Abstract
Long-chain n-3 polyunsaturated fatty acids (PUFAs) can derive from diet or from α-linolenic acid (ALA) by elongation and desaturation. We investigated the association of common genetic variation with plasma phospholipid levels of the four major n-3 PUFAs by performing genome-wide association studies in five population-based cohorts comprising 8,866 subjects of European ancestry. Minor alleles of SNPs in FADS1 and FADS2 (desaturases) were associated with higher levels of ALA (p = 3×10−64) and lower levels of eicosapentaenoic acid (EPA, p = 5×10−58) and docosapentaenoic acid (DPA, p = 4×10−154). Minor alleles of SNPs in ELOVL2 (elongase) were associated with higher EPA (p = 2×10−12) and DPA (p = 1×10−43) and lower docosahexaenoic acid (DHA, p = 1×10−15). In addition to genes in the n-3 pathway, we identified a novel association of DPA with several SNPs in GCKR (glucokinase regulator, p = 1×10−8). We observed a weaker association between ALA and EPA among carriers of the minor allele of a representative SNP in FADS2 (rs1535), suggesting a lower rate of ALA-to-EPA conversion in these subjects. In samples of African, Chinese, and Hispanic ancestry, associations of n-3 PUFAs were similar with a representative SNP in FADS1 but less consistent with a representative SNP in ELOVL2. Our findings show that common variation in n-3 metabolic pathway genes and in GCKR influences plasma phospholipid levels of n-3 PUFAs in populations of European ancestry and, for FADS1, in other ancestries.
Author Summary
Circulating long-chain n-3 polyunsaturated fatty acids (PUFAs) derive from fatty fish or from the conversion of the plant n-3 PUFA by elongation and desaturation. We looked for common genetic markers throughout the genome that might influence plasma phospholipid levels of the four major n-3 PUFAs in five large studies and pooled the results. We found that levels of all four n-3 PUFAs were associated with genetic markers in known desaturation and elongation genes. We also found evidence that conversion of the plant n-3 PUFA to longer chain n-3 PUFAs is less effective in people with certain desaturation-gene markers, which could be important for people who do not eat fish. We also found a marker in a gene involved in glucose metabolism, called the glucokinase regulator, to be associated with one intermediate n-3 PUFA. Some of these findings were seen across multiple race/ethnicities. Overall, these results have implications for how genes and the environment interact to influence circulating levels of fatty acids.
Introduction
High levels of n-3 polyunsaturated fatty acids (PUFA) in plasma phospholipids, cell membranes, and whole blood have been associated with lower risk of multiple diseases, including sudden cardiac death [1], fatal coronary heart disease [2], non-fatal myocardial infarction [3], heart failure [4], thickening of the carotid arteries [5], metabolic syndrome [6], breast cancer [7], chronic obstructive pulmonary disease [8], depression [9], [10] and dementia [11], [12].
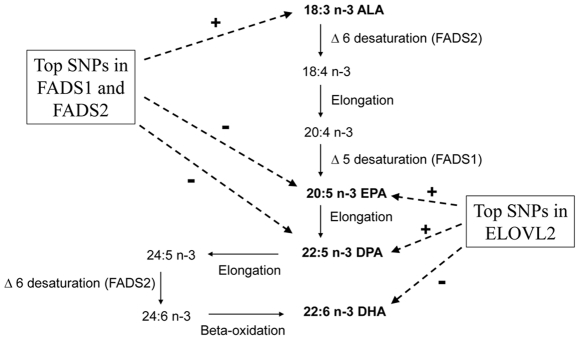
N-3 PUFAs are derived directly from the diet, including the plant-derived essential fatty acid α-linolenic acid (ALA, 18:3n3) and the seafood-derived long-chain n-3 PUFAs eicosapentaenoic acid (EPA, 20:5n3) and docosahexaenoic acid (DHA, 22:6n3) [13], [14]. Long-chain n-3 PUFAs can also be produced from ALA by the series of desaturation and elongation steps in the pathway shown in Figure 1; docosapentaenoic acid (DPA, 22:5n-3) can be produced from EPA. The pathway enzymes may be a major source of circulating long-chain n-3 PUFAs in people who consume very little or no seafood. However, the conversion of ALA to EPA and DHA has been shown to be generally low [15]–[17], and it is not known whether common genetic variation in the pathway affects this conversion. The n-6 essential fatty acid linoleic acid (LA) is elongated to long-chain n-6 PUFAs by the same pathway enzymes and could also compete with the conversion of ALA to EPA; it is not known whether genetic variation affects such competition.
Figure 1. N-3 polyunsaturated fatty acid metabolic pathway and summary of genome-wide associations in pathway.
The fatty acids indicated in bold were examined in this study. The genome-wide significant associations of two loci with each fatty acid are shown with dashed arrows. + and − signs indicate the direction of the associations.
There is evidence of co-heritability of EPA and DHA levels in erythrocyte membrane phospholipids [18]. Investigation of genetic factors influencing PUFA levels has largely focused on candidate genes, such as the desaturase genes FADS1 and FADS2, among participants of European ancestry [19]–[25]. Only one prior study reported a genome-wide association of n-3 PUFA levels evaluating total plasma n-3 PUFAs which includes triacylglycerols, phospholipids and free fatty acids, among 1075 participants [26]. The study found an association of EPA with variants in the FADS1 gene that reached genome-wide significance level; independent follow-up investigation showed associations of a selected FADS1 variant with erythrocyte membrane levels of EPA, ALA and DPA and of an ELOVL2 variant with DPA and DHA. These findings confirm an influence of FADS1 and ELOVL2 on selected n-3 PUFAs. However, statistical power may have been limited to confirm an influence of these genes on all four major n-3 PUFAs, of other genes in these pathways (e.g., FADS2), or of additional genes in other unknown biologic pathways. Prior studies have also not had adequate power to evaluate potential interaction between variation in these genes, EPA and DHA, and: (a) diet, (b) ALA levels, and (c) LA levels. In addition, there is limited information on genetic variation and n-3 PUFA levels in subjects of non-European ancestry.
To understand how common genetic variation affects n-3 PUFA phospholipid levels and potentially uncover novel associations, we conducted a meta-analysis of pre-planned genome-wide association analyses of plasma phospholipid n-3 PUFAs in 8,866 participants of European ancestry in five population-based studies, as part of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium [27]. We evaluated the four main n-3 PUFAs of the metabolic pathway, (ALA, EPA, DPA, and DHA) separately and accounted for the intercorrelations between these fatty acids. In addition, we investigated whether consumption of fatty fish or phospholipid levels of ALA and LA influenced the association of the identified genetic markers with EPA and DHA levels. Finally, we studied the most highly associated SNPs from the meta-analyses among samples of European ancestry in additional samples from African, Chinese and Hispanic ancestry.
Results
Meta-analysis of genome-wide associations of n-3 fatty acids
The study samples for the genome-wide association study (GWAS) comprised a total of 8,866 subjects of European ancestry. Table 1 shows sample size, demographic characteristics, fish consumption and phospholipid n-3 PUFA levels in the 5 cohorts that contributed to the study. Participants ranged from 21 to 102 years of age. Across the cohorts, mean levels of ALA varied from 0.14% to 0.44% of total fatty acids; EPA from 0.56% to 1.01%; DPA from 0.83% to 0.98%; and DHA from 2.29% to 5.09%. Relatively higher ALA levels were seen in the InCHIANTI cohort, likely reflecting differences in the composition of total plasma fatty acids (InCHIANTI) versus phospholipid fatty acids (ARIC, CARDIA, CHS, and MESA).[28]
Table 1. CHARGE cohorts descriptives.* .
| Plasma phospholipid concentration, % of total fatty acids | ||||||||
| Cohort/Ancestry | N | Age | Women % | Fish intake¥ | ALA† | EPA† | DPA† | DHA† |
| ARIC/European‡ | 3268 | 53.8 (5.6) | 51.0 | 1.2 (0–24) | 0.14 (0.05) | 0.56 (0.30) | 0.90 (0.17) | 2.82 (0.88) |
| CHS/European‡ | 2326 | 72.0 (5.1) | 61.3 | 2.0 (0–14) | 0.15 (0.05) | 0.59 (0.37) | 0.83 (0.17) | 2.97 (0.96) |
| CHS/African | 427 | 72.9 (5.5) | 68.3 | NA | 0.14 (0.05) | 0.56 (0.29) | 0.84 (0.17) | 3.52 (0.93) |
| InCHIANTI/European‡ | 1075 | 68.4 (15.5) | 54.9 | 1.6 (0–12) | 0.44 (0.25) | 0.61 (0.22) | NA | 2.29 (0.77) |
| CARDIA/European‡ | 1507 | 45.8 (3.4) | 53.3 | 1.0 (0–31) | 0.19 (0.09) | 0.85 (0.62) | 0.94 (0.21) | 3.09 (1.12) |
| CARDIA/African | 493 | 44.6 (3.8) | 60.7 | 1.2 (0–18) | 0.17 (0.08) | 0.67 (0.41) | 0.92 (0.21) | 3.27 (0.99) |
| MESA/European‡ | 690 | 61.6 (10.4) | 53.3 | 0.6 (0–11) | 0.18 (0.06) | 0.88 (0.58) | 0.95 (0.21) | 3.66 (1.38) |
| MESA/African | 631 | 61.6 (9.8) | 54.2 | 0.6 (0–14) | 0.16 (0.06) | 0.89 (0.56) | 0.98 (0.22) | 4.57 (1.42) |
| MESA/Chinese | 646 | 62.1 (10.4) | 50.8 | 0.8 (0–11) | 0.18 (0.07) | 1.01 (0.65) | 0.96 (0.24) | 5.09 (1.46) |
| MESA/Hispanic | 672 | 61.0 (10.0) | 53.7 | 0.2 (0–7) | 0.17 (0.06) | 0.64 (0.41) | 0.89 (0.19) | 3.34 (1.23) |
Descriptive characteristics of the cohorts of European ancestry included in the GWAS analyses and the cohorts of African, Chinese, and Hispanic ancestry included in the association analyses of selected SNPs.
*Values are mean (SD) unless specified otherwise.
‡Included in the meta-analyses of GWAS.
¥ Servings per week, median (range).
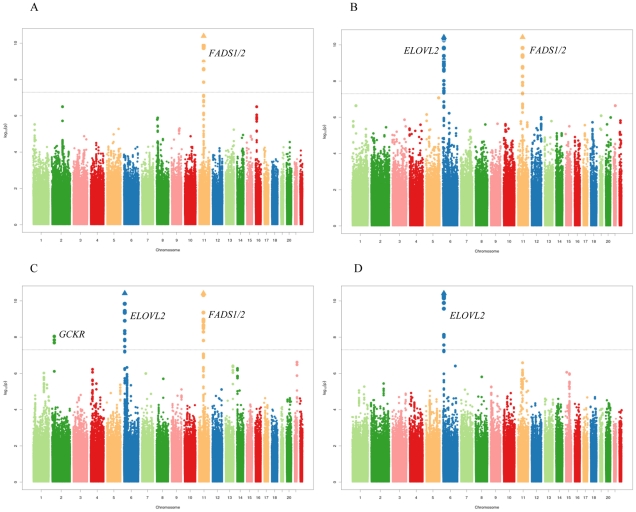
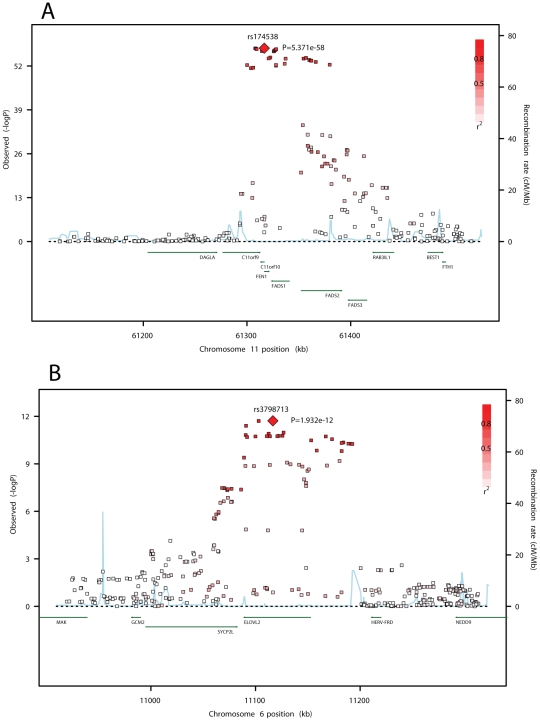
Figure 2A–2D show the meta-analysis of the genome-wide association results for ALA, EPA, DPA, and DHA. Variation in one or both of two major genetic loci was associated with plasma phospholipid levels of each n-3 PUFA at genome-wide levels of significance. The two loci are illustrated in association plots for EPA (Figure 3). One locus, on chromosome 11q12.2, contained the C11orf9/10, FEN1 and the desaturase genes FADS1, FADS2 and FADS3. The other locus, on chromosome 6p24.2, contained SYPC2L and the elongase gene ELOVL2. Many highly correlated SNPs reached genome-wide significance in the associations with n-3 PUFAs (Table 2 and Table S1A–S1D). Variant alleles at SNPs in the chromosome 11 locus were associated with higher levels of ALA and lower levels of EPA and DPA, and variant alleles at SNPs in the chromosome 6 locus were associated with higher levels of EPA and DPA and lower levels of DHA (Table 2, Table 3, Table 4). From the meta-analysis results, we estimated that the most highly associated SNPs on chromosome 11 explained 3.8% of the variance of ALA, 2.0% of the variance of EPA and 8.6% of the variance of DPA. The most highly associated SNPs on chromosome 6 explained 0.4% of the variance of EPA, 2.8% of the variance in DPA and 0.7% of the variance in DHA.
Figure 2. Meta-analysis of genome-wide associations with n-3 polyunsaturated fatty acids.
A: α-linolenic acid (ALA), B: eicosapentaenoic acid (EPA), C: docosapentaenoic acid (DPA), D: docosahexaenoic acid (DHA). Associations were graphed by chromosome position and –log10 (p-value) up to p-values of 10−10. Triangles indicate additional SNPs with p-values less than 10−10. Genes of interest in the regions with significantly associated SNP variants are indicated.
Figure 3. SNP association plots for EPA–associated regions.
Genome-wide association significance level is plotted against the y-axis as –log10 (p-value). Genetic coordinates are as per NCI build 36. A) FADS cluster region. LD is indicated by color scale in relationship to marker rs174538. B) ELOVL2 region. LD is indicated by color scale in relationship to marker rs3798713. The color scheme is red for strong linkage disequilibrium (LD; r2≥0.8) and fading color for lower LD.
Table 2. Loci associated with SNP markers with P-values <5.0×10−8.
| Most significant SNP | ||||||
| Fatty acid (region) | Analysis adjustments | Number of SNPs| | SNP*, allele | MAF | P-value | Parameter Coefficient (CI) |
| ALA (11) | Main model† | 75 | rs174547,C/T | 0.33 | 4×10−64 | 0.016 (0.014, 0.018) |
| ALA (11) | EPA‡ | 86 | rs174547,C/T | 0.33 | 7×10−84 | 0.019 (0.017, 0.021) |
| DPA (2) | Main model† | 3 | rs780094, T/C | 0.41 | 9×10−09 | 0.017 (0.011, 0.022) |
| DPA (6) | Main model† | 85 | rs3734398, C/T | 0.43 | 1×10−43 | 0.040 (0.035, 0.046) |
| DPA (6) | rs2236212, rs174547‡ | 22 | rs12662634, A/G | 0.18 | 3×10−10 | −0.023 (−0.039, −0.021) |
| DPA (11) | Main model† | 97 | rs174547,C/T | 0.33 | 4×10−154 | −0.075 (−0.069, −0.080) |
| EPA(6) | Main model† | 47 | rs3798713, C/G | 0.43 | 2×10−12 | 0.035 (0.025, 0.045) |
| EPA(6) | DHA‡ | 78 | rs3798713, C/G | 0.43 | 8×10−35 | 0.054 (0.045, 0.062) |
| EPA(6) | ALA‡ | 77 | rs3798713, C/G | 0.43 | 3×10−12 | 0.035 (0.025, 0.045) |
| EPA (11) | Main model† | 78 | rs174538, A/G | 0.28 | 5×10−58 | −0.083 (−0.094, −0.073) |
| EPA (11) | DHA‡ | 77 | rs174535, C/T | 0.31 | 2×10−59 | −0.072 (−0.063, −0.080) |
| EPA (11) | ALA‡ | 87 | rs174535, C/T | 0.31 | 1×10−84 | −0.097 (−0.088, −0.107) |
| DHA (6) | Main model† | 57 | rs2236212, C/G | 0.43 | 1×10−15 | −0.113 (−0.086, −0.141) |
| DHA (6) | EPA‡ | 76 | rs2236212, C/G | 0.43 | 7×10−39 | −0.162 (−0.138, −0.186) |
Results of the meta-analyses of n-3 fatty acid GWAS; main results with standard adjustments described in Methods, and additional results further adjusted for one other fatty acid or for selected SNPs.
†Adjustments of the cohort-specific GWAS; the main model included age, sex, and if appropriate study site and principal components to account for population structure.
‡Adjustment in addition to main model adjustments.
| Number of SNPs with p-values <5.0×10−8.
*The SNPs rs3798713, rs2236212 and rs3734398 are in perfect LD; rs174538, rs174535 and rs174547 are in LD with r2≥0.9. Positions for genome built 36.3 are 61327359 (rs174547), 27594741 (rs780094), 11090959 (rs3734398), 11182177 (rs12662634), 11116608 (rs3798713), 61316657 (rs174538), 61307932 (rs174535), 11103001 (rs2236212).
Table 3. Associations of rs174548 (G allele) with n-3 polyunsaturated fatty acids in samples of different ancestries.
| ALA | EPA | DPA | DHA | |||||||
| N | MAF | Beta* (SE) | p-value | Beta* (SE) | p-value | Beta* (SE) | p-value | Beta* (SE) | p-value | |
| European ancestry cohort † | ||||||||||
| ARIC | 3268 | 0.29 | 0.013 (0.001) | 7×10−24 | −0.085 (0.008) | 6×10−27 | −0.075 (0.004) | 1×10−72 | −0.095 (0.024) | 6×10−05 |
| CARDIA | 1507 | 0.30 | 0.019 (0.004) | 3×10−05 | −0.071 (0.030) | 0.02 | −0.069 (0.008) | 4×10−18 | −0.066 (0.046) | 0.15 |
| CHS | 2326 | 0.28 | 0.019 (0.002) | 2×10−25 | −0.102 (0.011) | 7×10−20 | −0.078 (0.005) | 2×10−48 | −0.075 (0.029) | 0.01 |
| InCHIANTI | 1075 | 0.24 | 0.033 (0.012) | 0.005 | −0.063 (0.009) | 2×10−11 | NA | −0.030 (0.037) | 0.41 | |
| MESA | 690 | 0.29 | 0.023 (0.003) | 5×10−11 | 0.077 (0.027) | 0.005 | −0.056 (0.0012) | 2×10−06 | −0.117 (0.071) | 0.10 |
| Overall‡ | 8866 | 0.29 | 0.016 (0.001) | 8×10−59 | −0.081 (0.005) | 8×10−53 | −0.074 (0.003) | 3×10−138 | −0.076 (0.016) | 1×10−06 |
| African ancestry cohort | ||||||||||
| CARDIA | 1493 | 0.21 | 0.007 (0.004) | 0.05 | −0.070 (0.018) | 9×10−05 | −0.042 (0.009) | 3×10−06 | −0.122 (0.044) | 0.005 |
| CHS | 426 | 0.20 | 0.009 (0.004) | 0.03 | 0.011 (0.299) | 0.71 | −0.099 (0.015) | 0.51 | 0.001 (0.088) | 0.99 |
| MESA | 628 | 0.22 | 0.006 (0.004) | 0.13 | −0.028 (0.036) | 0.44 | −0.033 (0.014) | 0.02 | −0.018 (0.111) | 0.87 |
| Overall‡ | 2547 | 0.21 | 0.007 (0.002) | 0.001 | −0.061 (0.016) | 1×10−4 | −0.051 (0.007) | 3×10−14 | −0.089 (0.037) | 0.02 |
| Chinese ancestry cohort | ||||||||||
| MESA | 633 | 0.58 | 0.019 (0.004) | 1×10−06 | −0.046 (0.033) | 0.17 | −0.048 (0.012) | 3×10−05 | −0.008 (0.072) | 0.91 |
| Hispanic ancestry cohort | ||||||||||
| MESA | 661 | 0.52 | 0.016 (0.003) | 3×10−07 | −0.083 (0.020) | 4×10−05 | −0.078 (0.012) | 3×10−11 | −0.128 (0.053) | 0.02 |
For each fatty acid, results are shown for the individual cohorts and for the meta-analyses among samples of European ancestry and samples of African ancestry.
*Beta-regression coefficient associated with one copy of the G allele.
†Included in the GWAS.
‡Meta-analysis of results of same ancestry cohorts.
Table 4. Associations of rs3734398 (C allele) with n-3 polyunsaturated fatty acids in samples of different ancestries.
| ALA | EPA | DPA | DHA | |||||||
| N | MAF | Beta* (SE) | p-value | Beta* (SE) | p-value | Beta* (SE) | p-value | Beta* (SE) | p-value | |
| European ancestry cohort † | ||||||||||
| ARIC | 3268 | 0.42 | 0.0002 (0.001) | 0.87 | 0.028 (0.008) | 2×10−04 | 0.042 (0.004) | 4×10−24 | −0.119 (0.022) | 8×10−08 |
| CARDIA | 1507 | −0.002 (0.003) | 0.52 | 0.055 (0.022) | 0.01 | 0.037 (0.008) | 1×10−06 | −0.090 (0.041) | 0.03 | |
| CHS | 2326 | 0.43 | 0.003 (0.002) | 0.08 | 0.044 (0.010) | 7×10−06 | 0.040 (0.005) | 1×10−13 | −0.144 (0.027) | 2×10−07 |
| InCHIANTI | 1075 | 0.43 | −0.0007 (0.009) | 0.94 | 0.037 (0.010) | 2×10−04 | NA | −0.092 (0.032) | 0.004 | |
| MESA | 690 | 0.42 | 0.0001 (0.003) | 0.98 | 0.014 (0.029) | 0.06 | 0.041 (0.011) | 2×10−04 | −0.065 (0.068) | 0.34 |
| Overall‡ | 8866 | 0.43 | 0.0008 (0.001) | 0.37 | 0.035 (0.005) | 4×10−12 | 0.040 (0.003) | 1×10−43 | −0.114 (0.014) | 2×10−15 |
| African ancestry cohort | ||||||||||
| CARDIA | 1493 | 0.25 | −0.002 (0.004) | 0.57 | 0.040 (0.017) | 0.02 | 0.045 (0.008) | 1×10−07 | −0.077 (0.041) | 0.06 |
| CHS | 426 | 0.25 | 0.003 (0.003) | 0.37 | −0.016 (0.022) | 0.46 | 0.043 (0.013) | 0.001 | −0.191 (0.071) | 0.008 |
| MESA | 628 | 0.25 | −0.004 (0.004) | 0.32 | 0.043 (0.035) | 0.02 | 0.030 (0.013) | 0.02 | −0.042 (0.107) | 0.69 |
| Overall‡ | 2547 | 0.25 | −0.0002 (0.002) | 0.93 | 0.022 (0.013) | 0.08 | 0.041 (0.006) | 7×10−12 | −0.099 (0.034) | 0.003 |
| Chinese ancestry cohort | ||||||||||
| MESA | 633 | 0.92 | −0.005 (0.008) | 0.51 | −0.097 (0.071) | 0.17 | −0.005 (0.026) | 0.85 | −0.200 (0.154) | 0.20 |
| Hispanic ancestry cohort | ||||||||||
| MESA | 661 | 0.57 | −0.002 (0.003) | 0.56 | −0.006 (0.021) | 0.79 | 0.033 (0.012) | 0.01 | −0.110 (0.055) | 0.05 |
For each fatty acid, results are shown for the individual cohorts and for the meta-analyses among samples of European ancestry and samples of African ancestry.
*Beta-regression coefficient associated with one copy of the G allele.
†Included in the GWAS.
‡Meta-analysis of results of same ancestry cohorts.
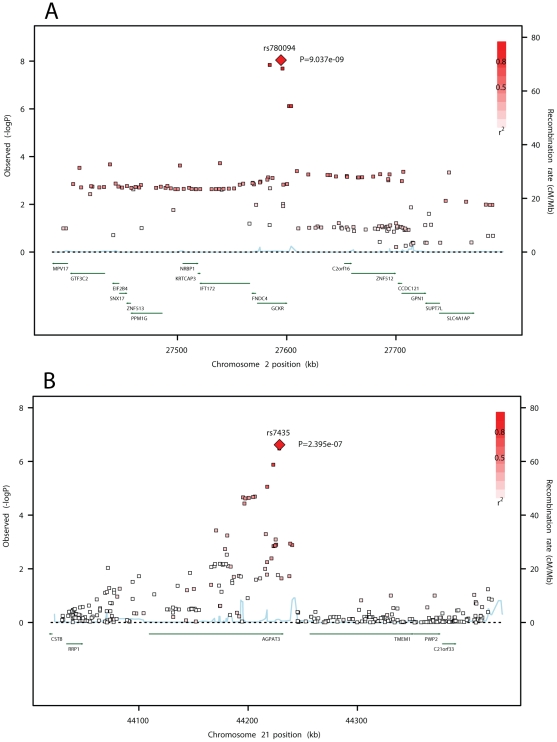
Another genome-wide significant association with DPA was observed with SNPs on chromosome 2 in the GCKR gene (most associated SNP: rs780094, p = 9.0×10−9, Table 2 and Figure 4A). In addition, DPA showed a possible association with SNPs in AGPAT3, a gene on chromosome 21 involved in phospholipid metabolism (most associated SNP: rs7453, p = 2.4×10−7; Figure 4B and Table S1C).
Figure 4. SNP association plots for DPA–associated regions.
Genome-wide association significance level is plotted against the y-axis as –log10 (p-value). Genetic coordinates are as per NCBI build 36. A) GCKR region. LD is indicated by color scale in relationship to marker rs780094. B) AGPAT3 region. LD is indicated by color scale in relationship to marker rs7435. The color scheme is red for strong linkage disequilibrium (LD; r2≥0.8) and fading color for lower LD.
Levels of EPA and DHA were correlated, as were levels of ALA and EPA. Spearman correlations in the 5 cohorts ranged from 0.26 to 0.57 for EPA-DHA and from 0.20 to 0.28 for ALA-EPA. When two outcomes are positively correlated but exhibit genetic associations in opposite directions, it is possible to increase the power of discovery efforts by adjusting the association between SNPs and one fatty acid trait for the other [29]. For this reason, we performed meta-analyses of genome-wide association results for ALA adjusted for EPA, EPA adjusted for ALA, EPA adjusted for DHA and DHA adjusted for EPA. These analyses did not reveal additional genome-wide significant loci, although the statistical significance of the adjusted associations was increased (Table 2). For example, the significance corresponding to the association of the SNP rs2236212 in ELOVL2 with DHA increased from p = 1.3×10−15 to p = 7.0×10−39 with adjustment for EPA, with many more genome-wide significant associations in that region. In addition, the association of rs4985167 on chromosome 16 (PDXDC1) with ALA approached genome-wide significance with adjustment for EPA (p = 7.6×10−8).
To explore whether the signals in the chromosome 6 and 11 loci could be explained by one SNP alone, we performed GWAS of DPA with adjustment for rs2236212 and rs174547 in the ARIC, CARDIA, CHS and MESA cohorts and meta-analyzed the results. No new association was found in the chromosome 11 locus where the adjustment reduced the association for all other SNPs, from a minimum p-value of 1.1×10−51 to p>10−6 (not shown). In contrast, 22 SNPs in the ELOVL2 region reached genome-wide significance, with ten of these SNPs previously undetected (Table 2, Table S1E). The A allele of rs12662634, the most highly associated SNP, had minor allele frequency of 0.18 and was associated with lower level of DPA (regression coefficient associated with one copy of A allele: −0.030, p = 2.7×10−10). This association was new, as the estimated effect in the unadjusted GWAS of DPA was 0.008 (p = 0.06). Among the 12 SNPs that had been detected before, rs9368564 showed the most significant association (regression coefficient and p-value associated with one copy of G allele: 0.027, p = 1.4×10−9, in the adjusted GWAS, and 0.048, p = 10−40, in the unadjusted GWAS). This result may reflect residual association in the adjusted analysis or yet another new association.
To explore if reducing the variability in EPA would reveal additional associations, we also performed GWAS of EPA adjusted for estimated fish intake. No additional associations beyond those previously seen for SNP in FADS1/2 and ELOVL2 were observed in these analyses (not shown). Finally, adjustment of the GWAS of the four n-3 PUFAs for levels of triglycerides, high density lipoprotein and low density lipoprotein did not materially change the results (not shown). For example, in the GWAS of DPA, the estimated effect of one copy of the T allele of rs780094 (GCKR) went from 0.0157 (p = 1.15×10−8) without adjustment to 0.0175 (p = 2.52×10−9) with adjustment, in meta-analyses that included 7663 participants.
Association of top SNPs with n-3 PUFAs in samples from participants of African, Chinese, and Hispanic ancestry
To determine whether the gene-fatty acid associations were consistent across different ethnicities, we examined the associations of genotype at two representative SNPs with phospholipid n-3 PUFA levels, in samples of African, Chinese and Hispanic ancestry. Results of these analyses for the selected SNPs are shown in Table 3 and Table 4, together with meta-analysis and cohort-specific results among the samples of European ancestry. Frequency of the G allele of rs174548 (FADS1) was 0.29, 0.21, 0.58, 0.52 and frequency of the C allele of rs3734398 (ELOVL2) was 0.43, 0.25, 0.92, 0.57 in samples of European, African, Chinese and Hispanic ancestry respectively. Associations of rs174548 with n-3 PUFA were generally similar across all ancestries, with the G allele associated with higher ALA and lower long-chain n-3 PUFA levels, although associations did not always reach statistical significance, perhaps due to limited sample sizes (Table 3). The associations of rs3734398 with EPA, DPA and DHA were similar for samples of African ancestry versus European ancestry (Table 4). Among samples of Chinese ancestry, SNP rs3734398 was not highly polymorphic (C allele frequency of 92%) and no significant associations were detected. In samples of Hispanic ancestry, the C allele of rs3734398 was associated with higher DPA and lower DHA, but it was not associated with EPA.
Interactions
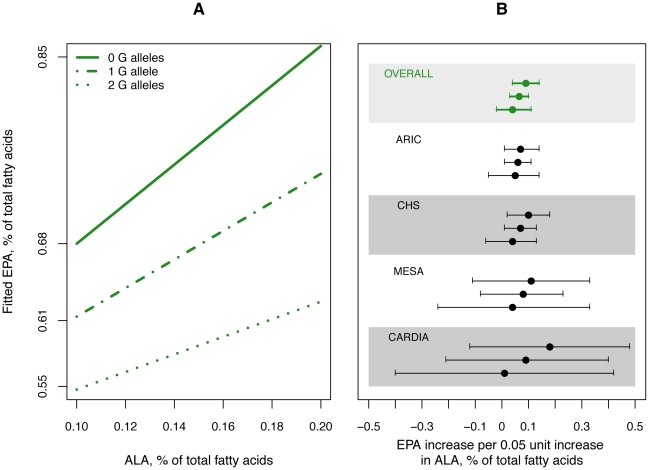
We evaluated several potential interactions in the samples of European ancestry, with statistical significance defined at alpha = 0.004 (0.05 divided by 13 tested interactions). We found little evidence that fatty fish consumption (≥vs. <0.6 servings/week) modified the associations of rs1535 (FADS2) or rs3734398 (ELOVL2) with levels of DHA or EPA. We also did not observe any interaction between plasma phospholipid levels of LA (continuous linear) and genotype at these two SNPs on the levels of DHA or EPA. Plasma phospholipid levels of ALA (continuous linear) also did not modify the association of genotype at these two SNPs with levels of DHA, or of genotype at rs3734398 with levels of EPA. However, there was a significant interaction of ALA with rs1535 genotype and EPA levels (meta-analyzed interaction coefficient p = 9.3×10−7), illustrated in Figure 5. Per one SD unit (0.05% of total fatty acids) increase in ALA, EPA levels increased by 0.086% of total fatty acids (23% of one SD) in the absence of the minor allele (G); by 0.063% (17% of one SD) in the presence of one copy (G-); and by 0.036% (10% of one SD) in the presence of two copies (GG).
Figure 5. Influence of variation in rs1535 on the estimated association of plasma phospholipid ALA levels with EPA levels.
Panel A illustrates the association of ALA with EPA in carriers of 0, 1, and 2 copies of rs1535 G allele with fitted lines constructed from meta-analyzed estimates. Panel B shows the % change in EPA corresponding to a 0.05 unit increase in ALA in carriers of 0 (top estimate), 1 (middle estimate), 2 (bottom estimate) copies of rs1535 G allele.
Discussion
We report here the results of the largest GWAS of plasma phospholipid n-3 PUFAs to date, with 8,866 participants of European ancestry. The associations of the two top hits on chromosomes 6 and 11 are shown in context of the n-3 PUFA pathway in Figure 1. Genetic variation in the desaturase genes FADS1 and FADS2 was associated with higher levels of ALA and lower levels of EPA and DPA suggesting genetic variants that affect the conversion of ALA to EPA and DPA. In the main analyses, genetic variation in the elongase gene ELOVL2 was associated with higher levels of EPA and DPA and lower levels of DHA, suggesting variants that decrease the conversion of EPA and DPA to DHA. These reciprocal associations support a role of genetic variation in the pathway for circulating levels of n-3 PUFAs in free-living populations.
The associations of FADS1/2 and ELOVL2 with n-3 PUFAs were generally consistent with the previous GWAS in InCHIANTI (total plasma n-3 PUFAs) and follow-up candidate replication in the Genetics of Lipid-Lowering Drugs and Diet Network (GOLDN) Study (erythrocyte n-3 PUFAs) [26]. In that prior study, only the association of variation in FADS1 with EPA reached genome-wide significance. Follow-up investigations in GOLDN suggested associations of FADS1 gene variation with ALA and DHA, and of ELOVL2 gene with DHA and DPA, such as the ones reported here [26]. In the present larger meta-analyses, these prior suggestive associations reached genome-wide significance, providing a definite picture of the association of genes in the PUFA pathway with phospholipid n-3 PUFAs. While the InCHIANTI cohort showed similar estimates of association with ALA, EPA and DHA as the other cohorts in the present study, further studies are needed to further explore genetic associations of n-3 PUFAs in triglycerides and other fatty acid compartments.
Using ratios of n-6 PUFAs as proxy of desaturase activities, Bokor et al reported that the minor allele of FADS1 rs174546 was associated with lower delta-5 desaturase activity and the minor allele of FADS2 rs968567 was associated with higher delta-6 desaturase activity [30]. We found both rs174546 and rs968567 were associated with higher levels of ALA and lower levels of EPA and DPA at genome-wide significance level (Table S1A–S1C). Furthermore, adjustment of the GWAS of DPA for the most associated SNP from the FADS1/2 genes and the most associated SNP from ELOVL2 did not reveal additional associations on chromosome 11. This result suggests that each associated SNP conveyed the information contained in the other SNPs of the same broad region on chromosome 11. In contrast, we found evidence for two independent associations, exemplified by rs2236212 and rs12662634, in the region of the ELOVL2 gene.
The associations of the FADS1/2 and ELOVL2 genes with the phospholipid levels of EPA and DHA did not vary depending on the frequency of fatty fish consumption, suggesting the genetic effects are independent of fish intake at the levels of consumption in the studied populations. The absence of interaction of FADS1/2 by fish consumption is consistent with findings from the Netherlands KOALA Birth Cohort Study [31], in which higher levels of fish consumption were associated with similar slopes of plasma phospholipid EPA and DHA levels among individuals with 0, 1, or 2 copies of minor FADS1/2 alleles. The associations of FADS1/2 and ELOVL2 genes with EPA and DHA also did not vary depending on phospholipid LA. Linoleic acid is desaturated and elongated to n-6 metabolites (e.g. arachidonic acid) using the same desaturases and elongase(s) as ALA, and in dietary trials, higher dietary LA reduces plasma phospholipid EPA [32]. Animal experiments suggest that high dietary LA inhibits FADS2 gene expression [33]. Our findings do not support an influence of LA on the association of genetic variation in the pathway with n-3 PUFA levels, at levels of LA consumption in these cohorts.
We report for the first time a GWAS of DPA, a central intermediate in the n-3 fatty acid pathway (Figure 1). While present in small quantities in fatty fish, DPA plasma levels appear unrelated to dietary intake [3], suggesting a primarily metabolic origin. Supporting this, we found that DPA exhibited stronger genetic associations than the other n-3 PUFAs.
In addition to its association with variants of desaturase and elongase genes, DPA was associated with variation in the glucokinase regulator gene GCKR, a pleiotropic gene associated with multiple outcomes in GWAS [34]. The T allele of rs780094 is associated with lower fasting glucose and insulin [35] and with higher triglycerides [36]–[38]; this allele was associated with higher DPA levels in the present study, and the association was independent of triglyceride levels. Given the known influence of long-chain n-3 PUFAs on hepatic triglyceride production [39] and possibly glucose-insulin homeostasis [40], the mechanism of potential pleiotropic effects of this allele on both DPA and these pathways merit further attention.
We found a potential association of DPA with AGPAT3, a gene encoding 1-acylglycerol-3-phosphate O-acyltransferase 3. DPA is a known substrate for the AGPAT3 protein, which transfers a fatty acid in sn-2 position of lysophosphatic acid, a step in the phospholipid biosynthesis pathway. A possible association of DPA with AGPAT3 variation supports an origin of phospholipid DPA from de novo phospholipid synthesis. In contrast, phospholipid EPA and DHA often originate to a greater extent from diet and are predominantly integrated into phospholipids by the process of acyl-chain remodelling [41]. The genetic associations reported here together with growing evidence of the association of DPA with lower risk of coronary heart disease [3], [42], [43] should stimulate further work on factors regulating this fatty acid.
The GWAS of ALA adjusted with EPA revealed a possible new association of phospholipid ALA with variation in PDXDC1. The PDXDC1 protein, a vitamin B6-dependent decarboxylase, is expressed preferentially in the intestine [44], but its function is not known. Animal studies support an influence of dietary vitamin B6 (pyridoxal) on serum and liver levels of ALA and other PUFAs [45], [46], which has been interpreted as an effect on desaturase enzymes activity. The association of PDXDC1 with ALA, if confirmed in other studies, raises the possibility of another vitamin B6-dependent protein affecting ALA, for example through involvement in intestinal ALA absorption.
In addition to an overall association of FADS1/2 variation with less ALA and more EPA, we found that the minor G allele of rs1535 was associated with a reduction of the magnitude of the association between ALA and EPA. In persons with two copies of the G allele, the association of ALA with EPA was less than half the association observed in persons with two copies of the A allele. These results suggest an influence of variation in FADS1/2 on the rate of conversion of ALA into EPA. This conversion is of great clinical and public health interest, given the evidence for importance of long-chain n-3 PUFAs (such as EPA) in many chronic diseases, their limited dietary supply worldwide, and the much greater potential supply of plant-derived ALA. On average, the conversion of ALA to EPA is quite low [47]. Prior tracer studies in humans have shown that the majority of dietary ALA is beta-oxidized for energy or directed into long-term storage as triglycerides, with less than 5% being incorporated into phospholipids where ALA is more readily converted to EPA [16], [17], [47]. Genetic variation that increases or decreases the rate of conversion of ALA to EPA could have implications for individual-based recommendations for consumption of plant- versus seafood-derived n-3 PUFA. The genetic variation may also indicate novel targets for drugs that may increase this conversion.
The associations of a representative SNP in the FADS1/2 genes observed in the meta-analyses of samples of European ancestry were generally similar in samples of African, Chinese, and Hispanic ancestry. Associations of ELOVL2 were less consistent in different ancestries. However, the frequency of the ELOVL2 rs3734398 G allele varied substantially with ancestry, from 25% in African samples to 92% in Chinese samples. Lack of association may be due to inadequate statistical power, chance, different background diet [14], or possible race/ethnic differences in the activity of elongases from the ELOVL2 and ELOVL5 genes.
Our study, the largest GWAS to-date of fatty acid biomarkers, demonstrates key associations of genetic variation with phospholipid n-3 PUFA levels, including genes in the n-3 PUFA metabolic pathway and, for DPA, novel pathways including the pleiotropic gene GCKR. Our results also imply that common variation may result in less efficient conversion of ALA to EPA.
Materials and Methods
Ethics statement
All cohort participants gave written informed consent, including consent to participate in genetic studies. All studies received approval from local ethical oversight committees.
Study samples
The data were obtained from 2 cohort studies in the CHARGE Consortium, the Atherosclerosis Risk in Communities (ARIC) Study and the Cardiovascular Health Study (CHS), and 3 additional cohort studies, the Coronary Artery Risk Development in Young Adults (CARDIA) Study, the Invecchiare in Chianti (InCHIANTI) Study, and the Multi-Ethnic Study of Atherosclerosis (MESA).
Fatty acid measurements
In all cohorts but InCHIANTI, plasma phospholipids were first isolated by thin layer chromatography; fatty acids were then separated by gas chromatography. In InCHIANTI, total plasma fatty acids were measured using a similar gas chromatography technique. Details of fatty acid measurements are provided in Text S1. Levels of EPA, DHA, ALA and DPA were expressed as % of total fatty acids.
Imputation and statistical analysis
Genotyping was done in each cohort separately using high-density SNP marker platforms (ARIC, CARDIA and MESA - Affymetrix 6.0, CHS - Illumina 370, InCHIANTI - Illumina 550). Samples with call rates below 95% (ARIC, CARDIA, MESA), or 97% (CHS, InCHIANTI) at genotyped markers were excluded. Genotypes were imputed to approximately 2.5 million HapMap SNPs by using either MACH [48] (ARIC, InCHIANTI), BIMBAM [49] (CHS), BEAGLE [50] (CARDIA) or IMPUTE [51] (MESA). SNPs for which testing Hardy Weinberg equilibrium resulted in p<10−5 (CHS) or p<10−6 (ARIC) were excluded from imputation. SNPs with minor allele frequency (MAF) ≤1% were excluded from the meta-analyses. Additional details on genotyping and imputation per cohort are provided in Text S1.
Association analysis between genotype and each fatty acid was done separately within each study cohort according to a pre-specified plan. All studies conducted linear regression analysis using an additive genetic model, i.e. regression of phenotype on the number of reference alleles, or equivalently the imputed dosage for imputed genotypes. All analyses were adjusted for age, sex, and site of recruitment where appropriate, and used robust standard errors. In addition, CARDIA, CHS and MESA analyses were adjusted for principal components to account for possible population genetic substructure. The results in InCHIANTI included in the present study have been previously published [26].
Meta-analysis of main effects
For each SNP and fatty acid, GWAS-specific results were combined using inverse-variance weighted meta-analysis in METAL (www.sph.umich.edu/csg/abecasis/metal). Genomic control correction was applied to each study prior to the meta-analysis. Genomic control correction factors ranged from 1.00 to 1.07 (ALA), 1.00–1.08 (EPA), 1.00–1.03 (DPA) and 1.01–1.13 (DHA). P-values less than 5×10−8 were considered significant. Because total plasma levels of ALA (measured in InCHIANTI) are higher than plasma phospholipid levels of ALA (measured in the other cohorts), we performed a z-score based meta-analysis of ALA with the 5 cohorts as a sensitivity analysis. Results did not differ from that of inverse-variance weighted meta-analysis, i.e. from those presented. The proportion of fatty acid variance explained by a particular variant allele was calculated from the formula.
(β2*2*MAF(1-MAF))/Var(Y), where β is the regression coefficient for one copy of the allele, MAF is the minor allele frequency and Var(Y) is the variance of the fatty acid.
Interaction analyses
We tested 13 interactions using cross-products in the linear regression models. Two of the most associated SNPs available in all cohorts (rs1535 in FADS2 and rs3734398 in ELOVL2) were chosen for investigation of interactions with a) fatty fish intake (dichotomized at 0.6 servings/week, a cut-point around the 25th percentile of fish consumption in the CHS and ARIC cohorts), b) plasma phospholipid ALA levels (continuous linear) and c) plasma phospholipid LA levels (continuous linear) on the outcomes of EPA and DHA. Additionally, we tested the interaction between rs1535 and plasma phospholipid EPA with DHA levels as the outcome. Interaction coefficients from cohort-specific analyses were meta-analyzed. For interactions of SNPs with ALA on the outcomes of EPA and DHA, we performed z-score meta-analysis with all the cohorts to assess statistical significance, and inverse-variance weighted meta-analysis excluding InCHIANTI to estimate the magnitude of the interaction. P-values less than 0.004 (0.05/13 tests) were considered significant for the interactions.
Analyses of selected SNPs in cohorts of African, Chinese, and Hispanic ancestry
We investigated the association of two selected SNPs which had been directly genotyped as part of candidate gene studies in the African American cohort in CARDIA and the African, Chinese and Hispanic American cohort in MESA, and which were available from genome-wide scans on African Americans in the CHS cohort. We used linear regression and additive models as described above. Results in the 3 African American cohorts were meta-analyzed using inverse-variance weighted meta-analysis.
Supporting Information
Details of participating cohorts.
(DOC)
Supplementary results from the CHARGE consortium. A. Comprehensive results for ALA with p<5*10−6. B. Comprehensive results for EPA with p<5*10−6. C. Comprehensive results for DPA with p<5*10−6. D. Comprehensive results for DHA with p<5*10−6. E. Results for DPA adjusted for rs2236212 and rs174547 with p<5*10−8.
(DOC)
Acknowledgments
The authors thank the other investigators, the staff and the participants of the ARIC study, the CARDIA study, the CHS study, the MESA study, and the InCHIANTI study for their important contributions. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. A full list of principal CARDIA investigators and institutions can be found at http://www.cardia.dopm.uab.edu/o_pain.htm. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. The authors acknowledge the essential role of the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) Consortium in development and support of this. CHARGE members include NHLBI's Atherosclerosis Risk in Communities (ARIC) Study, NIA's Iceland Age Gene/Environment Susceptibility Study, NHLBI's Cardiovascular Health Study (CHS) and Framingham Heart Study, and the Netherland's Rotterdam Study. The authors acknowledge the use of the SNAP server from the Broad Institute (http://www.broadinstitute.org/mpg/snap/) to construct regional association plots.
Footnotes
The authors have declared that no competing interests exist.
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, HHSN268201100012C, R01HL087641, R01HL59367, and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. The Cardiovascular Health Study research reported in this article was supported by the National Heart, Lung, and Blood Institute contract N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133 and grants HL075366, HL080295, HL087652, HL105756, and HL085251 with additional contribution from the National Institute of Neurological Disorders and Stroke. Additional support from the National Institute on Aging was provided through AG-023269, AG-15928, AG-20098, and AG-027058. DNA handling and genotyping was supported in part by the National Center for Research Resources grant M01RR00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and the National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The Coronary Artery Risk Development in Young Adults (CARDIA) study is funded by contracts N01-HC-95095, N01-HC-48047, N01-HC-48048, N01-HC-48049, N01-HC-48050, N01-HC-45134, N01-HC-05187, N01-HC-45205, and N01-HC-45204 from the National Heart, Lung, and Blood Institute to the CARDIA investigators. Genotyping of the CARDIA participants was supported by grants U01-HG-004729, U01-HG-004446, and U01-HG-004424 from the National Human Genome Research Institute and R01-HL-084099 from the National Heart, Lung, and Blood Institute to MF. The InCHIANTI Study baseline (1998-2000) was supported as a “targeted project” (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336) and was supported in part by the Intramural research program of the National Institute on Aging, National Institutes of Health, Baltimore, Maryland. The Multi-Ethnic Study of Atherosclerosis (MESA) and MESA SHARe were supported by contracts N01-HC-95159 through N01-HC-95169 and RR-024156 from the National Heart, Lung, and Blood Institute. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02HL64278. MESA Family is conducted and supported in collaboration with MESA investigators; support is provided by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071252, R01HL071258, R01HL071259. The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. JA Nettleton is supported by a K01 from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (5K01DK082729-02). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Siscovick DS, Raghunathan TE, King I, Weinmann S, Wicklund KG, et al. Dietary intake and cell membrane levels of long-chain n-3 polyunsaturated fatty acids and the risk of primary cardiac arrest. Jama. 1995;274:1363–1367. doi: 10.1001/jama.1995.03530170043030. [DOI] [PubMed] [Google Scholar]
- 2.Lemaitre RN, King IB, Mozaffarian D, Sotoodehnia N, Rea TD, et al. Plasma phospholipid trans fatty acids, fatal ischemic heart disease, and sudden cardiac death in older adults: the cardiovascular health study. Circulation. 2006;114:209–215. doi: 10.1161/CIRCULATIONAHA.106.620336. [DOI] [PubMed] [Google Scholar]
- 3.Sun Q, Ma J, Campos H, Rexrode KM, Albert CM, et al. Blood concentrations of individual long-chain n-3 fatty acids and risk of nonfatal myocardial infarction. Am J Clin Nutr. 2008;88:216–223. doi: 10.1093/ajcn/88.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamagishi K, Nettleton JA, Folsom AR. Plasma fatty acid composition and incident heart failure in middle-aged adults: the Atherosclerosis Risk in Communities (ARIC) Study. Am Heart J. 2008;156:965–974. doi: 10.1016/j.ahj.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sala-Vila A, Cofan M, Perez-Heras A, Nunez I, Gilabert R, et al. Fatty acids in serum phospholipids and carotid intima-media thickness in Spanish subjects with primary dyslipidemia. Am J Clin Nutr. 2010;92:186–193. doi: 10.3945/ajcn.2009.28807. [DOI] [PubMed] [Google Scholar]
- 6.Warensjo E, Sundstrom J, Lind L, Vessby B. Factor analysis of fatty acids in serum lipids as a measure of dietary fat quality in relation to the metabolic syndrome in men. Am J Clin Nutr. 2006;84:442–448. doi: 10.1093/ajcn/84.1.442. [DOI] [PubMed] [Google Scholar]
- 7.Shannon J, King IB, Moshofsky R, Lampe JW, Gao DL, et al. Erythrocyte fatty acids and breast cancer risk: a case-control study in Shanghai, China. Am J Clin Nutr. 2007;85:1090–1097. doi: 10.1093/ajcn/85.4.1090. [DOI] [PubMed] [Google Scholar]
- 8.Shahar E, Boland LL, Folsom AR, Tockman MS, McGovern PG, et al. Docosahexaenoic acid and smoking-related chronic obstructive pulmonary disease. The Atherosclerosis Risk in Communities Study Investigators. Am J Respir Crit Care Med. 1999;159:1780–1785. doi: 10.1164/ajrccm.159.6.9810068. [DOI] [PubMed] [Google Scholar]
- 9.Tiemeier H, van Tuijl HR, Hofman A, Kiliaan AJ, Breteler MM. Plasma fatty acid composition and depression are associated in the elderly: the Rotterdam Study. Am J Clin Nutr. 2003;78:40–46. doi: 10.1093/ajcn/78.1.40. [DOI] [PubMed] [Google Scholar]
- 10.Feart C, Peuchant E, Letenneur L, Samieri C, Montagnier D, et al. Plasma eicosapentaenoic acid is inversely associated with severity of depressive symptomatology in the elderly: data from the Bordeaux sample of the Three-City Study. Am J Clin Nutr. 2008;87:1156–1162. doi: 10.1093/ajcn/87.5.1156. [DOI] [PubMed] [Google Scholar]
- 11.Samieri C, Feart C, Letenneur L, Dartigues JF, Peres K, et al. Low plasma eicosapentaenoic acid and depressive symptomatology are independent predictors of dementia risk. Am J Clin Nutr. 2008;88:714–721. doi: 10.1093/ajcn/88.3.714. [DOI] [PubMed] [Google Scholar]
- 12.Schaefer EJ, Bongard V, Beiser AS, Lamon-Fava S, Robins SJ, et al. Plasma phosphatidylcholine docosahexaenoic acid content and risk of dementia and Alzheimer disease: the Framingham Heart Study. Arch Neurol. 2006;63:1545–1550. doi: 10.1001/archneur.63.11.1545. [DOI] [PubMed] [Google Scholar]
- 13.Sun Q, Ma J, Campos H, Hankinson SE, Hu FB. Comparison between plasma and erythrocyte fatty acid content as biomarkers of fatty acid intake in US women. Am J Clin Nutr. 2007;86:74–81. doi: 10.1093/ajcn/86.1.74. [DOI] [PubMed] [Google Scholar]
- 14.Chung H, Nettleton JA, Lemaitre RN, Barr RG, Tsai MY, et al. Frequency and type of seafood consumed influence plasma (n-3) fatty acid concentrations. J Nutr. 2008;138:2422–2427. doi: 10.3945/jn.108.089631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burdge GC, Wootton SA. Conversion of alpha-linolenic acid to palmitic, palmitoleic, stearic and oleic acids in men and women. Prostaglandins Leukot Essent Fatty Acids. 2003;69:283–290. doi: 10.1016/s0952-3278(03)00111-x. [DOI] [PubMed] [Google Scholar]
- 16.Goyens PL, Spilker ME, Zock PL, Katan MB, Mensink RP. Conversion of alpha-linolenic acid in humans is influenced by the absolute amounts of alpha-linolenic acid and linoleic acid in the diet and not by their ratio. Am J Clin Nutr. 2006;84:44–53. doi: 10.1093/ajcn/84.1.44. [DOI] [PubMed] [Google Scholar]
- 17.Goyens PL, Spilker ME, Zock PL, Katan MB, Mensink RP. Compartmental modeling to quantify alpha-linolenic acid conversion after longer term intake of multiple tracer boluses. J Lipid Res. 2005;46:1474–1483. doi: 10.1194/jlr.M400514-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Lemaitre RN, Siscovick DS, Berry EM, Kark JD, Friedlander Y. Familial aggregation of red blood cell membrane fatty acid composition: the Kibbutzim Family Study. Metabolism. 2008;57:662–668. doi: 10.1016/j.metabol.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, et al. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15:1745–1756. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 20.Rzehak P, Heinrich J, Klopp N, Schaeffer L, Hoff S, et al. Evidence for an association between genetic variants of the fatty acid desaturase 1 fatty acid desaturase 2 (FADS1 FADS2) gene cluster and the fatty acid composition of erythrocyte membranes. Br J Nutr. 2009;101:20–26. doi: 10.1017/S0007114508992564. [DOI] [PubMed] [Google Scholar]
- 21.Xie L, Innis SM. Genetic variants of the FADS1 FADS2 gene cluster are associated with altered (n-6) and (n-3) essential fatty acids in plasma and erythrocyte phospholipids in women during pregnancy and in breast milk during lactation. J Nutr. 2008;138:2222–2228. doi: 10.3945/jn.108.096156. [DOI] [PubMed] [Google Scholar]
- 22.Malerba G, Schaeffer L, Xumerle L, Klopp N, Trabetti E, et al. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 2008;43:289–299. doi: 10.1007/s11745-008-3158-5. [DOI] [PubMed] [Google Scholar]
- 23.Bokor S, Dumont J, Spinneker A, Gonzalez-Gross M, Nova E, et al. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J Lipid Res. 2010;51:2325–2333. doi: 10.1194/jlr.M006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zietemann V, Kroger J, Enzenbach C, Jansen E, Fritsche A, et al. Genetic variation of the FADS1 FADS2 gene cluster and n-6 PUFA composition in erythrocyte membranes in the European Prospective Investigation into Cancer and Nutrition-Potsdam study. Br J Nutr. 2010. pp. 1–12. [DOI] [PubMed]
- 25.Okada T, Sato NF, Kuromori Y, Miyashita M, Iwata F, et al. Thr-encoding allele homozygosity at codon 54 of FABP 2 gene be associated with impaired delta 6 desatruase activity and reduced plasma arachidonic acid in obese children. J Atheroscler Thromb. 2006;13:192–196. doi: 10.5551/jat.13.192. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka T, Shen J, Abecasis GR, Kisialiou A, Ordovas JM, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009;5:e1000338. doi: 10.1371/journal.pgen.1000338. doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodson L, Skeaff CM, Fielding BA. Fatty acid composition of adipose tissue and blood in humans and its use as a biomarker of dietary intake. Prog Lipid Res. 2008;47:348–380. doi: 10.1016/j.plipres.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Vittinghoff E, Glidden D, Shiboski S, McCulloch C. New York: Springer; 2005. Regression Methods in Biostatistics: Linear Logistic, Survival and Repeated Measures Models. [Google Scholar]
- 30.Bokor S, Dumont J, Spinneker A, Gonzalez-Gross M, Nova E, et al. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J Lipid Res. 2010. [DOI] [PMC free article] [PubMed]
- 31.Molto-Puigmarti C, Plat J, Mensink RP, Muller A, Jansen E, et al. FADS1 FADS2 gene variants modify the association between fish intake and the docosahexaenoic acid proportions in human milk. Am J Clin Nutr. 2010;91:1368–1376. doi: 10.3945/ajcn.2009.28789. [DOI] [PubMed] [Google Scholar]
- 32.Liou YA, King DJ, Zibrik D, Innis SM. Decreasing linoleic acid with constant alpha-linolenic acid in dietary fats increases (n-3) eicosapentaenoic acid in plasma phospholipids in healthy men. J Nutr. 2007;137:945–952. doi: 10.1093/jn/137.4.945. [DOI] [PubMed] [Google Scholar]
- 33.Tang C, Cho HP, Nakamura MT, Clarke SD. Regulation of human delta-6 desaturase gene transcription: identification of a functional direct repeat-1 element. J Lipid Res. 2003;44:686–695. doi: 10.1194/jlr.M200195-JLR200. [DOI] [PubMed] [Google Scholar]
- 34.Hindorff L, Junkins H, Hall P, Mehta J, Manolio T. A Catalog of Published Genome-Wide Association Studies. Available at: www.genome.gov/gwastudies.
- 35.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris WS, Bulchandani D. Why do omega-3 fatty acids lower serum triglycerides? Curr Opin Lipidol. 2006;17:387–393. doi: 10.1097/01.mol.0000236363.63840.16. [DOI] [PubMed] [Google Scholar]
- 40.Friedberg CE, Janssen MJ, Heine RJ, Grobbee DE. Fish oil and glycemic control in diabetes. A meta-analysis. Diabetes Care. 1998;21:494–500. doi: 10.2337/diacare.21.4.494. [DOI] [PubMed] [Google Scholar]
- 41.Lands WE. Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J Biol Chem. 1958;231:883–888. [PubMed] [Google Scholar]
- 42.Rissanen T, Voutilainen S, Nyyssonen K, Lakka TA, Salonen JT. Fish oil-derived fatty acids, docosahexaenoic acid and docosapentaenoic acid, and the risk of acute coronary events: the Kuopio ischaemic heart disease risk factor study. Circulation. 2000;102:2677–2679. doi: 10.1161/01.cir.102.22.2677. [DOI] [PubMed] [Google Scholar]
- 43.Oda E, Hatada K, Katoh K, Kodama M, Nakamura Y, et al. A case-control pilot study on n-3 polyunsaturated fatty acid as a negative risk factor for myocardial infarction. Int Heart J. 2005;46:583–591. doi: 10.1536/ihj.46.583. [DOI] [PubMed] [Google Scholar]
- 44.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertrandt J, Klos A, Debski B. Influence of vitamin B6 supplementation on polyunsaturated fatty acids concentration in serum and liver of rats fed a diet restricted in protein. Nahrung. 2004;48:99–103. doi: 10.1002/food.200300338. [DOI] [PubMed] [Google Scholar]
- 46.Bordoni A, Hrelia S, Lorenzini A, Bergami R, Cabrini L, et al. Dual influence of aging and vitamin B6 deficiency on delta-6-desaturation of essential fatty acids in rat liver microsomes. Prostaglandins Leukot Essent Fatty Acids. 1998;58:417–420. doi: 10.1016/s0952-3278(98)90163-6. [DOI] [PubMed] [Google Scholar]
- 47.Burdge GC, Finnegan YE, Minihane AM, Williams CM, Wootton SA. Effect of altered dietary n-3 fatty acid intake upon plasma lipid fatty acid composition, conversion of [13C]alpha-linolenic acid to longer-chain fatty acids and partitioning towards beta-oxidation in older men. Br J Nutr. 2003;90:311–321. doi: 10.1079/bjn2003901. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of participating cohorts.
(DOC)
Supplementary results from the CHARGE consortium. A. Comprehensive results for ALA with p<5*10−6. B. Comprehensive results for EPA with p<5*10−6. C. Comprehensive results for DPA with p<5*10−6. D. Comprehensive results for DHA with p<5*10−6. E. Results for DPA adjusted for rs2236212 and rs174547 with p<5*10−8.
(DOC)