Abstract
Type VI secretion systems (T6SS) are macromolecular machines of the cell envelope of Gram-negative bacteria responsible for bacterial killing and/or virulence towards different host cells. Here, we characterized the regulatory mechanism underlying expression of the enteroagregative Escherichia coli sci1 T6SS gene cluster. We identified Fur as the main regulator of the sci1 cluster. A detailed analysis of the promoter region showed the presence of three GATC motifs, which are target of the DNA adenine methylase Dam. Using a combination of reporter fusion, gel shift, and in vivo and in vitro Dam methylation assays, we dissected the regulatory role of Fur and Dam-dependent methylation. We showed that the sci1 gene cluster expression is under the control of an epigenetic switch depending on methylation: fur binding prevents methylation of a GATC motif, whereas methylation at this specific site decreases the affinity of Fur for its binding box. A model is proposed in which the sci1 promoter is regulated by iron availability, adenine methylation, and DNA replication.
Author Summary
DNA methylation plays an important role in the regulation of genes involved in assembly of cell surface adhesins or appendages. Methylation at a GATC motif by the Dam methylase influences binding of transcriptional regulators, leading to variation in the gene expression pattern. In several cases, this may lead to different cell subpopulations allowing a rapid adaptation to varying environments. In this work, we uncover the regulatory mechanism controlling expression of the sci1 Type VI secretion gene cluster in entero-aggregative Escherichia coli, which encodes a structure required for inter-bacterial interaction. We showed that this gene cluster is repressed by Fur in iron-replete conditions and that Fur binding on the promoter prevents methylation of a GATC motif. In iron-limited conditions, Fur is relieved from the promoter allowing expression of the gene cluster and methylation of the GATC motif. Methylation prevents de novo Fur binding allowing constitutive expression. Our findings support a model in which the expression of the Type VI secretion gene cluster is regulated by a non-stochastic epigenetic switch: switch from the OFF to ON phases depends on iron availability whereas the ON to OFF switch depends on DNA replication and competition between Dam-dependent methylation and Fur binding.
Introduction
Type VI secretion systems (T6SS) are macromolecular assemblies encoded within the genome of most Gram negative bacteria [1]–[3]. They are composed of at least 13 subunits, called core components, which are believed to form a trans-envelope apparatus from the cytoplasm to the outside of the cell [2]. Several subunits of this transport system have extensive homologies with Type IV secretion components, or functional homologues in envelope spanning complexes such as an outer membrane lipoprotein [4], a protein anchoring the system to the peptidoglycan layer [5], [6] and an AAA+ ATPase. The exciting discoveries that two core components exhibit remarkable structure conservation with two bacteriophage structural proteins reshaped this field [7]. Two T6SS proteins released in the environmental milieu, Hcp and VgrG, are structurally related to the tail tube and the cell-puncturing device of bacteriophage T4, gp19 and the gp27-gp5 complex respectively [8]–[11]. From these data, it has been suggested that T6SS will assemble a bacteriophage upside-down structure, anchored to the cell envelope through the bacteriophage-unrelated membrane or membrane-associated subunits. In this model, Hcp will assemble a tube-like structure resembling the bacteriophage tail, and displaying a VgrG trimer at the tip [7], [10]. Interestingly, a number of VgrG proteins are fused to an additional, C-terminal domain, carrying an effector function [12]. It has been initially proposed that T6SS are important virulence factors towards eukaryotic host cells [13]; however, although this turned to be true in several cases, most VgrG proteins do not carry the C-terminal extension.
Recently, several research groups demonstrated that a number of T6SS, including those of Pseudomonas aeruginosa, Burkholderia thailadensis and Vibrio cholerae are required for inter-bacterial competition, and anti-bacterial toxins secreted by the P. aeruginosa HSI-1 T6SS have been identified [14]–[17]. Indeed, mixed cultures between a T6SS-producing strain and a different species showed killing of the T6SS non-producing in a T6SS-dependent manner. Therefore, the role of T6SS in host pathogenesis is somehow limited to the competition towards other microorganisms, to gain access to a specific niche where additional virulence factors may act directly against host cells [16], [18]. However, the roles of T6SS are not limited to virulence towards host cells or towards surrounding bacteria, but several studies reported roles in resisting amoeba predation, stress sensing, or biofilm formation [4], [13], [19]. It appears that T6SS are adapted to the specific needs of each individual bacterium, and are therefore subjected to specific and precise regulatory modulations [20], [21]. Indeed, a wide array of different mechanisms have been reported: control by quorum sensing mechanisms, two-component systems, transcriptional factors, histone-like proteins or alternate sigma factors [20]–[22].
In this study, we sought to identify the regulatory mechanism underlying expression of the enteroaggregative Escherichia coli (EAEC) sci1 T6SS gene cluster. Using random mini-Tn mutagenesis of a strain carrying a translational reporter fusion to the sci1 promoter, we identified the Ferric uptake regulator Fur as the main repressor of the expression of this cluster. The Fur protein has been well characterized in several bacteria in which it acts as a transcriptional repressor of iron-regulated promoters [23]. In presence of iron, Fur represses the expression of these promoters, while in absence of iron, Fur is relieved from these promoters leaving access for the RNA polymerase [23]. The target promoters of Fur have been identified in E. coli K12 and a consensus Fur binding sequence (or Fur box) has emerged [23]. We identified two Fur boxes in the promoter region of the sci1 T6SS gene cluster, including one overlapping with the putative -10 box. The direct binding of Fur was further confirmed by in vivo Fur titration and in vitro gel shift assays. Interestingly, close analysis of the promoter elements and Fur binding boxes showed an overrepresentation of GATC motifs, which are targets for the DNA adenine methylase Dam. Dam catalyzes methylation at the N6 position of the adenine of the GATC motif. Dam methylation has been shown to be involved in a variety of processes, including the control of the timing of replication, mismatch repair or transcriptional regulation [24]–[27]. In several cases, Dam-dependent methylation modulates DNA-protein interactions [28]). The genes under the control of this mechanism display a phase variation expression pattern, in which binding of the transcriptional factor depends of the methylated state of the DNA [29]. Using a combination of in vivo and in vitro methylation assays, as well as in vitro Fur binding assays on nonmethylated and methylated sci1 promoter, we demonstrated that the sci1 promoter expression depends on the outcome of the competition between Fur binding and Dam-dependent methylation. We propose that the sci1 gene cluster expression undergoes an epigenetic switch, varying from an ON to an OFF state in response to iron availability and DNA replication.
Results
The sci1 T6SS gene cluster is weakly expressed in in vitro conditions
The first gene of the enteroaggregative E. coli (EAEC) sci1 T6SS gene cluster, sciH, is preceded by a 578-bp non-coding sequence, which is hereafter called sci1 promoter. To test the activity of the sci1 promoter, we constructed a lacZ transcriptional reporter fusion in which lacZ expression is controlled by the sci1 promoter region (from –578 to + 18 [relative to the sciH start codon]). The 596-pb DNA fragment corresponding to the sci1 promoter region has been inserted into the SmaI site of the pBR322-derived multicopy plasmid pGE593 [30], encoding a promoterless lacZ gene. Pilot studies performed in LB medium showed that the basal level of β-galactosidase produced from the promoterless lacZ gene was quite high (∼ 180 Miller units), whereas the sci1-lacZ fusion only displayed a 3- to 4-fold higher expression (∼600–800 Miller units). To avoid any artifactual effect on fusion activity due to the high copy number of the plasmid, we deleted the chromosomal pcnB gene, a gene involved in the regulation of the copy number of pBR322-derived vectors [31]. In this background, the activity of the promoterless fusion was considerably decreased (∼ 40 Miller units). We also noted that the sci1-lacZ fusion was poorly expressed in LB rich medium. We then tested the activity of the sci1-lacZ fusion under several conditions. We found that the activity of the sci1-lacZ fusion increased in the late stage of exponential growth phase and in stationary growth phase, after acid exposure or in minimal media (data not shown).
Random mini-Tn10 transposon mutagenesis identified Fur as a regulator of EAEC sci1 expression
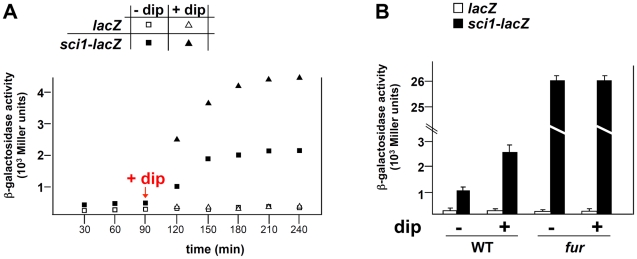
To gain further insight onto the sci1 regulatory mechanism, we performed a transposon mutagenesis to identify regulators. Cells carrying the sci1-lacZ fusion, which form white to light blue colonies on X-Gal LB (pH 8.0) plates, were transformed with the pNKBOR plasposon, a suicide vector carrying a mini-Tn10 transposon and its cognate transposase [32]. We screened ∼20,000 kanamycin-resistant clones for higher lacZ activity and obtained three dark blue clones with a ∼25-fold increase in β-galactosidase activity (data not shown). Sequencing the site of transposition revealed that insertions occurred at three independent positions within the fur gene, which encodes the master regulator of iron and pH homeostasis (data not shown). Since Tn insertions resulted in the disruption of the fur gene, these data suggest that the Fur protein may act as a negative regulator of sci1 gene cluster expression. To independently verify the role of Fur in sci1 expression, we measured the activity of the sci1-lacZ reporter fusion in presence of the iron chelator 2,2′-dipyridyl. As shown in Figure 1A, the activity of the reporter fusion increased upon exposure to dipyridyl. Construction of the EAEC fur null strain confirmed the transposon mutagenesis data (Figure 1B). By contrast, deletion of aggR, the gene encoding the AraC-like transcriptional activator of the EAEC sci2 T6SS gene cluster [33], had no effect on the promoterless and the sci1-lacZ fusions (data not shown).
Figure 1. The EAEC sci1 T6SS gene cluster is regulated by iron levels and the Fur repressor.
(A) β-galactosidase activity of a promoterless lacZ fusion (open symbols) and of the sci1-lacZ reporter fusion (closed symbols) upon addition of the iron chelator 2,2′-dipyridyl (dip; 100 µM, squares) in a EAEC wild-type (WT) strain (triangles: no dip added). (B) β-galactosidase activity of a promoterless lacZ fusion (white bars) and of the sci1-lacZ reporter fusion (black bars) after 120 minutes of culture (OD600nm = 0.8) upon a 30 min treatment with 2,2′-dipyridyl (+dip; 100 µM) or ethanol-carrier (-dip) in a WT strain or its isogenic fur mutant.
Sequence analysis of the sci1 promoter region
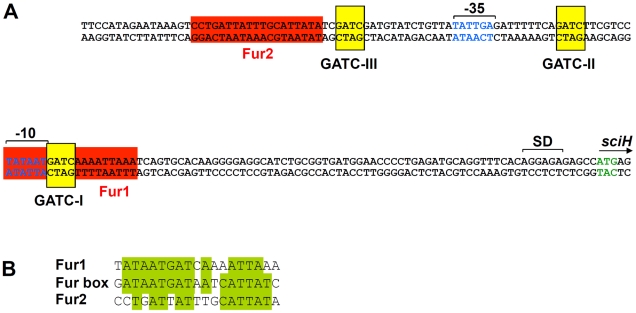
The Fur protein has been extensively studied. Fur acts as a dimer and participates in regulation of genes involved in iron homeostasis and tolerance to acid stresses [23]. Once complexed to iron, Fur binds to a well-defined 19-bp sequence (GATAATGATAATCATTATC), called the ‘Fur box’ [23]. To determine whether the effect of the fur mutation was direct, we first analyzed the sci1 promoter sequence. Interestingly, two putative Fur binding sites were identified (Figure 2A). The fur2 binding box (11 out of the 19 nucleotides of the consensus Fur box, see Figure 2B) is located upstream the putative -35 element of the putative σ70 promoter. fur1 (13 out of the 19 nucleotides of the consensus Fur box, see Figure 2B) overlaps with the putative -10 box. The position of the fur1 box suggests that Fur may prevent RNA polymerase (RNAP) binding to the -10 element, a characteristic commonly observed for transcriptional repressors, including the Fur protein [34], [35]. Overall, the in silico analysis of the sci1 promoter sequence suggests that Fur directly bind to the sci1 promoter.
Figure 2. In silico analysis of the sci1 proximal promoter region.
(A) The proximal sci1 promoter region. The ATG translational codon of sciH is indicated as well as the Shine Delgarno (SD). The putative -10 and -35 elements of the σ70 promoter (identified by the BProm algorithm) are indicated in blue, as well as Fur-binding sequences (red boxes) and GATC Dam-dependent methylation sites (yellow boxes). The two Fur-binding sequences and GATC sites are numbered from the start site. (B) Sequence alignment of the fur1 and fur2 sci1 boxes with the E. coli Fur box consensus sequence. Identical bases are framed in green.
The Fur protein binds to the sci1 promoter at fur1 and fur2
To test whether Fur binds the sci1 promoter in vivo, we used the Fur titration assay (FURTA [36]). In this assay, a chromosomal fhuF::lacZ fusion is derepressed if a Fur box is carried on a high copy plasmid. We thus cloned the sci1 promoter sequence as well as the two putative Fur boxes (the fur1 and fur2 sequences flanked by the natural downstream and upstream 3 bases) and controls (the Fur-dependent cir and Fur-independent sci2 promoters) into the high copy pT7.5 vector. Figure 3A shows that transformation of the reporter strain with pT7.5 derivatives carrying the sci1 promoter or of the two putative Fur boxes derepressed the fhuF::lacZ fusion, leading to a lac + phenotype on MacConkey agar plates supplemented with FeSO4.
Figure 3. Fur binds to the EAEC sci1 T6SS promoter in vivo and in vitro.
(A) Fur Titration assay (FURTA). H1717 reporter cells (fhuF-lacZ) carrying the empty vector or the vector bearing the sci1, sci2, or cir promoters, or the fur1 or fur2 sequences were spotted on MacConkey plates (upper panel) or on MacConkey plates supplemented with FeSO4 (30 µM; lower panel). A lacZ+ phenotype reports a derepression of the fhuF-lacZ reporter fusion by titration of the Fur protein bound to the fhuF promoter. (B) Electrophoretic mobility shift assay of the sci1 promoter (upper panel) or of the fur1 (middle panel) or fur2 (lower panel) sequences using purified Fur (lane 1, no protein; lane 2, 0.5 nM; lane 3, 2 nM, lane 4, 5 nM, lane 5, 20 nM) in presence of FeCl3 or in presence of EDTA (lane 6, Fur at 20 nM) or using purified NtrC transcriptional activator (lane 7, 50 nM). Controls include Fur shift assays of the Fur-dependent cir promoter (lane 8, no protein; lane 9, Fur at 5 nM) or of the Fur-independent sci2 promoter (lane 10, 20 nM). (C) Competition experiments for Fur binding (lane 1, no protein; lanes 2–6, Fur at 20 nM) with duplex consensus Fur- (lane 3, molecular ratio sci1:fur box 1∶2; lane 4, molecular ratio 1∶10) or σ54-binding sequence (lane 5, molecular ratio sci1:σ54-box 1∶2; lane 6, molecular ratio 1∶10). (D) Binding of the Eσ70 RNA polymerase holoenzyme (RNAP) (lanes 1, 4, 7 and 9, no RNAP; lanes 2 and 5, RNAP 0.5 unit; lanes 3, 6, 8 and 10, RNAP 2 units) on the sci1 or control cir promoter pre-incubated (+) or not (−) with Fur (20 nM). Fur-DNA, (Fur)2-DNA, and RNAP-DNA complexes are indicated by *, **, and ǂ respectively.
Fur binding to the sci1 promoter was then confirmed in vitro. The E. coli K12 Fur protein was purified to homogeneity by metal affinity chromatography and tested for its ability to bind the sci1 promoter by electrophoretic mobility shift assays (EMSA) (Figure 3B, upper panel). As expected, the cir promoter fragment was shifted by the Fur protein in a metal-dependent manner, whereas no shift was observed for the sci2 promoter fragment, even at high Fur concentration. The Fur protein bound the sci1 promoter, in a metal-dependent manner. Interestingly, two shifts were observed, suggesting formation of two distinct complexes. Digestion of the sci1 promoter fragment by SspI leads to two fragments, a 384-bp 5′ DNA fragment and a 212-bp 3′ DNA fragment which contains the two putative Fur boxes. Following SspI digestion and EMSA, we observed that the 212-bp fragment—but not the 396-bp fragment—was retarded in the presence of the Fur protein (data not shown). We further tested whether two fragments encompassing the fur1 or the fur2 box were retarded by Fur. Figure 3B shows that both fragments were retarded although the affinity of Fur was higher for the fur1 box. Specificity of Fur binding was further confirmed by using specific (a duplex consensus 19-bp Fur box flanked by 7 bases) or non-specific (a duplex consensus 22-bp σ54 box flanked by 7 bases) unlabelled competitors (Figure 3C). Overall, our data suggest that Fur binds the sci1 promoter, including at a position that overlaps with one of the RNAP-binding elements. We then tested whether Fur exerts a competitive effect to RNAP binding. As expected from the position of the fur1 box relative to the putative -10 element, pre-incubation of the sci1 promoter probe with Fur decreased the affinity of RNAP for the DNA fragment (Figure 3D).
The Fur protein prevents Dam-dependent methylation at GATC-I
Interestingly, sequence analysis of the sci1 promoter also revealed the existence of three GATC motifs over a 53-nucleotide region (see Figure 2A). The adenine of GATC motifs are recognized and methylated by the DNA adenine methylase (Dam). Two of these sites (GATC-II and GATC-III) flank the putative -35 element whereas the third site, GATC-I, is located at the 3′ of the putative -10 element and overlaps with the fur1 box (Figure 2A).
The observation that the fur1 box contains a GATC site (GATC-I) suggests that Fur and Dam-dependent methylation overlap for the regulation of the sci1 T6SS gene cluster. Several examples have been reported of interplays between the methylation at GATC sequences and binding of transcriptional regulators, including Lrp and OxyR, as a phenomenon known as “phase variation” [24]–[29]. We therefore tested whether Fur influences Dam-dependent methylation (Figure 4 and Figure 5) and vice-versa (Figure 6). We took advantage of the observation that each GATC site within the sci1 promoter was part of a 6-nucleotide palindrome sequence (see Figure S1) for which methylation-sensitive or –insensitive restriction endonucleases are commercially available. We then compared the digestion profile of a PCR-generated fragment encompassing the 596-bp of the sci1 promoter. The non-methylated PCR product was digested by all restriction nucleases with the exception of the methylated GATC-specific DpnI enzyme (Figure 4, upper panel). Upon in vitro methylation by Dam, all the sites were methylated and thus, restriction nucleases sensitive to Dam methylation (MboI, BspD1, Hpy188I, and BclI) were inactive on this fragment (Figure 4, middle panel). When Dam was added to a mixture containing the PCR product and an excess of the Fur protein, the Dam-sensitive BspD1 and Hpy188I nucleases were inactive, demonstrating that the GATC-II and –III sites were methylated (Figure 4, lower panel). By contrast, the methylation-sensitive BclI nuclease was active on the GATC-I site, revealing that the GATC-I site remained non-methylated in presence of Fur. These results thus demonstrate than Fur protects GATC-I from methylation in vitro.
Figure 4. Fur protects GATC-I from methylation in vitro.
A radiolabeled PCR product corresponding to the 596-bp sci1 promoter was digested by the restriction enzymes indicated on top (no, no digestion). Upper panel, untreated PCR product; middle panel, PCR product treated with the Dam methylase; lower panel, PCR product incubated with purified Fur (20 nM) prior to Dam methylation. The sizes of the digestion products (in bp) are indicated on the left. Red and blue frames emphasize the observation that incubation with Fur did not change the digestion profiles for GATC-II (-II) and GATC-III (-III) whereas the green frame emphasize the observation that GATC-I (-I) was not methylated upon Fur binding. Schematic representations of the conclusions of the left panels are shown on right. See Figure S1 for positions of restriction sites and sizes of DNA fragments.
Figure 5. Fur protects GATC-I from methylation in vivo.
The sci1 promoters purified from the EAEC wild-type strain (WT, upper panel) or its isogenic dam (second panel rom top) or fur (lower panel) mutant strains, or from the WT strain treated with 2,2′-dipyridyl (WT + dip; third panel from top) were digested by the restriction enzymes indicated on top (no, no digestion). The sizes of the digestion products (in bp) are indicated on the left. Arrows indicate the position of the digestion product obtained with the BclI restriction enzyme emphasizing the observation that GATC-I was not methylated in a WT strain but was methylated in a fur mutant strain (or in a WT strain treated with 2,2′-dipyridyl). Schematic representations of the conclusions of the left panels are shown on right. See Figure S1 for positions of restriction sites and sizes of DNA fragments.
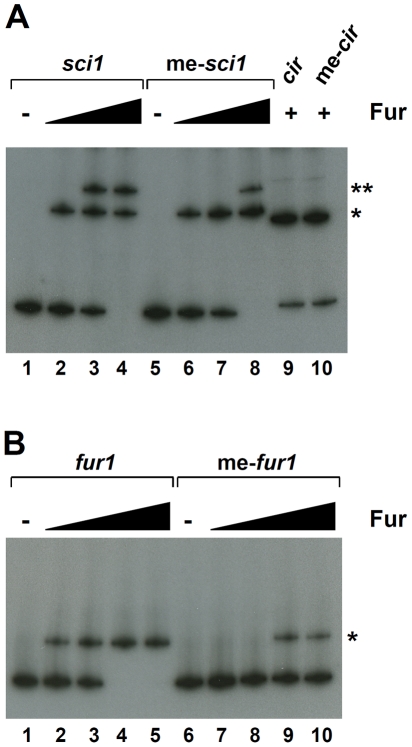
Figure 6. GATC methylation influences Fur binding on fur1.
(A) Electrophoretic mobility shift assay of the non methylated or Dam-methylated (me-) sci1 or cir promoter using purified Fur (lanes 1 and 5, no protein; lanes 2 and 6, 2 nM; lanes 3 and 7, 5 nM; lanes 4 and 8-10, 20 nM). (B) Electrophoretic mobility shift assay of the non methylated or Dam-methylated (me-) fur1 sequence using purified Fur (lanes 1 and 6, no protein; lanes 2 and 7, 0.5 nM; lanes 3 and 8, 2 nM; lanes 4 and 9, 5 nM; lanes 5 and 10, 20 nM). Fur-DNA, (Fur)2-DNA complexes are indicated by *, and ** respectively.
We then tested the methylation state of each GATC sequence of the sci1 promoter in vivo. The plasmid carrying the sci1-lacZ fusion was extracted from various genetic backgrounds and each GATC site was analyzed using the restriction assay. In the dam derivative, none of the sites was methylated (Figure 5, second panel from top). Interestingly, the GATC-II and –III sites were methylated in a wild-type background as shown by the absence of activity of the methylation-sensitive BspDI and Hpy188I nucleases on the promoter substrate (Figure 5, top panel). However, GATC-I remained nonmethylated, suggesting it is protected from Dam-dependent methylation in vivo. This protection was due to the presence of the Fur protein bound to the fur1 box since the GATC-I site was fully methylated in fur mutant cells (Figure 5, lower panel) or when wild-type cells were exposed to 2,2′-dipyridyl (Figure 5, third panel from top). Overall, the results showed in Figure 4 and Figure 5 demonstrate that Fur binding prevents DNA adenine methylation at the GATC-I site.
GATC-I methylation decreases affinity of Fur for the fur1 binding site
Reciprocally, we tested the effect of DNA adenine methylation on Fur binding. A radiolabelled PCR product was methylated in vitro by Dam, and then used as probe in electrophoretic mobility shift assays. As shown in Figure 6, gel shift assays demonstrated that methylation of the sci1 promoter fragment decreased the affinity of Fur whereas had no effect on the control cir promoter (Figure 6A). However, only one of the two Fur binding boxes seemed affected since methylation of the sci1 promoter affected the formation of the (Fur)2-DNA complex. We therefore tested whether methylation of the fur1 box affected Fur binding. As shown in panel (B), methylation of the fur1 box had a negative impact on Fur binding (Figure 6B).
Discussion
In this study, we have shown that the sci1 Type VI secretion gene cluster is poorly expressed under the laboratory conditions. Using a combination of random mutagenesis, reporter fusion, titration and gel mobility shift assays, we have shown that Fur represses the expression of this cluster. We also demonstrated that expression of this cluster is modulated by DNA adenine methylation at the GATC-I site by the Dam methylase. Using in vivo and in vitro methylation protection assays, we showed that Fur binding prevents methylation at the GATC-I site, whereas methylation at GATC-I decreases affinity of Fur for its binding sequence (see Figure 7 and below).
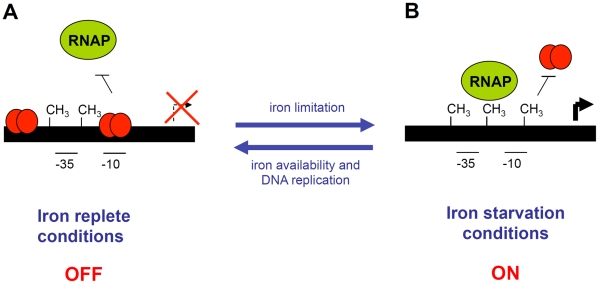
Figure 7. Schematic representation of the EAEC sci1 T6SS gene cluster epigenetic switch regulatory mechanism.
(A) In iron-replete conditions, Fur (red balls) represses the expression of the sci1 gene cluster by binding to specific boxes overlapping the putative -10 transcriptional element. The expression of the sci1 gene cluster is in the OFF phase. (B) In iron starvation conditions, Fur is relieved from the putative -10 element, leaving the promoter available for RNAP binding and transcription. Dam-dependent methylation at the GATC-I site prevents Fur binding. The expression of the sci1 gene cluster is in the ON phase. Transition to the OFF phase requires both iron replete conditions and hemi-methylation of GATC-I after DNA replication. Methylated GATC sites are indicated by CH3.
Fur regulation
Although suggested for the regulation of the Edwardsiella tarda Evp and the distantly-related Francisella tularensis FPI Type VI secretion systems, the role of the Fur protein has not yet been characterized in the regulation of these clusters [20]. In the case of the sci1 gene cluster, we identified two Fur boxes, including one—the fur1 box—overlapping with the putative -10 box of the putative σ70-dependent promoter. Indeed, using competition gel shift experiments we have shown that Fur prevents RNA polymerase binding at the sci1 promoter. The fur2 box being located upstream the promoter, its role is not yet clear, but one may hypothesize that this second, lower affinity, box has a role in cooperativity, i.e., increasing the local concentration of the Fur protein around the sci1 promoter. Further experiments are therefore required to clearly understand the specific role of each Fur binding box.
Dam regulation
We also observed a role of Dam methylation in the regulation of the EAEC sci1 gene cluster. Interestingly, Dam methylation site are over-represented in the sci1 promoter: whereas a GATC sequence should statistically be present every ∼250 bases, three GATC sequences are present within a 53-nucleotide sequence flanking the putative -35 and -10 elements of the promoter. This represents a 10-fold increase over random average. Dam methylation has been shown to be involved in a variety of processes such as mismatch repair, temporal regulation of the replication, as well as transcriptional regulation [25], [28]. In this latter case, it is noteworthy that most Dam-dependent gene regulations have been identified in the case of genes encoding cell surface structures such as conjugation machines, Type III secretion systems, several pili and fimbriae, adhesines, or enzymes required for the modification of the antigen O of the lipopolysaccharide [25], [37]. In several of these cases, methylation may cause biphase or phase variation, allowing that bacteria produce variable structures at the cell surface and to escape host immunogenicity. Although T6SS-dependent structures have not been yet observed at the cell surface, the presence of both Hcp and VgrG in cell culture supernatant and the homologies of these proteins with tail tube and syringe components of the bacteriophage T4 suggest that these proteins might form an extracellular appendice. It is noteworthy that if this were the case, the regulation of this cell surface structure will be dependent on Dam, which has been previously shown to be involved in the regulation of various pili, fimbriae or adhesines. More striking, these cell surface structures are involved in adhesion or biofilm formation such as the EAEC sci1 Type VI secretion system [4].
Fur and Dam interplay
The most interesting data generated in this study concerns the competitive effect of the Fur and Dam proteins at the -10 box. Using in vivo and in vitro methylation protection assays, we found that Fur binding prevents methylation at the GATC-I site. We observed that GATC-I was unmethylated in the WT strain, whereas became methylated upon exposure of this strain to 2,2′-dipyridyl or in a fur mutant strain. It is noteworthy that these experiments were done using a low copy plasmid, and that it remains to test whether identical methylation patterns occur on the chromosome. Methylation of a GATC site depending on the presence of a regulatory protein has been exemplified in several cases (see [25]) such as the regulation of the gut, carAB or agn43 operons, in which binding of the GutR, CarP or OxyR transcriptional factors on the promoter prevents Dam methylation [38]–[40]. In reciprocal experiments, using gel mobility shift assays, we showed that Fur had a lower affinity for the fur1 box upon GATC-I methylation. Therefore, the addition of a methyl group on the adenine of the GATC-I motif is sufficient to diminish the Fur affinity for the fur1 box. This nucleotide is conserved in the consensus Fur binding motif (see Figure 2B) and the adjacent thymine residue is directly engaged in interaction with the repressor [23]. Consistent with these findings, Dam-dependent methylation abrogating binding of transcriptional activators had been reported [41], [42]. It has been suggested that methylation interferes with transcriptional factors binding by direct steric occlusion or by the local modification in the DNA conformation [28].
From these data, we propose that the expression of the sci1 Type VI secretion gene cluster is under the control of a regulatory mechanism controlling transitions between ON and OFF expression states (Figure 7). In this model, the switch will be controlled by the level of iron and by an epigenetic mechanism involving the Dam methylase. In iron rich conditions, Fur will prevent RNAP binding at the -10 box, as well as GATC-I site methylation: the expression of the sci1 gene cluster will be stably maintained in a repressed state (OFF phase). To switch to the ON state, Fur must be displaced from the fur1 box. In iron-limited conditions, Fur will be relieved from the promoter, allowing expression of the gene cluster. Fur displacement will leave the GATC-I site available for Dam-dependent methylation, which will in turn prevent de novo Fur binding, allowing the expression of the sci1 gene cluster to be maintained in a stable ON phase. Dam thereby acts as a positive regulator by stabilizing the ON expression state. In this model, fur mutant cells are locked in the ON state, whereas dam mutant cells are locked in the OFF phase, which is consistent with our reporter fusion studies.
Although different, this elegant mechanism resembles phase variation, the better studied examples being the regulation of agn43 which involves the OxyR repressor and Dam methylation [40]–[43], and the Pap (pyelonephritis-associated pili) switch, which involves the Leucine-responsive protein Lrp and Dam methylation (for a review, see [44]). In both cases, competition between the transcriptional factor and Dam methylation allows the transition between the OFF and ON transcriptional states [25]. In phase variation mechanisms, the stochastic passage from the ON to the OFF phase leads to the formation of subpopulations. In the mechanism described in this study, the expression of the sci1 T6SS is controlled by iron levels and hence, no subpopulations have been observed (data not shown). Although epigenetic switches resulting from competition between Dam methylation and transcriptional activators have been reported, this is the first study demonstrating competitive effects between Dam and the Fur repressor. It is noteworthy that no GATC site is found in the putative Fur-binding boxes of the E. tarda or F. tularensis T6SS gene clusters, suggesting that the Fur/Dam interplay mechanism is not widely distributed for the expression of T6SS gene clusters.
A model of the sci1 epigenetic switch
In the case of the Pap switch, it has been shown that transition to the ON phase requires the PapI protein, the expression of which is induced by PapB. The OFF-ON transition is therefore controlled by PapB and PapI. It has been proposed that this modulation is a stochastic event, although environmental factors may somehow influence this modulation [44]. In the case of the sci1 promoter, the passage of the OFF to the ON phase is probably less random, and is essentially dependent upon the iron availability; however maintenance in a stable ON phase requires methylation of the GATC-I site. How the bacteria manage the passage from the ON to the OFF phase is an interesting question. The ON to OFF switch requires a change from a methylated to nonmethylated state of the GATC-I sequence, which can only occurs upon DNA replication. In the case of agn43, it has been shown that OxyR can bind to the hemimethylated operator upon DNA replication [45]. However, it is noteworthy that GATC-I methylation decreases Fur affinity for fur1 (and does not abrogate it). It remains possible that Fur binds to a hemimethylated fur1 box, therefore facilitating the ON to OFF switch during DNA replication in iron replete conditions. In this model, the absence of DNA replication should maintain the expression of the sci1 gene cluster in an ON state, irrespective of iron levels, a hypothesis that remains to be tested. Further studies using quantitative competition experiments between Fur and Dam methylation, as well as Fur binding on hemimethylated templates will probably provide important details on this mechanism. In sum, we suggest that Dam methylation will therefore serve two roles: (i) preventing de novo Fur binding in absence of DNA replication (irrespective of iron levels), and (ii) slowing down de novo Fur binding upon DNA replication. This mechanism will timely regulate expression of the sci1 T6SS gene cluster.
On a physiological perspective, it is difficult to reconcile the regulatory mechanism dissected in this study and the role of the EAEC T6SS in the timing of biofilm formation. One may hypothesize that iron limitation occurs in the digestive track due to high competition between micro-organisms or by the activation of host mechanisms to sequester iron at the mucosal surface [46]. Several regulatory mechanisms have thus been developed by pathogens to induce virulence genes expression in response to iron starvation [46]. Iron starvation may thus have been hijacked by EAEC to initiate the switch of the population to the ON phase, and thus to timely regulate biofilm formation.
Materials and Methods
Bacterial strains, media, growth condition, and chemicals
Strains used in this study and their relevant characteristics are listed in Table S1. Escherichia coli K12 DH5α was used for cloning procedures. The enteroaggregative E. coli strain 17-2 (kindly provided by Arlette Darfeuille-Michaud, University of Clermont-Ferrand, France) was used for this study. The FURTA reporter strain (H1717, fhuF::lacZ [36]) was generously provided by Klaus Hantke (Tuebingen Universitat, Germany). The E. coli K12 BW25113furΩkan and damΩkan strains from the KEIO collection [47] were obtained through Patrice L. Moreau (LCB, Marseille). Strains were routinely grown in LB broth at 37°C, with aeration. When required, M9 minimal medium supplemented with glucose 0.4% or MacConkey agar (purchased from Difco) were used. Plasmids and cassettes were maintained by the addition of ampicillin (100 µg/ml for K12, 200 µg/ml for EAEC), kanamycin (50 µg/ml for K12, 50 µg/ml for chromosomal insertion on EAEC, 100 µg/ml for plasmid-bearing EAEC), or chloramphenicol (40 µg/ml). Bromo-chloro-indolyl-β-D-galactopyranoside (X-Gal), Iso-propyl-thio-galactopyranoside (IPTG) and 2,2′-dipyridyl were purchased from Fluka. RNA polymerase holoenzyme (saturated with σ70) was purchased from Epicentre Biotechnologies. Custom oligonucleotides were synthesized by Eurogentec. With the exception of the Pfu Turbo Taq polymerase (Stratagen), restrictions and modification enzymes were purchased from New England Biolabs.
Strain constructions
Deletion of the lacZ gene into the wild-type EAEC 17-2 strain was performed using the modified one-step inactivation procedure [48] with the pKOBEG plasmid [49] and oligonucleotides carrying 50-nucleotide extensions homologous to regions adjacent to the target gene (oligonucleotides are listed in Table S1). White colonies were screened on kanamycin LB plates supplemented with X-Gal (40 µg/ml) and IPTG (100 µM). The kanamycin cassette was then excised using plasmid pCP20. Deletion of the pcnB gene was performed into the 17-2ΔlacZ strain using the same procedure. The corresponding strain, 17-2ΔlacZΔpcnB, is considered as the WT reporter strain throughout the study. Deletions of the fur and of the aggR genes were done similarly into the reporter strain, whereas deletion of the dam gene was done into the reporter strain and its Δfur derivative (oligonucleotides listed in Table S1).
Plasmid constructions
Plasmids used in this study are listed in Table S1. Plasmids pKOBEG [49] and pCP20 [48] were provided by Arlette Darfeuille-Michaud and Barry Wanner respectively. Plasmid pBT4-1 [50], carrying the E. coli K12 fur gene was kindly provided by Sam Dukan and Maialene Chabalier through the Danièle Touati's strain collection. Plasmid pNKBOR [32] was kindky provided by Daniel Vinella. Plasmids encoding the transcriptional fusions were constructed by cloning the promoters from the sci1 and sci2 T6SS gene clusters (amplified by PCR using the Pfu Turbo [Stratagene] polymerase and corresponding oligonucleotides; sci1, from –578 to +18 relative to the initiation start codon of sciH (EC042_4524) [nucleotides 4852298-4852892; 596-bp]; sci2, from –421 to + 46 relative to the initiation start codon of aaiA (EC042_4562) [nucleotides 4892656-4893121; 467-bp]) upstream the 'lacZ gene into the blunt SmaI site of the dephosphorylated pGE593 vector [30]. In these constructions, lacZ is under the control of the promoter of the corresponding gene. pT7.5 [51] derivatives carrying the sci1, sci2, and cirA promoters were engineered by a double PCR technique, allowing amplification of the DNA sequence of interest flanked by extensions annealing to the target vector. The product of the first PCR was then used as oligonucleotides for a second PCR using the target vector as template [4], [52]. pT7.5 derivatives carrying the fur1 and fur2 boxes of the sci1 promoter were obtained by insertion using insertion quick change mutagenesis and complementary pairs of oligonucleotides. All constructs have been verified by restriction analyses and DNA sequencing (Genome Express).
Beta-galactosidase assay
β-galactosidase activity was measured on whole cells by the method of Miller [55]. Reported values represent the average of at least three independent triplicates with a variation of less than 10% from the mean (standard deviation shown on graphics).
Random mutagenesis using pNKBOR plasposon
The EAECΔlacZΔpcnB strain carrying the reporter fusion plasmid pGE593-sci1 was randomly mutagenized using the pNKBOR plasposon [32]. 350 ng of pNKBOR were transformed into EAEC ΔlacZΔpcnB electro-competent cells and the cell suspension was spread on fifty 20-cm LB agar plates (pH 8.0) supplemented with 100 µg/ml of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) and 50 µg/ml of kanamycine. Blue colonies were picked and streaked on identical plates. After validation, chromosomal DNAs were purified from overnight culture using the DNeasy Blood and Tissue kit (Qiagen), and digested by BglII. Upon ligation (T4 DNA ligase, Promega), fragments were transformed into CC118λpir electro-competent cells and spread on LB agar plates supplemented with kanamycine. Plasmids carrying the pNKBOR insert were extracted, verified by restriction analyses, and the chromosome-pNKBOR junction site was sequenced.
In vivo Fur binding assay: Fur titration assay (FURTA)
The Fur titration assay relies on the activity of the fhuF::lacZ chromosomal transcriptional fusion in presence of a sequence carried on a multicopy plasmid [36]. In absence of plasmid or in presence of the pT7.5 vector, Fur represses the fhuF::lacZ fusion, leading to a lac− phenotype. The presence of a Fur binding site on the multicopy plasmid leads to a de-repression of the fhuF::lacZ transcriptional fusion, and a lac+ phenotype. Briefly, 5 µl of an exponential culture of the H1717 strain bearing the pT7.5 vector or derivatives were spotted on a MacConkey plate supplemented with ampicillin and FeS04 30 µM. Controls to verify that the fhuF::lacZ fusion was active and responsive to iron levels in all the strains tested were done in parallel by spotting the same cultures on MacConkey plates supplemented with ampicillin (iron starved medium leading to de-repression [i.e., lac+ phenotype] in all cases).
Fur purification
This purification procedure is based on the ability of the native, non-recombinant Fur protein to bind metal affinity beads. The E. coli K12 Fur proteins (100% identical to the EAEC Fur protein) was purified by ion-metal affinity chromatography from DH5α carrying the pBT4-1 plasmid after induction with IPTG (200 µM) for 3 hours at 37°C as previously described [50]. 1011 cells were harvested, resuspended in buffer A (Tris-HCl 20 mM, NaCl 100 mM, Imidazole 5 mM, pH 8.0) supplemented with MgCl2 10 mM, DNase 100 µg/ml and RNase 100 µg/ml, and disrupted by French Press. The cleared cell lysate containing the soluble fraction was loaded on a cobalt column (Talon, Clontech) equilibrated with buffer A, and after extensive washing with buffer A, the Fur protein was eluted in buffer A supplemented with 400 mM imidazole. Fractions were pooled, dialysed against buffer B (Tris-HCl 10 mM, NaCl 100 mM, pH 7.2) and 10-fold concentrated using the Amicon technology (Millipore, cut-off of 3,000 Da) before storage at -80°C. The final concentration of the Fur protein was 1.30 mg/ml.
Electrophoretic mobility shift assay—EMSA
PCR products were generated using a mix of dNTPs supplemented with [α-32P]dGTP (5 µCi per PCR in a total volume of 50 µl; Perkin-Elmer), and purified using the Wizard Gel and PCR clean-up kit (Promega). Dam methylated substrates were prepared as described below except that the column-purified PCR products were digested by MboI (to hydrolyze un-methylated substrates) and full-length fragments were gel-purified using the Wizard gel and PCR clean-up kit (Promega). EMSAs with Fur and RNA polymerase holo-enzyme (RNAP) were adapted from previously published protocols [53], [54]. PCR products were incubated in a final volume of 10 µl in Fur EMSA binding buffer (10 mM Tris-borate, 40 mM KCl, 5% glycerol, 1 mM MgCl2, 100 µM MnCl2, 2 mM DTT, BSA 100 µg/ml and sonicated salmon sperm DNA 1 µg/ml, pH 7.5) at the concentration of 2 nM with increasing concentrations of Fur, of RNA polymerase holo-enzyme (saturated in σ70, Epicentre Biotechnologies), or of both Fur and RNAP. In competition experiments, Fur was added 5 min prior to RNAP addition. The mixture was incubated for 30 minutes at 25°C and then loaded on a pre-run 8% non denaturing polyacrylamide (Tris-borate) gel, and DNA and DNA-complexes were separated at 100 V in Tris-Borate buffer (45 mM Tris base, 45 mM boric acid, 100 µM MnCl2 buffer). Gels were fixed in 10% trichloro-acetic acid for 10 minutes, and exposed to Kodak BioMax MR films.
In vitro Dam methylation and endonuclease restriction assays
The in vitro methylation protection assay was done as previously published [38] with modifications. Briefly, purified radio-labelled PCR products were in vitro methylated by the Dam methylase (New England Biolabs) in methylation buffer (50 mM Tris-HCl pH 7.5, 5 mM dithiothreitol (DTT), 5% glycerol, 20 mM KCl, 1 mM MgCl2, 100 µM MnCl2, bovine serum albumine (BSA) 100 µg/ml, in presence of 80 µM of S-adenosylmethionine (SAM)), as recommended by the manufacturer at 37°C for 4 hours. For competition experiments, the PCR products were first incubated with purified the Fur protein in methylation buffer for 30 min at 37°C before addition of the Dam methylase. Half of the mixture was loaded on an acrylamide gel to verify Fur binding by EMSA. The remaining was treated for 20 min at 65°C to heat-inactivate the Dam methylase, and column-purified. 5 nmoles of the PCR products were then digested by the indicated restriction endonucleases (all purchased from New England Biolabs) in the buffers recommended by the manufacturer. DNA fragments were resolved on a pre-run denaturing 12%-acrylamide gel at 200 V in TBE buffer. Gels were fixed in 10% trichloro-acetic acid for 10 minutes, and exposed to Kodak BioMax MR films.
In vivo Dam methylation assay
The promoter-fusion vector pGE593-sci1 was extracted from 5×109 exponentially-growing cells from various backgrounds (wild-type, Δdam, Δfur) treated or not with 200 µM 2,2′-di-pyridyl using the Wizard Miniprep kit (Promega). Plasmids were digested by EcoRI and BamHI and the 600-bp promoter fragments were gel-purified (Wizard Gel and PCR clean-up kit, Promega). 200 ng of the fragment were then subjected to digestion by the indicated restriction endonucleases in the buffers recommended by the manufacturer. DNA fragments were resolved on a pre-run denaturing 12%-acrylamide gel at 200 V in TBE buffer and visualized under ultra-violet after GelRed staining as recommended by the manufacturer (FluoProbes).
Supporting Information
Schematic representation of the sci1 promoter region. The position of the fur1 and fur2 boxes and of the GATC sites are indicated (GATC-dis, distal GATC). Each GATC is part of a palindrome sequence recognized by specific methylation-sensitive (underlined name), methylation-insensitive (plain name) or methylation-dependent (italicized name) restriction enzymes. The size of the digestion products obtained for each enzyme (if accessible for digest) is indicated. Please note that Hpy188I has a palindromic penta-nucleotide recognition sequence, and therefore is only sensitive to methylation of top strand.
(PPT)
Strains, plasmids, and oligonucleotides used in this study. a sequence adjacent to the target gene underlined, sequence annealing on pKD4 italicized. b consensus sequence underlined. c sequence complementary to target vector underlined. d sequence annealing on the target vector underlined. e sequence complementary to B oligonucleotide italicized.
(PDF)
Acknowledgments
We thank Arlette Darfeuille-Michaud (Université de Clermont-Ferrand, France) for strain 17-2; Daniel Vinella, Julia Bos and Laurent Loiseau (LCB, Marseille, France) for advice and protocols for transposon mutagenesis; Klaus Hantke (Tuebingen Universitat, Germany) for the FURTA reporter strain; Danièle Touati, Sam Dukan, and Maïalène Chabalier for plasmid pBT4-1; and members of the Lloubès, Bouveret and Sturgis research groups for discussions and encouragements. A special thanks to Mireille Ansaldi for strains, plasmids, protocols, and discussions; Frédéric Barras for the early observation of Dam methylation sites within the sci1 promoter sequence, fruitful discussions, and advice; Pepe for critical reading of the manuscript and encouragement; Patrick Stragier for discussions; and Sophie Fonphec for encouragement.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the Institut National des Sciences Biologiques of the Centre National de la Recherche Scientifique through a PEPS grant (Projet Exploratoire - Premier Soutien, SDV.2009-1935) and an Agence Nationale de la Recherche grant (ANR-10-JCJC-1303-03) to EC. YRB is supported by a doctoral fellowship from the French Ministère de la Recherche et de la Technologie. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner's guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Cascales E. The Type VI secretion toolkit. EMBO Rep. 2008;9:735–41. doi: 10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 2008;154:1570–83. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- 4.Aschtgen MS, Bernard CS, De Bentzmann S, Lloubès R, Cascales E. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol. 2008;190:7523–31. doi: 10.1128/JB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aschtgen MS, Gavioli M, Dessen A, Lloubès R, Cascales E. The SciZ protein anchors the enteroaggregative Escherichia coli Type VI secretion system to the cell wall. Mol Microbiol. 2010;75:886–99. doi: 10.1111/j.1365-2958.2009.07028.x. [DOI] [PubMed] [Google Scholar]
- 6.Aschtgen MS, Thomas MS, Cascales E. Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP… what else? Virulence. 2010;1:535–40. doi: 10.4161/viru.1.6.13732. [DOI] [PubMed] [Google Scholar]
- 7.Kanamaru S. Structural similarity of tailed phages and pathogenic bacterial secretion systems. Proc Natl Acad Sci USA. 2009;106:4067–8. doi: 10.1073/pnas.0901205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–30. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci USA. 2009;106:4160–5. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106:4154–9. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–7. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 12.Pukatzki S, McAuley SB, Miyata ST. The type VI secretion system: translocation of effectors and effector-domains. Curr Opin Microbiol. 2009;12:11–7. doi: 10.1016/j.mib.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–33. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwarz S, West TE, Boyer F, Chiang W-C, Carl MA, et al. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010;6:e1001068. doi: 10.1371/journal.ppat.1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends Microbiol. 2010;18:531–7. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci USA. 2010;107:19520–4. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jani AJ, Cotter PA. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe. 2010;8:2–6. doi: 10.1016/j.chom.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber B, Hasic M, Chen C, Wai SN, Milton DL. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ Microbiol. 2009;11:3018–28. doi: 10.1111/j.1462-2920.2009.02005.x. [DOI] [PubMed] [Google Scholar]
- 20.Bernard CS, Brunet YR, Gueguen E, Cascales E. Nooks and crannies in type VI secretion regulation. J Bacteriol. 2010;192:3850–60. doi: 10.1128/JB.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung KY, Siame BA, Snowball H, Mok YK. Type VI secretion regulation: crosstalk and intracellular communication. Curr Opin Microbiol. 2011;14:9–15. doi: 10.1016/j.mib.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Bernard CS, Brunet YR, Gavioli M, Lloubès R, Cascales E. Regulation of Type VI secretion gene clusters by σ54 and cognate enhancer binding proteins. J Bacteriol. 2011;193:2158–2167. doi: 10.1128/JB.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escolar L, Pérez-Martín J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–9. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barras F, Marinus MG. The great GATC: DNA methylation in E. coli. Trends Genet. 1989;5:139–43. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- 25.Casadesús J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–56. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marinus MG, Casadesus J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol Rev. 2009;33:488–503. doi: 10.1111/j.1574-6976.2008.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Løbner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Curr Opin Microbiol. 2005;8:154–60. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Low DA, Weyand NJ, Mahan MJ. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun. 2001;69:7197–204. doi: 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Woude MW. Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol. 2011;14:205–211. doi: 10.1016/j.mib.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Eraso JM, Weinstock GM. Anaerobic control of colicin E1 production. J Bacteriol. 1992;174:5101–9. doi: 10.1128/jb.174.15.5101-5109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–90. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 32.Rossignol M, Basset A, Espéli O, Boccard F. NKBOR, a mini-Tn10-based transposon for random insertion in the chromosome of Gram-negative bacteria and the rapid recovery of sequences flanking the insertion sites in Escherichia coli. Res Microbiol. 2001;152:481–5. doi: 10.1016/s0923-2508(01)01221-9. [DOI] [PubMed] [Google Scholar]
- 33.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:1267–82. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 34.Escolar L, de Lorenzo V, Pérez-Martín J. Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol Microbiol. 1997;26:799–808. doi: 10.1046/j.1365-2958.1997.6211987.x. [DOI] [PubMed] [Google Scholar]
- 35.Escolar L, Pérez-Martín J, de Lorenzo V. Coordinated repression in vitro of the divergent fepA-fes promoters of Escherichia coli by the iron uptake regulation (Fur) protein. J Bacteriol. 1998;180:2579–82. doi: 10.1128/jb.180.9.2579-2582.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stojiljkovic I, Bäumler AJ, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J Mol Biol. 1994;236:531–45. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 37.Broadbent SE, Davies MR, van der Woude MW. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol Microbiol. 2010;77:337–53. doi: 10.1111/j.1365-2958.2010.07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Woude M, Hale WB, Low DA. Formation of DNA methylation patterns: nonmethylated GATC sequences in gut and pap operons. J Bacteriol. 1998;180:5913–20. doi: 10.1128/jb.180.22.5913-5920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charlier D, Gigot D, Huysveld N, Roovers M, Piérard A, et al. Pyrimidine regulation of the Escherichia coli and Salmonella typhimurium carAB operons: CarP and integration host factor (IHF) modulate the methylation status of a GATC site present in the control region. J Mol Biol. 1995;250:383–91. doi: 10.1006/jmbi.1995.0384. [DOI] [PubMed] [Google Scholar]
- 40.Kaminska R, van der Woude MW. Establishing and maintaining sequestration of Dam target sites for phase variation of agn43 in Escherichia coli. J Bacteriol. 2010;192:1937–45. doi: 10.1128/JB.01629-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haagmans W, van der Woude M. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol Microbiol. 2000;35:877–87. doi: 10.1046/j.1365-2958.2000.01762.x. [DOI] [PubMed] [Google Scholar]
- 42.Waldron DE, Owen P, Dorman CJ. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol Microbiol. 2002;44:509–20. doi: 10.1046/j.1365-2958.2002.02905.x. [DOI] [PubMed] [Google Scholar]
- 43.Henderson IR, Owen P. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J Bacteriol. 1999;181:2132–41. doi: 10.1128/jb.181.7.2132-2141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernday A, Braaten B, Low D. The intricate workings of a bacterial epigenetic switch. Adv Exp Med Biol. 2004;547:83–9. doi: 10.1007/978-1-4419-8861-4_7. [DOI] [PubMed] [Google Scholar]
- 45.Correnti J, Munster V, Chan T, Woude M. Dam-dependent phase variation of Ag43 in Escherichia coli is altered in a seqA mutant. Mol Microbiol. 2002;44:521–32. doi: 10.1046/j.1365-2958.2002.02918.x. [DOI] [PubMed] [Google Scholar]
- 46.Carpenter BM, Whitmire JM, Merrell DS. This is not your mother's repressor: the complex role of Fur in pathogenesis. Infect Immun. 2009;77:2590–601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaveroche MK, Ghigo JM, d'Enfert C. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 2000;28:E97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tardat B, Touati D. Iron and oxygen regulation of Escherichia coli MnSOD expression: competition between the global regulators Fur and ArcA for binding to DNA. Mol Microbiol. 1993;9:53–63. doi: 10.1111/j.1365-2958.1993.tb01668.x. [DOI] [PubMed] [Google Scholar]
- 51.Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–8. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Ent F, Löwe J. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods. 2006;67:67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Dubrac S, Touati D. Fur-mediated transcriptional and post-transcriptional regulation of FeSOD expression in Escherichia coli. Microbiology. 2002;148:147–56. doi: 10.1099/00221287-148-1-147. [DOI] [PubMed] [Google Scholar]
- 54.Lithgow JK, Haider F, Roberts IS, Green J. Alternate SlyA and H-NS nucleoprotein complexes control hlyE expression in Escherichia coli K-12. Mol Microbiol. 2007;66:685–98. doi: 10.1111/j.1365-2958.2007.05950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller J. Cold Spring Harbor Laboratory, NY; 1972. Experiments in Molecular Genetics, p.352-355. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of the sci1 promoter region. The position of the fur1 and fur2 boxes and of the GATC sites are indicated (GATC-dis, distal GATC). Each GATC is part of a palindrome sequence recognized by specific methylation-sensitive (underlined name), methylation-insensitive (plain name) or methylation-dependent (italicized name) restriction enzymes. The size of the digestion products obtained for each enzyme (if accessible for digest) is indicated. Please note that Hpy188I has a palindromic penta-nucleotide recognition sequence, and therefore is only sensitive to methylation of top strand.
(PPT)
Strains, plasmids, and oligonucleotides used in this study. a sequence adjacent to the target gene underlined, sequence annealing on pKD4 italicized. b consensus sequence underlined. c sequence complementary to target vector underlined. d sequence annealing on the target vector underlined. e sequence complementary to B oligonucleotide italicized.
(PDF)