Abstract
Just as animal monozygotic twins can experience different environmental conditions by being reared apart, individual genetically identical trees of the genus Populus can also be exposed to contrasting environmental conditions by being grown in different locations. As such, clonally propagated Populus trees provide an opportunity to interrogate the impact of individual environmental history on current response to environmental stimuli. To test the hypothesis that current responses to an environmental stimulus, drought, are contingent on environmental history, the transcriptome- level drought responses of three economically important hybrid genotypes—DN34 (Populus deltoides × Populus nigra), Walker [P. deltoides var. occidentalis × (Populus laurifolia × P. nigra)], and Okanese [Walker × (P. laurifolia × P. nigra)]—derived from two different locations were compared. Strikingly, differences in transcript abundance patterns in response to drought were based on differences in geographic origin of clones for two of the three genotypes. This observation was most pronounced for the genotypes with the longest time since establishment and last common propagation. Differences in genome-wide DNA methylation paralleled the transcriptome level trends, whereby the clones with the most divergent transcriptomes and clone history had the most marked differences in the extent of total DNA methylation, suggesting an epigenomic basis for the clone history-dependent transcriptome divergence. The data provide insights into the interplay between genotype and environment in the ecologically and economically important Populus genus, with implications for the industrial application of Populus trees and the evolution and persistence of these important tree species and their associated hybrids.
Keywords: epigenetics, forest trees, poplar
There has been a longstanding interest in the impact of previous individual experience on current ability to respond to a given stimulus. Classical studies on human monozygotic (MZ) twins have provided key insights into the respective roles of genetics and personal history in shaping current responses to developmental and environmental cues (1). Recently, comparison of MZ twins supported the hypothesis that the personal history experienced by each MZ twin resulted in a divergence in genome expression between pairs of MZ twins over time (2). This divergence was more pronounced if the MZ twins were raised apart (2). These findings support the notion that an individual's personal environmental history shapes the extent to which different regions of the genome can be expressed, with concomitant impact on an individual's capacity to respond to future prevailing environmental conditions.
Forest trees in the genus Populus provide an excellent opportunity to explore the lasting impact of personal environmental history on an individual's capacity to respond to subsequent environmental stimuli. In nature, Populus trees can reproduce through vegetative propagation, where genetically identical individuals, or ramets, arise clonally from roots or from dispersed fragments of branches. Multiple taxa in the genus Populus (including cottonwoods, poplars, and their hybrids) are frequently propagated asexually in a commercial context by using stem cuttings of branches containing dormant buds in early spring (3). Vegetatively propagated poplar clones are planted in multiple locations, thereby creating several populations of ramets, each with their own local environment and history.
Here we examine the persistent influence of recent individual history on the Populus transcriptome-level response to an important environmental stress, drought. Drought is a crucial determinant of survival and growth of Populus trees (4–7), and is readily manipulated under experimental conditions. Given this, in the present study, Populus trees were established and grown under common environmental conditions, and then examined at the physiological and transcriptome levels for their response to water withdrawal. Ramets with distinct histories were obtained from two different geographic regions for each of three different Populus genotypes, and assessed under common, controlled environmental conditions.
Notably, when assessed for their response to drought, the source of the ramet (i.e., from one location vs. another) influenced the nature of the drought transcriptome for two of the three genotypes examined. Consideration of the relationship between transcriptome responsiveness, its mechanistic underpinnings, and the distinctiveness of personal history revealed striking parallels between human MZ twins and Populus ramets. The data emphasize the importance of individual history in shaping the environmentally responsive transcriptome. The findings of this study also have important implications for the capacity of long-lived, widely distributed species to contend with diverse environmental conditions, as well as considerable practical implications for afforestation and reforestation efforts.
Results and Discussion
This study empirically tested the extent of transcriptome remodeling induced by a drought stimulus in ramets sourced from different locations. Three commercially important hybrid Populus genotypes were used in the study: DN34 (Populus deltoides × Populus nigra), Walker [P. deltoides var. occidentalis × (Populus laurifolia × P. nigra)], and Okanese [Walker × (P. laurifolia × P. nigra)]. The genotypes each have a distinct propagation history. The DN34 genotype has been propagated since the early 1900s, whereas Walker was first propagated as a clonal population in 1946 and Okanese since 1986.
Half the DN34 genotype ramets were sourced from a nursery in the Canadian province of Manitoba (MB) and the other half were from a nursery in Saskatchewan (SK), whereas half the Walker and Okanese ramets were sourced from the same nursery in SK and the other half from a nursery in Alberta (AB; Fig. 1 A, C, and E). Each of the nursery sites was geographically and climatically distinct (Tables S1 and S2). All ramets were propagated under stereotypical “common garden” conditions in the same climate-controlled growth room. Ramets were propagated from equivalent hardwood cuttings and grown to statistically equivalent heights and stem diameters (Fig. S1). Drought was then imposed on half the ramets of a given genotype/source combination, whereas the other half continued to receive water to field capacity.
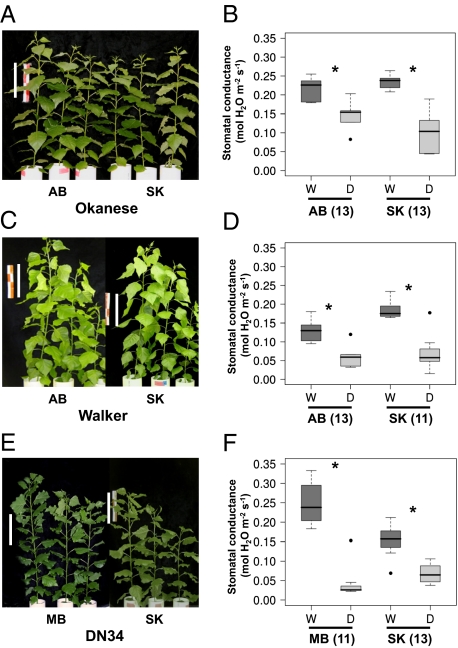
Fig. 1.
Tree appearance and midday stomatal conductance changes of hybrid Populus clones. Tree images correspond to a subsample of (A) Okanese, (C) Walker, and (E) DN34 sourced from different locations before the water-withholding experiment. (Scale bar: 20 cm.) Box plots indicating stomatal conductance measures for well watered samples (dark gray) and waterdeficient samples (light gray) of (B) Okanese, (D) Walker, and (F) DN34 from each location. Asterisks indicate a significant difference between well watered and water-deficient sample leaves (P < 0.05, Welch unpaired t test; n = 5–6). The number of days taken for treatments to exhibit a significant difference is shown in parentheses.
Genotype and Clone History Shape Physiological Responses to Drought.
To ensure physiological comparability between genotypes after drought, ramets were sampled when stomatal conductance was statistically significantly different between well watered and water-deficient samples at midday for two consecutive days. Stomatal closure is one of the earliest responses to shoot or root dehydration (8); therefore, stomatal conductance was used as a measure of stomatal aperture regulation in leaves.
The Okanese hybrid populations from AB and SK took equal numbers of days to exhibit significant decreases in stomatal conductance levels in the water-deficient samples (Fig. 1B). In contrast, the AB and SK populations of Walker had slightly different drought responses, with the former taking two more days than the latter to exhibit significant differences due to water limitation (Fig. 1D). A similar trend was observed in both DN34 populations, whereby significant differences in stomatal conductance were observed in the MB population 2 d earlier than the SK population (Fig. 1F). Variable stomatal behavior has been observed at the intraspecific level in poplars originating from different locations (9, 10), and the results observed here show that even genetically identical clones may have slightly varied stomatal behavior under drought conditions in relation to their site of origin.
Whole Transcriptome Divergence Is Shaped by Time of Day and Location.
Plant responses to stresses such as drought, including changes in stomatal behavior, are underpinned by major transcriptome reconfigurations to adjust metabolism, growth, and development to the prevailing environment (11–13). In diverse plant species, including Populus, transcriptome responses to drought are shaped by the time of day, such that different drought-response transcriptomes arise at different times of the day (14–16). To date, the impact of clone history (e.g., nursery source) on the drought transcriptome is unknown. To determine the influence of clone history on Populus drought transcriptomes, microarray analysis was used to compare drought-mediated reconfiguration of transcriptomes at two different times of day, across three commercially important Populus hybrid genotypes, with the clones obtained from two different nursery sources for each poplar genotype.
For each genotype, the Pearson correlation coefficient (PCC) was calculated for all pairs of arrays for all detectable transcripts (Fig. 2 and Fig. S2). Unsupervised, hierarchical cluster analysis was then used to group together the treatments with most similar transcript profiles (Fig. 3 and Fig. S3). Microarray transcript abundance data were independently validated by quantitative RT-PCR (qRT-PCR) for a sample of genes (Fig. S4 and Table S3). Hypothetically, for each genotype, microarray data should cluster on the basis of time of day and/or by drought treatment only, and not by clone history (i.e., nursery source). That is, clone history should have no impact on the relatedness of transcript abundance profiles at a given time of day or for a given treatment, as the clones obtained were genetically identical across the two nurseries.
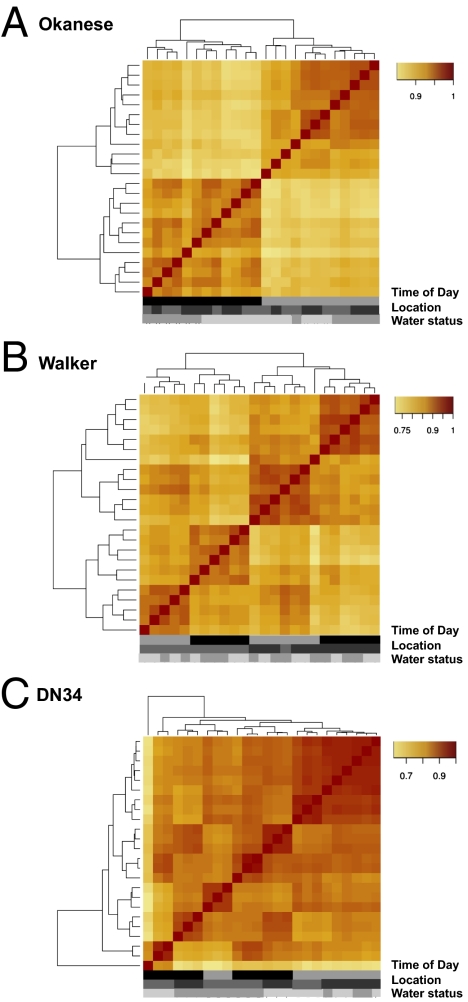
Fig. 2.
Clustering poplar transcriptomes. PCC heat maps of whole transcriptome profiles of (A) Okanese, (B) Walker, and (C) DN34 sourced from different locations. Samples are clustered in the same order on both axes. The color of each cell corresponds to the PCC for the compared samples. Samples are indicated by time of sample collection (predawn, black; midday, light gray), location of origin (AB, light gray; SK, dark gray), and water status (well watered, gray; water-deficient, light gray).
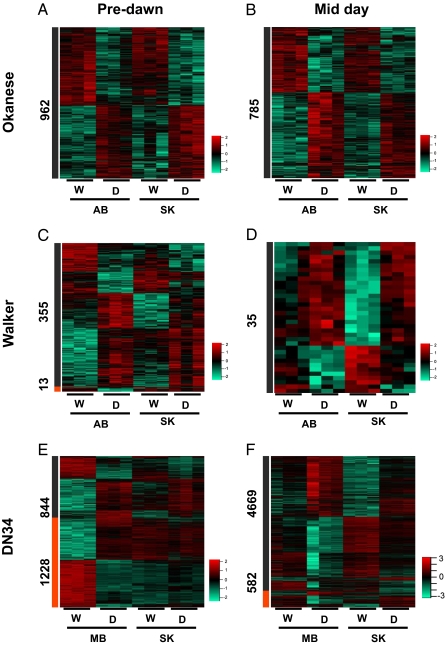
Fig. 3.
Treatment and treatment:location interaction effects for each hybrid. Heat maps representing the relative abundance of drought responsive transcripts for (A and B) Okanese, (C and D) Walker, and (E and F) DN34 obtained from two locations at predawn (PD) and midday (MD) time points. The number indicated to the side of the heat map corresponds to transcripts with significant treatment main effects only [gray bar, Benjamini–Hochberg (BH) adjusted P < 0.05] and to transcripts with significant treatment:location interactions (orange bar, BH adjusted P < 0.05) at the specified time point. Each column represents a discrete biological sample, and all treatments are presented as biological triplicate replicates. Red indicates higher, and green indicates lower, levels of transcript abundance. Expression levels are row-normalized; W, well watered samples; D, water-deficient samples.
Consistent with expectations for genetically identical material, for the transcript abundance data derived from the Okanese genotype, the two locations were indistinguishable. The data clustered by time of day, with predawn and midday transcript abundance profiles clustered into two distinct groups (Fig. 2A). This indicated that the transcript abundance profiles at a particular time point were most similar to each other. In the genotype Okanese, transcript abundance profiles clustered primarily based on time of day, followed by water status (Fig. 2A). Clone history had no impact. The findings were consistent regardless of whether stringent or more inclusive microarray analyses were applied to the data (Fig. 2A and Fig. S2A).
In contrast to Okanese, the history of the clones influenced the relatedness of transcript abundance patterns for both DN34 and Walker (Fig. 2 B and C). For the Walker genotype, as was the case with Okanese, time of day greatly influenced transcriptome relatedness. In contrast to Okanese, however, clone history was also a strong determinant in shaping Walker transcriptomes. For the DN34 genotype, the nursery source (i.e., clone history) and time of day had a very strong impact on transcript abundance profiles, with water status having a lower impact on transcriptome relatedness. The findings were consistent regardless of whether stringent or more inclusive microarray analyses were applied to the data (Fig. 2 B and C and Fig. S2 B and C). These findings are also borne out by principal component analyses (PCA) of all transcripts present, among all populations of hybrids, whereby separation by genotype was influenced by the time of day, and in the case of Walker and DN34, also by clone history (Figs. S4, S5, and S6). Taken together, the data suggest that clone history can have a profound effect on the nature of the transcriptomes that arise because of a change in water status, or in response to the time of day. The microarray data were subjected to greater scrutiny to dissect the extent to which clone history impacted the drought transcriptome of each of the poplar hybrid genotypes.
Interplay Between Clonal History and Drought Response Within Identical Genotypes Reflected in Specific Transcriptome Reconfigurations.
The extent to which clone history influenced specific transcriptome alterations in response to drought was examined by ANOVA (Tables S5 and S6). Linear Models for MicroArrays package (LIMMA)-based ANOVA identified probe sets with differential transcript accumulation attributable to water deficit independent of other factors (i.e., treatment main effect), as well as the identification of probe sets with transcript accumulation patterns that were dependent on the interaction between drought and location (i.e., treatment:location interaction) (17). This identified genes whose transcript abundance profiles responded differently to drought depending on the location from which the source materials were derived. Genes for which the treatment:location interaction was significant showed, for example, no difference in drought-responsive transcript abundance in the ramets from one location and a clear change in transcript abundance in the clones derived from the other location, or increased transcript abundance in response to drought in the ramets from one location and decreased transcript abundance in the clones from the other location. Importantly, genes comprising the treatment:location interaction group would not normally be expected in a stereotypical experiment when genetically identical individuals were treated under identical conditions. To determine the impact of clone history on the transcriptome, each genotype and time of day was assessed individually.
For Okanese, the drought treatment main effect was significant for 962 and 785 probe sets at predawn and midday, respectively (Table S5 and Fig. 3 A and B). A very small proportion of probe sets showed transcript abundance changes that were attributable to treatment:location interaction in this genotype (0 probe sets at predawn, 1 probe set at midday). This suggests that clone history had little effect on drought-responsive transcriptome remodeling in Okanese.
Relative to Okanese, the Walker genotype had a greater number of probe sets (13) for which treatment:location interaction was significant predawn (Table S5 and Fig. 3 C and D). In contrast to Okanese, Walker clone history had a greater impact on the drought transcriptome at one time point, highlighting that identifiable groups of genetically identical ramets can respond differently to a common stress when those groups have had distinct histories. This indicates that location-derived effects may persist in populations of genetically identical plants and influence future responses to a common stress.
An even more pronounced clone history-dependent trend was observed for transcriptome remodeling in the DN34 genotype. In addition to transcriptome alterations attributable to drought alone (treatment main effect only, 844 probe sets at predawn, 4,669 at midday; Table S5 and Fig. 3 E and F), a large number of probe sets comprised the treatment:location interaction group at both time points. At predawn and midday, there were 1,228 and 582 probe sets, respectively, that reported on statistically significant differences in transcript abundance contingent on a combination of water deficit and the location from which the clone was derived (Table S4 and Fig. 3 E and F). The strong location-dependent trends in the drought transcriptome in DN34 compared with the other hybrids provides strong evidence for the variable influence of an individual's site of origin and environmental history in this particular hybrid.
Notably, when the microarray analysis was conducted with probe set transcript abundance preprocessing that was more inclusive, the trends reported earlier were qualitatively equivalent. In fact, the trends were accentuated by the inclusion of a greater number of probe sets (Fig. S3 and Table S5). The analyses emphasize the role of individual history in shaping an environmental response.
Causes of Transcriptome Divergence.
The discovery of genes that define a treatment:location interaction group within a given genotype is consistent with the hypothesis that an individual's history can play a role in subsequent transcriptome remodeling. Strikingly, across the three genotypes used in these experiments, there was a gradient in the numbers of probe sets in the treatment:location interaction groups. Okanese showed the smallest impact of location of origin on drought transcriptome remodeling, whereas Walker and DN34 showed intermediate and strong effects of clone history on drought transcriptome remodeling, respectively. This is interesting relative to the respective “ages” of the three genotypes, whereby age is defined by the time since original selection and propagation as a unique clone. In this regard, the “oldest” poplar hybrid clone in these experiments was DN34, whereas the “youngest” clone was Okanese. Given this, it may be that that time since last common propagation provides some clues why clones with divergent histories show divergent transcript abundance profiles.
For clonally propagated individuals, like the poplar hybrids in the present study, somatic mutations that arise in the meristematic cells of a given ramet can give rise to distinct clonal lineages harboring specific mutations. Such somatic mutations can give rise to lineages with distinct phenotypes and environmental responses (18–21). Consequently, somatic mutation could explain the differences in transcriptome responses observed between the clones derived from two different locations within a given genotype. Moreover, consistent with the findings here, divergence in response caused by somatic mutation would be expected to increase as a function of time since the ramets last shared a common origin. Consequently, two approaches were used to determine if genetic divergence between clones from the two locations might account for the differences in the drought transcriptome observed between locations.
Each poplar hybrid genotype/location combination was genotyped by using nine unlinked simple sequence repeat (SSR) markers. SSRs are typically neutral and are used as a tool to detect inter- and intraspecific variation. Previously, unique clones of a single poplar species and hybrid poplar cultivars could be differentiated by using between two and nine SSR loci (22, 23). This is consistent with the number of loci examined to support monozygosity of human twins (2, 24). Thirty individuals (n = 3–6 ramets per genotype and location) were genotyped by using nine primers pairs, and information on a total of 1,050 SSR amplicons was recorded (Table S7). For each poplar genotype, all individuals analyzed showed the same multilocus genotype regardless of geographic origin. All told, the microsatellite evidence does not indicate that extensive somatic mutation has occurred between locations for any given hybrid poplar genotype.
In addition to SSR genotyping, highly variable regions of two unlinked nuclear loci were sequenced, providing information on 600 and 1,000 bp of noncoding sequence, respectively, for each of the two loci (Fig. S7). The heterozygous nature of the hybrid genotypes was reflected in the high number of polymorphisms detected for each locus; however, in all but one instance, no more than two allelic variants (i.e., haplotypes) per genotype and location were detected. Notably, for each genotype, the same polymorphisms were detected in ramets from different locations. The possibility of a third allelic variation in one instance could have arisen through a rare somatic mutation or could possibly reflect a PCR artifact. Importantly, this single occurrence of possible somatic mutation appeared in the Okanese genotype only, which is noteworthy because this genotype had the lowest level of divergence in transcriptome reconfiguration. Taken together, the SSR data and targeted sequencing data show that somatic mutations between clones derived from two different locations are rare, if they exist at all, in DN34 and Walker. It is unlikely that such a low somatic mutation rate would account for the observed large number of transcripts varying in response to water deficit between the two locations in DN34 and Walker.
Variation in Global DNA Methylation Is Dependent on Location in Some Genotypes.
An alternative and easily testable cause for divergent drought transcriptomes of both DN34 and Walker populations could be a result of epigenome influences. Epigenome modifications alter genome packaging and accessibility to the transcriptional machinery, thereby shaping transcriptome-level responses (25). In keeping with this, epigenome alterations in DNA methylation have been suspected to partly contribute to somaclonal variation (26, 27). Additionally, stress responses in plants were affected by past experiences in a manner that subsequent phenotypes were shaped by epigenome modifications at specific loci (28). Altered DNA methylation levels can lead to differences apparent in gene regulation under conditions of stress (28–30).
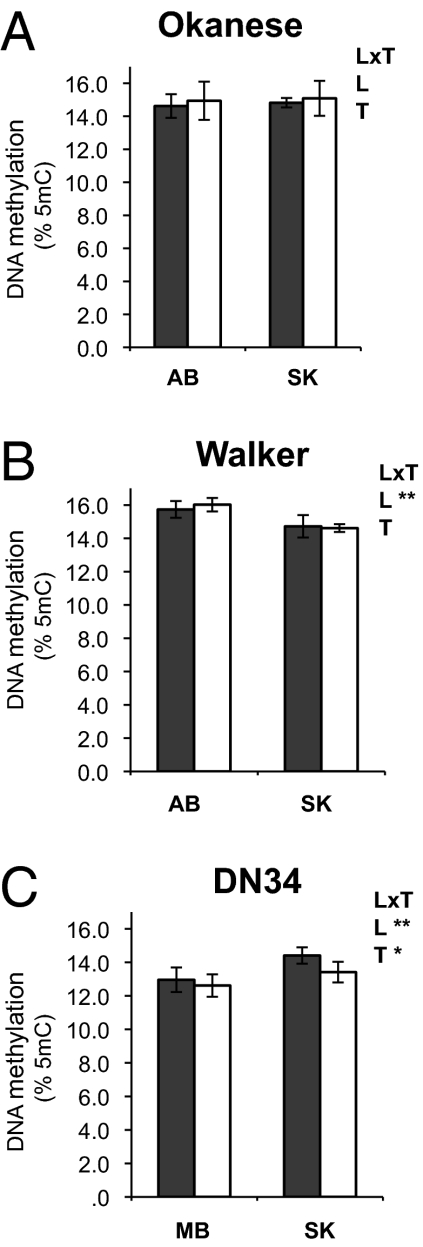
To study the possible role of epigenome marks in shaping the divergent drought transcriptome responses observed in Populus trees of the same genotype, total DNA methylation (global 5-methylcytosine levels as percentage of total cytosine) was investigated (Fig. 4). Although global DNA methylation levels in the genotype Okanese were independent of location or treatment (Fig. 4A), significant location effects were observed in the Walker and DN34 genotypes (Fig. 4 B and C). Thus, differences in DNA methylation in Populus hybrids might represent a possible mechanistic link between individual histories of Walker and DN34 plants propagated at different locations and divergent drought transcriptome responses observed in a common garden experiment. It is tempting to speculate that location-specific epigenome modifications might be of special significance for long-lived organisms such as poplar trees.
Fig. 4.
Global DNA methylation levels. Percentage of 5mC residues is given for plants grown under well watered (shaded bars) and water-limited (white bars) conditions for the Populus genotypes (A) Okanese, (B) Walker, and (C) DN34. For each graph, L indicates the location effect, T the treatment effect, and L×T the location:treatment interaction term (*P < 0.05, **P < 0.01). Mean values (n = 6; biological triplicates × technical duplicates) and SD bars are represented.
Studies of genetically identical human MZ twins similarly uncovered divergent epigenome patterns, including differences in global DNA methylation content, and highlighted that these differences were more pronounced in older MZ twins and in those twin pairs that were raised apart (2). Further indications that the environment influences DNA methylation patterns in plants are suggested by studies that focused on genetic and epigenome variation between populations or species growing in contrasting environments (31–33).
In addition to a significant location effect, an effect of drought treatment on DNA methylation was observed in ramets of the genotype DN34 (Fig. 4C). Although the total levels of DNA methylation in leaves measured in the present study were higher than previously detected for the shoot apex (34), the detected treatment effect of drought was consistent with the finding that differences in DNA methylation levels occurred after drought treatment in only some poplar genotypes.
The extent and distribution of DNA methylation within plant genes, and more generally within plant genomes, is highly variable, and is dependent on tissue type, plant age, developmental stage, and environmental factors (30, 35–37). Given this variation, it would be premature at this stage to speculate precisely where the modest changes in total DNA methylation documented herein may reside in the genome. This said, given the correlation between history-dependent differences in transcriptome activity, and global DNA methylation, it is tempting to hypothesize that the methylation differences, potentially within gene regulatory regions, may shape history-dependent transcriptome reconfiguration.
Conclusions
Mechanisms to adapt to a local environment are of key importance for plant species, and especially for long-lived organisms like forest trees (38). Here we report on the persistent influence of the geographic origin (i.e., nursery effect) on a stress response in a common controlled environment for three economically important poplar hybrid genotypes. Our findings support the hypothesis that the transcriptome-level drought response of a given poplar genotype can be shaped by the history of that clone. In keeping with this, for the genotypes tested here, the evidence suggests that the divergence in transcriptome-level response between ramets is a function of the time since the ramets last shared a common environment. The older the clone, the more likely ramets from different locations had a divergent history, and, consequently, divergent drought transcriptomes. Differences in total DNA methylation observed in older genotypes may hint at a mechanism that underpins the differences in drought transcriptomes that are shaped by clone history. These mechanisms may also be influenced by the unique genetic backgrounds that contribute to the hybrid genotypes. DN34 is a cross-sectional hybrid, whereas Walker is a cross-sectional hybrid between a pure and a within-section hybrid and Okanese is a backcrossed complex hybrid. These diverse genetic backgrounds may contribute to the patterns of transcriptome and epigenome divergence noted here.
The impact of clone history on subsequent response to stress could have profound implications for natural forests and tree plantations. Forests consisting of Populus trees can comprise patchworks of large contiguous blocks of genetically identical ramets (39–41). Individuals from different parts of the same natural monoclonal stand, or clonal individuals grown at different geographic sites, could have divergent “histories” on account of differences in parameters such as water availability, prevailing wind, soil conditions, or exposure to pests or pathogens. Consequently, individuals within these clonal groups could express divergent transcriptomes on account of molecular mechanisms, such as epigenome reprogramming, that arise from their distinct histories. This would provide a layer of diversity in gene expression responses that could buffer individuals within a clonal population from the deleterious effects associated with absence of genetic diversity in that population (29) and enable them to acclimate to environmental fluctuations over long life cycles. In an applied context, foresters should be aware that sourcing plantation material from different nurseries means that they are drawing on stock that may exhibit greater phenotypic diversity than would be suggested by genotype, which may respond differently to prevailing conditions contingent on nursery source, despite genetic identity. Together, these implications underscore the fact that long-lived organisms like trees contend with stresses through mechanisms that are more complex than hitherto expected.
Materials and Methods
Plant Material.
Unrooted dormant stem cuttings of three hybrid Populus hybrid clones DN34 [Populus × canadensis Moench var. eugenei (P. deltoides × P. nigra)], Walker [P. deltoides var. occidentalis × (P. laurifolia × P. nigra)], and Okanese [Walker × (P. laurifolia × P. nigra)] were each obtained from nurseries in two different Canadian provinces for a total of six hybrid clone populations. Okanese and Walker were each sourced from AB and SK. DN34 was obtained from MB and SK. Details on plant source, genotyping, and growth conditions are described in SI Materials and Methods.
Water-Withholding Experiment.
Details on water-withholding experiment are provided in SI Materials and Methods. In brief, rooted cuttings were grown without water limitation for a minimum of 9 wk. One half of each population was grown without further input of water, whereas the other half was well watered. For each hybrid population, the onset of a physiological response (i.e., change in stomatal conductance) to water deficit was monitored daily on the first fully expanded leaf. A statistically significant difference in leaf stomatal conductance in water-deficient plants, for two consecutive days at midday, compared with well watered plants, was used as an indicator of water stress. On the third day, the first fully expanded leaves were harvested from nine to 12 trees in each population, both well watered and water-deficient, at two time points: predawn (1 h before lights were turned on) and midday (middle of the light period). The leaves were pooled to generate biological triplicate samples, and flash-frozen in liquid nitrogen for further analyses.
Transcript Abundance Analysis.
RNA analysis, transcript abundance detection with Affymetrix GeneChip Poplar Genome Array, qRT-PCR, and microarray analysis were performed largely as previously described (14, 15). Details are provided in SI Materials and Methods. The transcript abundance analyses were conducted independently for each hybrid. The microarrays were analyzed as a 2 × 2 × 2 factorial complete randomized ANOVA design (two locations, two time points, two treatments).
Global Methylation Analysis.
Global DNA methylation quantification was performed by isocratic cation-exchange HPLC as described (42) by using proportions of the same tissue that has been used for RNA extraction and microarray hybridization. Foliar tissue samples used for total DNA methylation analysis were identical to the foliar samples collected for RNA isolation and microarray hybridization. The effect of the independent factors—treatment and location—were analyzed by using a 2 × 2 factorial ANOVA. Details are described in SI Materials and Methods.
Sequencing of Noncoding Regions.
Promoter and noncoding regions for two unlinked nuclear loci, POPTR_0003s11430 and POPTR_0012s13180, were selected to study potentially highly variable regions, and to assess the influence of location on sequence diversity. PCR-amplified DNA fragments were subcloned, sequenced, and assessed for polymorphic sites (SI Materials and Methods).
Supplementary Material
Acknowledgments
We are grateful for the excellent input provided by three anonymous reviewers. We also express our gratitude to Bruce Hall and Andrew Petrie for greenhouse assistance, John McCarron for the experimental setup, and Joan Ouellette for technical assistance. Research infrastructure and technical support was provided by the Centre for Analysis of Genome Evolution and Function. O.W. was supported by a Natural Science and Engineering Research Council of Canada (NSERC) Canadian Graduate Scholarship. S.D.M. is a Canada Research Chair. This work was supported by funding from NSERC (to M.M.C., A.L.P., and S.D.M.), the Canada Foundation for Innovation, and the University of Toronto (M.M.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE27693).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103341108/-/DCSupplemental.
References
- 1.Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet. 2002;3:872–882. doi: 10.1038/nrg932. [DOI] [PubMed] [Google Scholar]
- 2.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanturf JA, van Oosten C, Netzer DA, Coleman MD, Portwood CJ. Ecology and silviculture of poplar plantations. In: Dickmann DI, Isebrands JG, Eckenwalder JE, Richardson J, editors. Poplar Culture in North America. Ottawa: NRC Research Press; 2001. pp. 153–206. [Google Scholar]
- 4.Ceulemans R, Impens I, Steenackers V. Genetic-variation in aspects of leaf growth of Populus clones, using the leag plastochron index. Can J Forest Res. 1988;18:1069–1077. [Google Scholar]
- 5.Tschaplinski TJ, Tuskan GA, Gebre GM, Todd DE. Drought resistance of two hybrid Populus clones grown in a large-scale plantation. Tree Physiol. 1998;18:653–658. doi: 10.1093/treephys/18.10.653. [DOI] [PubMed] [Google Scholar]
- 6.Griffin DH, Schaedle M, DeVit MJ, Manion PD. Clonal variation of Populus tremuloides responses to diurnal drought stress. Tree Physiol. 1991;8:297–304. doi: 10.1093/treephys/8.3.297. [DOI] [PubMed] [Google Scholar]
- 7.Pallardy SG, Kozlowski TT. Water relations of populus clones. Ecology. 1981;62:159–169. [Google Scholar]
- 8.Chaves MM, Maroco JP, Pereira JS. Understanding plant responses to drought - from genes to the whole plant. Funct Plant Biol. 2003;30:239–264. doi: 10.1071/FP02076. [DOI] [PubMed] [Google Scholar]
- 9.Sparks JP, Black RA. Regulation of water loss in populations of Populus trichocarpa: The role of stomatal control in preventing xylem cavitation. Tree Physiol. 1999;19:453–459. doi: 10.1093/treephys/19.7.453. [DOI] [PubMed] [Google Scholar]
- 10.Bassman JH, Zwier JC. Gas exchange characteristics of Populus trichocarpa, Populus deltoides and Populus trichocarpa × P. deltoides clones. Tree Physiol. 1991;8:145–159. doi: 10.1093/treephys/8.2.145. [DOI] [PubMed] [Google Scholar]
- 11.Harb A, Krishnan A, Ambavaram MMR, Pereira A. Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 2010;154:1254–1271. doi: 10.1104/pp.110.161752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues FA, de Laia ML, Zingaretti SM. Analysis of gene expression profiles under water stress in tolerant and sensitive sugarcane plants. Plant Sci. 2009;176:286–302. [Google Scholar]
- 13.Matsui A, et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- 14.Hamanishi ET, et al. Intraspecific variation in the Populus balsamifera drought transcriptome. Plant Cell Environ. 2010;33:1742–1755. doi: 10.1111/j.1365-3040.2010.02179.x. [DOI] [PubMed] [Google Scholar]
- 15.Wilkins O, Waldron L, Nahal H, Provart NJ, Campbell MM. Genotype and time of day shape the Populus drought response. Plant J. 2009;60:703–715. doi: 10.1111/j.1365-313X.2009.03993.x. [DOI] [PubMed] [Google Scholar]
- 16.Wilkins O, Bräutigam K, Campbell MM. Time of day shapes Arabidopsis drought transcriptomes. Plant J. 2010;63:715–727. doi: 10.1111/j.1365-313X.2010.04274.x. [DOI] [PubMed] [Google Scholar]
- 17.Doncaster CP. Analysis of Variance and Covariance: How to Choose and Construct Models for the Life Sciences. Cambridge, UK: Cambridge Univ Press; 2007. [Google Scholar]
- 18.Moncada X, Pelsy F, Merdinoglu D, Hinrichsen P. Genetic diversity and geographical dispersal in grapevine clones revealed by microsatellite markers. Genome. 2006;49:1459–1472. doi: 10.1139/g06-102. [DOI] [PubMed] [Google Scholar]
- 19.Mock KE, Rowe CA, Hooten MB, Dewoody J, Hipkins VD. Clonal dynamics in western North American aspen (Populus tremuloides) Mol Ecol. 2008;17:4827–4844. doi: 10.1111/j.1365-294X.2008.03963.x. [DOI] [PubMed] [Google Scholar]
- 20.Ally D, Ritland K, Otto SP. Can clone size serve as a proxy for clone age? An exploration using microsatellite divergence in Populus tremuloides. Mol Ecol. 2008;17:4897–4911. doi: 10.1111/j.1365-294X.2008.03962.x. [DOI] [PubMed] [Google Scholar]
- 21.Heinze B, Fussi B. Somatic mutations as a useful tool for studying clonal dynamics in trees. Mol Ecol. 2008;17:4779–4781. doi: 10.1111/j.1365-294X.2008.03964.x. [DOI] [PubMed] [Google Scholar]
- 22.Rahman MH, Rajora OP. Microsatellite DNA fingerprinting, differentiation, and genetic relationships of clones, cultivars, and varieties of six poplar species from three sections of the genus Populus. Genome. 2002;45:1083–1094. doi: 10.1139/g02-077. [DOI] [PubMed] [Google Scholar]
- 23.Bekkaoui F, Mann B, Schroeder B. Application of DNA markers for the identification and management of hybrid poplar accessions. Agrofor Syst. 2003;59:53–59. [Google Scholar]
- 24.Becker A, et al. Twin zygosity. Automated determination with microsatellites. J Reprod Med. 1997;42:260–266. [PubMed] [Google Scholar]
- 25.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 26.Kaeppler SM, Kaeppler HF, Rhee Y. Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol. 2000;43:179–188. doi: 10.1023/a:1006423110134. [DOI] [PubMed] [Google Scholar]
- 27.Rodríguez López CM, Wetten AC, Wilkinson MJ. Progressive erosion of genetic and epigenetic variation in callus-derived cocoa (Theobroma cacao) plants. New Phytol. 2010;186:856–868. doi: 10.1111/j.1469-8137.2010.03242.x. [DOI] [PubMed] [Google Scholar]
- 28.Finnegan EJ. Epialleles - a source of random variation in times of stress. Curr Opin Plant Biol. 2002;5:101–106. doi: 10.1016/s1369-5266(02)00233-9. [DOI] [PubMed] [Google Scholar]
- 29.Boyko A, Kovalchuk I. Epigenetic control of plant stress response. Environ Mol Mutagen. 2008;49:61–72. doi: 10.1002/em.20347. [DOI] [PubMed] [Google Scholar]
- 30.Chinnusamy V, Zhu J-K. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol. 2009;12:133–139. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrera CM, Bazaga P. Epigenetic differentiation and relationship to adaptive genetic divergence in discrete populations of the violet Viola cazorlensis. New Phytol. 2010;187:867–876. doi: 10.1111/j.1469-8137.2010.03298.x. [DOI] [PubMed] [Google Scholar]
- 32.Lira-Medeiros CF, et al. Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS ONE. 2010;5:e10326. doi: 10.1371/journal.pone.0010326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paun O, et al. Stable epigenetic effects impact adaptation in allopolyploid orchids (Dactylorhiza: Orchidaceae) Mol Biol Evol. 2010;27:2465–2473. doi: 10.1093/molbev/msq150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gourcilleau D, et al. DNA methylation and histone acetylation: genotypic variations in hybrid poplars, impact of water deficit and relationshios with productivity. Ann For Sci. 2010;67:208. [Google Scholar]
- 35.Furner IJ, Matzke M. Methylation and demethylation of the Arabidopsis genome. Curr Opin Plant Biol. 2011;14:137–141. doi: 10.1016/j.pbi.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kvaalen H, Johnsen O. Timing of bud set in Picea abies is regulated by a memory of temperature during zygotic and somatic embryogenesis. New Phytol. 2008;177:49–59. doi: 10.1111/j.1469-8137.2007.02222.x. [DOI] [PubMed] [Google Scholar]
- 39.Dickmann DI, Kuzovkina J. Poplars and Willows of the World, with Emphasis on Silviculturally Important Species. FAO Forest Management Division Working Paper IPC/9-2. Rome: FAO; 2008. [Google Scholar]
- 40.Kemperman JA, Barnes BV. Clone size in american aspens. Can J Bot. 1976;54:2603–2607. [Google Scholar]
- 41.Mitton JB, Grant MC. Genetic variation and the natural history of quaking aspen. Bioscience. 1996;46:25–31. [Google Scholar]
- 42.Rozhon W, Baubec T, Mayerhofer J, Mittelsten Scheid O, Jonak C. Rapid quantification of global DNA methylation by isocratic cation exchange high-performance liquid chromatography. Anal Biochem. 2008;375:354–360. doi: 10.1016/j.ab.2008.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.