Abstract
Cohesin is a member of the Smc family of protein complexes that mediates higher-order chromosome structure by tethering different regions of chromatin. We present a new in vitro system that assembles cohesin-DNA complexes with in vivo properties. The assembly of these physiological salt-resistant complexes requires the cohesin holo-complex, its ability to bind ATP, the cohesin loader Scc2p and a closed DNA topology. Both the number of cohesin molecules bound to the DNA substrate and their distribution on the DNA substrate are limited. Cohesin and Scc2p bind preferentially to cohesin associated regions (CARs), DNA sequences with enriched cohesin binding in vivo. A subsequence of CARC1 promotes cohesin binding to neighboring sequences within CARC1. The enhancer-like function of this sequence is validated by in vivo deletion analysis. By demonstrating the physiological relevance of these in vitro assembled cohesin-DNA complexes, we establish our in vitro system as a powerful tool to elucidate the mechanism of cohesin and other Smc complexes.
Keywords: cohesion, DNA binding, structural maintenance of chromosomes, yeast
Cohesin mediates higher-order chromosome structure by binding and tethering different regions of chromatin. These cohesin activities are important for sister chromatid cohesion, condensation, transcription, and DNA repair (recently reviewed in refs. 1 and 2). Despite the importance of cohesin to genome integrity and chromosome dynamics, the molecular mechanism by which cohesin binds and tethers chromatin fibers remains to be elucidated. The complex architecture of cohesin and the complex features of its in vivo chromatin binding have impeded attempts to elucidate its molecular function.
At the core of cohesin’s architecture are the Smc1p and Smc3p subunits, which are orthologs of the structural maintenance of chromosomes (SMC) ATPases family. Each Smc protein contains a central hinge domain flanked by two long coiled-coil domains that culminate in a head domain. The Smc proteins form a large ring by dimerization of their hinge domains at one end and the their head domains at the other end. The dimerization of the heads also generates two ABC-like ATPase domains. The third subunit, Mcd1p/Scc1p/Rad21p, links the heads together, whereas a fourth subunit Scc3p/Irr1p/SAp binds to Mcd1p (3, 4). This complex architecture coupled with the fact that none of the cohesin subunits have known DNA binding motifs indicated early on that cohesin binds DNA by a mechanism distinct from other DNA binding proteins.
Additional evidence for a different mechanism of binding came from in vivo studies of cohesin function. First, its DNA binding is not attributable to a single subunit but rather requires the assembly and function of all the subunits. In vivo assembled cohesin/chromosome complexes can be very stable as evidenced by the inability to elute cohesin from isolated chromosomes with 1.6 M salt (5). Binding to chromosomes also requires ATP binding to both Smc1p and Smc3p (6–8). In all organisms, cohesins bind to specific regions on chromosome arms as well as to large domains in the pericentric region (9, 10). In budding and fission yeast, cohesins bind to highly AT-rich intergenic regions covering 0.5–2.0 Kb (9–12). These broad binding sites are called cohesin associated regions (CARs) (9). The broad size of CARs and their AT richness have led researchers to assume that cohesin binding to chromosomes is not dictated by DNA sequence. Finally, the association of cohesin with chromosomes requires the Scc2p/Scc4p complex that loosely interacts with cohesin but whose molecular function is unknown (5). For example, Scc2p/Scc4p complex may act directly on cohesin or recruit chromatin remodeling factors to allow cohesin access to DNA. Taken together, the unusual features of CARs, the requirement for ATP and multiple subunits, and the extreme salt resistance all make cohesin different from canonical DNA binding proteins.
Many of the unusual properties of cohesin binding to chromosomes can be explained by the ring or embrace model. In this model chromosomes become topologically entrapped within the large ring of cohesin through an ATP-dependent mechanism (13–15). This model is consistent with the requirement for all subunits of cohesin for DNA binding. Indeed, cleavage of the protein backbone of cohesin subunits or the DNA backbone of isolated circular minichromosomes can release cohesin from chromosomes. The topological binding rather than direct contact with DNA would also allow it to bind to anywhere on chromosomes and yet be enriched at CARs by indirect mechanisms of chromosome metabolism. For example, one in vivo study suggests that cohesin is loaded at Scc2p/Scc4p binding sites distinct from CARs and then is pushed to CARs by transcription (16). Although the simplicity of the embrace model is attractive, a number of studies suggest that the simple entrapment of DNA and its derivative hypotheses may need to be modified. For example, in vivo cleavage of the DNA backbone next to a CAR fails to release cohesin from the DNA (17) suggesting cohesin may bind chromosomes by additional or alternative mechanisms than topological entrapment. Indeed, in vivo FRAP studies suggest cohesin can bind to chromosomes in two distinct modes that differ in residence time on chromosomes (18–21). A recent in vivo study indicates that Scc2p/Scc4p binding sites lie within CARs, indicating that cohesin enrichment at CARs may not be indirect (22). Colocalization of cohesin and its loader was also detected in human and drosophila (23, 24). Finally in the embrace model, the thinness of the ring and the absence of direct contact with DNA infer that the number of cohesins bound to chromosomes should be limited only by the number of cohesins in the cell, a concept that has been assumed but not tested.
Clearly many of these uncertainties about cohesin binding to DNA could be resolved by in vitro biochemical studies. Indeed, a number of studies have utilized cohesin assembled from purified subunits to analyze its interaction with generic DNA. These studies document cohesin’s ability to bind linear DNA independent of DNA sequence and its high affinity for DNA secondary structures (25–27). However, the physiological relevance of these observations is not clear as these cohesin/DNA complexes fail to mimic known properties of in vivo assembled complexes like ATP and Scc2p dependence, salt resistance, topology dependence, or preferential binding to different DNA regions. Furthermore, the biological relevance of these in vitro properties has not been validated by a diagnostic in vivo test. Biochemical studies of other SMC complexes had similar drawbacks (28–31). Therefore, the study of cohesin and all Smc proteins have suffered from the absence of a physiologically relevant in vitro system, coupled with the failure to assess the physiological relevance of in vitro observations. With this in mind, we developed an in vitro system that recapitulates the critical properties of in vivo assembled complexes. Furthermore, we successfully employ this system to address fundamental questions about cohesin-CAR interactions in vitro and demonstrate its relevance in vivo.
Results
Assaying Cohesin Binding to DNA in Vitro.
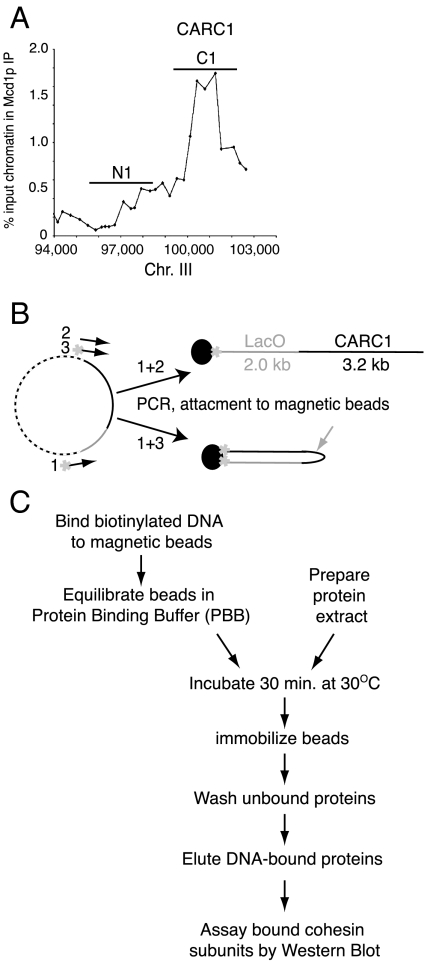
The starting DNA substrate in our assay was prepared by amplifying a 5-kb fragment that has a 3.2-kb sequence containing CARC1 (Fig. 1A). One or both ends of the PCR product were labeled with 5′ biotin (Fig. 1B). A PCR product labeled at one end should bind streptavidin-coated beads with linear topology (C1-L), whereas a PCR product labeled at both ends should allow both ends to bind to the same bead generating a closed topology (C1-C) (Fig. 1B). Analyses of C1-L and C1-C with restriction endonucleases confirm that greater than 93% have the linear and closed topology, respectively (SI Text). By elution of the bound DNA from C1-L and C1-C, we estimate that approximately 1,000–2,000 DNA molecules are bound per bead.
Fig. 1.
A new system to study cohesin/DNA binding activity. (A) Representative ChIP of CARC1. The positions of the substrates C1 and N1 are indicated. (B) Substrates were prepared by PCR using one or two primers labeled with biotin (*). DNA was attached to streptavidin-coated magnetic beads (dark balls) creating either a linear (C1-L, Top) or closed (C1-C, Bottom) structure. The gray arrow indicates the Pst I site. (C) Flowchart of the standard binding assay.
We assembled cohesin onto C1-L and C1-C, as described in Fig. 1C. We incubated the substrate with crude extracts from yeast cells arrested in S phase using hydroxyurea. We choose S-phase extracts for two reasons. First, in vivo experiments showed that S-phase cells have fully assembled cohesin complexes, are competent for cohesin loading onto DNA, and also are competent to convert chromatin-bound cohesin to its cohesive state, the latter being a future goal of our approach. Second, cohesin from a crude extract seemed more likely than purified or recombinant cohesin to have all the activities needed for loading cohesin onto DNA in a physiologically relevant manner. After incubation, beads were immobilized using a magnet, and then unbound proteins were removed by multiple washes. The bound fraction was eluted from DNA, and the presence of bound cohesin subunits were analyzed by Western blot with antibodies against cohesin subunits. For most analyses we used antibodies directed against the Mcd1p subunit of cohesin as a marker for cohesin binding to the beads. Previous in vivo analyses have shown that Mcd1p binds chromatin/DNA only in the context of the other subunits of cohesin. In addition, the binding of all the other subunits to chromatin/DNA requires functional Mcd1p (4, 7).
Cohesin Binding in Our in Vitro Assay Recapitulates Its Properties in Vivo.
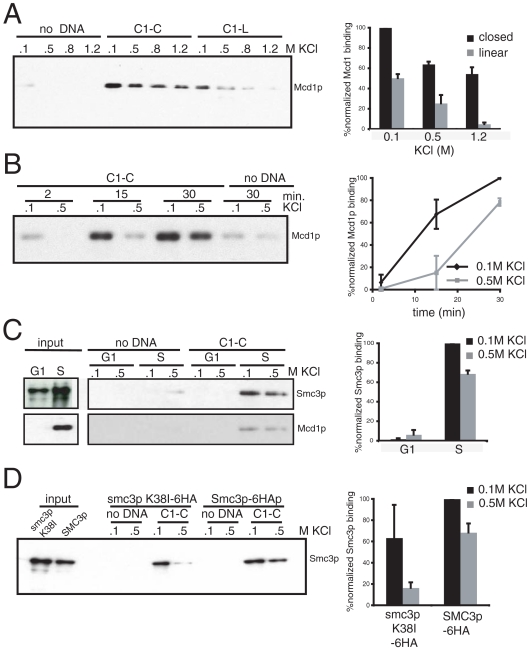
We first asked whether cohesin would associate with C1-L, and if so, whether this DNA binding would mimic the salt-resistant binding of in vivo assembled complexes. C1-L was incubated in S-phase yeast extracts; then the beads were washed with buffer containing 0.1, 0.5, 0.8, or 1.2 M KCl to remove unbound cohesin and to assess cohesin binding under conditions of increasing stringency (Fig. 2A). Western blot analyses show that Mcd1p binds to beads only when beads contain DNA. However Mcd1p is readily removed at salt concentrations above 0.1 M. Therefore, linear DNA does not allow the assembly of physiologically stable cohesin-DNA complexes.
Fig. 2.
In vitro cohesin binding recapitulates that of cohesin bound in vivo. (A) Protein extract was prepared from 3131-V13 cells arrested in S and mixed with C1-L or C1-C under the standard assay conditions with the following modification: Unbound proteins were washed with 0.1, 0.5, 0.8, or 1.2 M KCl. Bound proteins were analyzed with antibody against Mcd1p. Error bars represent SD of two independent experiments. (B) Protein extract was prepared from 3131-V13 cells arrested in S and mixed with C1-C under the standard assay conditions with the following modification: Samples were incubated for 2, 15. or 30 min before the unbound fraction was washed. Bound proteins were analyzed with antibody against Mcd1p. Error bars represent SD of two independent experiments. (C) Protein extract was prepared from YIO71 cells arrested in G1 or S and mixed with C1-C under the standard assay conditions. Bound proteins were analyzed with antibodies against HA (Smc3-6HA, Upper) and Mcd1p (Lower). Error bars represent SD of two independent experiments. (D) Protein extract was prepared from YIO5031 and YIO5032 cells arrested in S and mixed with C1-C under the standard assay conditions. Bound proteins were analyzed with antibodies against HA (Smc3p-6HA and smc3p K38I-6HA). Error bars represent SD of two independent experiments.
Substrate topology has been implicated as an important feature of cohesin binding to DNA based on in vitro assays of circular minichromosomes where cohesin had been preassembled in vivo. Therefore, we wondered whether our in vitro assay would recapitulate salt-resistant cohesin binding if we used the topologically closed C1-C substrate. Incubation of equal amounts of C1-C and C1-L with the S-phase extract led to roughly twice as much Mcd1p binding as with C1-L in 0.1 M salt, suggesting that topology enhanced overall binding. Moreover, 60% of the C1-C Mcd1p remained bound to C1-C even at 1.2 M KCl, in contrast to the quantitative depletion we observed from C1-L. To further characterize the relationship between the salt-sensitive and salt-resistant cohesin/C1-C complexes, we measured the kinetics of their formation (Fig. 2B). Salt-sensitive complexes form first, followed by the salt-resistant complexes. This difference in kinetics supports distinct modes of binding. Hereafter we define salt-resistant cohesin-DNA complexes as those that remain intact after washes with 0.5 M KCl or greater. The topologically modulated assembly of a salt-resistant Mcd1p/DNA complex in our system mimics two important parameters of cohesin/chromosome complexes assembled in vivo, indicating that our in vitro assembled complexes are physiologically relevant.
Two alternative explanations might explain the preferred binding of cohesin to C1-C over C1-L. First, C1-L may be degraded by the extract. However, we observe no loss of either C1-C or C1-L DNA after 30 min of incubation with our extracts (SI Text). Alternatively, the less constrained DNA of C1-L may allow the binding of an inhibitor that precludes cohesin binding like the assembly of nucleosomes. The assembly of nucleosomes seemed unlikely because it had been shown that chromatin assembly in yeast extract requires specific conditions that are not present in our assay (32). Nevertheless, we compared the presence of nucleosomes on C1-L and C1-C by detecting the presence of histones H2B and H3 in the bound fraction. Neither template binds H2B or H3, indicating that neither assembles nucleosomes (SI Text). Although we cannot rule out some other unforeseen inhibitor, the preferential binding of cohesin to the C1-C is consistent with the topological requirements observed in in vivo studies.
To assess further the physiological relevance of our in vitro system, we explored whether the in vivo regulation of cohesin binding to DNA is also recapitulated. In G1 cells, Mcd1p is degraded and its gene expression repressed, so very little if any Mcd1p is present (33, 34). Consequently, only a partial cohesin complex of the Smc1p/Smcp3p heterodimers forms, and this subcomplex is not bound to chromosomes. To test whether this was also true in our system, we incubated extracts from G1 or S-phase arrested cells with C1-C and followed the binding of an epitope-tagged Smc3p to the beads (Fig. 2C). Almost no Smc3p from a G1 extract is detected in the DNA-bound fraction, whereas both Smc3p and Mcd1p from the S-phase extract are recovered in the salt-resistant DNA-bound fraction. Furthermore, in the S-phase extract, the ratio of the salt-sensitive to salt-resistant fraction of Smc3p and Scc3p is similar to the ratio observed for Mcd1p (Fig. 2C and SI Text). These results have three major implications that validate our in vitro assay. First, we recapitulate with our extracts the normal cell cycle control of cohesin binding to chromosomes. Second, we demonstrate that binding of Smc3p to DNA requires Mcd1p as it does in vivo. Third, the cofractionation of Smc3p, Scc3p, and Mcd1p in the high salt-resistant fraction validates Mcd1p as a marker for the cohesin complex and strongly supports the assembly of cohesin/DNA complexes in our in vitro system.
Another important in vivo parameter of cohesin binding to chromosomes is its dependency on binding and hydrolysis of ATP by Smc1p and Smc3p (6–8). To test whether our in vitro system also mimicked this dependency, we reduced cohesin affinity to ATP with a K38I mutation in the ATP-binding cassette of Smc3p. This mutation inhibits both ATP binding to Smc3p and in vivo cohesin binding to chromosomes (6, 8). Yet, it does not affect the integrity of the complex (6, 8). We prepared S-phase extracts from cells expressing Smc3p and either wild-type Smc3p-6HA or smc3p-6HA K38I, and assayed C1-C substrate binding via Western blot using HA antibodies. Although smc3p-6HA K38I binding to C1-C is comparable to the Smc3p-6HA in low salt, its ability to form salt-resistant complexes is reduced fivefold compared to the Smc3p-6HA (Fig. 2D). Our data suggest that ATP binding to Smc3p is essential for the formation of the high-affinity nucleo-protein complex consistent with in vivo analyses. However, the ability to bind at low salt in an ATP-independent manner may represent an undescribed intermediate of cohesin binding. In summary, our in vitro assembled cohesin–DNA complexes recapitulate key attributes of in vivo assembled complexes, including salt resistance, dependence on DNA topology, and ATP binding of Smc3p.
In Vitro Assembled Cohesin–DNA Complexes Recapitulate Preference for CARs.
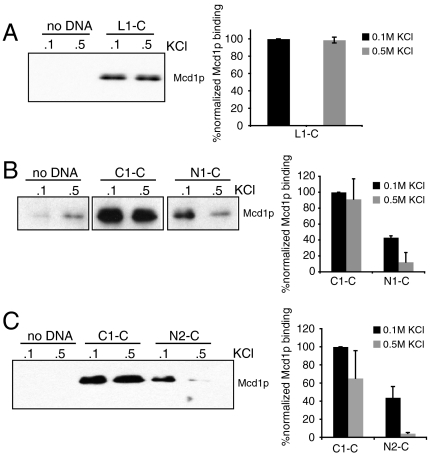
Previous studies used generic DNA sequences to study cohesin–DNA binding properties. However, cohesin is known to enrich at CARs in vivo. We tested whether the assembly of our physiological cohesin–DNA complexes also recapitulates this key in vivo property. First, we tested if we can assemble salt-resistant cohesin/DNA complexes by using a different CAR sequence. We prepared a circular substrate L1-C that contains 3.1 kb of CAR L1 from chromosome XII (9). Similar to C1-C, cohesin showed high affinity to L1-C suggesting that cohesin association with CAR DNA in our system is not a unique property of C1-C (Fig. 3A).
Fig. 3.
Cohesin has higher affinity to CAR DNA than non-CAR DNA in vitro. (A) Protein extract was prepared from YIO71 cells arrested in S phase and mixed with L1-C. Reaction was done under the standard assay conditions. Bound proteins were analyzed with antibodies against Mcd1p. Error bars represent SD of two independent experiments. (B) Protein extract was prepared from 3131-V13 cells arrested in S phase and mixed with C1-C and N1-C. Reaction was done under the standard assay conditions. Bound proteins were analyzed with antibodies against Mcd1p. Proteins were normalized to the DNA on the beads. Error bars represent SD of two independent experiments. (C) Protein extract was prepared from 3131-V13 cells arrested in S phase and mixed with C1-C and N2-C. Reaction was done under the standard assay conditions. Bound proteins were analyzed with antibodies against Mcd1p. Proteins were normalized to the DNA on the beads. Error bars represent SD of two independent experiments.
Next, we replaced the CAR sequences with 3-kb sequences with chromosomal sequences that exhibit low cohesin binding in vivo. These non-CAR sequences, N1-C and N2-C, lie immediately adjacent to CARC1 or the MAT locus, respectively (Fig. 1A). Then we compared the ability of C1-C, N1-C, and N2-C to form salt-sensitive and salt-resistant complexes with cohesin (Fig. 3 B and C). Salt-sensitive complexes with N1-C and N2-C are reduced 2-fold compared to C1-C, and salt-resistant complexes are reduced even further, 7.5-fold for N1-C and 15-fold for N2-C. The results show that CARs have a cis element that promotes the formation of high-affinity cohesin/DNA complex in vitro.
The Number and Position of Cohesins Bound to C1-C Is Limited.
In the simple ring model where the cohesin ring entraps but does not directly bind DNA, the potential number of cohesins that could load on DNA should be very large because the ring is very thin and it does not interact with specific DNA sequences. Furthermore, once loaded the cohesins should diffuse along DNA. However, in vivo, both the location and number of cohesins on chromosomes appear limited as they are enriched at CARs with approximately 3–20 cohesins per CAR (7, 10, 35). This finite ratio may simply reflect that almost all of the active cohesin in the cell is chromosome bound. Furthermore, cohesin enrichment at CARs has been proposed to be a consequence of transcription. Alternatively features of the complex itself may limit the inherent capacity of cohesins to bind CARs and to spread.
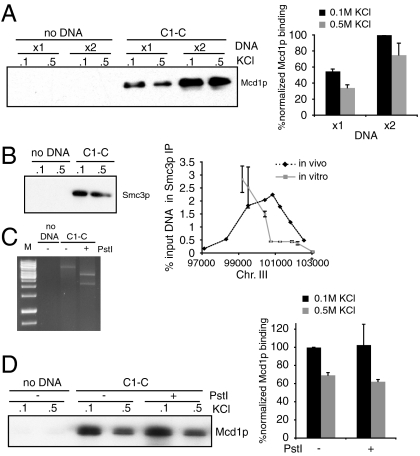
Our in vitro assay allowed us to address whether cohesin binding to DNA is limited by active cohesin molecules. First the ratio of cohesin molecules per DNA molecule in our extract is about 500∶1. Incubation of C1-C in the extract longer than 30 min shows no significant increase in the total amount of cohesin/C1-C complexes. At this steady state only about 2% of the Smc3p and Mcd1p molecules in the extract are bound to C1-C beads. With this and the knowledge of the number of DNA molecules on each bead, we calculate that the ratio of bound cohesin per DNA molecule is only between 1 to 10 despite the vast excess of cohesin (see Materials and Methods). Thus the number of cohesin molecules that can bind to DNA in vitro is limited. This limitation is not a result of limiting active cohesin molecules. When we double the amount of C1-C in the reaction, it doubles the amount of cohesin binds to DNA (Fig. 4A). This limitation is likely physiologically relevant given the striking similarity between ratio of cohesin per CAR in our in vitro system and that measured in vivo.
Fig. 4.
The number and position of cohesins bound to C1-C is limited. (A) Protein extract was prepared from 3131-V13 cells arrested in S phase and mixed with circular substrates under the standard assay conditions with the following modification: Reaction contained 20 or 40 μL beads. Bound proteins were analyzed with antibodies against Mcd1p. Error bars represent SD of two independent experiments. (B) Protein extract was prepared from YIO71 cells arrested in S and mixed with C1-C under the standard assay conditions. Bound proteins were analyzed with antibodies against HA (Smc3-6HA, Left). Samples washed with 0.5 M salt were analyzed by ChIP as described in Materials and Methods (Right). The asterisk shows binding to lacO sequence. Error bars represent SD of two independent experiments. (C) Protein extract was prepared from 3131-V13 cells arrested in S and mixed C1-C under the standard assay conditions with the following modification. After the 30-min assembly, 100 U Pst I were added to the reaction mix and incubated for additional 10 min at 30 °C. Bound proteins were analyzed with antibodies against Mcd1p. (D) Protein extract was prepared from YI971 cells arrested in S and mixed with C1-C. The reaction was done as described in Materials and Methods. Bound proteins were analyzed with antibodies against HA. Error bars represent SD of two independent experiments.
To test further whether the number and the position of cohesins bound to C1-C are limited, we mapped cohesin binding to C1-C. We used a chromatin immunoprecipitation protocol similar to that used to map in vivo cohesin binding to chromosomes (Fig. 4B). First, cohesin loading is highly enriched on CARC1 and absent from neighboring vector sequences. Poor binding to bacterial sequences has been observed in vivo (9, 36). Second, a peak of cohesin binding occurs in a subregion of CARC1 on the C1-C substrate as it does in in vivo chromosomes. The maximal peak level of cohesin cross-linked to CARC1 on C1-C (3%) is very similar to that seen in vivo (2.5–5%) consistent with the assembly of a physiological relevant cohesin/chromosome complex. The limited distribution of cohesin binding to C1-C strongly supports our conclusion based upon average stoichiometry of cohesin to DNA molecule that only one or a few cohesin can bind to C1-C.
The limited distribution of cohesin on C1-C is unlikely due to transcription or exclusion by chromatin. As shown above, C1-C does not assemble into chromatin. Transcription is unlikely to be supported by our extracts as no rNTPs are added. Furthermore, when transcription in the extract is blocked by the addition of transcription inhibitor thiolutin, no significant difference in cohesin/DNA complex assembly is observed (SI Text). Interestingly, the distribution of cohesin binding of CARC1 in our extracts is not identical to that observed in vivo. This suggests that our in vitro system may be missing some cis boundary elements or activities that control the distribution of cohesin binding within CARC1.
The limited distribution of cohesin on C1-C suggests that the diffusion of cohesin along C1-C, if it occurs at all, is constrained after forming the salt-resistant complex with C1-C. To test this possibility we performed a standard binding reaction to allow cohesin to form a salt-stable complex with C1-C, then added Pst I restriction endonuclease to the reaction. When > 99% of the DNA was digested, beads were immobilized and washed with low or high salt buffer as before (Fig. 1B). No significant difference in cohesin binding is detected after cutting the DNA, indicating that the salt-resistant complexes remain stably bound to the now linear C1-L-like substrate (Fig. 4 C and D). The ability of cohesin to maintain a stable complex with the linear DNA C1-C after it is cut but not with C1-L infers that the complex matures. This maturation may involve more intimate interactions with C1-C during or after the formation of the salt-resistant complex preventing cohesin from disengaging from or diffusing along C1-C once it becomes linear.
Dissection of CARC1 Identifies Subregions That Modulate Cohesin Affinity to DNA.
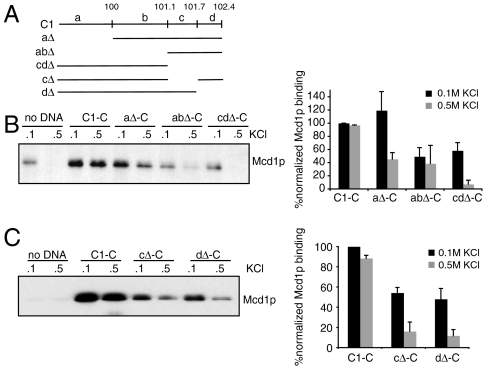
To begin to identify elements in CARC1 that contribute to formation of salt-resistant cohesin we divided CARC1 DNA into four regions marked a–d (Fig. 5A). We constructed a panel of deletions of these four regions. Formation of salt-sensitive complexes is only moderately affected by these deletions. The formation of salt-resistant complexes is reduced by about 2-fold for the aΔ-C and abΔ-C substrates compared to full-length C1-C. However, it is reduced 13-fold for cdΔ-C substrate, suggesting that a critical element for cohesin binding lies within region cd (Fig. 5B). Interestingly, region cd lies to the right of the binding peak identified by the in vitro ChIP (Fig. 4B). This suggests that sequences within CARC1 lead to asymmetric assembly of cohesin. Individual deletions of c and d regions significantly reduce cohesin binding, identifying at least two regions important for assembly of salt-resistant complex (Fig. 5C).
Fig. 5.
Cohesin binding is affected by subdomains within C1-C1. (A) Schematic of deletions in C1. (B) Protein extract was prepared from YIO71 cells arrested in S and mixed with aΔ-C, abΔ-C, or cdΔ-C. The reaction was conducted under the standard assay conditions. Bound proteins were analyzed with antibodies against Mcd1p. Proteins were normalized to the DNA on the beads. Error bars represent SD of two independent experiments. (C) Protein extract was prepared from YIO71 cells arrested in S and mixed with cΔ-C, dΔ-C. The reaction was conducted under the standard assay conditions. Bound proteins were analyzed with antibodies against Mcd1p. Proteins were normalized to the DNA on the beads. Error bars represent SD of two independent experiments.
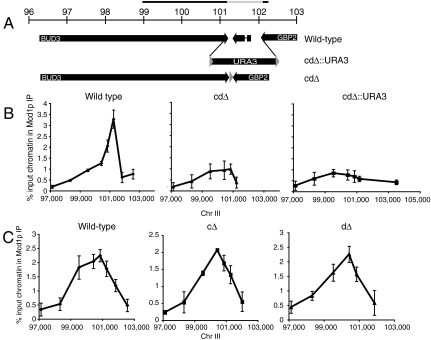
We next tested the in vivo relevance of the CARC1 sequences that modulate cohesin binding in vitro. We constructed deletions in chromosome III in two steps. First, we deleted c, d or cd and replaced them with the URA3 gene flaked by bacterial sequences. Then we selected for loss of the URA3 gene by recombination between flanking loxP sites leaving the desired deletion with just one loxP site (Fig. 6A). We compared in vivo enrichment of cohesin at the wild-type CARC1 and deletion derivatives by chromatin immunoprecipitation (Fig. 6C).
Fig. 6.
CARC1 subdomains modulate in vivo cohesin binding. (A) Genomic organization of CARC1 and deletion of cd by LoxP-URA3 integration (cdΔ∷URA3) and Cre recombinase (cdΔ). The bar above the ruler shows the location of the in vitro substrate C1. The gray bar indicates the 1-kb fragment missing from cdΔ. (B) ChIP analysis of CARC1 from 2185-6B (WT), YIO6041 (cdΔ), and YIO604 (cdΔ∷URA3). Coordinates were adjusted to the chromosomal deletion or insertion Error bars represent SD of three independent experiments. (C) ChIP analysis of CARC1 from 2185-6B (WT), YIO6051 (cΔ), and YIO6052 (dΔ). Coordinates were adjusted to the chromosomal deletions. Error bars represent SD of three independent experiments.
Region cd of CARC1 lies to the right of the peak of in cohesin binding to CARC1 both in vitro and in vivo. Deleting the corresponding sequence in chromosome III (cdΔ) reduces this peak by 3.5-fold compared to wild-type cells. To exclude the possibility that reduction in cohesin association with the cdΔ is due to shortening of the distance between flanking elements such as transcription terminators, we tested the strain in which we replaced the cd region with the URA3 gene (Fig. 6A, cdΔ∷URA3). Note, the URA3 sequence we used has low cohesin binding in vivo. The similar size of URA3 and the cd region preserves the distance between the potential flanking elements. Cohesin occupancy of the CARC1 region cdΔ∷URA3 is reduced to levels similar to that of cdΔ. As a control, we monitored cohesin enrichment on a second CAR located about 80 kb from CARC1. We find its cohesin binding is identical in cdΔ, cdΔ∷URA3, and wild-type strains, indicating that deletions reduce cohesin binding only at the CARC1 locus as expected of a cis element (SI Text). Thus, two critical in vitro traits of cd are conserved in vivo: its ability to increase cohesin binding to neighboring sequences and the magnitude of this enhancement.
However, deletion of c and d regions individually did not impact cohesin binding in vivo, indicating that the cd region has at least two elements, either of which is sufficient for cohesin binding (Fig. 6C). In contrast, our in vitro analyses also identified two elements in cd but both are necessary (Fig. 5C). It would appear that the two elements in c and d regions are more redundant in vivo for assembly of stable cohesin complexes presumably reflecting a current limitation in our in vitro system. However, we are strongly encouraged by the fact both the in vivo and in vitro data suggest that CARC1 contains elements within both c and d that regulate cohesin assembly on DNA.
In Vitro Assembly of Cohesin/DNA Complex Is Scc2 Dependent.
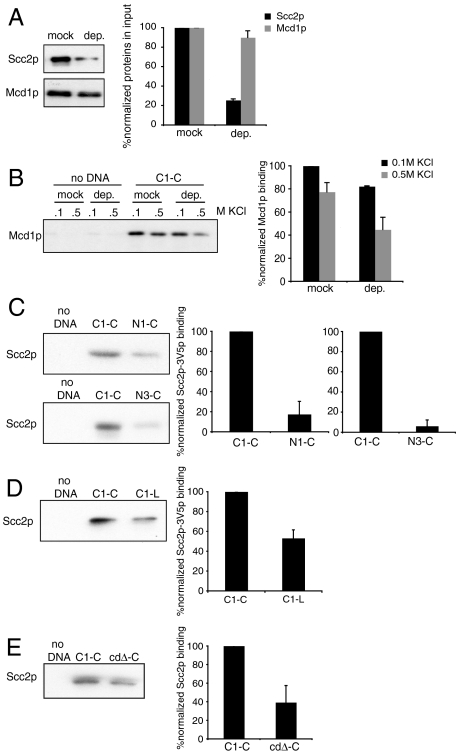
Cohesin loading on chromosomes depends on the Scc2p-Scc4p complex, yet the exact role of Scc2p is unclear. First, we tested if the in vitro assembled cohesin/DNA complexes in our system depend on Scc2p. We immunodepleted Scc2p from the extract to a about 25% of its initial level (Fig. 7A). This immunodepletion does not reduce the amount of cohesin as monitored through Mcd1p. Mock-depleted and depleted extracts showed no difference in the assembly of cohesin/DNA complex in low salt (Fig. 7B). However the formation of salt-resistant C1-C cohesin complexes is reduced about 50% in the Scc2p depleted extract. Complementation of this defect by the addition of recombinant Scc2p is not currently feasible because we and others have been unable to purify Scc2p either directly from yeast or in a recombinant system. Because of this limitation we conclude that Scc2p and/or an associated factor is required for the efficient formation of the salt-resistant cohesin/DNA complex but not for the formation of the salt-sensitive complex.
Fig. 7.
In vitro assembly of cohesin/DNA complexes depends on Scc2p. (A) Scc2p-3V5 was either mock or immunodepleted from protein extract prepared from JH5257 cells arrested in S. About 25% Scc2p-3V5 remained in the depleted extract (d) in comparison with the mock-depleted extract (m). Error bars represent SD of two independent experiments. (B) Protein extract was prepared from JH5257. Scc2p-3V5 was either mock or immunodepleted and the extract was mixed with C1-C under the standard assay conditions. Bound proteins were analyzed with antibodies against Mcd1p. Statistical difference between mock and depleted samples was confirmed by p < 0.05, t test. Error bars represent SD of two independent experiments. (C) Protein extract was prepared from JH5257 cells arrested in S phase and mixed with C1-C or NC3-C. The reaction was done as described. Bound proteins were analyzed with antibodies against V5. Proteins were normalized to the DNA on the beads. Error bars represent SD of two independent experiments. (D) Protein extract was prepared from JH5257 cells arrested in S phase and mixed with C1-C or C1-L. The reaction was done as describes. Bound proteins were analyzed with antibodies against V5. Proteins were normalized to the DNA on the beads. Error bars represent SD of two independent experiments. (E) Protein extract was prepared from JH5257 cells arrested in S phase and mixed with C1-C or cdΔ. The reaction was done as described. Bound proteins were analyzed with antibodies against V5. Proteins were normalized to the DNA on the beads. Error bars represent SD of two independent experiments.
We then asked whether the ability of different substrates to bind Scc2 might explain their relative proficiency to assemble salt-resistant complexes with cohesin. Substrates were mixed with extract made from cells containing Scc2p-3V5 and arrested in S phase, as before. After 30 min unbound proteins were washed with 0.1 M KCl and the amount of Scc2p-3V5 in the bound fraction was determined using antibodies against V5. Scc2p-3V5 is bound to beads with C1-C but not to beads lacking DNA. The binding of Scc2p-3V5 by C1-C is consistent with the functional requirement of Scc2p for formation of the stable complex (Fig. 7B). Compared to C1-C, the binding of Scc2p-3V5 to non-CAR substrates NC1-C and NC3-C is reduced by 6- and 20-fold, respectively (Fig. 7C). Indeed, this decrease in Scc2p-3V5 binding is very similar to the reduced ability of NC sequences to form salt-resistant DNA complexes with cohesin. (Fig. 3 B and C). In contrast, the binding of Scc2-3V5 to the linear C1-L and the cdΔ-C is reduced 2-fold (Fig. 7 D and E). This reduction is not sufficient to cause the 5- to 10-fold reduction in formation of the salt-resistant cohesin complex on C1-L and even greater reduction on cdΔ-C (Figs. 2A and 5B). Thus these two substrates reveal the existence of additional steps beyond Scc2p recruitment to assemble stable cohesin-DNA complexes.
Discussion
Here we describe the development of a biochemical system to study the interaction of cohesin with DNA in vitro. The in vitro assembled cohesin/DNA complex shares many physiological properties with cohesin /chromosome complexes assembled in vivo. Both in vivo and in vitro complexes exhibit resistance to high salt (5). As predicted from in vivo analyses, the formation of stable complexes in vitro is dependent upon the topology of the DNA substrate, the presence of Mcd1p, cell cycle stage of the extract, and ATP binding of Smc3p (reviewed in ref. 1). Also as predicted from in vivo studies, cohesin–DNA complexes form preferentially on CARs (9, 10, 36). By recapitulating physiological properties, our DNA–cohesin complexes are the only in vitro assembled DNA-Smc complex to be validated as biologically relevant. Clearly this biological relevance provides a critical foundation to perform advanced mechanistic studies with high-resolution structural and single molecule methodology.
Our studies also reveal that cohesin associates with DNA by more than a single mode of DNA binding. As indicated above one mode, the salt-resistant complex exhibits all the properties of the stable in vivo complex. The slow formation of the salt-resistant complex is consistent with significant rearrangement of the cohesin structure during DNA binding. Indeed, in vivo studies infer rearrangements of the head and distal hinge domains of Smc1p and Smc3p for proper DNA binding (37, 38). The second mode, the salt-sensitive complex, forms much more rapidly and independent of substrate, topology, substrate sequence (CAR), and Scc2p. Recent studies have identified distinct populations of cohesin that have dramatically different residence time on chromosomes in vivo (18, 21). Our results suggest that the low residence time complexes may in fact be due to our low-salt nonspecific complex. Thus in the future, it will be interesting to determine which of the cohesin chromosome complexes, defined by different binding half lives, are Scc2p dependent.
The binding of cohesin to our substrate contradicts two predictions of the simple embrace model. First, the number of cohesins that can bind to our substrate is limited to an average of 2–10 even when active cohesin is in vast excess in the extract. Second, cohesin is unable to bind or subsequently to diffuse randomly along our substrate as evidenced by our ChIP analysis. In agreement with the ChIP study, once the slow formation of the salt-resistant complex between cohesin and the topologically closed DNA substrate is complete, cohesin remains bound even upon linearization of the DNA.
In vivo studies to assess cohesin’s ability to diffuse along DNA have yielded different results. Introduction of a double-strand break proximal to CARs on chromosome III does not release chromatin-bound cohesin, consistent with a constraint on diffusion (17). Furthermore, the ability to constrain cohesin diffusion on chromosomes and our substrate is attractive as more and more studies implicate cohesin function in transcription regulation where regional specific modulation of chromatin is essential for specificity (23, 39). However, cleavage of the DNA of an isolated minichromosome releases about 80% of the cohesin associated with the minichromosome’s pericentric region implying that at least some cohesin can diffuse along DNA (15, 40).
There are a number of possible differences to explain this apparent discrepancy. Cohesin may slowly transit from simple topological binding to a more intimate interaction as part of its maturation to a tethering complex. The closed, but not the linear, DNA substrate prevents cohesin diffusion off the ends before the maturation of the complex. Such nontopological binding could involve a surface within the cohesin ring, similar to the mechanism of DNA binding for the Smc-like Msh2 and RecR proteins (41, 42). In vitro DNA binding assays with recombinant Smc hinge domain from bacteria and mouse condensin showed nontopological DNA binding by these domains (28, 43). The fraction of cohesin in the more mature state may vary between CARs and pericentric regions. Alternatively all cohesin may have the same mode of binding but cohesin’s diffusion along DNA may be constrained specifically at euchromatic CARs by its association with conventional DNA binding proteins to regulate its function in transcription (44–46). In this model the assembly of cohesin with DNA binding proteins must also proceed slowly to explain the difference between the linear and closed substrate. Further analysis of complex assembly with this in vitro system should provide a means to distinguish between these possibilities.
Finally, we use our in vitro system to elucidate two factors that are responsible for the preference to form physiological cohesin–DNA complexes on CARs. Although cohesin itself forms low-salt complexes with both CAR and non CAR DNA, Scc2p binds preferentially to CARs. Furthermore, depletion of Scc2p impedes the formation of salt-resistant cohesin–CAR complexes. Together these results suggest that the specificity for the formation of high salt complexes on CARs may result from the preferential binding of Scc2p to CARs. Neither Scc2p nor its partner, Scc4p, have known DNA binding motifs. Mechanisms of targeting Scc2p are suggested from studies implicating histone modifications or auxiliary factors like Sir2, Ctcf, and Atrx, in the targeting of cohesin to specific chromosomal sites or regions (17, 44–49). Because our substrates lack histones, we suggest that a transcription-like factor, to be discovered, may target Scc2p to CARs in yeast, which in turn acts on cohesin to promote the formation of the salt-resistant cohesin–CAR.
A second factor that promotes formation of physiological cohesin-CAR complexes is the 1 Kb cd sequence of CARC1. We show that deletion of cd dramatically reduces the ability of CARC1 to form salt-resistant complexes with cohesin in vitro and to bind cohesin in vivo. In both cases, cd deletion reduces cohesin binding to CARC1 sequences adjacent to cd as well as cd itself. The discovery of cd suggests that sequences important for cohesin loading to CARs lie within the CARs themselves. Interestingly the deletion of cd does partially reduce Scc2p binding. These observations are consistent with a recent study localizing Scc2p-Scc4p complexes to CARs as opposed to a previous study that concluded cohesins migrate to CARs from distal loading sites (16, 22). The cd sequence may contain specific protein binding motifs or structures (like cruciform) necessary for Scc2p binding. Unfortunately, identification of a small sequence with potential bioinformatic utility has been confounded by our subsequent deletion analysis of cd that has revealed functional redundancy. Clearly the reduction in Scc2p binding by the cd deletion is not sufficient to explain the dramatic effect on formation of the salt-resistant cohesin-CAR complex (this study). This result implies that additional factors beyond simple binding of Scc2p regulate formation of a stable cohesin-CAR complex. The existence of these factors has eluded previous in vivo analyses and therefore, identifying the cd-dependent, Scc2p-independent factor will be an exciting future direction.
Materials and Methods
Detailed information about reagents and molecular methods used in this work are presented in SI Materials and Methods. We describe growth conditions for yeast, the preparation of DNA substrates, and the preparation of the yeast extracts from cells harboring wild-type and smc3-K38Ip as well as extracts from cells staged in G1. We provide detailed protocols for assembly and detection of cohesin-DNA complexes on magnetic beads. We describe the immunodepletion of Scc2p, the in vitro and in vivo deletion of CARC1 sequences, and the detection of cohesin binding to these DNA variants both in vitro and in vivo by chromatin immunoprecipitation.
Supplementary Material
Acknowledgments.
We thank Vinny Guacci, Jill Heidinger-Pauli, Fred Tan, and all other members of the Koshland lab for constructive comments and technical support. This work was funded by Howard Hughes Medical Institute and National Iinstitutes of Health Grant GM092813.
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected in 2010.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107504108/-/DCSupplemental.
References
- 1.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: A simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 2.Barbero JL. Cohesins: Chromatin architects in chromosome segregation, control of gene expression and much more. Cell Mol Life Sci. 2009;66:2025–2035. doi: 10.1007/s00018-009-0004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 4.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 5.Ciosk R, et al. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 6.Arumugam P, et al. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 7.Weitzer S, Lehane C, Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 8.Heidinger-Pauli JM, Onn I, Koshland D. Genetic evidence that the acetylation of the Smc3p subunit of cohesin modulates its ATP-bound state to promote cohesion establishment in Saccharomyces cerevisiae. Genetics. 2010;185:1249–1256. doi: 10.1534/genetics.110.116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glynn EF, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:e259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 13.Nasmyth K, Haering CH. Cohesin: Its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 14.Haering CH, et al. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 15.Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Lengronne A, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unal E, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 18.Bernard P, et al. Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J. 2008;27:111–121. doi: 10.1038/sj.emboj.7601955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gause M, Misulovin Z, Bilyeu A, Dorsett D. Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5 and Wapl. Mol Cell Biol. 2010;30:4940–4951. doi: 10.1128/MCB.00642-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Austin C, Novikova N, Guacci V, Bellini M. Lampbrush chromosomes enable study of cohesin dynamics. Chromosome Res. 2009;17:165–184. doi: 10.1007/s10577-008-9015-9. [DOI] [PubMed] [Google Scholar]
- 21.Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol. 2006;16:1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 22.Kogut I, Wang J, Guacci V, Mistry RK, Megee PC. The Scc2/Scc4 cohesin loader determines the distribution of cohesin on budding yeast chromosomes. Genes Dev. 2009;23:2345–2357. doi: 10.1101/gad.1819409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misulovin Z, et al. Association of cohesin and nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akhmedov AT, et al. Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J Biol Chem. 1998;273:24088–24094. doi: 10.1074/jbc.273.37.24088. [DOI] [PubMed] [Google Scholar]
- 26.Akhmedov AT, Gross B, Jessberger R. Mammalian SMC3 C-terminal and coiled-coil protein domains specifically bind palindromic DNA, do not block DNA ends, and prevent DNA bending. J Biol Chem. 1999;274:38216–38224. doi: 10.1074/jbc.274.53.38216. [DOI] [PubMed] [Google Scholar]
- 27.Losada A, Hirano T. Intermolecular DNA interactions stimulated by the cohesin complex in vitro: Implications for sister chromatid cohesion. Curr Biol. 2001;11:268–272. doi: 10.1016/s0960-9822(01)00066-5. [DOI] [PubMed] [Google Scholar]
- 28.Hirano M, Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirano M, Hirano T. Opening closed arms: Long-distance activation of SMC ATPase by hinge-DNA interactions. Mol Cell. 2006;21:175–186. doi: 10.1016/j.molcel.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 30.Cui Y, Petrushenko ZM, Rybenkov VV. MukB acts as a macromolecular clamp in DNA condensation. Nat Struct Mol Biol. 2008;15:411–418. doi: 10.1038/nsmb.1410. [DOI] [PubMed] [Google Scholar]
- 31.Onn I, Aono N, Hirano M, Hirano T. Reconstitution and subunit geometry of human condensin complexes. EMBO J. 2007;26:1024–1034. doi: 10.1038/sj.emboj.7601562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultz MC, Hockman DJ, Harkness TAA, Garinther WI, Altheim BA. Chromatin assembly in a yeast whole-cell extract. Proc Natl Acad Sci USA. 1997;94:9034–9039. doi: 10.1073/pnas.94.17.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 35.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 36.Megee PC, Koshland D. A functional assay for centromere-associated sister chromatid cohesion. Science. 1999;285:254–257. doi: 10.1126/science.285.5425.254. [DOI] [PubMed] [Google Scholar]
- 37.Gruber S, et al. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 38.Mc Intyre J, et al. In vivo analysis of cohesin architecture using FRET in the budding yeast Saccharomyces cerevisiae. EMBO J. 2007;26:3783–3793. doi: 10.1038/sj.emboj.7601793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorsett D, et al. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–4753. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanov D, Nasmyth K. A physical assay for sister chromatid cohesion in vitro. Mol Cell. 2007;27:300–310. doi: 10.1016/j.molcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Drotschmann K, Yang W, Brownewell FE, Kool ET, Kunkel TA. Asymmetric recognition of DNA local distortion. Structure-based functional studies of eukaryotic Msh2-Msh. J Biol Chem. 2001;276:46225–46229. doi: 10.1074/jbc.C100450200. [DOI] [PubMed] [Google Scholar]
- 42.Honda M, Fujisawa T, Shibata T, Mikawa T. RecR forms a ring-like tetramer that encircles dsDNA by forming a complex with RecF. Nucleic Acids Res. 2008;36:5013–5020. doi: 10.1093/nar/gkn471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griese JJ, Witte G, Hopfner KP. Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res. 2010;38:3454–3465. doi: 10.1093/nar/gkq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 45.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 46.Kernohan KD, et al. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev Cell. 2010;18:191–202. doi: 10.1016/j.devcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 47.Wu CS, Chen YF, Gartenberg MR. Targeted sister chromatid cohesion by Sir2. PLoS Genet. 2011;7:e1002000. doi: 10.1371/journal.pgen.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stedman W, et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi TS, Basu A, Bermudez V, Hurwitz J, Walter JC. Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008;22:1894–1905. doi: 10.1101/gad.1683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.