell Biol. 2004;24:8994–9005. doi: 10.1128/MCB.24.20.8994-9005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielewoy N, Guaderrama M, Wohlschlegel JA, Ashe M, Yates JR, 3rd, Wittenberg C. Npr2, yeast homolog of the human tumor suppressor NPRL2, is a target of Grr1 required for adaptation to growth on diverse nitrogen sources. Eukaryot Cell. 2010;9:592–601. doi: 10.1128/EC.00192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud RM, Kossiakoff AA, Chambers JL. Mechanisms of zymogen activation. Annu Rev Biophys Bioeng. 1977;6:177–193. doi: 10.1146/annurev.bb.06.060177.001141. [DOI] [PubMed] [Google Scholar]
- Tumusiime S, Zhang C, Overstreet MS, Liu Z. Differential regulation of transcription factors Stp1 and Stp2 in the Ssy1-Ptr3-Ssy5 amino acid sensing pathway. J Biol Chem. 2011;286:4620–4631. doi: 10.1074/jbc.M110.195313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Vancura A, Sessler A, Leichus B, Kuret J. A prenylation motif is required for plasma membrane localization and biochemical function of casein kinase I in budding yeast. J Biol Chem. 1994;269:19271–19278. [PubMed] [Google Scholar]
- Wang PC, Vancura A, Mitcheson TG, Kuret J. Two genes in Saccharomyces cerevisiae encode a membrane-bound form of casein kinase-1. Mol Biol Cell. 1992;3:275–286. doi: 10.1091/mbc.3.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielemans K, Jean C, Vissers S, Andre B. Amino acid signaling in yeast: post-genome duplication divergence of the Stp1 and Stp2 transcription factors. J Biol Chem. 2010;285:855–865. doi: 10.1074/jbc.M109.015263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Wolf DH. From lysosome to proteasome: the power of yeast in the dissection of proteinase function in cellular regulation and waste disposal. Cell Mol Life Sci. 2004;61:1601–1614. doi: 10.1007/s00018-004-4134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Ottow K, Poulsen P, Gaber RF, Albers E, Kielland-Brandt MC. Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J Cell Biol. 2006;173:327–331. doi: 10.1083/jcb.200602089. [DOI] [PMC free article] [PubMed] [Google Scholar]