Abstract
Fc receptors transport maternal antibodies across epithelial cell barriers to passively immunize newborns. FcRY, the functional counterpart of mammalian FcRn (a major histocompatibility complex homolog), transfers IgY across the avian yolk sac, and represents a new class of Fc receptor related to the mammalian mannose receptor family. FcRY and FcRn bind immunoglobulins at pH ≤6.5, but not pH ≥7, allowing receptor–ligand association inside intracellular vesicles and release at the pH of blood. We obtained structures of monomeric and dimeric FcRY and an FcRY–IgY complex and explored FcRY's pH-dependent binding mechanism using electron cryomicroscopy (cryoEM) and small-angle X-ray scattering. The cryoEM structure of FcRY at pH 6 revealed a compact double-ring “head,” in which the N-terminal cysteine-rich and fibronectin II domains were folded back to contact C-type lectin-like domains 1–6, and a “tail” comprising C-type lectin-like domains 7–8. Conformational changes at pH 8 created a more elongated structure that cannot bind IgY. CryoEM reconstruction of FcRY dimers at pH 6 and small-angle X-ray scattering analysis at both pH values confirmed both structures. The cryoEM structure of the FcRY–IgY revealed symmetric binding of two FcRY heads to the dimeric FcY, each head contacting the CH4 domain of one FcY chain. FcRY shares structural properties with mannose receptor family members, including a head and tail domain organization, multimerization that may regulate ligand binding, and pH-dependent conformational changes. Our results facilitate understanding of immune recognition by the structurally related mannose receptor family and comparison of diverse methods of Ig transport across evolution.
Transfer of immunoglobulin (Ig) from mother to offspring is important for passive acquisition of immunity. In mammals, transport of maternal IgG by the neonatal Fc receptor (FcRn) occurs in utero or after birth through uptake of IgG in ingested milk (1). Critical to the function of FcRn in IgG transport is the strong pH dependence of its interaction with IgG: FcRn binds IgG with an nM affinity at the acidic pH of intracellular endosomes and releases it at the slightly basic pH of the blood (1). Surprisingly, FcRn shares sequence and structural similarity with class I major histocompatibility complex (MHC) molecules, which present antigenic peptides to T cells (2, 3), rather than to other Ig receptors, such as the FcγRs, FcαRI, and FcεRI, which are members of the Ig gene superfamily with two or three Ig-like domains arranged in tandem (4, 5).
Although FcRn has been characterized in many mammalian species, including human and nonhuman primates (6, 7), rodents (8), ruminants (9), and marsupials (10), homologs have not been found outside of mammals. However, nonmammalian species, including birds and some reptiles, transfer maternal Ig to offspring. For example, IgY, the avian and reptilian counterpart of IgG, is packaged into egg yolk and then transported across the yolk sac membrane into the embryonic bloodstream during late embryonic development (11). The yolk sac membranes of chicks express an IgY binding receptor with functional characteristics similar to FcRn: i.e., high-affinity binding at pH 6, and no binding at pH 7.4 (12, 13).
Affinity purification of the IgY binding protein from chicken yolk sac, subsequently named FcRY, and molecular cloning of its gene (14), revealed it to be a new class of Fc receptor lacking sequence and architectural similarity to FcRn or the Ig superfamily Fc receptors that recognize mammalian IgG, IgA, IgE (4, 5), or avian IgY (15). Instead, FcRY is the avian homolog of the mammalian secretory phospholipase A2 receptor (PLA2R), a member of the mannose receptor (MR) family (14). FcRY is the only member of the MR family known to function as an Ig receptor, although other members participate in immune recognition. For example, MR binds pathogens via recognition of carbohydrates rarely found in mammalian glycoproteins, and the dendritic cell receptor DEC-205, another MR family member, functions in the immune system by regulating antigen presentation (16, 17).
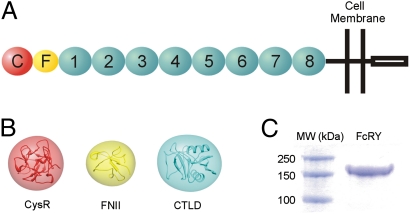
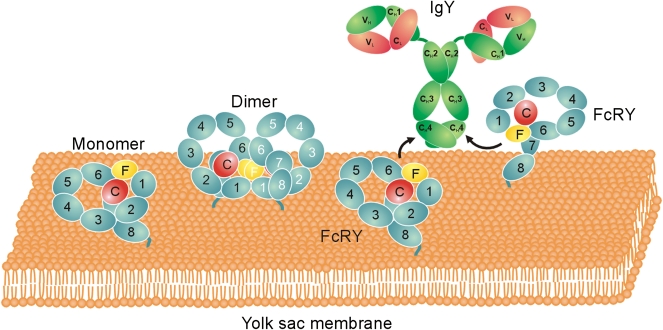
In common with PLA2R and other MR family members (16), FcRY is a type I membrane glycoprotein with a large ectodomain comprising 10 domains of known structure: an N-terminal cysteine-rich (CysR) domain, a fibronectin type II (FNII) repeat, and eight C-type lectin-like domains (CTLDs; Fig. 1 A and B). A recombinant form of the FcRY ectodomain was shown to bind IgY and the FcY fragment of IgY with high affinity at acidic, but not basic, pH (14), and full-length FcRY expressed in polarized mammalian epithelial cells functioned in endocytosis, bidirectional transcytosis, and recycling of chicken FcY/IgY (18), analogous to the functions of FcRn in epithelial and endothelial cells (1).
Fig. 1.
Composition and characterization of FcRY. (A) Schematic model of FcRY showing individual domains: CysR (red sphere labeled C), FNII (yellow oval labeled F), CTLDs 1–8 (cyan ovals labeled 1–8), the transmembrane region, and the cytoplasmic tail. (B) Ribbon diagrams of structures related to FcRY domains (CysR, PDB ID code 1DQO; FNII, PDB ID code 2FN2; CTLD, PDB ID code 2CL8). Electron density calculated to 25 Å from the coordinates is superimposed upon each ribbon diagram. (C) SDS/PAGE analysis of the purified FcRY ectodomain used for cryoEM and SAXS studies.
Here we used single-particle electron cryomicroscopy (cryoEM) to investigate the structures of FcRY alone, both in its monomeric and dimeric forms, and the FcRY–IgY complex. We also used small-angle X-ray scattering (SAXS) analysis to confirm the compact structure of FcRY at pH 6 derived by cryoEM and to investigate the conformational change to a more elongated structure at pH 8 that presumably prevents IgY binding. The results are compared with structural studies of mammalian MR members and to pH-dependent IgG recognition by FcRn.
Results
3D Reconstruction of FcRY at pH 6.
The ectodomain of FcRY was purified from the supernatants of baculovirus-infected insect cells as described (14) (Fig. 1C), and its structure investigated by cryoEM single-particle analysis, which can be used to generate 3D structures of proteins or assemblies that would be difficult or impossible to crystallize (19). Single-particle analysis is most easily done with assemblies or very large proteins (19), thus the relatively small size of the FcRY ectodomain (∼150 kDa; ∼180 kDa including carbohydrate) presented challenges for visualization and alignment of particles. Cryo-images of FcRY generally showed low contrast, but individual molecules could be visualized when frozen under conditions that yielded very thin ice (Fig. S1A). Several cryoEM data sets were collected for FcRY at pH 6, and a total of 11,745 particles were selected for reconstruction of a structure at ∼23 Å resolution. This is a relatively low resolution for a cryoEM structure derived from this number of particles, likely resulting from conformational flexibility within the protein, as speculated to be the case for a 33-Å single-particle EM structure of mannose receptor in negative stain (20) and a 24-Å cryoEM single-particle structure of the anaphase-promoting complex (APC) (21).
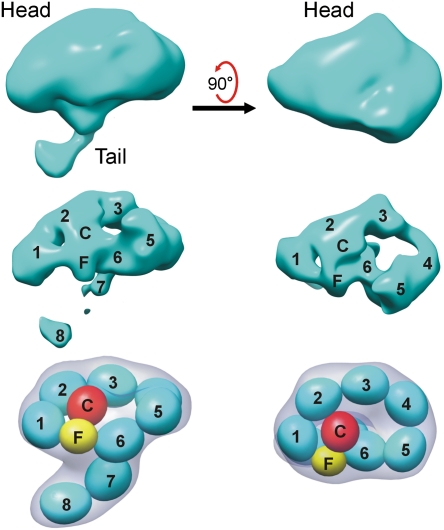
The 3D reconstruction of FcRY revealed that its 10 extracellular domains were arranged to form a compact “head” region and an extended “tail” (Fig. 2). By adjusting the density contour levels, fitting structures for individual domains from related proteins (Fig. 1 A and B) into the density, and using knowledge from previous biochemical experiments (14), we constructed a model for the likely domain arrangement of the FcRY ectodomain at pH 6 (Fig. 2). Because the N-terminal CysR and FNII domains fold back to contact the CTLDs at pH 6 (14), we concluded that the thin tail did not involve the N-terminal domains, but instead represented the two C-terminal domains arranged in tandem, consistent with its width and length. This left the head region to contain the CysR-FNII-CTLD1-6 domains. Using low-resolution (25 Å) filtered versions of individual domain structures (Fig. 1B), we were able to place all 10 extracellular domains to essentially fill the entire density volume. Tracing the domains through the density at high contour levels revealed that the best arrangement of the domains was achieved when we placed the CysR and FNII domains plus CTLDs 1 and 2 into a ring-like structure with CysR contacting CTLD2, and CTLDs 3–6 into a second, larger ring-like structure formed through an interaction between CTLD6 and the FNII domain (Fig. 2). These contacts suggested that the head region of FcRY would be relatively rigid, whereas the tail, which comprised CTLDs 7 and 8, would be more flexible. Although there was more than one way to place the FcRY domains within the head region density, the domain placement shown in Fig. 2 is the only one consistent with reconstructions of an FcRY dimer and an FcRY–IgY complex, subsequently determined independently (see below), the biochemical data (14), and with structures of other MR family members (20).
Fig. 2.
FcRY structure at pH 6. Electron density of the FcRY monomer from a reconstruction at 23 Å resolution is shown at low (Top) and high (Middle) contour levels. The domain arrangement of the FcRY monomer (derived by fitting low-resolution filtered structures into the cryoEM density and applying knowledge of the results of biochemical experiments; Fig. S3) is shown with numbers and letters corresponding to the domains in the Middle row and schematically with low resolution calculated envelopes (gray) representing each of the domains in the Bottom row. The individual domains of FcRY are color coded as in Fig. 1.
Similar procedures were used to investigate the structure of FcRY at pH 8, but attempts to reconstruct the pH 8 structure from 1,829 particles were unsuccessful, presumably because FcRY ectodomain was flexible at basic pH and existed in multiple conformations. However, the average diameter of boxed particles from the cryo-images at pH 8 was ∼20% larger than the average diameter of selected particles at pH 6 (∼180 Å vs. ∼142 Å), suggesting a more open conformation at basic pH, consistent with the slower migration of FcRY by size exclusion chromatography at pH 6 than at pH 8, and with previous analytical ultracentrifugation results (14).
SAXS Analysis of FcRY at Acidic and Basic pH.
To obtain independent structural information for the structure of FcRY at acidic pH and to assess potential conformational changes at basic pH, we performed analyses using small-angle X-ray scattering (SAXS), which provides information about the size and shape of molecules in solution (22). The scattering curves derived from FcRY samples at pH 6 and pH 8 showed significant differences (Fig. S2). Guinier plot analyses yielded radius of gyration (Rg) values of 45 ± 2 Å at pH 6 and 55 ± 2 Å at pH 8, consistent with a pH-dependent conformational change to a more open structure at basic pH. Pair distribution plots were calculated and used for 3D structural modeling using ab initio calculations. Independent calculations were repeated at least 10 times for the structures at each pH value. The resulting models at acidic pH (example shown in Fig. S2) showed an overall shape with similarities to the cryoEM structure, containing recognizable head and tail portions. Although it was not possible to unambiguously identify domains within the SAXS model, the domain assignment derived from the cryoEM single-particle analysis fit the SAXS models reasonably well, revealing structures consistent with the CysR and FnII domains contacting the CTLDs. By contrast, the models obtained at pH 8 showed a more open structure (Fig. S2), consistent with a large conformational change occurring at pH 8. In these elongated structures, it is plausible that the CysR and FNII domains were not folded back to contact the CTLD domains as in the pH 6 structure (Fig. S2).
3D Reconstruction of an FcRY Dimer at pH 6.
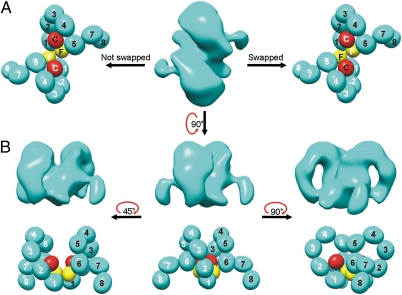
In addition to particles corresponding to FcRY monomers, we identified a minor subset of larger particles in cryoEM images of FcRY at pH 6. Because the FcRY ectodomain had been shown to exist as a mixture of monomer and dimer at high concentrations (14), we assumed the larger particles corresponded to FcRY dimers. The two species could not always be distinguished during particle selection because the size difference was not obvious at some orientations. However, dimer particles were sometimes dominant in certain regions of some of the grids (Fig. S1B), which could correspond to local regions of high concentration. To characterize the FcRY dimer, which may be relevant to reported oligomers of MR family members (17, 20), and to obtain an independent verification of the structure of the FcRY monomer, we conducted a single-particle reconstruction of the FcRY dimer. A total of 1,883 particles were selected to reconstruct the dimer at ∼28 Å resolution. A variety of symmetry operators for averaging were tested using the same data set. Only twofold symmetry averaging yielded stable structural features, consistent with the identification of the larger FcRY species as a dimer (14). The resulting reconstruction, determined without using information from the monomer structure, revealed two associated ring-like features, each with a protruding tail perpendicular to the plane of the ring (Fig. 3 A and B). The size of the reconstructed dimer corresponded to two FcRY monomers, and each monomer resembled the FcRY monomer structure derived from the independent reconstruction, thereby validating the monomer reconstruction. Using the domain assignment of the FcRY monomer, each of the rings in the FcRY dimer corresponded to the larger ring in the FcRY monomer that contained CTLDs 3–6. The smaller ring observed in the FcRY monomer (CysR-FNII, CTLDs 1–2) was not revealed as clearly as the larger ring, perhaps because it was involved in dimer contacts. Also, the tails (CTLDs 7–8) in the FcRY dimer were slightly shorter than their counterparts in the FcRY monomer, perhaps due to flexibility that averaged out some of the tail density during the averaging performed in the reconstruction.
Fig. 3.
FcRY dimer structure. (A) Electron density (Middle) and possible domain arrangements for the FcRY dimer (Left, a non–domain-swapped dimer; Right, a domain-swapped dimer) from a reconstruction at 28 Å resolution. The domain arrangement was derived as described in Fig. 2. (B) Three different views of the FcRY dimer. Electron density is shown above the corresponding schematic domain arrangement in each view. The CysR (red) and FnII (yellow) domains were not labeled to indicate that the same domain arrangement would apply to a domain-swapped vs. non–domain-swapped dimer.
Applying the domain assignment for the FcRY monomer structure at pH 6 to the EM density of the dimer yielded a structure in which two FcRY monomers associated through N-terminal regions, including the FNII and CTLD1 domains. Dimer formation could result from direct association between these domains or through 3D domain swapping (23) in which the CysR and FNII domains from one monomer associate with the CTLDs of the partner monomer (Fig. 3A). Although the resolution of the dimer structure was too low to distinguish swapped from nonswapped dimers, the observation that the CysR-FNII fragment does not bind the CTLDs at pH 8 (14), taken together with the more extended pH 8 conformation (Fig. S2), is consistent with inherent flexibility in FcRY that could facilitate domain swapping involving the CysR and FNII domains.
3D Reconstructions of the FcRY–IgY Complex.
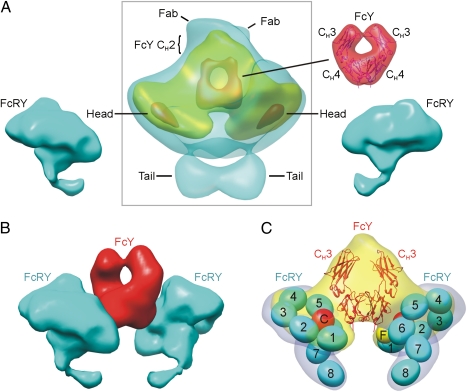
FcRY–ligand complexes were formed at pH 6 by incubating purified FcRY with IgY or the FcY fragment of IgY at a 2:1 molar ratio (Fig. S1C), based on previous studies demonstrating that FcRY formed a 2:1 complex with IgY (14). A total of 4,572 particles were picked to reconstruct the FcRY–IgY complex, and 3,835 particles were picked for the FcRY–FcY complex. Twofold symmetry averaging was applied in initial model building and refinement of complexes to take advantage of the twofold symmetry of dimeric FcY. The core portions of the FcRY–FcY and FcRY–IgY reconstructions were similar, but at low contour levels, the FcRY–IgY complex showed extra density with two short elongated densities emanating from it, which likely represented the N-terminal CH2 domains of FcY (absent in the FcY fragment used for the FcRY–FcY reconstruction) and the C-terminal portions of two Fab fragments. Complete density for the Fabs was not present, probably due to flexibility of the Fabs relative to FcY, which would result in disorder of the N-terminal regions of the Fabs. To obtain a more accurate reconstruction of the complex, we combined the FcRY–IgY and FcRY–FcY particles to obtain a final reconstruction from 8,407 particles at ∼26 Å resolution. Weak densities for the Fab and CH2 regions of IgY were visible in the averaged reconstruction (Fig. 4A), which we will subsequently refer to as FcRY–IgY.
Fig. 4.
FcRY–IgY structure. (A, Middle) Electron density of the FcRY–IgY complex from a reconstruction at 25 Å resolution. Low, medium, and high contour levels (cyan, yellow, and red, respectively) are shown with the approximate locations of different portions of the complex structure indicated by lines. (Left and Bottom Right) electron density for the pH 6 FcRY monomer structure shown on both sides of the complex in the orientation in which it would interact with the FcY portion of IgY. (Top Right) FcY coordinates (PDB code 2W59) enclosed in low-resolution (25 Å) filtered electron density. (B) Fitting of the FcRY–IgY complex density with density from the FcRY structure and the calculated FcY density from its crystal structure as shown in A. (C) FcRY–IgY electron density (medium contour level) fit with FcY coordinates and the domain model of the pH 6 FcRY structure (as shown in Fig. 2 with a low-resolution calculated envelope in gray).
Having established that the core portion of the FcRY–IgY reconstruction corresponded to IgY, we interpreted the two paddle-shaped densities on either side of the IgY as FcRY monomers. Consistent with this interpretation, the paddle-shaped densities were each similar in size to one FcRY monomer, as observed in the monomer and dimer reconstructions, and each contained a head region and density observed at low contour levels (Fig. 4A) that resembled the tails in the FcRY monomer (Fig. 2) and dimer (Fig. 3). As in the monomeric and dimeric FcRY reconstructions, the tail densities disappeared at higher contour levels (Fig. 4A), consistent with flexibility of this region of FcRY. Facilitating the fitting of FcRY into the complex density, the relative position of the head region plane and the tail was a consistent feature of FcRY in the monomer, dimer, and FcRY–IgY complex structures: the tail was always bent toward the small ring (CysR-FNII-CTLD1–2) within the plane of the head region. The central portion of the FcRY–IgY complex density was a triangular shape into which the crystal structure of the CH3–CH4 domains of FcY (24) could be fit (Fig. 4 B and C). The docking of FcY was aided by a circular feature at high contour levels in the density, which corresponded to the space between the CH3 and CH4 domains of FcY (Fig. 4 A–C). The orientation of FcY docked into the complex density was consistent with the assignment of the weak densities on one end of the FcY as Fabs.
The docking of FcRY and FcY into the FcRY–IgY complex structure allowed interpretation of the mode of interaction between the receptor and its ligand: each FcRY used the side of its head region at the junction between FNII and CTLD6 to contact the CH4 domain of one chain of the dimeric FcY (Fig. 4C). The binding interface on FcRY therefore required the compact configuration involving the conjunction of both rings within the FcRY head, a conformation that was lost upon forming the extended structure at pH 8 (Fig. S2). The FcRY domains in direct contact with FcY are likely to be the FNII and CTLD6 domains, with possible involvement of the CTLD5 domain (Fig. 4C). The resulting complex structure was consistent with an upright orientation of IgY on a cell membrane (Fig. 5), with the FcRY tail corresponding to the C-terminal portion of the receptor to which the transmembrane region would be attached. Fig. 5 also depicts the likely orientations of FcRY monomer and dimer on a membrane, indicating that dimerization via the FcRY interface found in the dimer reconstruction would be possible when attached to a membrane, and that the IgY binding interface on FcRY would be occluded upon dimerization.
Fig. 5.
Likely orientations of FcRY and FcRY–IgY on a membrane. Schematic structures for the FcRY monomer and dimer are shown as the full-length proteins would be oriented on a membrane bilayer. The two FcRY monomers on the Right are shown in an orientation that would allow formation of a 2:1 FcRY–IgY complex.
Discussion
Here we investigated the structure of avian FcRY, an IgY receptor, and its mechanism of pH-dependent binding to IgY. Similar to mammalian MR and other members of the MR family (16, 17), the FcRY ectodomain contains multiple domains arranged in tandem, including an N-terminal CysR domain, a FNII domain, and eight CTLDs (14). We used cryoEM single-particle analysis to derive structures of FcRY and FcRY–IgY at ∼25-Å resolution and investigated the pH-dependent conformational change of the FcRY ectodomain by cryoEM and SAXS. Although the low resolution of our structures precluded placing domains unambiguously based on the experimental density alone, there was only one domain trace that was consistent with creating a surface for IgY binding that accounts for the biochemical data (14) (Fig. S3), explains the elongated structure of FcRY at pH 8 in which CysR-FNII is not folded back upon the CTLDs, rationalizes how FcRY dimers may form, and resembles the structures of other MR family members, which were reconstructed from negatively stained samples at 25–33 Å resolution (20).
A 3D cryoEM reconstruction of FcRY at pH 6, an acidic pH permissive for IgY binding, revealed that it adopted a compact conformation containing a large, double-ringed N-terminal head and a C-terminal tail. The first ring in the head was formed by the FcRY CysR and FNII domains, which were folded back to form a structure in which the CysR domain contacted CTLD2, as also seen in the neutral pH structure of Endo180 (25). A variation of this ring-like structure was found in MR, such that its CysR domain interacted with CLTDs 4 and 5 (20). Endo180 CTLD2 and MR CTLD4 are the only CTLDs in their respective receptors that contain the conserved amino acids found in functional (i.e., carbohydrate-binding) C-type lectins (17, 20). By contrast, all eight CTLDs in FcRY lack residues that are critical for carbohydrate recognition by C-type lectins (14). In our pH 6 FcRY structure, the CTLDs following CTLD2 participated in a second ring-like structure in which CTLDs 3 and 4 were extended away from CTLD2, with CTLDs 5 and 6 turning back toward the first ring-like structure to form an apparent contact between CTLD6 and the FNII domain. Thus, the FcRY head contained the N-terminal eight domains of the ectodomain, with the two C-terminal domains forming the tail. Although MR and Endo180 had longer tails, all three receptors shared common features in their compact forms, including a C-terminal tail of two or more CTLDs and an N-terminal head in which the CysR-FNII region made contact with one or more of the CTLDs. An independent verification of the cryoEM structure of FcRY at pH 6 was obtained by reconstruction of an FcRY dimer, which revealed a twofold symmetric association of two FcRY monomers with the FNII and CTLD1 domains forming the dimer interface.
The cryoEM structures of the FcRY–IgY complex allowed insight into how FcRY recognizes IgY. Previous experiments demonstrated that FcRY bound to IgY and FcY with approximately equal affinities (14), which narrowed down the binding interface on IgY to the CH3 and CH4 domains of the FcY protein used for the experiments. Consistent with this observation, the complex structure, which had twofold symmetry with a 2:1 FcRY–IgY stoichiometry, revealed the FcRY binding site to be localized primarily to the FcY CH4 domain. The low resolution of the complex structure prohibits precise location of the FcRY binding site on FcY, but it appeared to be near the counterpart of the FcRn binding site on IgG, which comprises the IgG CH2–CH3 domain interface (26) that corresponds to IgY CH3–CH4. Another common feature of FcRY–IgY and FcRn–IgG recognition is their 2:1 receptor–ligand stoichiometry, which is in contrast to the 1:1 receptor–ligand stoichiometry of the FcγR and FcεRI receptors (4, 5).
The binding site for IgY on FcRY involved the double-ring structure of the FcRY head, with apparent contacts between FcY and the FNII portion of CysR-FNII region (part of the first ring in which CysR contacted CTLD2), and with CTLD6 (part of the second ring in which the intermediate CTLDs were folded back to create a CTLD6-FNII contact). The IgY-binding interface on the FcRY monomer would be occluded by formation of FcRY dimers because the dimer interface involved the FNII and CTLD1 domains, suggesting that dimerization at acidic pH is part of a mechanism by which FcRY regulates binding of IgY at locations of high receptor density on the membrane. Likewise, multimerization of mammalian MR family members (20) has been suggested to regulate binding of their ligands (17). Multimerization of mammalian and avian MR family members has been observed at pH values permissive for ligand binding; thus, multimers of MR were found at basic pH (20), whereas dimers of FcRY were found only at acidic pH (14).
The structures of FcRY and the FcRY–IgY complex derived by single-particle cryoEM were consistent with other data, including models from SAXS experiments at pH 6, and with previous binding data (Fig. S3) showing that two FcRY fragments, one comprising the CysR and FNII domains and the other comprising CTLDs 1–8, did not bind to IgY in isolation, but bound to each other and to IgY at pH 6 (14). The double-ring arrangement of domains in the pH 6 cryoEM structure rationalized the results of the binding experiments in that the interaction of the CysR-FNII fragment with the CTLDs, specifically CysR with CTLD2 and FNII with CTLD6, would reconstitute the intact conformation of FcRY effectively, thereby regaining the mode of IgY recognition seen in the FcRY–IgY complex structure (Figs. S3 and S4). Other truncation mutants, CysR-FNII-CTLD1–4 and CysR-FNII-CTLD5–8, showed no binding to IgY (14), rationalized by the cryoEM model because neither of these fragments would maintain the compact conformation of FcRY found at acidic pH. In addition, a mixture of two fragments, the CysR domain alone (not including the FNII domain) plus CTLDs 1–8, could not reconstitute IgY binding, as predicted from the double-ring structure of FcRY at acidic pH, which would require the FNII contact with CTLD6. A truncation mutant that might be expected to maintain binding to IgY would be CysR-FNII-CTLD1–6, which lacks the tail domain CTLDs that did not contact FcY in the complex structure. However, we were unable to express adequate amounts of this fragment for binding studies, perhaps because some or all of the CTLDs in the tail and/or at junction with the tail are tightly associated with each other, as previously suggested for some of the CTLDs in MR family members (27), such that improper truncation would interfere with the folding process during expression.
We used both cryoEM and SAXS to investigate the conformational change leading to a more open structure of FcRY at pH 8 that does not bind IgY. Although a 3D reconstruction by single-particle EM methods was not possible for the pH 8 structure, presumably due to conformational flexibility, pH 8 FcRY particles were more extended than their pH 6 counterparts, compatible with the model derived from the SAXS data. Evidence for pH-dependent fluctuations of mammalian MR family members between compact and more open forms has also been described (17, 20, 25). Interestingly, both the mammalian and avian MR family members adopted their more compact conformations at their active pH rather than at the pH that is nonpermissive for binding their ligands; thus Endo180 and MR exhibited compact bent structures at neutral/basic pH and more open structures at acidic pH, opposite from the pH-dependent properties of FcRY (17).
The lack of interaction between the FcRY CysR-FNII and CTLD1–8 fragments at pH 8 (14) was consistent with the more open conformation at basic pH revealed by cryoEM and SAXS, and previously by size exclusion chromatography and analytical ultracentrifugation (14). These results suggested that the CysR-FNII region could be pivoted away from the rest of the ectodomain in the basic pH structure, similar to structural models for the open conformations of mammalian MR family members (17). For FcRY, it appears that movement of the CysR-FNII region away from the CTLDs destroyed the IgY binding site, which required the double-ring structure of the FcRY head: in particular, the folding back of the CysR-FNII region to contact CTLD2 and CTLD6. The propensity for displacement of the CysR-FNII region in the transition from the bent structure at pH 6 to the extended structure at pH 8 suggests that this portion of FcRY could exist in a conformational equilibrium even at pH 6, which might facilitate receptor dimerization via 3D domain swapping.
The pH-dependent affinity transition for the FcRY–IgY interaction can be described as primarily involving a pH-dependent conformational change in the receptor. By contrast, the pH 6.5 and pH 8 structures of FcRn, the mammalian IgG receptor that is the functional counterpart of FcRY, did not differ significantly (28). Instead of a conformational change, the pH-dependent affinity transition for the FcRn–IgG interaction involves histidines on the IgG Fc, which form salt bridges with negatively charged residues on FcRn only at acidic pH when protonated (26). Protonation/deprotonation of FcRY histidines over the narrow pH range of the FcRY–IgY affinity transition might be responsible for the pH-dependent conformational change of FcRY that leads to its pH-dependent ligand-binding activity. For example, interdomain salt bridge(s) between positively charged histidine(s) and negatively charged residue(s) could be responsible for some of the favorable interactions that create the compact form of FcRY at acidic pH. Upon deprotonation of the histidines at basic pH, the negative charge(s) would be uncompensated, leading to the conformational transition to the more extended structure.
Like its mammalian counterpart FcRn, FcRY evolved from a protein fold whose original function was of no apparent relation to Ig binding or transport. As the only MR family member with an Fc receptor function, and one of only a few Fc receptors that are not members of the Ig gene superfamily, FcRY is an intriguing model for the evolution and function of Ig and MR family receptors. Here we have shown how FcRY recognizes IgY at acidic pH and the nature of the conformational change that prevents binding at basic pH. These structural studies are relevant not only for evolutionary comparisons of mechanisms of Ig recognition and transport, but also for facilitating understanding of the diverse ligand recognition properties, pH-dependent conformational changes, and multimerization of mammalian MR family members.
Materials and Methods
The FcRY ectodomain (residues 1–1,370 of the mature protein plus a C-terminal 6×-His tag) was expressed in baculovirus-infected insect (Hi5) cells and purified from infected cell supernatants by nickel-nitrilotriacetic acid and size exclusion chromatography as previously described (14). The purified protein was buffer exchanged and concentrated in 50 mM Bis-Tris (pH 6.0), 150 mM NaCl, or 50 mM Tris (pH 8.0), and 150 mM NaCl. Chicken IgY was purchased from Sigma, and FcY was purchased from Jackson Immunoresearch. The Fc region of IgY contains three constant domains (CH2, CH3, and CH4). The region analogous to IgG Fc is a dimer of the IgY CH3 and CH4 domains. N-terminal sequencing of FcY revealed that it started at the CH3 domain (14).
CryoEM data collection, reconstructions, and SAXS experiments are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Anthony West for advice and sharing of experimental data; Inderjit Nangiana, Michael Anaya, Jost Vielmetter, and the Caltech Protein Expression Center for expression of proteins; Alasdair McDowell, Jian Shi, and Grant Jensen for help with microscopy; Thomas Weiss at the Stanford Synchrotron Radiation Lightsource for assistance with SAXS data collection; and Beth Stadtmueller and Anthony West for critical reading of the manuscript. This work was supported by National Institutes of Health Grant 2 R37 AI041239-06A1 (to P.J.B.) and gifts to the California Institute of Technology to support electron microscopy from the Gordon and Betty Moore Foundation and the Agouron Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The cryoEM maps reported in this paper have been deposited in the Research Collaboratory for Structural Bioinformatics Electron Microscopy Data Bank, www.emdatabank.org/. (accession nos. EMD5316, EMD5317, and EMD5318 for the FcRY monomer, dimer, and FcRY-IgY complex, respectively).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106925108/-/DCSupplemental.
References
- 1.Roopenian DC, Akilesh S. FcRn: The neonatal Fc receptor comes of age. Nat Rev Immunol. 2007;7:715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 2.Burmeister WP, Gastinel LN, Simister NE, Blum ML, Bjorkman PJ. Crystal structure at 2.2 A resolution of the MHC-related neonatal Fc receptor. Nature. 1994;372:336–343. doi: 10.1038/372336a0. [DOI] [PubMed] [Google Scholar]
- 3.Simister NE, Mostov KE. An Fc receptor structurally related to MHC class I antigens. Nature. 1989;337:184–187. doi: 10.1038/337184a0. [DOI] [PubMed] [Google Scholar]
- 4.Nimmerjahn F, Ravetch JV. Fcgamma receptors: Old friends and new family members. Immunity. 2006;24:19–28. doi: 10.1016/j.immuni.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Woof JM, Burton DR. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat Rev Immunol. 2004;4:89–99. doi: 10.1038/nri1266. [DOI] [PubMed] [Google Scholar]
- 6.Leach JL, et al. Isolation from human placenta of the IgG transporter, FcRn, and localization to the syncytiotrophoblast: Implications for maternal-fetal antibody transport. J Immunol. 1996;157:3317–3322. [PubMed] [Google Scholar]
- 7.Spiekermann GM, et al. Receptor-mediated immunoglobulin G transport across mucosal barriers in adult life: Functional expression of FcRn in the mammalian lung. J Exp Med. 2002;196:303–310. doi: 10.1084/jem.20020400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simister NE, Rees AR. Isolation and characterization of an Fc receptor from neonatal rat small intestine. Eur J Immunol. 1985;15:733–738. doi: 10.1002/eji.1830150718. [DOI] [PubMed] [Google Scholar]
- 9.Mayer B, et al. Localization of the sheep FcRn in the mammary gland. Vet Immunol Immunopathol. 2002;87:327–330. doi: 10.1016/s0165-2427(02)00059-4. [DOI] [PubMed] [Google Scholar]
- 10.Western AH, et al. Expression of the FcRn receptor (alpha and beta) gene homologues in the intestine of suckling brushtail possum (Trichosurus vulpecula) pouch young. Mol Immunol. 2003;39:707–717. doi: 10.1016/s0161-5890(02)00260-2. [DOI] [PubMed] [Google Scholar]
- 11.Kowalczyk K, Daiss J, Halpern J, Roth TF. Quantitation of maternal-fetal IgG transport in the chicken. Immunology. 1985;54:755–762. [PMC free article] [PubMed] [Google Scholar]
- 12.Linden CD, Roth TF. IgG receptors on foetal chick yolk sac. J Cell Sci. 1978;33:317–328. doi: 10.1242/jcs.33.1.317. [DOI] [PubMed] [Google Scholar]
- 13.Tressler RL, Roth TF. IgG receptors on the embryonic chick yolk sac. J Biol Chem. 1987;262:15406–15412. [PubMed] [Google Scholar]
- 14.West AP, Jr, Herr AB, Bjorkman PJ. The chicken yolk sac IgY receptor, a functional equivalent of the mammalian MHC-related Fc receptor, is a phospholipase A2 receptor homolog. Immunity. 2004;20:601–610. doi: 10.1016/s1074-7613(04)00113-x. [DOI] [PubMed] [Google Scholar]
- 15.Viertlboeck BC, et al. The chicken leukocyte receptor complex encodes a primordial, activating, high-affinity IgY Fc receptor. Proc Natl Acad Sci USA. 2007;104:11718–11723. doi: 10.1073/pnas.0702011104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.East L, Isacke CM. The mannose receptor family. Biochim Biophys Acta. 2002;1572:364–386. doi: 10.1016/s0304-4165(02)00319-7. [DOI] [PubMed] [Google Scholar]
- 17.Llorca O. Extended and bent conformations of the mannose receptor family. Cell Mol Life Sci. 2008;65:1302–1310. doi: 10.1007/s00018-007-7497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tesar DB, Cheung EJ, Bjorkman PJ. The chicken yolk sac IgY receptor, a mammalian mannose receptor family member, transcytoses IgY across polarized epithelial cells. Mol Biol Cell. 2008;19:1587–1593. doi: 10.1091/mbc.E07-09-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Böttcher B, Hipp K. Single-particle applications at intermediate resolution. Adv Protein Chem Struct Biol. 2010;81:61–88. doi: 10.1016/B978-0-12-381357-2.00003-7. [DOI] [PubMed] [Google Scholar]
- 20.Boskovic J, et al. Structural model for the mannose receptor family uncovered by electron microscopy of Endo180 and the mannose receptor. J Biol Chem. 2006;281:8780–8787. doi: 10.1074/jbc.M513277200. [DOI] [PubMed] [Google Scholar]
- 21.Dube P, et al. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol Cell. 2005;20:867–879. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Mertens HD, Svergun DI. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J Struct Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor AI, Fabiane SM, Sutton BJ, Calvert RA. The crystal structure of an avian IgY-Fc fragment reveals conservation with both mammalian IgG and IgE. Biochemistry. 2009;48:558–562. doi: 10.1021/bi8019993. [DOI] [PubMed] [Google Scholar]
- 25.Rivera-Calzada A, et al. Three-dimensional interplay among the ligand-binding domains of the urokinase-plasminogen-activator-receptor-associated protein, Endo180. EMBO Rep. 2003;4:807–812. doi: 10.1038/sj.embor.embor898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin WL, West AP, Jr, Gan L, Bjorkman PJ. Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: Mechanism of pH-dependent binding. Mol Cell. 2001;7:867–877. doi: 10.1016/s1097-2765(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 27.Napper CE, Dyson MH, Taylor ME. An extended conformation of the macrophage mannose receptor. J Biol Chem. 2001;276:14759–14766. doi: 10.1074/jbc.M100425200. [DOI] [PubMed] [Google Scholar]
- 28.Vaughn DE, Bjorkman PJ. Structural basis of pH-dependent antibody binding by the neonatal Fc receptor. Structure. 1998;6:63–73. doi: 10.1016/s0969-2126(98)00008-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.