Abstract
SNARE proteins are crucial for membrane fusion in vesicular transport. To ensure efficient and accurate fusion, SNAREs need to be sorted into different budding vesicles. This process is usually regulated by specific recognition between SNAREs and their adaptor proteins. How different pairs of SNAREs and adaptors achieve their recognition is unclear. Here, we report the recognition between yeast SNARE Vti1p and its adaptor Ent3p derived from three crystal structures. Surprisingly, this yeast pair Vti1p/Ent3p interacts through a distinct binding site compared to their homologues vti1b/epsinR in mammals. An opposite surface on Vti1p_Habc domain binds to a conserved area on the epsin N-terminal homology (ENTH) domain of Ent3p. Two-hybrid, in vitro pull-down and in vivo experiments indicate this binding interface is important for correct localization of Vti1p in the cell. This previously undescribed discovery that a cargo and adaptor pair uses different binding sites across species suggests the diversity of SNARE-adaptor recognition in vesicular transport.
Keywords: cargo recogniton, Vti1p sorting, crystallography
Secretion and intracellular transport rely on vesicular traffic in eukaryotic cells. Vesicle transport can be divided into multiple steps: budding, targeting, tethering, and fusion (1). During the fusion process, the vesicles fuse with the target membrane and release the cargo proteins. The key players in membrane fusion are SNAREs (soluble N-ethylmaleimide-sensitive factor attachment receptor) (2, 3). SNARE proteins are defined by their characteristic heptad repeat SNARE domains. During the fusion process, four SNARE domains (usually one on the vesicle membrane and three on the target membrane) associate to form a stable twisted four-helix bundle, bringing the two opposing membranes in close contact and facilitating fusion (3). Thus, SNAREs are regarded as the basic element in membrane fusion (2–4).
Vesicle fusions are specific processes that require the specific localization of SNAREs on different membranes. There are 24 SNAREs in yeast and about 40 SNAREs in mammals with specific subcellular localization (5–7) (Fig. S1). SNAREs are recruited as cargo proteins to different subcellular membranes by adaptor proteins (8). The maintenance of compartment identity requires highly specific recognition between SNAREs and their adaptor proteins during vesicle budding. Distinct from many cargo proteins, most SNAREs do not contain the short, linear sorting motifs that are recognized by common adaptors like AP or GGA for sorting into vesicles (there are few exceptions including Vamp4, which contains the signal motif dileucine recognized by AP) (9–11). It has been reported, however, that the N-terminal three-helix bundle (Habc domain) of many SNAREs is recognized through interaction with the ENTH (epsin N-terminal homology) domain of a few epsin-like family proteins (8, 12, 13).
The epsin-like family is characterized by a conserved N-terminal ENTH domain (11, 14, 15) and consists of four epsin proteins (epsin1, epsin2, epsin3, epsinR) in humans and four proteins (Ent1p, Ent2p, Ent3p, Ent4p) in yeast. The epsin1 ENTH domain contains an additional N-terminal hydrophobic α0 helix. When binding to phosphoinositides, ENTH causes membrane curvature by inserting this α0 into the membrane (16). EpsinR is an adaptor for the late endosomal SNARE vti1b in mammals. Similarly, Ent3p is the adaptor for SNARE Vti1p in yeast (which is homologous to human vti1b) (8, 12, 13). Both Vti1p and vti1b are required for several transport steps between TGN, endosomes, and lysosomes (17–19).
The vast amount of SNAREs and the necessity of specific recognition of each SNARE to its adaptor suggest that SNARE proteins have diverse ways to recognize their adaptor. Thus, the SNARE–adaptor protein interaction at the molecular level has been the subject of interest. Four surface-peptide interacting structures have been reported (signal peptide of syntaxin5, Bet1, Sed5 being recognized by COP II Sec23/24, SNARE VAMP7_Longin domain interacting with adaptor Hrb’s unstructured C-terminal tail) (20–22). In addition, two domain–domain interacting complexes that impart more understanding of the specific recognition were reported. One is SNARE Sec22 being recognized by COP II Sec23/24. The other is the Habc domain of mouse vti1b in complex with human epsinR_ENTH domain (23, 24). Habc domains are found or predicted at the N termini of more than 10 yeast SNAREs (Fig. S1).
Here, we report three crystal structures in the yeast system: Habc domain of yeast Vti1p, ENTH domain of yeast Ent3p, and their complex. Comparison with the corresponding mammalian structure reveals that yeast Vti1p uses an opposite surface on its Habc domain to bind Ent3p. We further designed mutations on this interface that disrupt the interaction of yeast Vti1p-Ent3p. These mutations affected the sorting of Vti1p in vivo by both sucrose density gradient centrifugation and size exclusion chromatography. Thus, this interface is responsible for Vti1p-Ent3p recognition in yeast. This result suggests the SNARE–adaptor recognition could gain versatility by using distinct interfaces on the same scaffold.
Results
Structure of Yeast Vti1p_Habc and Ent3p_ENTH.
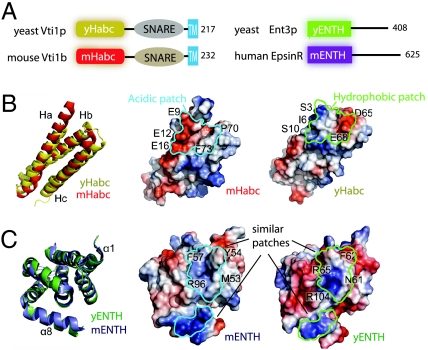
In addition to the SNARE motif, Vti1 proteins have a C-terminal transmembrane domain and an N-terminal Habc domain (Fig. 1A). The Habc domain in both mammalian vti1b and yeast Vti1p is responsible for interaction with the ENTH domains (8). We first solved the structure of the yeast Vti1p_Habc domain (designated as yHabc) at 1.9 Å. The yHabc contains approximately 100 residues with a three-helix bundle fold similar to vti1b and many other SNAREs (2, 24) (Fig. 1B). Three helices (Ha, Hb, and Hc) of the helix bundle closely resemble the structure of mammalian vti1b_Habc (designated as mHabc) with an rmsd of 2.17 Å. However, because of the low similarity of their amino acid sequences (16% identity on this region), yHabc has a different electrostatic potential distribution at the surface. The difference is especially apparent on the region where mHabc binds to the mammalian epsinR_ENTH (designated as mENTH). An acidic patch in mHabc, which is important for interacting with mENTH, is absent in the yHabc structure (Fig. 1B).
Fig. 1.
Structures of yHabc and yENTH. (A) Schematic of the SNARE proteins yeast Vti1p, mouse vti1b, and their adaptor proteins yeast Ent3p, human EpsinR. (B) Structural superimposition of Habc domains. (C) Structural superimposition of ENTH domains. The Habc/ENTH binding interface in the mammalian complex is circled in cyan; the corresponding region on yeast is circled in green. The mENTH and yENTH share a hydrophobic-basic patch and a basic α8 helix. In contrast, yHabc shows a surface electrostatic distribution different from mHabc.
The difference between yHabc and mHabc raised the question of whether their ENTH binding partners also differ in the binding interface. We further solved the crystal structure of the yeast Ent3p_ENTH domain (designated as yENTH) at 2.1-Å resolution. The ENTH domains of Ent3p and epsinR are highly homologous in amino acid sequence (45% identity). Accordingly, the yENTH domain has a conserved tertiary structure of eight α helices (α1–α8) closely resembling the ENTH domain of its mammalian homologue epsinR (rmsd = 1.33 Å, Fig. 1C). The yENTH also shares a similar electrostatic potential distribution on the potential Habc-binding site with mENTH. This Habc-binding site on mENTH, including a basic charged helix (α8) and a hydrophobic-basic patch that interacts with the acidic patch in mHabc, is also seen in yENTH (Fig. 1C). The discrepancy between the similarity of the ENTH domains and the differences between Habc domains raised a question of whether a human-like ENTH-Habc complex forms in yeast.
Complex Structure of Yeast Habc and ENTH.
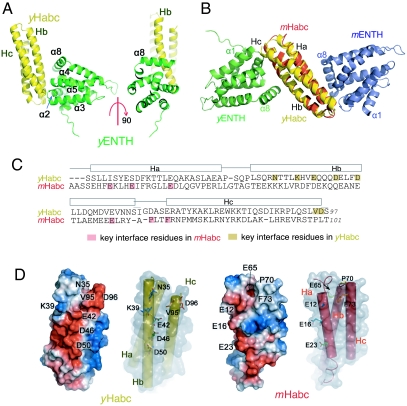
To answer this question, we further investigated the interaction of yHabc and yENTH. We first determined that yHabc binds to yENTH with a dissociation constant (Kd) of 12 μM (Fig. S2), an affinity close to that of the mammalian mHabc–mENTH pair (22 μM) (24). Although direct mixing of the two purified proteins yielded crystals of the complex, they had no sufficient diffraction quality. To improve the crystals, we further isolated the complex peak on a size exclusion column and used the purified complex for crystallization. This significantly improved the crystals, and the complex structure was solved at 2.2-Å resolution (Fig. 2A).
Fig. 2.
Complex structure of yHabc-yENTH. (A) Overall structure of yHabc–yENTH complexes. (B) yENTH and mENTH bind to the opposite sides of Habc domain when yHabc (yellow) was superimposed onto mHabc (red). (C) Sequence comparison of Habc domains shows that yHabc and mHabc use distinct residues for binding to ENTH domains. (D) Distinct ENTH-binding interface on yHabc and mHabc.
In the crystals of yHabc–yENTH complex, each yHabc molecule packed with two yENTH molecules and together they formed the minimal asymmetric unit. When combined with other packing units, each yHabc has contacts with five surrounding yENTH domains in the crystals (Fig. S3). Surprisingly, none of these five contacts was observed in the previous mammalian mHabc–mENTH complex structure. Among them, one contact is dominant. It has the largest interface (670 Å2) and occupies 11% of the total surface area of yHabc and 8% of the surface area of yENTH. This yHabc–yENTH contact is formed by a cross-attachment of α2–α3 loop, α4–α5 loop, and α8 in the yENTH domain with the helix Hb and the C terminus of Hc of the three-helix bundle of yHabc (Fig. 2A). In contrast, the second packing interface is much smaller with only 6% of yHabc and 4% of yENTH. Thus, this largest interface is likely to represent the native complex of yHabc and yENTH and was later confirmed (see below).
Two Opposite Sides of Habc Bind to ENTH.
To clarify the difference between these two Habc–ENTH complexes from yeast and mammals, we aligned their structures by superimposing the Habc domains. Surprisingly, ENTH domains from human epsinR and yeast Ent3p bind to two opposite sides of the three-helix bundle of Habc (Fig. 2B). While the interface on yHabc domain is formed by helix Hb and the C terminus of Hc, the interface on mHabc domain is formed by Ha and a loop region between Hb and Hc (Fig. 2B).
In addition to the distinct assembly, these two complexes also differ in their binding surface on the Habc domains. The interface on yHabc is extended and covered with strong acidic residues. In contrast, the interface on mHabc is formed by a few acidic residues and a new hydrophobic cavity close to F73 (Fig. 2D). Significantly, the two interfaces are formed by two sets of different residues that are located at distinct positions on both sequence and the tertiary structure of Habc. Three acidic residues (E42, D46, and D50) forming the yHabc interface are located on the helix Hb, while three other different acidic residues (E12, E16, and E23) forming the mHabc interface are from the helix Ha (Fig. 2C). In addition, two residues (P70, F73) that form the hydrophobic cavity in mHabc are absent in yHabc (Fig. 2C). These differences indicate that the yHabc interacts with ENTH domain through a distinct site that is not present in its mammalian homologue mHabc.
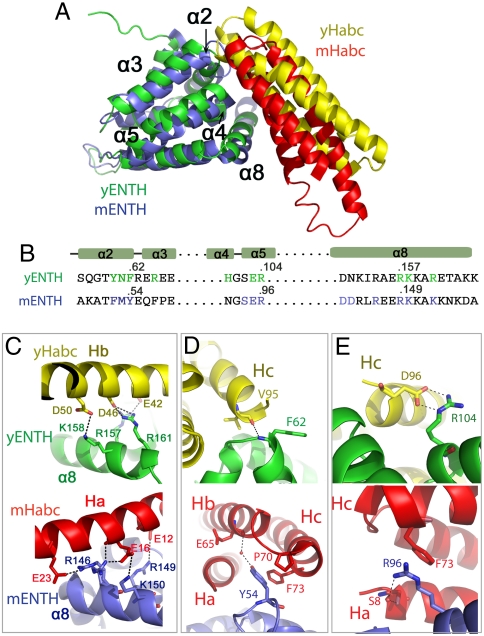
Same Site on ENTHs Binds to Two Habc Domains.
To further compare the difference between two Habc–ENTH complexes, we superimposed their structures using the ENTH domains. Interestingly, the two ENTH domains use the same site to bind to the Habc domains. In both complexes, an interface involving α2–α3 loop, α4–α5 loop, and α8 in ENTH binds to Habc (Fig. 3A).
Fig. 3.
Conserved site on ENTH domains binds to Habc domains. (A) Same site on ENTH domain binds to two Habc domains when yENTH was superimposed onto mENTH. (B) Sequence comparison of ENTH domains shows that interface residues are mostly conserved between yeast and mammals. (C) Conserved basic residues on ENTH α8 bind to different acidic residues of Habc domains. Acidic residues on mHabc are from Ha, while those on yHabc are from Hb. (D and E) Conserved residues on ENTH use different conformations to bind to different site on Habc domains. F62 in yENTH corresponds to Y54 in mENTH (D). R104 in yENTH correspond to R96 in mENTH (E).
We further checked sequence conservation of this shared binding site to identify whether different residues are used in ENTHs to bind to the two distinct sites on Habc. Surprisingly, many of the interface residues in ENTH domains are identical between yeast and human (4 out of a total of 10 shared in yeast and mammals; Fig. 3B). For other interface residues, changes are small (Y60 to F52, F62 to Y54, R161 to K153, from yeast to human). In contrast, all of these conserved residues are used to recognize the different interfaces on Habc. For example, three basic residues in α8 of both yENTH and mENTH (R157, K158, R161 in yENTH and R146, R149, K150 in mENTH) form salt bridges but with different acidic residues on Habc domains (E42, D46, and D50 on the helix Hb in yHabc and E12, E16, and E23 on Ha in mHabc) (Fig. 3C). Two conserved residues, F62 and R104 in yENTH (corresponding to Y54 and R96 in mENTH, respectively), form different types of interactions to Habc domains by adopting alternative conformations. The hydrophobic side chain of F62 in the α2–α3 loop of yENTH is flipped away from the binding interface. The flipped F62 interacts with yHabc through a new hydrogen bond between the main-chain NH- group of F62 and the main-chain C = O of V95 on Hc of yHabc (Fig. 3D). In contrast, its homologous residue Y54 in mENTH forms a hydrophobic patch and binds to the cavity formed by P70, F73, and E65 on the loop between helix Hb and Hc of mHabc (Fig. 3D). The side chain of R104 in the α4–α5 loop of yENTH binds to yHabc through a salt bond with D96 at the C terminus of Hc (Fig. 3E), while its homologous residue R96 in mENTH forms a stacking interaction with the phenol ring of F73 on Hb-Hc loop of mHabc and a water-bridged hydrogen bond to the side chain of S8 on Ha (Fig. 3E). Thus, the same sites on ENTHs form distinct interactions with different sites on Habc domains in yeast and mammals.
Structural-Based Mutagenesis Disrupts the yHabc-yENTH Interaction.
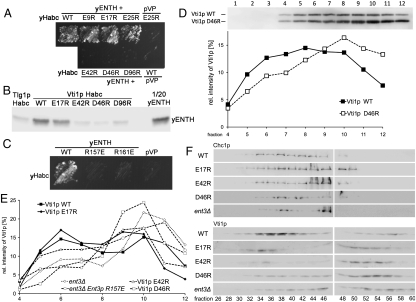
To confirm that the observed yHabc–yENTH complex in crystal represents the true complex in vivo, we carried out two-hybrid experiments in yeast. We first made mutations based on the mammalian complex. We chose α8 of ENTH domain as a target because it contains conserved residues in both yeast and mammals (Fig. 3C). In mammals, these basic residues form salt bridges with three acid residues on the helix Ha in mHabc. Mutagenesis of one acid residue (E12 to R) was sufficient to disrupt the interaction in mammals (24). To test for the interactions in yeast, we mutated each acidic residue (E9, E17, E25) in Ha of yHabc to arginine and paired them with yENTH in two-hybrid assays. Growth of yHabc mutants E9R, E17R, or E25R with yENTH was comparable to wild-type yHabc (WT) on selective plates (Fig. 4A) indicating that these three acid residues do not form the yHabc–yENTH interface. We then designed mutations on yHabc based on the yeast yHabc–yENTH complex structure. Three important contacts are observed in the crystal, and each of them was mutated (Fig. S4A). E42 and D46 in the helix Hb form two salt bridges with R157 and R161 in the helix α8 of yENTH, respectively. Introduction of the mutations yHabc_E42R or yHabc_D46R abolished their ability to grow on selective plates and therefore disrupted the interaction between yENTH and yHabc (Fig. 4A). Second, D96 on Hc binds directly to R104 in yENTH (Fig. 3E and Fig. S4C). The mutation yHabc_D96R also disrupted the interaction (Fig. 4A). To further confirm if the effects of these mutations in two-hybrid are direct, we studied their interactions in vitro. Purified yENTH with a Strep-tag was used to interact with WT and mutant His6-tagged yHabc. While His6-yHabc WT and E17R pulled down yENTH–Strep, His6-yHabc_E42R, D46R, and D96R could not bind to yENTH–Strep (Fig. 4B). Thus the effects of the point mutations are direct and consistent in both two-hybrid and in vitro pull-down assays. Moreover, we mutated two interacting residues on the yENTH domain and found each point mutation (R157E and R161E) could also disrupt yENTH’s interaction with yHabc in two-hybrid assays (Fig. 4C). Taken together, these results support that yHabc of yeast Vti1p binds to the yENTH domain of Ent3p in the way as observed in our crystal structure.
Fig. 4.
The yHabc/yENTH interface is required for the correct sorting of Vti1p. (A) Yeast two-hybrid assays of yHabc WT and mutants cotransformed with wild-type yENTH. VP16 (pVP) was used as a negative control. Interaction of yHabc/yENTH was disrupted by mutations designed from the yHabc/yENTH interface (E42R, D46R on Hb, D96R on Hc) but unaffected by mutations based on the mammalian interface (E9R, E17R, E25R on Ha). (B) In vitro pull-down of Strep-tagged yENTH by His-tagged yHabc (Vti1p_Habc) WT and mutants. His-tagged Tlg1p_Habc was used as a negative control. All proteins were bacterially expressed and purified. Antibodies against Ent3p were used for detection after SDS/PAGE. (C) Yeast two-hybrid assays of yENTH wt and two mutants cotransformed with wild-type yHabc. VP16 (pVP) was used as a negative control. (D) Different sucrose density gradient distribution of Vti1p and Vti1p_D46R in the same ent5▵ cells. Fractions were analyzed by immunoblotting for Vti1p. Vti1p_D46R could be distinguished from Vti1p WT by a higher mobility. Relative intensities of Vti1p in the gradient fractions were plotted against the fraction number. Representative gradients of three independent experiments are shown. (E) Disruption of Vti1p interaction to Ent3p changed the subcellular localization of yeast Vti1p. Homogenates of vti1▵ cells expressing Vti1p, Vti1p_E17R, Vti1p_E42R or Vti1p_D46R or ent3▵ cells in the absence or presence of Ent3p_R157E were analyzed as described in D. Representative gradients of three independent experiments are shown. (F) Mislocalization of Vti1p_D46R to smaller organelles was detected by size exclusion chromatography. Fractions were analyzed by immunoblotting for Vti1p (lower panels). The control (clathrin heavy chain Chc1p) was blotted using the same samples (upper panels). Loss of Vti1p_E42R from fractions 34–38 was detectable but less pronounced compared to Vti1p_D46R.
Disruption of the Interaction Affects the Sorting of Vti1p in Vivo.
To determine whether disruption of the interaction between Ent3p and Vti1p influences in vivo sorting of Vti1p, we compared the distribution of wild-type and mutant Vti1p in yeast cells. First, we monitored the distribution of Vti1p by sucrose density gradient centrifugation. Vti1p mislocalization in the absence of Ent3p and Ent5p was previously shown to result in a shift of Vti1p to denser organelles (13). Although Ent5p itself does not bind to Vti1p (8), Ent5p might have synergistic effects with Ent3p in the sorting of SNARE proteins. For example, the mislocalization of the endosomal SNARE Pep12p is aggravated in ent3Δent5Δ cells, while absence of Ent3p alone has only slight phenotypes (25). In order to evaluate specific effects between Ent3p and Vti1p in yeast, we used cells with ent5Δ background. We detected more Vti1p_D46R and slightly more Vti1p_E42R in the denser fractions (10–12) than wild-type Vti1p that was assayed in parallel (Fig. 4D and Fig. S5). The increased presence in denser fractions corresponds to a mislocalization of Vti1p_D46R and Vti1p_E42R to denser organelles in the absence of the Ent3p interaction, thus suggesting that the point mutation affects the distribution of Vti1p between organelles in yeast. A shift of endogenous wild-type Vti1p to denser fractions was also observed in ent3Δ cells (Fig. 4E and Fig. S4C). Furthermore, distribution of Vti1p mutants was tested in vti1Δ cells. The distribution of mammalian complex-derived mutant Vti1p_E17R is similar to Vti1p, consistent with its full ability to bind to yeast Ent3p in two-hybrid and pull-down assays. In contrast, the yeast complex-derived mutants show more (Vti1p_D46R) or slightly more (Vti1p_E42R) abundance in denser fractions than wild-type Vti1p (Fig. 4E and Fig. S4B). Finally, when an interacting residue in yeast Ent3p was mutated (Ent3p_R157E in ent3Δ cells), the endogenous Vti1p was also mislocalized (Fig. 4E and Fig. S4C).
To further confirm these mutant effects, we carried out another independent assay. Organelles in yeast cells were fractioned by size exclusion chromatography using a Sephacryl S-1000 column. Wild-type Vti1p was found to be less abundant in larger organelles (fractions 32–38) in ent3Δ cells compared to in wild-type cells (Fig. 4F). Consistently, the interaction-disrupting mutant Vti1p_D46R (derived from the yeast complex structure) was less abundant in larger organelles in vti1Δ cells and displayed a fractionation similar to wild-type Vti1p in ent3Δ cells. By contrast, the distribution of mammalian complex-derived mutant Vti1p_E17R was similar to that of the wild-type Vti1p (Fig. 4F). Taken together, these experiments show that the yeast Vti1p_Habc uses a distinct site to recognize the ENTH of its adaptor protein Ent3p in yeast, and that this new interaction between Ent3p and Vti1p is important for the sorting of Vti1p in vivo.
Discussion
Specific recognitions of cargo and adaptor proteins are central to vesicular transport in eukaryotic cells. More than 10 complex structures of cargo-adaptor pairs have been reported (20–24, 26–32). However, no switch of the binding surfaces in cargo-adaptor interaction has been found. Our results show that the Habc domains of yeast and mammalian Vti1 proteins bind their ENTH adaptors by opposite faces. Point mutagenesis of the interface residues verified that this new interaction is responsible for the formation of the yeast Vti1p–Ent3p complex. In contrast, mutations of Vti1p residues based on the mammalian Habc–ENTH interaction did not disrupt the yeast complex. Furthermore, a set of mutations unique to the yeast complex affects the distribution of Vti1p in yeast indicating the correct sorting of Vti1p between organelles requires this new interaction. The partial sorting of Vti1p_E42R into budding vesicles may be caused by its weak interaction with Ent3p.
In contrast to the single Vti1 protein (Vti1p) that is encoded in fungi and serves in several endosomal trafficking steps (5, 18, 19), mammals contain two different Vti1p homologues, vti1a and vti1b, with more specialized functions (6, 17, 33). Two distinct features are observed when the Vti1_Habc sequences from yeast to human are aligned (Fig. S6, and also seen in ref. 6). First, lower eukaryotic Vti1_Habc domains have a conserved acidic residue (D96) on helix Hc that is used to bind the ENTH domain as seen in our yeast complex. All mammalian vti1a and vti1b proteins have lost this acidic residue, suggesting that they do not bind the ENTH domain through the yeast interface. The vti1b binding site involves a large acidic surface patch in the helix Ha and another hydrophobic region between Hb and Hc (F73 in mouse). This newly evolved acidic-hydrophobic sequence accounts for the specific binding between the vti1b_Habc and epsinR_ENTH in the mammalian complex (24). This feature is conserved among mammalian vti1b sequences but not in vti1a. This lineage-specific sequence variance may be a signature for the differential roles of Vti1 proteins: Vti1a functions at early endosomes while vti1b functions at late endosomes (13, 17), and vti1a is not able to bind epsinR (8, 12).
One consequence of the distinct interacting modes between yeast and mammals is the orientation of the SNARE proteins. As the α0 helices of ENTH domains are inserted into membranes for anchoring (34), both epsinR and Ent3p orient their N termini toward the membrane. However, due to the opposite binding orientation of Habc domains, the ENTH-bound yeast Vti1p and mammalian vti1b will be placed oppositely (Fig. S7). This difference may have functional consequences for cargo sorting because the rest of vti1b will be directed away from the membrane and may allow for more flexibility in sorting of SNARE complexes than Vti1p. In fact, vti1b has been shown to sort at least two more SNARE partners, syntaxin 8 and VAMP8 (35, 36). Superimposition of the yeast Habc–ENTH with the closed four-helix bundle form of Habc–SNARE structure of Sso1 (37) shows steric clashes between ENTH and the closed SNARE motif. The binding of Ent3p to Vti1p may help Vti1p to form an “open state” to free its SNARE motif and further regulate the SNARE-mediated membrane fusion process. Consistently, deletion of the Habc domain from Vti1p was found to stimulate the SNARE-mediated fusion of liposomes in vitro (38). The ability to form two opposite binding sites on the triple-helix Habc domain also implicates the potential diversity of SNAREs and epsin adaptors recognition. For example, it is known that Ent3p_ENTH interacts with the SNAREs Pep12p and Syn8p (39) that possess Habc domains. However, the ENTH interaction was disrupted in Pep12p by a F20L mutation or a deletion of residues 19–26 (13). This indicates that the helix Ha of Habc domain in Pep12p is important for interaction with Ent3p_ENTH and these residues are far from the ENTH-binding interface of yeast Vti1p. Thus, yeast Pep12p may have a similar ENTH-binding site as mammalian vti1b or it may contain a third ENTH-binding site.
Methods
Protein Preparation.
Ent3p_ENTH domain (uniprotKB ID: P47160; amino residues 28–170) and Vti1p_Habc domain (uniprotKB ID: Q04338; amino residues 3–99) were subcloned and expressed in Rossetta (DE3). Purified proteins (yENTH, yHabc, and the complex) were concentrated to 10 mg/mL for crystallization.
Crystallization and Structure Determination.
Crystallization of the proteins was performed at 288 K using the hanging-drop vapor diffusion method. All diffraction data were processed with HKL2000 program (40). Structural models were built and refined by Molrep, Coot, CNS, and Refmac5 (41–44). All current models have good geometry.
Sucrose Density Gradient Centrifugation and Size Exclusion Chromatography.
Subcellular fractionation of osmotically lysed spheroplasts was performed as described on a density gradient with steps of 22%–55% sucrose (13). Size exclusion chromatography was performed as described (45) with modifications. Each strain was analyzed twice with comparable results with exception of variable clathrin signals in fraction 48 (see details in SI Text).
Supplementary Material
Acknowledgments.
We thank Prof. Min Guo and Xuebiao Yao for helpful discussions. We also thank Jana Jesuthas, Anne Muschinski, and Milena Herbort for excellent technical assistance. Jaqueline Alonso Lunar is acknowledged for the construction of plasmids, Sonja Habelt for protein purification. We thank S. Lemmon (University of Miami) for the generous gift of Chc1p antibodies. We appreciate the assistance from Shanghai Synchrotron Radiation Facility. Financial support for this project was provided by research grants from the Chinese National Natural Science Foundation (30025012 and 10979039), the Chinese Ministry of Science and Technology (2006CB806500 [973], 2006CB910200 and 2006AA02A318), the Chinese Academy of Sciences (KSCX2-YW-R-60), the Chinese Ministry of Education (20070358025), and the Natural Science Foundation of Anhui Province (090413081).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3ONJ (Vti1p_Habc), 3ONK (Ent3p_ENTH), and 3ONL (Vti1p_Habc–Ent3_ENTH complex)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013101108/-/DCSupplemental.
References
- 1.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 2.Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 3.Sudhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickner W. Membrane fusion: Five lipids, four snares, three chaperones, two nucleotides, and a rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–135. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- 5.Kienle N, Kloepper TH, Fasshauer D. Phylogeny of the SNARE vesicle fusion machinery yields insights into the conservation of the secretory pathway in fungi. BMC Evol Biol. 2009;9:19. doi: 10.1186/1471-2148-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kloepper TH, Kienle CN, Fasshauer D. SNAREing the basis of multicellularity: Consequences of protein family expansion during evolution. Mol Biol Evol. 2008;25:2055–2068. doi: 10.1093/molbev/msn151. [DOI] [PubMed] [Google Scholar]
- 7.Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744:120–144. doi: 10.1016/j.bbamcr.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Chidambaram S, Mullers N, Wiederhold K, Haucke V, von Mollard GF. Specific interaction between SNAREs and epsin N-terminal homology (ENTH) domains of epsin-related proteins in trans-Golgi network to endosome transport. J Biol Chem. 2004;279:4175–4179. doi: 10.1074/jbc.M308667200. [DOI] [PubMed] [Google Scholar]
- 9.Peden AA, Park GY, Scheller RH. The Di-leucine motif of vesicle-associated membrane protein 4 is required for its localization and AP-1 binding. J Biol Chem. 2001;276:49183–49187. doi: 10.1074/jbc.M106646200. [DOI] [PubMed] [Google Scholar]
- 10.Aridor M, Traub LM. Cargo selection in vesicular transport: The making and breaking of a coat. Traffic. 2002;3:537–546. doi: 10.1034/j.1600-0854.2002.30804.x. [DOI] [PubMed] [Google Scholar]
- 11.Owen DJ, Collins BM, Evans PR. Adaptors for clathrin coats: Structure and function. Annu Rev Cell Dev Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- 12.Hirst J, Miller SE, Taylor MJ, von Mollard GF, Robinson MS. EpsinR is an adaptor for the SNARE protein Vti1b. Mol Biol Cell. 2004;15:5593–5602. doi: 10.1091/mbc.E04-06-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chidambaram S, Zimmermann J, von Mollard GF. ENTH domain proteins are cargo adaptors for multiple SNARE proteins at the TGN endosome. J Cell Sci. 2008;121:329–338. doi: 10.1242/jcs.012708. [DOI] [PubMed] [Google Scholar]
- 14.Duncan MC, Payne GS. ENTH/ANTH domains expand to the Golgi. Trends Cell Biol. 2003;13:211–215. doi: 10.1016/s0962-8924(03)00076-x. [DOI] [PubMed] [Google Scholar]
- 15.Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- 16.Ford MG, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 17.Antonin W, et al. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19:6453–6464. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Mollard GF, Nothwehr SF, Stevens TH. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J Cell Biol. 1997;137:1511–1524. doi: 10.1083/jcb.137.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer von Mollard G, Stevens TH. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol Biol Cell. 1999;10:1719–1732. doi: 10.1091/mbc.10.6.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller EA, et al. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- 21.Mancias JD, Goldberg J. Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO J. 2008;27:2918–2928. doi: 10.1038/emboj.2008.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pryor PR, et al. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell. 2008;134:817–827. doi: 10.1016/j.cell.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mancias JD, Goldberg J. The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol Cell. 2007;26:403–414. doi: 10.1016/j.molcel.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 24.Miller SE, Collins BM, McCoy AJ, Robinson MS, Owen DJ. A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature. 2007;450:570–574. doi: 10.1038/nature06353. [DOI] [PubMed] [Google Scholar]
- 25.Copic A, Starr T, Schekman R. Ent3p and Ent5p exhibit cargo-specific functions in trafficking proteins between the trans-Golgi network and the endosomes in yeast. Mol Biol Cell. 2007;18:1803–1815. doi: 10.1091/mbc.E06-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson LP, et al. A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell. 2010;141:1220–1229. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edeling MA, et al. Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev Cell. 2006;10:329–342. doi: 10.1016/j.devcel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly BT, et al. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 2008;456:976–979. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olesen LE, et al. Solitary and repetitive binding motifs for the AP2 complex alpha-appendage in amphiphysin and other accessory proteins. J Biol Chem. 2008;283:5099–5109. doi: 10.1074/jbc.M708621200. [DOI] [PubMed] [Google Scholar]
- 31.Misra S, Puertollano R, Kato Y, Bonifacino JS, Hurley JH. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature. 2002;415:933–937. doi: 10.1038/415933a. [DOI] [PubMed] [Google Scholar]
- 32.Shiba T, et al. Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport. Nat Struct Biol. 2003;10:386–393. doi: 10.1038/nsb920. [DOI] [PubMed] [Google Scholar]
- 33.Kreykenbohm V, Wenzel D, Antonin W, Atlachkine V, von Mollard GF. The SNAREs vti1a and vti1b have distinct localization and SNARE complex partners. Eur J Cell Biol. 2002;81:273–280. doi: 10.1078/0171-9335-00247. [DOI] [PubMed] [Google Scholar]
- 34.Horvath CA, Vanden Broeck D, Boulet GA, Bogers J, De Wolf MJ. Epsin: Inducing membrane curvature. Int J Biochem Cell Biol. 2007;39:1765–1770. doi: 10.1016/j.biocel.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Atlashkin V, et al. Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8. Mol Cell Biol. 2003;23:5198–5207. doi: 10.1128/MCB.23.15.5198-5207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon DE, Mirza M, Sahlender DA, Jakovleska J, Peden AA. Coiled-coil interactions are required for post-Golgi R-SNARE trafficking. EMBO Rep. 2009;10:851–856. doi: 10.1038/embor.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munson M, et al. Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nat Struct Biol. 2000;7:894–902. doi: 10.1038/79659. [DOI] [PubMed] [Google Scholar]
- 38.Paumet F, et al. Concerted auto-regulation in yeast endosomal t-SNAREs. J Biol Chem. 2005;280:21137–21143. doi: 10.1074/jbc.M500841200. [DOI] [PubMed] [Google Scholar]
- 39.Zimmermann J, Chidambaram S, Fischer von Mollard G. Dissecting Ent3p: The ENTH domain binds different SNAREs via distinct amino acid residues while the C-terminus is sufficient for retrograde transport from endosomes. Biochem J. 2010;431:123–134. doi: 10.1042/BJ20100693. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Vagin A, Teplyakov A. Molrep: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov G, Vagin A, Dodson E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez GE, Payne GS. Laa1p, a conserved AP-1 accessory protein important for AP-1 localization in yeast. Mol Biol Cell. 2006;17:3304–3317. doi: 10.1091/mbc.E06-02-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.