Abstract
Metabolic changes in cancer have been observed for almost a century. The mechanisms underlying these changes have begun to emerge from the recent studies implicating the tumor suppressor p53 in multiple metabolic pathways. The ability of p53 to regulate metabolism may also play important roles in the physiology of normal cells and organs. Here we demonstrate that p53 lowers bile acid (BA) levels under both normal and stressed conditions primarily through up-regulating expression of small heterodimer partner, a critical inhibitor of BA synthesis. Our results uncover a unique metabolic regulatory axis that unexpectedly couples p53 to BA homeostasis. Our results also warrant future studies to investigate a possible role of this axis in the tumor suppression by p53, because excessive quantities of BAs are cytotoxic and can cause liver damage and promote gastrointestinal cancers.
In the 1920s, Warburg found that cancers show a metabolic switch from oxidative phosphorylation to glycolysis (1). Recent findings reveal that glycolysis benefits cancer cells by providing a constant supply of building blocks to facilitate the construction of new daughter cells and demonstrate that the tumor suppressor p53 is an integral player in the Warburg effect (2–5). Suggesting that p53 is a key regulator of a wider range of metabolic processes than previously appreciated, p53 has also been shown to regulate other metabolic pathways, including autophagy and oxidative stress (2–5). Importantly, the metabolic roles of p53 have consequences beyond cancer and likely influence various aspects of normal and diseased life (2–5). The regulation of metabolic pathways is clearly an important aspect of p53 function that may eventually provide unique therapeutic targets for cancer and likely other diseases (5). However, the role of p53 in metabolism appears to be far from straightforward and remains to be fully elucidated. In this paper, we present evidence supporting a direct role of p53 in bile acid (BA) homeostasis, a surprising previously undescribed addition to the list of metabolic functions of p53.
BAs, synthesized from cholesterol molecules, play a critical role in eliminating excess cholesterol from the body and process dietary fat by facilitating the formation of micelles (6, 7). However, as excessive quantities of BAs are cytotoxic (8) and can cause liver damage (9) and may even contribute to the development of gastrointestinal cancers (10), it is important to tightly regulate BA synthesis. The nuclear receptor SHP (small heterodimer partner) regulates diverse physiological processes, including BA homeostasis (6, 7). BAs serve as a ligand of the nuclear receptor farnesoid X receptor (FXR), which induces the expression of SHP in hepatocytes (11, 12). In turn, SHP suppresses the expression of cholesterol 7α-hydroxylase (Cyp7A1) and sterol 12α-hydroxylase (Cyp8B1), two crucial enzymes that orchestrate the conversion of cholesterol to BAs (11, 12). Cyp7A1 catalyzes the synthesis of two primary BAs, cholic acid (CA) and chenodeoxycholic acid, and Cyp8B1 catalyzes CA synthesis (6, 7). In the intestine, FXR induces an intestinal hormone, FGF15 (FGF19 in human), which activates hepatic FGF receptor type 4 (FGFR4) signaling to inhibit BA synthesis, likely through a JNK-dependent signaling cascade (13–18). BAs can also activate PKC or induce the synthesis of inflammatory cytokines (TNFα, and IL-1β) and their release from Kupffer cells (15, 19–22). Both PKC and these cytokines can suppress Cyp7A1 expression in hepatocytes (15, 19–22). Thus, BAs antagonize their own synthesis via multiple regulatory pathways.
We recently found that inactivation of a histone H3-lysine-4-methyltransferase MLL3, a transcriptional coactivator of p53 and FXR (23, 24), results in enlarged gallbladders and increased BA levels (24). These findings, combined with the linkage of p53 to metabolism (2–5), prompted us to investigate whether p53 regulates BA synthesis in the liver. Our results in this paper reveal that p53 is indeed an intrinsic regulator of BA homeostasis under both normal and stressed conditions and that this regulation appears to involve at least in part a dual ability of p53 to directly activate expression of SHP via p53-response elements (p53REs) in SHP as well as to enhance the stability of the SHP protein.
Results
Down-Regulation of BA Levels by p53.
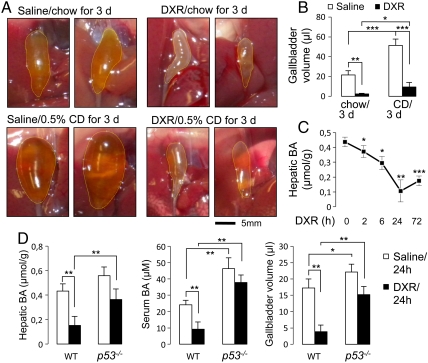
To test whether p53 controls BA homeostasis, we subjected wild-type mice to intraperitoneal injections of saline or doxorubicin (DXR), an activator of p53 signaling. DXR-injected mice showed dramatically shrunk gallbladders relative to saline treated mice (Fig. 1 A and B). DXR injection also suppressed the enlargement of gallbladders in mice fed a diet containing 0.5% CA (Fig. 1 A and B). DXR treatment led to a decrease in hepatic and serum BA levels (Fig. 1 C and D). Demonstrating that p53 is an effector of DXR, the DXR-dependent responses were significantly blunted in p53-null mice (25) (Fig. 1D and Fig. S1). Relative to wild-type mice, p53-null mice also showed an increased BA pool size (Fig. S2). Interestingly, serum BA levels and gallbladder size/volume were significantly higher in p53-/- mice than in wild-type mice even without DXR-dependent p53 activation (Fig. 1D and Fig. S1), suggesting that p53 is an intrinsic suppressor of BA levels under both normal and acutely stressed (e.g., DXR-treated) conditions. Consistently, low but readily detectable levels of p53 protein were found in normal liver tissue (26).
Fig. 1.
DXR regulates BA homeostasis through p53. (A) Representative pictures of gallbladders of wild-type male mice intraperitoneally injected with saline or DXR and then fed a regular chow diet or a diet containing 0.5% CA (CD) for 3 d. Similar results were obtained with female mice. (B) Quantitation of gallbladder volume for the mice in A. n = 4. (C) Quantitation of hepatic BA levels of wild-type male mice (n = 3–4) over 72 h after intraperitoneal injection of DXR. Similar results were obtained with female mice. (D) Measurement of hepatic and serum BA levels and gallbladder volume of wild-type and p53-null male mice (n = 5), 24 h after intraperitoneal injection of saline or DXR. Similar results were obtained with female mice.
Control of Expression of BA Synthesis Regulators by p53.
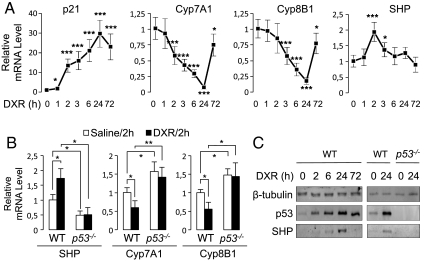
To elucidate the molecular basis of DXR action to decrease BA levels, we monitored the expression of various players in BA homeostasis over 72 h post DXR treatment. Expression of p21 mRNA, a well-defined target of p53, was induced (Fig. 2A). Interestingly, expression of Cyp7A1 and Cyp8B1 mRNAs was reduced with a maximum reduction at 24 h post DXR treatment (Fig. 2A). In the livers of p53-null mice (25), levels of Cyp7A1/Cyp8B1 were substantially elevated and failed to respond to DXR (Fig. 2B). These data suggest that p53 suppresses BA synthesis by inhibiting the expression of Cyp7A1/Cyp8B1 enzymes. Of note, our analysis of mouse and human Cyp7A1/Cyp8B1 genes failed to identify any motif similar to the consensus p53REs (4), suggesting that p53 may not directly regulate the expression of Cyp7A1/Cyp8B1.
Fig. 2.
DXR controls expression of critical regulators of BA homeostasis through p53. (A) qRT-PCR measurement of levels of mRNAs for p21, Cyp7A1, Cyp8B1, and SHP in wild-type male mice (n = 3–4) over 72 h after intraperitoneal injection of DXR. Similar results were obtained with female mice. (B) qRT-PCR measurement of levels of mRNAs for SHP, Cyp7A1, and Cyp8B1 of wild-type and p53-null male mice (n = 3), 2 h after intraperitoneal injection of saline or DXR. Similar results were obtained with female mice. (C) Western blotting analysis of hepatic expression of β-tubulin (loading control), p53, and SHP in wild-type and p53-null mice at 0, 2, 6, 24, and 72 h post DXR treatment.
Next, we examined expression of known regulators of Cyp7A1/Cyp8B1. Interestingly, hepatic expression of SHP, a negative regulator of Cyp7A1/Cyp8B1, was sharply induced at 2 h after DXR treatment and then gradually normalized to the basal level by 72 h post DXR treatment (Fig. 2A). The DXR-dependent induction of SHP at 2 h post DXR treatment was abolished in p53-null mice (Fig. 2B), indicating that p53 is required for the acute induction of SHP by DXR. Importantly, the basal levels of SHP without DXR injection were significantly reduced in p53-null mice, suggesting that p53 also maintains SHP expression under normal conditions.
Intriguingly, SHP mRNA levels in the liver were highest at 2 h post DXR treatment and almost normalized to the basal level at 24 h post DXR treatment, the time point in which Cyp7A1/Cyp8B1 mRNA levels were lowest (Fig. 2A). This argues against the possibility that p53 suppresses expression of Cyp7A1/Cyp8B1 via inducing expression of SHP, unless SHP protein levels are also highest at 24 h post DXR treatment. Indeed, SHP protein levels were readily observed at 24 h but significantly less at 2, 6, and 72 h post DXR treatment (Fig. 2C). One possible explanation for this observation is a role for DXR/p53 signaling to stabilize SHP proteins. Of note, it has been shown that SHP proteins are rapidly degraded in hepatocytes and that bile acids and bile acid-induced FGF19 signaling pathways increase SHP stability by inhibiting its ubiquitination and proteasomal degradation (27). In support of the idea that p53 signaling stabilizes SHP proteins, SHP proteins transiently expressed in HepG2 cells treated with DXR were significantly more stable than those expressed in HepG2 cells treated with vehicle alone (Fig. S3). The molecular basis underlying this intriguing action of p53 signaling to stabilize SHP proteins remains to be determined.
SHP as a Direct Transcriptional Target Gene of p53.
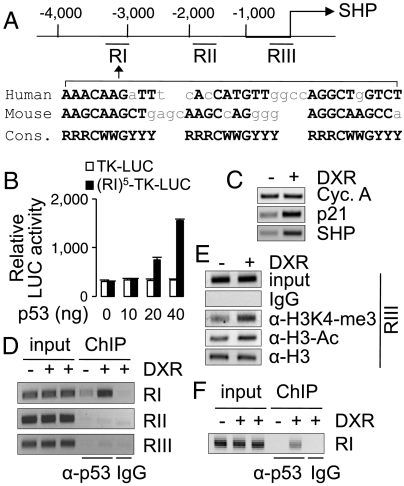
Our data raise the possibility that p53 suppresses BA synthesis, at least in part, by acting as a positive transcriptional regulator of SHP as well as by enhancing the protein stability of SHP. To investigate whether SHP is a direct transcriptional target of p53, we analyzed mouse and human SHP genes for motifs similar to the consensus p53REs (4). We found that highly conserved p53REs are present 2.7 kb and 3.1 kb upstream of mouse and human SHP-coding regions, respectively (Fig. 3A). Five copies of the distal region of mouse SHP (RI in Fig. 3A) were constructed into a luciferase reporter driven by a minimal thymidine kinase (TK) promoter. In HEK239 cells, p53 directed the dose-dependent transactivation of this reporter (Fig. 3B). Additionally, DXR treatment of a human liver carcinoma cell line, HepG2, increased expression of SHP and the positive control, p21 (4) (Fig. 3C), faithfully recapitulating the DXR-mediated up-regulation of SHP observed in the mouse liver. In ChIP experiments, p53 was recruited to the distal region of SHP (RI in Fig. 3A) in HepG2 cells, and this binding was enhanced by DXR treatment (Fig. 3D). In contrast, p53 was found neither in the proximal promoter region (RIII in Fig. 3A) containing the FXR response element (6) nor the region between the distal and promoter regions (RII in Fig. 3A) (Fig. 3D). DXR treatment resulted in transcriptionally active chromatin in the proximal promoter region of SHP in HepG2 cells, as shown by enrichment of two active chromatin marks, histone H3-lysine 4-trimethylation and acetylated H3 (Fig. 3E). Validating p53 occupancy of SHP in vivo, p53 was recruited to the distal region of SHP (RI in Fig. 3A) in the mouse liver exposed to DXR (Fig. 3F). Collectively, these data suggest that SHP is a direct transcriptional target gene of p53 and that recruitment of p53 to the distal region of SHP triggers the establishment of active chromatin in the proximal promoter region of SHP, resulting in up-regulation of SHP expression.
Fig. 3.
Direct binding of a distal enhancer of SHP by p53. (A) Schematic representation of a distal enhancer region of SHP with sequences similar to p53-response elements (RI), the promoter region containing FXR-response elements (RIII), and a nonspecific intervening region (RII). (B) A luciferase reporter driven by a minimal thymidine kinase promoter (TK) fused to five copies of SHP-p53REs, (RI)5-TK-LUC, but not the parental reporter with TK alone, TK-LUC, responded to p53 in a dose-dependent manner. (C) RT-PCR analysis of HepG2 cells treated with vehicle or DXR for 12 h to measure expression of p21, SHP, and cyclophilin A (control). (D–F) ChIP analysis of HepG2 cells (D and E) and mouse liver (F) treated with DXR for 2 h to examine the recruitment of p53 (D and F) and establishment of active chromatin (E). All experiments were repeated more than three times with similar results.
Overall, our results suggest that p53 signaling mobilizes at least two distinct mechanisms (i.e., transcriptional activation of SHP promoter and subsequently increasing the stability of SHP proteins) to accumulate highest levels of hepatic SHP proteins at 24 h post DXR treatment. In support of this idea, SHP proteins were not observed in the livers of p53-null mice even at 24 h post DXR treatment (Fig. 2C).
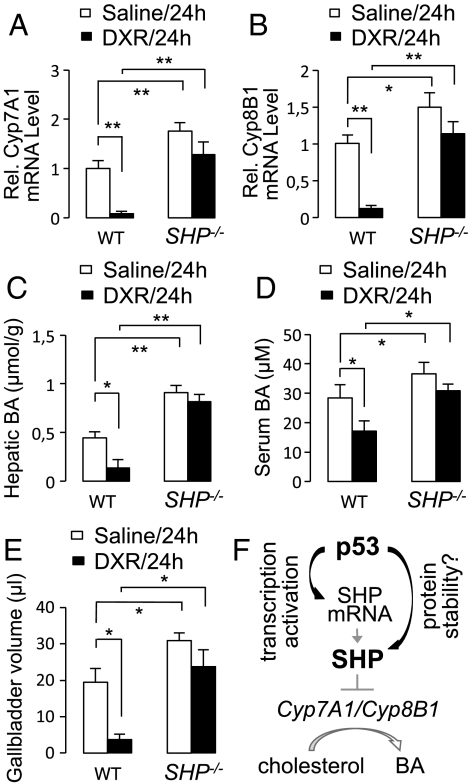
If SHP is required for p53-regulated BA homeostasis, SHP-null mice should be relatively refractory to DXR-triggered suppression of both Cyp7A1/Cyp8B1 and BA levels. Interestingly, DXR-dependent repression of Cyp7A1 and Cyp8B1 was not evident in SHP-null mice (Fig. 4 A and B). As reported (16), SHP-null mice exhibited increased BA levels and enlarged gallbladders (Fig. 4 C–E). Despite the increased BA levels in SHP-null mice, however, DXR failed to suppress hepatic and serum BA levels and gallbladder volume as readily as it did in wild-type mice (Fig. 4 C–E). These results demonstrate that SHP is indeed an essential mediator of p53-regulated BA homeostasis.
Fig. 4.
Involvement of SHP in DXR-mediated control of BA homeostasis. (A and B) qRT-PCR measurement of levels of mRNAs for Cyp7A1 (A) and Cyp8B1 (B) of wild-type and SHP-null male mice (n = 3), 24 h after intraperitoneal injection of saline or DXR. Similar results were obtained with female mice. (C–E) Measurement of hepatic and serum BA levels (C and D) and gallbladder volume (E) of wild-type and SHP-null male mice (n = 5), 24 h after intraperitoneal injection of saline or DXR. Similar results were obtained with female mice. (F) A working model for p53-SHP-Cyp7A1/B1 axis in BA homeostasis.
Impaired Intestinal Lipid Absorption by p53.
One major physiological function of BAs is to promote dietary lipid absorption from the intestine. Activation of p53 signaling by DXR injection reduced BA levels (Figs. 1 C and D and 4C–E). To test whether this leads to impaired dietary lipid absorption, we intraperitoneally injected saline or DXR into wild-type mice. Three days later, these mice were fed “1,2-dioleoyl-3-(pyren-1-yl)-decanoyl-rac-glycerol (DPG)” (28) using oral gavage. Small intestines of these mice were compared for levels of their absorption of DPG. Positive Oil-Red-O staining (i.e., absorption of DPG) was largely absent in the small intestines of DXR-treated mice, whereas it was evident in saline-treated mice (Fig. S4). These results demonstrate that activation of p53 signaling inhibits lipid absorption from intestines.
Discussion
Our results reveal a unique regulatory axis in BA homeostasis in which p53 utilizes at least two distinct mechanisms to up-regulate SHP, which in turn serves as a key negative regulator of BA synthesis (Fig. 4F). Although p53 is known to mobilize repair and cell death during times of cellular stress (4), there is also increasing evidence that p53 regulates metabolic homeostasis at normal physiological conditions and that these activities are important for cancer prevention (2–5). Our data show that p53-null mice have significantly higher levels of BAs than wild-type animals both in the absence and the presence of DXR (Fig. 1D). Thus, p53 appears to control BA homeostasis not only in response to acute stresses such as DXR but also under normal physiological conditions. Of note, BAs antagonize their own synthesis via multiple regulatory pathways (6, 7, 11–22). Interestingly, SHP-null mice fail to support DXR-dependent repression of Cyp7A1 and Cyp8B1 (Fig. 4 A and B) and of hepatic and serum BA levels and gallbladder volume (Fig. 4 C–E). These results strongly suggest that SHP is a key mediator of p53-regulated BA homeostasis. Correspondingly, we failed to observe any significant change in intestinal expression of FGF15, another key inhibitor of BA synthesis (13–18), upon DXR treatment (Fig. S5). Although it is possible that p53 may also impinge on bile acid homeostasis via other pathways (6, 7, 13–22), our results with SHP-null mice suggest that SHP likely lies downstream of those additional pathways targeted by p53. For instance, we found that bile salt export pump (BSEP), a protein involved in the canalicular secretion of BAs, is also down-regulated by DXR in the mouse liver (Fig. S6A). Considering that BSEP is a target of liver receptor homolog 1 (LRH-1) (29) and thus a potential negative target of SHP, it is possible that p53 also controls bile salt export via SHP.
Inactivation of our p53-SHP axis, such as mutations in p53 or SHP, may lead to chronically elevated levels of BAs. Of note, mice deficient in SHP or FXR, which show chronically elevated levels of BAs, are susceptible to liver and colorectal tumor formation (30–34), and BAs have also been suggested to play a causative role in human gastrointestinal cancers (10). Thus, it will be interesting to further investigate whether the p53-triggered decrease in BA levels represents a previously unrecognized avenue that p53 utilizes to exert its tumor suppression activity against liver and gastrointestinal cancers (35, 36).
Interestingly, BAs have been suggested to ameliorate insulin resistance (37, 38). Thus, our p53-SHP axis may also contribute to insulin resistance by lowering BA levels. In support of this possibility, the loss of a copy of p53 improved the insulin resistance of Ay mice, which are obese and diabetic (39).
An intriguing finding was that the induction of SHP mRNA by DXR peaked at 2 h post DXR treatment and then gradually adjusted back to the normal level at 72 h, whereas expression of p21 continued to increase and peaked at 24 h (Fig. 2A). Notably, the induction pattern of p21 mRNA was identical to the levels of p53 protein in the liver (compare Fig. 2 A and C). The unique, acute induction of SHP mRNAs by p53 may involve at least three distinct mechanisms. First, SHP down-regulates its own expression, because two nuclear receptors, HNF4 and LRH-1, up-regulate expression of SHP that forms a heterodimer with HNF4 and LRH-1 and suppress their transcriptional activity (12, 40–43). This negative autoregulatory feedback of SHP likely contributes to the normalization of SHP mRNA levels following its acute induction by p53. Second, as BAs function as ligands of FXR, a positive regulator of SHP, a decrease in BA levels by DXR may also lead to attenuation of FXR-dependent up-regulation of SHP, further contributing to the normalization of SHP mRNA levels. Finally, SHP has been recently shown to inhibit p53 transactivation of the miR-34a promoter (44). Consistent with these results, SHP repressed p53-dependent transactivation of the luciferase reporter directed by five copies of SHP p53REs (Fig. S7). This may also contribute to the normalization of SHP mRNA levels. Overall, these results suggest that p53 and SHP are regulated through multiple autoregulatory loops.
SHP mRNA levels peaked at 2 h post DXR treatment and were rapidly normalized to the basal level, whereas expression of Cyp7A1/Cyp8B1 mRNA levels continued to decline and were lowest at 24 h post DXR treatment (Fig. 2A). These results led to our finding that SHP protein is likely stabilized by p53 signaling (Fig. S3). Accordingly, SHP protein levels in the mouse liver became highest at 24 h post DXR treatment (Fig. 2C), explaining why Cyp7A1/Cyp8B1 mRNA levels were lowest at this time point (Fig. 2A). In addition, Cyp7A1 is also a target of HNF4 (40, 41) and LRH-1 (12, 42, 43). Interestingly, we found that expression of HNF4 and LRH-1 begins to be down-regulated by DXR at 6 h post DXR treatment (Fig. S6B). This may also contribute to the continuous decline in Cyp7A1 levels.
In summary, we found a unique metabolic regulatory axis of p53, which plays critical roles in BA homeostasis. Future studies will continue to uncover the multiple facets of this interesting p53-SHP pathway (Fig. 4F), including the molecular basis for the stabilization of SHP protein by p53 signaling, in fat and glucose homeostasis as well as tumor suppression. Given our finding of p53 as a key regulator of BA homeostasis, p53 modulators may have therapeutic values in treating diverse metabolic diseases involving impaired BA homeostasis, ranging from cholestasis and fatty liver to diabetes. Finally, DXR is not particularly an ideal tool to specifically study the hepatic p53 signaling. It will be interesting to test whether hepatic expression of SHP (and thus bile acid homeostasis) is modulated by p53 signaling under physiologically more relevant condition, such as hepatocellular injury associated with hepatosteatosis (45).
Materials and Methods
Animals.
Age-matched groups of 7- to 8-wk-old C57BL/6 mice (wild-type, SHP-null and p53-null) were fed either a standard rodent chow diet or a chow diet supplemented with 0.5% (wt/wt) CA. Then 10 mg/kg of DXR was administered by intraperitoneal injection in a constant volume of saline. Sample collections were done as described (46).
Measurements of Hepatic and Serum BA Levels and Gallbladder Volume.
The BA content of the liver and serum was measured using the BA assay kit (Diagnostic Chemicals Ltd). For measurement of the volume of the gallbladder, the weight of contents squeezed out from each gallbladder was measured.
Western Analysis.
Western assays for mouse livers were carried out using antibodies against β-tubulin (sc-5274, Santa Cruz), p53 (FL-393, Santa Cruz), and SHP (SC-30169, Santa Cruz).
ChIP Analysis.
ChIP assays for HepG2 cells and mouse livers were performed as described (47, 48). The antibodies used for ChIP assays were as follows: α-53 antibody (Pab 1801 and DO-1, Santa Cruz), α-H3K4-me3 antibody (05–745, Millipore), α-H3K9/K14-Ac (06–599, Upstate), and α-H3 antibody (Ab1791, Abcam). Sequences of the primers used for ChIP assays are available upon request.
RT-PCR/quantitative RT-PCR (qRT-PCR) Analysis.
RT-PCR and qRT-PCR assays were done as described (47). Sequences of the primers used for ChIP assays are available upon request.
Statistical Analysis.
Statistical differences were determined by Student’s t test. Statistical significance is displayed as P < 0.05 (one asterisk), P < 0.01 (two asterisks), or P < 0.001 (three asterisks).
Supplementary Material
Acknowledgments.
We thank Drs. Larry Donehower and David Moore (Baylor College of Medicine, Houston, TX) for p53- and SHP-null mice and Soo Lee and Karen Thiebes for critical reading of this manuscript. This work was supported by National Institutes of Health Grant DK064678 (to J.W.L).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019678108/-/DCSupplemental.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: A recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 4.Olovnikov IA, Kravchenko JE, Chumakov PM. Homeostatic functions of the p53 tumor suppressor: Regulation of energy metabolism and antioxidant defense. Semin Cancer Biol. 2009;19:32–41. doi: 10.1016/j.semcancer.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottlieb E, Vousden KH. p53 regulation of metabolic pathways. Cold Spring Harb Perspect Biol. 2010;2:a001040. doi: 10.1101/cshperspect.a001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chanda D, Park JH, Choi HS. Molecular basis of endocrine regulation by orphan nuclear receptor Small Heterodimer Partner. Endocr J. 2008;55:253–268. doi: 10.1507/endocrj.k07e-103. [DOI] [PubMed] [Google Scholar]
- 7.Chiang JY. Bile acids: Regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann AF. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med. 1999;159:2647–2658. doi: 10.1001/archinte.159.22.2647. [DOI] [PubMed] [Google Scholar]
- 9.Perez MJ, Briz O. Bile-acid-induced cell injury and protection. World J Gastroenterol. 2009;15:1677–1689. doi: 10.3748/wjg.15.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005;589:47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Chiang JYL, Kimmel R, Weinberger C, Stroup D. FXR responds to bile acids and represses cholesterol 7a-hydroxylase gene (CYP7a1) transcription. J Biol Chem. 2000;275:10918–10924. doi: 10.1074/jbc.275.15.10918. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin B, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 13.Holt JA, et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inagaki T, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–25. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001;276:15816–15822. doi: 10.1074/jbc.M010878200. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, et al. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 17.Yu C, Wang F, Jin C, Huang X, McKeehan WL. Independent repression of bile acid synthesis and activation of c-Jun N-terminal kinase (JNK) by activated hepatocyte fibroblast growth factor receptor 4 (FGFR4) and bile acids. J Biol Chem. 2005;280:17707–17714. doi: 10.1074/jbc.M411771200. [DOI] [PubMed] [Google Scholar]
- 18.Li T, Jahan A, Chiang JY. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 2006;43:1202–1210. doi: 10.1002/hep.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stravitz RT, et al. Hepatocellular protein kinase C activation by bile acids: Implications for regulation of cholesterol 7 α-hydroxylase. Am J Physiol. 1996;271:G293–G303. doi: 10.1152/ajpgi.1996.271.2.G293. [DOI] [PubMed] [Google Scholar]
- 20.De Fabiani E, et al. The negative effects of bile acids and tumor necrosis factor-α on the transcription of cholesterol 7α-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: A novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J Biol Chem. 2001;276:30708–30716. doi: 10.1074/jbc.M103270200. [DOI] [PubMed] [Google Scholar]
- 21.Feingold KR, Spady DK, Pollock AS, Moser AH, Grunfeld C. Endotoxin TNF, and IL-1 decrease cholesterol 7 α-hydroxylase mRNA levels and activity. J Lipid Res. 1996;37:223–228. [PubMed] [Google Scholar]
- 22.Miyake JH, Wang SL, Davis RA. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7α-hydroxylase. J Biol Chem. 2000;275:21805–21808. doi: 10.1074/jbc.C000275200. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, et al. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl Acad Sci USA. 2009;106:8513–8518. doi: 10.1073/pnas.0902873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim DH, Lee J, Lee B, Lee JW. ASCOM controls farnesoid X receptor transactivation through its associated histone H3 lysine 4 methyltransferase activity. Mol Endocrinol. 2009;23:1556–1562. doi: 10.1210/me.2009-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 26.Tsai W-W, Nguyen TT, Shi Y, Barton MC. p53-targeted LSD1 functions in repression of chromatin structure and transcription in vivo. Mol Cell Biol. 2008;28:5139–5146. doi: 10.1128/MCB.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao J, et al. Bile acid signaling pathways increase stability of small heterodimer partner (SHP) by inhibiting ubiquitin-proteasomal degradation. Genes Dev. 2009;15:986–996. doi: 10.1101/gad.1773909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galla HJ, Theilen U, Hartmann W. Transversal mobility in bilayer membrane vesicles: use of pyrene lecithin as optical probe. Chem Phys Lipids. 1979;23:239–251. [Google Scholar]
- 29.Song X, Kaimal R, Yan B, Deng R. Liver receptor homolog 1 transcriptionally regulates human bile salt export pump expression. J Lipid Res. 2008;49:973–984. doi: 10.1194/jlr.M700417-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, et al. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology. 2008;48:289–298. doi: 10.1002/hep.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang F, et al. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 32.Kim I, et al. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maran RR, et al. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J Pharmacol Exp Ther. 2009;328:469–477. doi: 10.1124/jpet.108.145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modica S, Murzilli S, Salvatore L, Schmidt DR, Moschetta A. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 2008;68:9589–9594. doi: 10.1158/0008-5472.CAN-08-1791. [DOI] [PubMed] [Google Scholar]
- 35.Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: Insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166–2176. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- 36.Zaika AI, El-Rifai W. The role of p53 protein family in gastrointestinal malignancies. Cell Death Differ. 2006;13:935–940. doi: 10.1038/sj.cdd.4401897. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe M, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe M, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 39.Minamino T, et al. A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat Med. 2009;15:1082–1087. doi: 10.1038/nm.2014. [DOI] [PubMed] [Google Scholar]
- 40.Shih DQ, et al. Loss of HNF-1alpha function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes. 2001;50:2472–2480. doi: 10.2337/diabetes.50.11.2472. [DOI] [PubMed] [Google Scholar]
- 41.Crestani M, Sadeghpour A, Stroup D, Galli G, Chiang JY. Transcriptional activation of the cholesterol 7alpha-hydroxylase gene (CYP7A) by nuclear hormone receptors. J Lipid Res. 1998;39:2192–2200. [PubMed] [Google Scholar]
- 42.Lu TT, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 43.Lee YK, Parker KL, Choi HS, Moore DD. Activation of the promoter of the orphan receptor SHP by orphan receptors that bind DNA as monomers. J Biol Chem. 1999;274:20869–20873. doi: 10.1074/jbc.274.30.20869. [DOI] [PubMed] [Google Scholar]
- 44.Lee J, et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J Biol Chem. 2010;285:12604–12611. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yahagi N, et al. p53 involvement in the pathogenesis of fatty liver disease. J Biol Chem. 2004;279:20571–20575. doi: 10.1074/jbc.M400884200. [DOI] [PubMed] [Google Scholar]
- 46.Sinal CJ, et al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;101:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 47.Chen WD. Farnesoid X receptor alleviates age-related proliferation defects in regenerating mouse livers by activating forkhead box m1b transcription. Hepatology. 2010;51:953–962. doi: 10.1002/hep.23390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.