Abstract
Paracoccidioides brasiliensis is a thermodimorphic fungus and the causative agent of paracoccidioidomycosis (PCM). The ability of P. brasiliensis to uptake nutrients is fundamental for growth, but a reduction in the availability of iron and other nutrients is a host defense mechanism many pathogenic fungi must overcome. Thus, fungal mechanisms that scavenge iron from host may contribute to P. brasiliensis virulence. In order to better understand how P. brasiliensis adapts to iron starvation in the host we compared the two-dimensional (2D) gel protein profile of yeast cells during iron starvation to that of iron rich condition. Protein spots were selected for comparative analysis based on the protein staining intensity as determined by image analysis. A total of 1752 protein spots were selected for comparison, and a total of 274 out of the 1752 protein spots were determined to have changed significantly in abundance due to iron depletion. Ninety six of the 274 proteins were grouped into the following functional categories; energy, metabolism, cell rescue, virulence, cell cycle, protein synthesis, protein fate, transcription, cellular communication, and cell fate. A correlation between protein and transcript levels was also discovered using quantitative RT-PCR analysis from RNA obtained from P. brasiliensis under iron restricting conditions and from yeast cells isolated from infected mouse spleens. In addition, western blot analysis and enzyme activity assays validated the differential regulation of proteins identified by 2-D gel analysis. We observed an increase in glycolytic pathway protein regulation while tricarboxylic acid cycle, glyoxylate and methylcitrate cycles, and electron transport chain proteins decreased in abundance under iron limiting conditions. These data suggest a remodeling of P. brasiliensis metabolism by prioritizing iron independent pathways.
Introduction
Iron is an essential nutrient for cellular function, but iron overload can be as detrimental as iron depletion. Thus, microorganisms use a complex network of systems to control iron levels in order to prevent free radical damage to proteins, ribonucleic acids, and cell membranes, keeping iron unavailable [1]. The absence of free iron in host tissues and host iron restriction mechanisms demand that pathogens develop an efficient iron uptake system in order to compete with the host for iron [2].
Iron deprivation responses in several fungi have been studied by transcriptional and proteomic analyses [3], [4], [5]. Iron homeostasis mechanisms including iron uptake, storage, and regulation have been extensively characterized especially in the fungal prototype Saccharomyces cerevisiae. Iron levels in S. cerevisiae are controlled by two major transcriptional factors identified as Aft1p and Aft2p. These two orthologues are missing in the majority of fungal species [6], [7], [8].
The human fungal pathogen Aspergillus fumigatus also has a well characterized iron acquisition system. Under iron starvation conditions A. fumigatus employs two high affinity iron uptake systems that include siderophore-assisted and reductive iron uptake [9]. A. fumigatus iron acquisition under iron-depleted conditions is controlled by transcriptional factors encoded by the genes sreA and hapX [10], [11]. Under low iron conditions, the expression of the GATA factor SreA is reduced resulting in desrepression of the bZip transcription factor HapX. Genes involved in reductive iron assimilation, siderophore biosynthesis and uptake are also affected [11]. This regulatory circuit is largely conserved and orthologs to sreA and hapX are found in most fungal species [10], [12]. Moreover, the A. fumigatus zinc cluster transcription factor AcuM suppresses sreA and induces hapX to stimulate expression of genes involved in reductive and siderophore mediated iron uptake [13].
Paracoccidoides brasiliensis is a dimorphic fungus and the etiologic agent of paracoccidioidomycosis (PCM). The disease is restricted to Latin America, and is the most prevalent systemic mycosis in the region [14]. PCM is a major public health concern in rural areas. Moreover, this infection can lead to potentially disabling injuries. Several studies indicate that the majority of PCM cases are reported in Brazil [15] with an annual mortality rate of 148 deaths per year [16].
Searching analyses on P. brasiliensis genome database (http://www.broadinstitute.org/annotation/genome/paracoccidioides_brasiliensis/MultiHome.html) showed that this fungus possess genes that encode proteins with sequence similarity to host iron uptake systems which utilize heme while also employing siderophore-assisted iron uptake and reductive iron assimilation [17]. The fungal genome of P. brasiliensis contains sequences that potentially encode all the necessary enzymes for siderophore biosynthesis; sidA, sidF, sidC and sidD orthologs of A. fumigatus [17]. Putative P. brasiliensis siderophore transporters were identified as orthologs to genes encoding the S. cerevisiae Sit1p and the A. nidulans MirB and MirC [17]. Further investigation of the P. brasiliensis genome revealed that it contains orthologs to hapx and sreA. Despite these advances the identity and total number of genes and proteins involved in P. brasiliensis response to iron deprivation remains unclear.
Different reports indicate that iron depletion in fungi promotes the metabolic remodeling of iron-dependent processes including oxidative respiration, amino acid biosynthesis, and fatty acid metabolism. [3], [4], [18]–[20]. Altered expression of heat shock proteins has also been found in fungi during iron limiting conditions most likely due to the accumulation of unfolded proteins in the cytoplasm [21].
In this study we utilized 2D gel electrophoresis coupled to mass spectrometry to identify P. brasiliensis proteins sensitive to low iron levels. We discovered 274 differentially regulated proteins and 96 of these were identified, rendering an integrated view of metabolic and cellular processes reorganization during iron deprivation. Moreover, in vivo analysis confirmed the expression pattern of selected genes. These data suggest that the remodeling of P. brasiliensis metabolism is one way the fungus adapts and survives in a nutrient deficient environment.
Results
Iron overload exacerbates P. brasiliensis infection in BALB/c mice
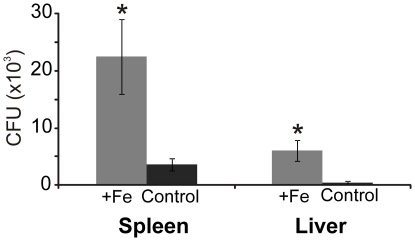
To investigate the effect of iron availability on P. brasiliensis colonization we infected BALB/c mice that were iron supplemented. We observed an increased CFU recovery from liver and spleen of mice treated with an iron supplement (Figure 1). The results suggest that iron availability increases the susceptibility of mice to P. brasiliensis infection.
Figure 1. Iron supplementation increases mice susceptibility to P. brasiliensis infection.
Colony forming units recovered from the spleen and liver of mice infected with P. brasiliensis. Mice were supplemented with iron (grey bars) or 0.9% (w/v) NaCl (black) for 15 days prior and during infection. Error bars represent standard deviation from four biological replicates while * represents p≤0.05.
Expression of P. brasiliensis iron acquisition genes during iron starvation
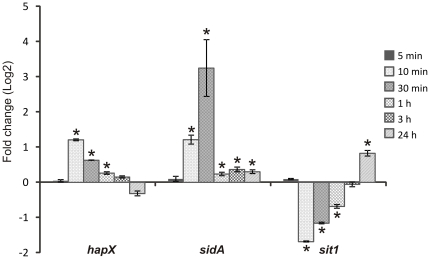
Before we began our proteomic analysis, we first sought to verify that our iron limiting conditions were restricting iron to the fungus by analyzing P. brasiliensis gene expression. To investigate the transcriptional profile of iron responsive genes in P. brasiliensis we used real-time RT-PCR. These genes included P. brasiliensis orthologs to the transcriptional regulator hapX, siderophore transporter sit1, and the siderophore biosynthesis gene sidA, encoding L-ornithine N5 –monooxygenase (Figure 2 and Table S1). In the first 10 minutes of iron deprivation, we observed an increased expression of the hapX and sidA orthologs. The level of hapX increased 2.3 times at 10 minutes of iron deprivation and the transcript level remained increased up to 1 hour in this condition. The sidA ortholog presented an expression level increased nine times compared to control following 30 minutes of iron starvation; transcript level remained at significant high levels in 24 hours of iron deprivation. The sit1 ortholog was over expressed following 24 hours of iron deprivation. These results indicate that P. brasiliensis was subjected to iron restriction conditions because the known siderophore iron uptake system is active upon iron restriction.
Figure 2. Induction of P. brasiliensis iron acquisition genes following iron restriction.
Quantitative RT-PCR determined P. brasiliensis transcript levels of hapX, sidA and sit1 during iron deprivation. Data were normalized to the L34 protein transcript. Student's t test was used for statistical comparisons. Error bars represent standard deviation from three biological replicates while * represents p≤0.05.
P. brasiliensis cell viability
Because we did not want to measure a proteome response which would represent cell death, we next sought to determine whether iron deprivation influenced P. brasiliensis cell viability. Using trypan blue staining we observed no significant differences in yeast cell viability up to 48 hours in iron deprivation (Figure S1). We did not observe a decrease in P. brasiliensis cell viability until 72 hours of exposure to iron limiting conditions (data not shown). These data show that the iron limiting incubation times selected for proteomic analysis did not influence P. brasiliensis cell viability.
2D-gel analysis of P. brasiliensis during iron starvation
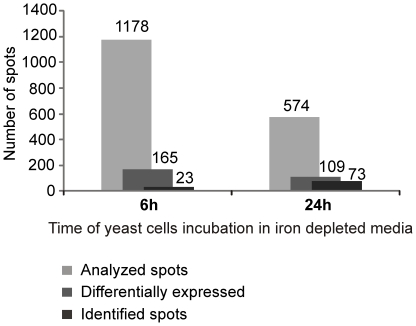
Based on the results described above 6 and 24 hours iron depletion incubation times were selected for proteomic analysis of P. brasiliensis yeast cells. Two-dimensional gel analysis was used to separate cytosolic fungal proteins while image analysis allowed for the quantification of proteins. Three independent experiments generate three replicates which included: 6 hours in iron depletion, 6 hours control, 24 hours in iron depletion, and 24 hours control. Using the gel image software a total of 1752 spots were successfully matched between control and iron depletion conditions (Figure 3 and Figure 4). Statistical analysis revealed a difference in protein abundance for 165 and 109 protein spots following 6 and 24 hours of iron deprivation respectively. This yielded a total of 274 differentially regulated proteins (Figure 3). A magnified gel region allows for visualization of representative spots with differential abundance (Figure 5).
Figure 3. Graphic summation of P. brasiliensis iron restriction proteomic analysis.
Number of spots differentially expressed were determined using 2D-gel image analysis software. Statistical analysis of the matched spots were performed using ANOVA.
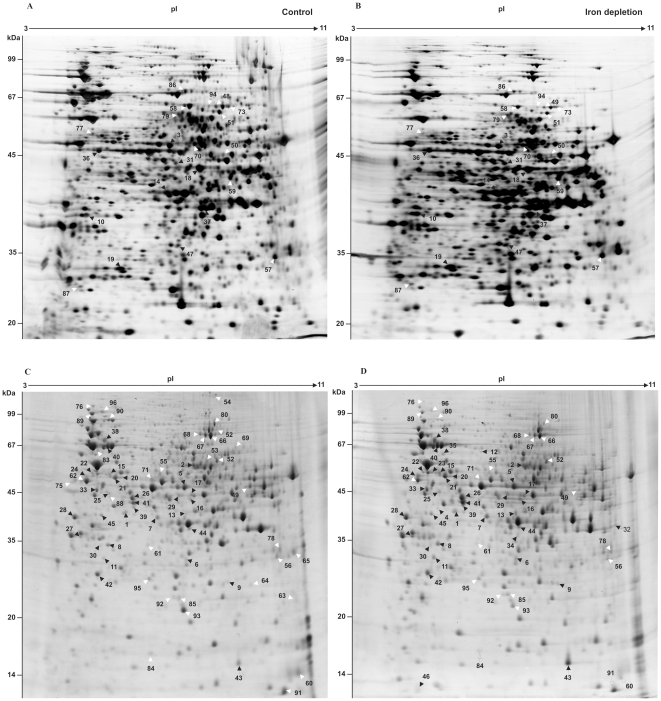
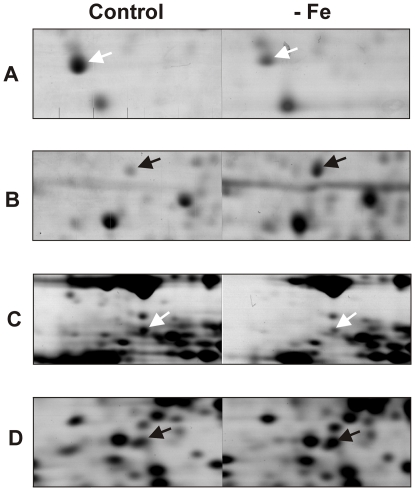
Figure 4. P. brasiliensis protein regulation during iron limiting conditions detected using 2D-gel analysis.
2D-gel analysis of P. brasiliensis proteins extracted from yeast cells grown in iron depleted media for 6 hours (B) and 24 hours (D). Gels A and C represent iron rich conditions. Black and white arrows indicate up-regulated and down-regulated proteins respectively. Identified protein spots are numbered and listed in Tables 1 and 2.
Figure 5. Magnified regions of the protein spots exhibiting protein abundance changes in the 2D gels shown in Figure 4 .
Protein spots correspond to the following proteins identified by mass spectrometry; Y20 protein (A), ATP synthase subunit beta (B), Tubulin alpha-1 chain (C), Conserved hypothetical protein (D). Black and white arrows indicate up-regulated and down-regulated proteins respectively.
Identification iron-regulated proteins
In order to determine the identities of the protein spots determined to be differentially regulated following 2D-gel analysis protein spots were subjected to in-gel digestion using trypsin and analyzed by mass spectrometry (MS). MS analysis following by protein database sequence matching resulted in the identification of 96 differentially expressed proteins (Figures 3 and 4). Seventy three proteins/isoforms were identified by peptide mass fingerprinting (PMF) and confirmed by MS/MS analysis while 23 of protein spots yielded identification by PMF only.
Among the 96 identified proteins 49 increased in abundance and 47 decreased. The p-values associated with each spot comparison are listed in Tables S2 and S3. Differences in protein spot intensity ranged from 1.2 to 3.6 (Tables 1 and 2). GenBank general information identifiers (gi), Broad Institute P. brasiliensis database accession numbers (PAAG), PMF and MS/MS mascot scores, protein molecular weights, and isoelectric points (pI) of each spot are also listed in Tables 1 and 2.
Table 1. P. brasiliensis identified proteins with increased expression during iron starvation.
| Spot1 | Time2 | GenBank gi3 | Protein identification | PMF score | MS/MS score | MW4 | pI 5 | ≥fold change6 |
| METABOLISM | ||||||||
| Nucleotide metabolism | ||||||||
| 1 | 24 h | gi|226280544 | Adenosine kinase | 193 | 49 | 36.6/41.3 | 5.4/5.5 | 1.3 |
| 2 | 24 h | gi|226283962 | GMP synthase | 133 | - | 60.3/55.2 | 6.2/7.4 | 3.6 |
| C-compound and carbohydrate metabolism | ||||||||
| 3 | 6 h | gi|295664927 | ATP-citrate-lyase | 159 | 303 | 52.8/46.6 | 5.9/6.9 | 1.2 |
| Amino acid, nitrogen and sulfur metabolism | ||||||||
| 4 | 24 h | gi|226280080 | 2-nitropropane dioxygenase | 163 | 107 | 38.8/41.9 | 5.4/5.1 | ** |
| 5 | 24 h | gi|27368050 | Formamidase | 108 | 100 | 46.1/46.1 | 6.1/7.1 | 1.3 |
| 6 | 24 h | gi|226284927 | Hydroxyacylglutathione hydrolase | 66 | 86 | 28.9/33.1 | 6.1/7.1 | 1.7 |
| 7 | 24 h | gi|226282479 | L-threonine 3-dehydrogenase | 105 | 129 | 38.0/40.3 | 5.8/6.4 | 1.9 |
| 8 | 24 h | gi|226293104 | Spermidine synthase | 93 | 32 | 33.6/36.0 | 5.3/5.1 | 1.6 |
| Lipid, fatty-acid and isoprenoid metabolism | ||||||||
| 9 | 24 h | gi|226279101 | Carbonic anhydrase | 90 | 104 | 32.6/27.4 | 9.1/8.2 | 1.8 |
| Phosphate metabolism | ||||||||
| 10 | 6 h | gi|295672504 | Inorganic pyrophosphatase | 141 | 67 | 33.3/38.0 | 5.1/4.8 | 1.3 |
| ENERGY | ||||||||
| Electron transport and membrane-associated energy conservation | ||||||||
| 11 | 24 h | gi|226282053 | ATP synthase subunit beta | 199 | 118 | 55.1/33.3 | 5.2/5.0 | 2.5 |
| Glycolysis and gluconeogenesis | ||||||||
| 12 | 24 h | gi|146762537 | Enolase | 110 | 173 | 47.4/63.4 | 5.6/6.2 | ** |
| 13 | 24 h | gi|29826036 | Fructose 1,6-biphosphate aldolase | 163 | 174 | 39.7/41.1 | 6.1/7.1 | 2.6 |
| 14 | 6 h | PAAG_01995 | Fructose 1,6-biphosphate aldolase | 77 | - | 39.6/40.3 | 6.0/6.8 | 1.4 |
| 15 | 24 h | gi|226279559 | Glucokinase | 113 | 49 | 55.7/52.2 | 5.3/5.0 | 2.8 |
| 16 | 24 h | gi|226285327 | Phosphoglycerate kinase | 108 | 72 | 45.3/43.4 | 6.4/7.3 | 1.6 |
| 17 | 24 h | gi|295669690 | Phosphoglycerate kinase | 149 | 72 | 45.3/44.1 | 6.4/7.3 | 1.8 |
| 18 | 6 h | gi|295669690 | Phosphoglycerate kinase | 82 | 53 | 45.3/42.9 | 7.8/7.7 | 1.2 |
| 19 | 6 h | gi|295670663 | Triosephosphate isomerase (Tpi) | 185 | 87 | 27.1/30.3 | 5.3/5.6 | 1.4 |
| Tricarboxylic-acid pathway | ||||||||
| 20 | 24 h | gi|295673931 | Pyruvate dehydrogenase protein X component | 110 | 41 | 52.7/50.6 | 6.4/5.4 | 1.5 |
| 21 | 24 h | gi|226280161 | Pyruvate dehydrogenase protein X component | 100 | 131 | 52.7/50.3 | 6.4/5.3 | 1.4 |
| CELL CYCLE AND DNA PROCESSING | ||||||||
| 22 | 24 h | gi|225683196 | Tubulin alpha-2 chain | 105 | - | 50.5/52.4 | 5.0/4.7 | 1.2 |
| 23 | 24 h | gi|225683196 | Tubulin alpha-2 chain | 117 | 57 | 50.5/53.2 | 5.0/4.9 | ** |
| 24 | 24 h | gi|226285902 | Tubulin beta chain | 146 | 65 | 50.3/51.1 | 4.8/4.5 | 1.8 |
| 25 | 24 h | gi|154705473 | Septin-1 | 123 | 87 | 44.1/44.3 | 5.2/5.1 | 2.0 |
| 26 | 24 h | gi|226294796 | Actin | 78 | 59 | 38.1/43.5 | 7.1/5.7 | 1.6 |
| 27 | 24 h | gi|38569374 | 14-3-3-like protein 2 | 120 | 84 | 29.7/37.1 | 4.6/4.4 | 1.7 |
| 28 | 24 h | gi|226282286 | DNA damage protein rad24 (14-3-3 protein) | 147 | 252 | 32.4/40.1 | 4.7/4.2 | 1.4 |
| TRANSCRIPTION | ||||||||
| 29 | 24 h | gi|226280907 | mRNA binding post-transcriptional regulator (Csx1) | 92 | 44 | 42.6/44.2 | 6.3/6.9 | 2.0 |
| 30 | 24 h | gi|226284577 | Type 2A phosphatase activator tip41 | 90 | - | 35.9/27.9 | 5.3/4.9 | 2.3 |
| 31 | 6 h | PAAG_06891 | mRNA binding post-transcriptional regulator (Csx1) | 96 | - | 42.4/43.4 | 6.3/7.1 | 2.0 |
| PROTEIN SYNTHESIS | ||||||||
| 32 | 24 h | gi|226280659 | 60S ribosomal protein L5 | 104 | - | 34.3/39.7 | 9.0/9.7 | ** |
| 33 | 24 h | gi|226282202 | ATP-dependent RNA helicase eIF4a | 130 | 60 | 45.0/45.1 | 5.1/4.8 | 1.3 |
| 34 | 24 h | gi|226280705 | Elongation factor 2 | 103 | 110 | 92.6/37.8 | 6.4/7.2 | ** |
| 35 | 24 h | gi|226283670 | Translation initiation factor eIF3 | 84 | 56 | 66.0/62.3 | 5.1/5.2 | ** |
| PROTEIN FATE (folding, modification, destination) | ||||||||
| 36 | 6 h | PAAG_05417 | Mitochondrial-processing peptidase | 140 | - | 53.0/45.2 | 5.8/7.1 | 1.3 |
| 37 | 6 h | gi|295661059 | G-protein complex beta subunit cpcb | 102 | 228 | 35.4/38.5 | 6.5/7.9 | 3.3 |
| CELL RESCUE, DEFENSE AND VIRULENCE | ||||||||
| 38 | 24 h | gi|60656557 | Heat shock protein 90 | 148 | 91 | 80.3/73.7 | 4.9/5.0 | 1.4 |
| 39 | 24 h | gi|226285144 | Hsp90 co-chaperone AHA1 | 150 | 48 | 36.5/42.0 | 5.4/5.7 | 1.4 |
| 40 | 24 h | gi|31324921 | Heat shock protein SSC1 (70 kDa) | 135 | 225 | 73.8/67.1 | 5.9/5.1 | 1.5 |
| 41 | 24 h | gi|295659116 | Hsp70-like protein | 244 | 126 | 70.9/42.7 | 5.0/5.7 | 1.6 |
| 42 | 24 h | gi|226286087 | Mitochondrial peroxiredoxin PRX1 | 197 | 97 | 24.8/29.2 | 5.2/4.8 | 1.2 |
| 43 | 24 h | gi|17980998 | Y20 protein | 92 | 82 | 21.6/15.5 | 6.0/8.5 | 2.2 |
| UNCLASSIFIED PROTEINS | ||||||||
| 44 | 24 h | gi|226278304 | Conserved hypothetical protein | 103 | 40 | 38.0/38.5 | 6.0/7.2 | 1.4 |
| 45 | 24 h | gi|226286114 | Conserved hypothetical protein | 92 | 114 | 34.7/40.6 | 5.3/4.9 | 1.6 |
| 46 | 24 h | gi|226279849 | Conserved hypothetical protein | 86 | 75 | 11.8/13.0 | 5.0/4.3 | ** |
| 47 | 6 h | PAAG_06617 | Conserved hypothetical protein | 113 | - | 30.5/34.9 | 6.2/7.1 | 1.3 |
**Spots visualized only in iron-depleted condition;
Spots numbers refers to Figure 3;
Time in iron starvation condition;
GenBank general information identifier;
Molecular weight (theoretical/experimental);
pI (theoretical/experimental).
≥fold in iron depletion.
Table 2. P. brasiliensis identified proteins with reduced expression during iron starvation.
| Spot1 | Time2 | GenBank gi3 | Protein identification | PMF score | MS/MS score | MW4 | pI 5 | ≤fold change6 |
| METABOLISM | ||||||||
| Nucleotide metabolism | ||||||||
| 48 | 6 h | gi|295672652 | Bifunctional purine biosynthesis protein ADE17 | 186 | 52 | 66.7/61.6 | 6.7/8.1 | 2.0 |
| C-compound and carbohydrate metabolism | ||||||||
| 49 | 24 h | gi|226277934 | 2-methylcitrate synthase (2-Mcs) | 254 | 153 | 51.5/46.2 | 9.0/8.8 | 3.1 |
| 50 | 6 h | PAAG_04550 | 2-methylcitrate synthase (2-Mcs) | 96 | - | 51.4/44.5 | 9.0/8.3 | 2.1 |
| 51 | 6 h | gi|295666177 | Mitochondrial 2-methylisocitrate lyase | 140 | 55 | 67.2/59.1 | 8.7/8.2 | 1.5 |
| 52 | 24 h | gi|116561512 | Isocitrate lyase (Icl) | 301 | 95 | 60.1/58.6 | 6.7/7.9 | 1.3 |
| 53 | 24 h | gi|116561512 | Isocitrate lyase (Icl) | 233 | 47 | 60.1/57.8 | 6.7/7.7 | ** |
| Amino acid, nitrogen and sulfur metabolism | ||||||||
| 54 | 24 h | gi|226281118 | Pentafunctional AROM polypeptide | 189 | - | 166/122 | 6.3/7.9 | ** |
| 55 | 24 h | gi|226277980 | Acetamidase | 177 | - | 59.2/52.7 | 5.8/6.4 | 1.6 |
| 56 | 24 h | gi|226285914 | Adenylyl-sulfate kinase | 172 | 41 | 23.8/33.5 | 8.6/9.5 | 1.9 |
| 57 | 6 h | gi|225679649 | Adenylyl-sulfate kinase | 125 | 55 | 23.8/32.7 | 9.1/9.7 | 1.1 |
| 58 | 6 h | gi|295663176 | Sulfate adenylyltransferase | 307 | 552 | 64.0/61.9 | 6.2/7.4 | 1.8 |
| 59 | 6 h | gi|295662426 | Aspartate aminotransferase | 121 | 137 | 50.8/41.8 | 8.3/8.5 | 1.6 |
| ENERGY | ||||||||
| Electron transport and membrane-associated energy conservation | ||||||||
| 60 | 24 h | gi|226279655 | Cytochrome c | 75 | 71 | 12.2/14.5 | 9.2/10.1 | 2.6 |
| 61 | 24 h | gi|225677786 | ATP synthase gamma chain | 87 | - | 32.4/36.0 | 6.7/6.1 | 1.6 |
| 62 | 24 h | gi|226282053 | ATP synthase subunit beta | 249 | 376 | 55.1/48.3 | 5.2/4.5 | 1.5 |
| 63 | 24 h | gi|226279593 | ATP synthase subunit 5 | 112 | 76 | 24.6/23.5 | 9.7/10.1 | ** |
| 64 | 24 h | gi|226291052 | ATP synthase subunit 4 | 94 | 69 | 26.7/26.2 | 9.4/8.8 | ** |
| 65 | 24 h | gi|226278316 | Electron transfer flavoprotein subunit beta | 103 | 136 | 21.6/34.3 | 9.3/10 | ** |
| Tricarboxylic-acid pathway | ||||||||
| 66 | 24 h | gi|226293399 | Aconitase | 172 | 211 | 79.1/77.7 | 6.4/7.8 | 1.7 |
| 67 | 24 h | gi|225684009 | Aconitase | 146 | 184 | 85.0/79.0 | 7.2/7.7 | 1.7 |
| 68 | 24 h | gi|225684009 | Aconitase | 139 | 55 | 85.0/80.1 | 7.2/7.5 | 1.6 |
| 69 | 24 h | gi|226278535 | ATP-citrate synthase subunit 1 | 84 | - | 72.4/65.9 | 8.0/8.4 | ** |
| 70 | 6 h | PAAG_03330 | Dihydrolipoyl dehydrogenase | 86 | - | 55.6/47.0 | 8.2/7.6 | 1.3 |
| Oxidation of fatty acids | ||||||||
| 71 | 24 h | gi|226278634 | Aldehyde dehydrogenase (Aldh) | 217 | 197 | 54.5/49.7 | 5.8/6.2 | 2.2 |
| 72 | 24 h | gi|226286163 | 3-hydroxybutyryl CoA dehydrogenase | 165 | 101 | 34.3/36.4 | 8.4/7.3 | 1.5 |
| 73 | 6 h | PAAG_06224 | Carnitine O-acetyltransferase | 89 | - | 69.4/61.7 | 8.2/8.5 | 2.1 |
| CELL CYCLE AND DNA PROCESSING | ||||||||
| 74 | 24 h | gi|154705473 | Septin-1 | 151 | 105 | 44.1/44.3 | 5.2/5.1 | ** |
| 75 | 24 h | gi|38569374 | 14-3-3-like protein 2 | 90 | 36 | 29.7/46.8 | 4.6/4.2 | ** |
| 76 | 24 h | gi|226278903 | Cell division cycle protein | 311 | 114 | 90.5/118 | 4.9/4.7 | 3.5 |
| 77 | 6 h | PAAG_01647 | Tubulin alpha-1 chain | 103 | - | 50.0/50.7 | 5.0/5.0 | 1.7 |
| TRANSCRIPTION | ||||||||
| 78 | 24 h | gi|226277842 | Prohibitin-1 | 180 | - | 30.9/35.1 | 8.7/9.6 | 1.6 |
| 79 | 6 h | gi|295673504 | cwfJ domain-containing protein | 88 | - | 61.6/58.3 | 6.1/7.2 | 1.4 |
| PROTEIN SYNTHESIS | ||||||||
| 80 | 24 h | gi|226280705 | Elongation factor 2 | 283 | 244 | 92.6/92.2 | 6.4/7.9 | 3.0 |
| 81 | 24 h | gi|226280705 | Elongation factor 2 | 189 | - | 92.6/85.0 | 6.4/7.9 | ** |
| 82 | 24 h | gi|28395450 | 40S ribosomal S12 protein | 95 | 63 | 16.8/19.3 | 5.0/4.4 | 1.7 |
| 83 | 24 h | gi|226282202 | ATP-dependent RNA helicase eIF4a | 142 | 63 | 45.0/64.1 | 5.1/4.8 | ** |
| PROTEIN FATE (folding, modification, destination) | ||||||||
| 84 | 24 h | gi|226285231 | Ubiquitin-conjugating enzyme variant MMS2 | 126 | 153 | 15.7/16.4 | 6.1/6.2 | 1.5 |
| 85 | 24 h | gi|34979129 | Peptidyl-prolyl cis/trans isomerase | 108 | 69 | 20.9/24.4 | 6.0/9.7 | 1.4 |
| 86 | 6 h | gi|295663176 | Dipeptidyl-peptidase | 201 | 84 | 86.2/70.0 | 7.9/7.3 | 1.7 |
| CELLULAR TRANSPORT AND TRANSPORT MECHANISMS | ||||||||
| 87 | 6 h | PAAG_03137 | Vacuolar protein sorting-associated protein | 72 | - | 22.1/26.3 | 5.0/4.6 | 1.2 |
| CELLULAR COMMUNICATION/SIGNAL TRANSDUCTION MECHANISM | ||||||||
| 88 | 24 h | gi|226285275 | Stomatin family protein | 89 | - | 48.7/43.4 | 6.5/5.1 | ** |
| CELL RESCUE, DEFENSE AND VIRULENCE | ||||||||
| 89 | 24 h | gi|60656557 | Heat shock protein 90 | 173 | 60 | 80.3/104 | 4.9/4.6 | 2.3 |
| 90 | 24 h | gi|31324921 | Heat shock protein SSC1 (70 kDa) | 214 | 263 | 73.8/96.8 | 5.9/5.1 | 2.0 |
| 91 | 24 h | gi|226278527 | 10 kDa heat shock protein, mitochondrial | 161 | 108 | 11.1/12.9 | 8.7/9.7 | 1.4 |
| 92 | 24 h | gi|226282384 | 30 kDa heat shock protein | 142 | - | 28.6/24.3 | 6.8/6.9 | 1.6 |
| 93 | 24 h | gi|17980998 | Y20 protein | 160 | 302 | 21.6/22.1 | 6.0/7.1 | 2.9 |
| 94 | 6 h | gi|24528587 | Peroxissomal catalase | 71 | - | 70.7/61.7 | 6.3/8.0 | 2.0 |
| UNCLASSIFIED PROTEINS | ||||||||
| 95 | 24 h | gi|226285365 | Conserved hypothetical protein | 135 | 194 | 26.0/28.2 | 5.8/6.1 | 1.2 |
| 96 | 24 h | gi|226286445 | Conserved hypothetical protein | 154 | 37 | 115/109 | 5.1/4.9 | 2.0 |
**Spots visualized only in iron-replete condition;
Spots numbers refers to Figure 3;
Time in iron starvation condition;
GenBank general information identifier;
Molecular weight kDa (theoretical/experimental);
pI (theoretical/experimental);
≤fold in iron depletion.
Identified proteins were grouped together based on their functionality according to the Munich Information Center for Protein Sequences (MIPS). Pie charts representing the distribution of the 96 identified spots according to their biological function are shown in Figure S2. P. brasiliensis differentially regulated proteins were involved in several cellular processes including energy (26%), metabolism (22.9%), cell rescue and virulence (12.5%), cell cycle (11.5%), protein synthesis (10.4%), transcription (5.2%), protein fate (3.1%), cellular communication (1%) and cell transport (1%). Unclassified proteins represented only a small fraction (6.2%) of the 96 protein spots analyzed by MS.
Twenty six percent of the P. brasiliensis identified proteins were grouped into energy processes. Glycolytic enzymes including enolase, fructose 1,6-biphosphate aldolase, glucokinase, phosphoglycerate kinase and triosephosphate isomerase were up regulated ranging in fold change from 1.2 to 2.8. The increased regulation of glycolytic enzymes was observed at 6 and 24 hours of iron deprivation (Table 1). The abundance of hydroxyacylglutathione hydrolase was also increased correlating with the detected increase in glycolytic enzyme regulation because this enzyme plays role in detoxification of glycolysis products.
While the abundance of P. brasiliensis glycolytic pathway enzymes increased during iron deprivation the tricarboxylic acid cycle (TCA) enzymes were decreased. TCA enzyme included aconitase, one subunit of ATP-citrate synthase, and dihydrolipoyl dehydrogenase (Figure S2, Table 2) depicted a decreased abundance during iron starvation. Four subunits of the electron transport ATP-synthase complex also decreased in abundance during iron restriction. Two other proteins related to electron transport were also down-regulated in P. brasiliensis during iron deprivation. These proteins included cytochrome c and electron transfer flavoprotein beta-subunit.
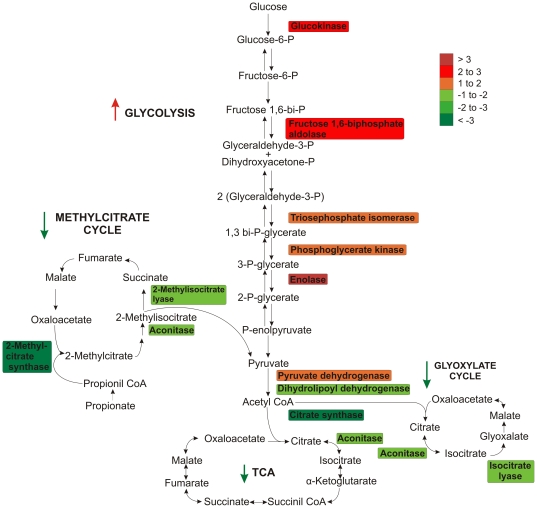
P. brasiliensis proteins involved in pathways using alternative sources of carbon, such as glyoxylate and methylcitrate cycles were also decreased in abundance during iron starvation. Two isoforms of the glyoxylate cycle enzyme isocitrate lyase, were decreased in abundance after 24 hours in iron depletion (Figure S2, Table 2). These isoforms play a regulatory role in glyoxylate cycle allowing fungi to use fatty acids to synthesize carbohydrates. Two P. brasiliensis enzymes involved in the methylcitrate cycle were decreased in abundance following iron restriction. These enzymes were 2-methylisocitrate lyase and 2-methylcitrate synthase. Figure 6 summarizes the P. brasiliensis metabolic proteins regulated during iron starvation in this study.
Figure 6. Overview of P. brasiliensis metabolic processes regulated during iron starvation revealed by proteomic analysis.
Schematic representation of proteins involved in P. brasiliensis carbon metabolism (glycolysis/TCA cycle/glyoxylate cycle/methylcitrate cycle). Proteins are colored according to their differences in abundance as detected by 2D-gel analysis of P. brasiliensis following iron starvation. Arrows indicate up or down regulation of metabolic pathways.
We also detected a decreased regulation of P. brasiliensis enzymes related to fatty acids metabolism including aldehyde dehydrogenase, 3-hydroxybutyryl CoA dehydrogenase, and carnitine-O-acetyltransferase (Table 2). Enzymes related to amino acid metabolism were also decreased in abundance during iron starvation, such as the pentafunctional AROM polypeptide, related to aromatic amino acids biosynthesis, and aspartate aminotransferase, the enzyme responsible for the amination of oxaloacetate to form aspartate. In contrast L-threonine 3-dehydrogenase was 1.9 times fold higher in abundance following iron deficiency (Tables 1 and 2). This enzyme is involved in threonine catabolism which produces glycine.
Another P. brasiliensis cellular process affected by iron starvation was cell rescue. Eight proteins, including isoforms, belonging to the heat shock protein family were altered in abundance following iron limitation (Tables 1 and 2).
P. brasiliensis mRNA and proteomic analysis correlate with in vivo infection
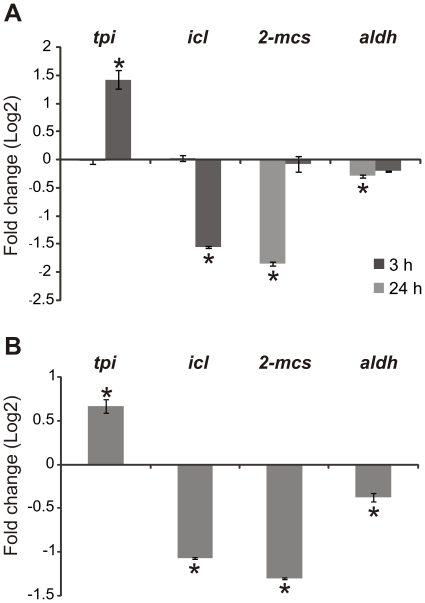
In order to validate the significance of our proteomic results we next sought to determine if changes in protein levels could be correlated with changes in transcript levels in vivo. We determined that the differences observed in proteomic assay are in agreement with transcriptional findings, using quantitative RT-PCR to measure isocitrate lyase (icl), aldehyde dehydrogenase (aldh) and 2-methylcitrate synthase (2-mcs) transcripts (Figure 7A). Protein and transcript levels of these enzymes decreased during iron limitation as depicted in Figure 7A and Table 2. The triosephosphate isomerase (tpi) transcript level was correlated with protein levels (Figure 7A and Table 1). The same oligonucleotides were next used to evaluate the transcriptional levels of these genes in P. brasiliensis isolated from mouse spleens. We observed that the changes in transcriptional profile of the genes tpi, icl, 2-mcs and aldh in yeast cells isolated from mice are similar to those observed in response to iron deprivation (Figure 7 B).
Figure 7. P. brasiliensis transcripts of yeast cells isolated from infected mice correspond to P. brasiliensis proteins differentially regulated under iron limiting conditions.
P. brasiliensis transcript levels of genes encoding enzymes involved in energy processes; triosephosphate isomerase (tpi), isocitrate lyase (icl), 2-methylcitrate synthase (2-mcs) and aldehyde dehydrogenase (aldh), isolated from yeast cells following iron starvation (A), and from yeast cells recovered from the spleens of infected BALB/c mice (B). Transcript levels were measured using quantitative RT-PCR. Data were normalized to the L34 protein transcript and presented as log2 (fold change). Student's t test was used for statistical comparisons. Error bars represent standard deviation from three biological replicates while * represents p≤0.05.
Western blot analysis and enzymatic activity correlates with proteomic data
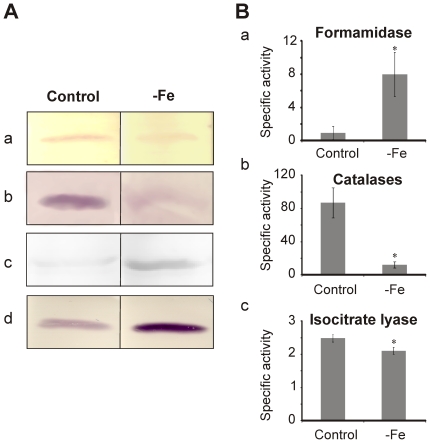
Western blot analysis was performed to confirm the P. brasiliensis protein level changes detected in the proteomic analysis. This was accomplished using antibodies specific to four proteins affected by iron levels as determined by 2D gel analysis. P. brasiliensis aconitase and isoctrate lyase (Figure 8 A, panels a and b) levels following iron depletion were observed to decrease in abundance by western blot and 2D-gel analysis. Formamidase and triosephosphate isomerase also yielded similar increases in abundance following western blot analysis to that described by 2D-gel analysis (Figure 8 A panels c and d).
Figure 8. Western blot and enzymatic activity analysis validate proteomic data of P. brasiliensis yeast cells during iron-limiting condition.
A) Western blot analysis of proteins probed with antibodies to (a) aconitase, (b) isocitrate lyase, (c) formamidase and (d) triosephosphate isomerase. B) Activity assay results from (a) formamidase, (b) catalases, (c) isocitrate lyase. Control represents yeast cells incubated for 24 hours in MMcM medium. −Fe represents yeast cells incubated for 24 hours in MMcM medium depleted of iron. Error bars represent standard deviation from three biological replicates while * represents p≤0.05.
Moreover, we performed enzymatic assays for formamidase, catalases, and isocitrate lyase using P. brasiliensis protein extracts derived from yeast cells grown under iron deprivation in order further validate our proteomic data. We observed a statistically significant increase in formamidase activity (Figure 8 B, panel a) and a decrease in catalases and isocitrate lyase activity during iron starvation (Figure 8 B, panels b and c, respectively). All data again are in agreement with the 2D-gel analysis.
Discussion
In this study, the treatment of mice with an iron supplement made them more susceptible to P. brasiliensis infection. Similar trends were also observed for the pathogenic fungi C. neoformans [22] and A. fumigatus [23]. Moreover, we found that P. brasiliensis regulated the abundance of 274 proteins following iron depletion. Host iron restriction is thought to be one part of the host defense mechanism. Our data suggest that a remodeling of P. brasiliensis metabolism by limiting the role of iron dependent pathways could be one mechanism in which the fungus adapts to the host environment.
Because iron is not abundantly available in host tissues pathogenic microorganisms have developed mechanisms which enable them to survive in low iron environments [24]. These mechanisms include iron uptake from host sources, and metabolic switching to iron independent pathways [25]–[31]. It is known that a component of the fungal cellular response to iron deficiency operates through the induction of a number of transcripts [18], [32]–[34]. Under our iron restriction conditions it was observed the induction of P. brasiliensis orthologs to the known iron dependent transcripts, such as the transcriptional regulator hapX, the siderophore biosynthetic enzyme sidA and the siderophore transporter sit1 [17]. These results suggest the activation of the siderophore pathway of P. brasiliensis under iron limiting conditions. Moreover, the induction of hapX suggests that this ortholog in P. brasiliensis is also integral to the fungus response to iron starvation as previously described in other organisms [34]. These data support our P. brasiliensis iron restriction model for further analysis using proteomic technologies.
Proteomic analysis revealed that during iron deprivation the majority of P. brasiliensis regulated proteins were related to energy and metabolic function. Among the pathways potentially repressed in P. brasiliensis during iron depletion was the tricarboxylic acid cycle (TCA). TCA oxidative reactions are mediated by enzymes containing Fe/S clusters including aconitase, succinate dehydrogenase and fumarase [35]. We observed that TCA enzymes: aconitase, an ATP-citrate synthase subunit and dihydrolipoyl dehydrogenase, were regulated in P. brasiliensis by iron availability. These data taken together suggest metabolic switching to pathways independent of enzymes with Fe/S clusters. The decreased expression of TCA enzymes observed in P. brasiliensis during iron deprivation has also been reported in other pathogenic bacteria and fungi [3], [4], [20], [36], [37]. The evidence of TCA repression during iron deprivation is further emphasized by a decreased expression of the P. brasiliensis enzyme aspartate aminotransferase. A substrate for aspartate aminotransferase is the TCA intermediate metabolite oxaloacetate [38]. A reduction in the amount of this substrate due to TCA repression may influence the decreased expression of aspartate aminotransferase. The correlation between aspartate biosynthesis and TCA was also reported in mice given acetate. The authors of this study observed an increase in TCA flux concurrent with an increase in aspartate aminotransferase [39].
In addition to TCA cycle enzymes, a decrease in the abundance of P. brasiliensis glyoxylate cycle enzymes were also observed during iron depletion. The glyoxylate pathway provides cells with an alternative source of carbon allowing for the consumption of acetate and/or ethanol in place of glucose [40]. The reduced expression of enzymes involved in the glyoxylate pathway correlates with the decreased expression of TCA enzymes because these cycles are coupled. Two of the five reactions of the TCA cycle are catalyzed by enzymes unique to the glyoxylate cycle [41]. Reduced levels of isocitrate lyase, an enzyme with a key role in the glyoxylate cycle, correlate with the negative regulation of this pathway during iron deprivation. We could speculate that during iron limitation and with glucose as the carbon source, P. brasiliensis reduces the glyoxylate cycle activity due to the requirement of aconitase which is an iron-dependent enzyme of this pathway. This speculation is proven stronger because of the induction of hapX. This transcription factor negatively regulates iron dependent energy metabolism.
Two P. brasiliensis enzymes involved in the methylcitrate cycle were also decreased in abundance following iron restriction. The methylcitrate cycle is another system that provides an alternative source of carbon through pyruvate production [42]. Similar to the glyoxylate cycle this pathway functions using enzymes that are part of the TCA cycle [43]. These data add further support to the overall metabolic adjustment of P. brasiliensis under iron deprivation.
P. brasiliensis proteins involved in oxidative phosphorylation also decreased in abundance following iron limitation. Oxidative phosphorylation promotes ATP formation from NADH and FADH2 produced by the TCA cycle [44]. The suppression of pathways such as TCA that feed into oxidative phosphorylation may be associated with the decreased expression levels of enzymes that are part of oxidative phosphorylation. Moreover, P. brasiliensis enzyme complexes part of the electron transport chain utilize iron as a prosthetic group. Thus, the maintenance of this pathway is iron dependent and with iron levels being low their production may be down regulated as we have detected in this study.
Our data suggest that the suppression of oxidative pathways during iron deprivation in P. brasiliensis is linked to an increase of glycolytic activity as demonstrated by the induction of most enzymes in the glycolytic pathway. The glucose concentration of the medium for all of our proteomic analysis remained above 0.8% (w/v) providing high glucose availability to the yeast cells (data not shown). Increased glycolysis activity from iron limitation has been observed in other bacteria and fungi including Staphylococcus aureus and S. cerevisiae [20], [45]. Moreover, the increased expression of P. brasiliensis hydroxyglutatione hydrolase observed in this study is also linked to glycolysis induction. Hydroxyglutatione hydrolase is involved in methylglyoxal detoxification which is a toxic molecule generated from the degradation of glycolytic products [46]. Hydroxyglutatione hydrolase prevents methylglyoxal accumulation by converting it to D-lactate in presence of glutathione [47]. These results suggest that P. brasiliensis remodels its energy metabolism in response to iron starvation by increasing glycolytic activity to compensate for the decrease of aerobic pathways, that are iron dependent.
It is important to note that the abundance changes detected by proteomic analysis for enzymes involved in energy processes during iron depletion were also observed following transcriptional analysis of selected genes of P. brasiliensis isolated from mouse spleens. Similar to what has been observed for other bacterial and fungal pathogens, these data suggest that iron deprivation treatment may mimic the environment P. brasiliensis encounters in the host. To further support this theory, the induction of genes involved with glucose utilization under host conditions have been reported for other fungal pathogens. C. albicans over expresses pfk2 that encodes for phosphofructokinase and pyk1 that encodes for pyruvate kinase when growing within the kidney [48].
Conclusions
Proteomic analysis of P. brasiliensis revealed that the major cellular response affected by iron restriction was related to energy production. Our data suggest that under iron limiting condition glycolysis was the more favored energy pathway over oxidative pathways that are dependent on enzymes containing Fe-S groups. Thus, P. brasiliensis relies more on the energy generated from glycolysis in order to compensate for the decrease in ATP production from oxidative phosphorylation. In addition to the activation of iron uptake systems, P. brasiliensis metabolic adjustment may be an indispensable survival mechanism the fungus requires when in a nutrient deficient environment.
Materials and Methods
All animal experiments were performed in accordance with the international rules for animal experimentation. The animal protocol was approved by the Universidade Federal de Goiás committee of the ethical treatment of animals (Number: 131/2008).
P.brasiliensis strain maintenance
P. brasiliensis Pb 01 (ATCC MYA-826) was used in all experiments. The yeast phase was maintained in vitro by sub culturing at 36°C in Fava Netto's semisolid medium [49] every 7 days. Fava Netto's semisolid medium components were as follows; 1% (w/v) peptone, 0.5% (w/v) yeast extract, 0.3% (w/v) proteose peptone, 0.5% (w/v) beef extract, 0.5% (w/v) NaCl, 4% (w/v) glucose, 1.2% (w/v) agar, pH 7.2.
BALB/c mice infection
Thirty day old female BALB/c mice (n = 4) were injected intraperitoneally with a solution containing Ferrodex diluted in 0.9% NaCl (w/v; 50 mg/kg; Tortuga Companhia Zootécnica Agrária, Brazil). Ferrodex is a ferric hydroxide solution containing dextran commonly used for animal supplementation experiments. The iron source was given every 2 days for 15 days prior to and during infection. For control experiments (n = 4) sterile 0.9% NaCl (w/v) was injected into mice at the same time intervals as iron supplementation. Mice were inoculated intraperitonially with 107 P. brasiliensis yeast cells as previously described [50]. After 2 weeks of harboring the organism, mouse spleens and livers were removed and homogenized in 5 mL of sterile 0.9% (w/v) NaCl. The homogenized sample was plated in brain heart infusion agar supplemented with 4% (v/v) fetal calf serum and 2% (w/v) glucose. The plates were incubated at 36°C and colony forming units (CFUs) were determined after 20 days.
Gene expression analyses of P. brasiliensis from infected mice were performed by isolating yeast cells from spleens as previously described with minor modifications [51]. The spleens of infected mice were homogenized in water using a grinder. To remove large pieces of animal tissue the homogenate was then filtered using a nylon membrane. The sample was frozen in liquid nitrogen and then centrifuged at 500× g for 5 minutes to remove any remaining animal tissue. Next, the sample was centrifuged at 7000× g for 15 minutes in order to isolate fungal cells. The samples containing fungal cells were checked under the microscope to make sure that they were not cross-contaminated by spleen cells. RNA was extracted from the fungal cells by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. It is important to note here that P. brasiliensis yeast cells were not cultured in vitro prior RNA extraction.
Iron depletion experiments
P. brasiliensis yeast cells were grown in McVeigh/Morton medium (MMcM) [52]. Iron depleted medium was supplemented with the iron chelator bathophenanthrolinedisulfonate (BPS; 50 µM; Sigma-Aldrich, Germany). P. brasiliensis yeast cells were incubated at 36°C with shaking at 150 rpm. Using trypan blue the number of viable cells was determined at time intervals of 2, 4, 6, 24 and 48 hours.
Protein extracts from yeast cells were prepared by inoculating 50 mL of Fava Neto's liquid medium with 108 cells/mL. Cultures were incubated overnight at 36°C under gentle shaking for 16 hours. Cells were centrifuged at 5000× g for 5 minutes and washed 5× in MMcM media [52] containing limited iron. Control cells were incubated in MMcM supplemented with 3.5 µM Fe(NH4)2(SO4)2, for 6 and 24 hours. Cells subjected to iron starvation were incubated in MMcM medium containing 50 µM of BPS with no iron supplementation.
Quantitative real time reverse transcription PCR (qRT-PCR) analysis
Following P. brasiliensis growth in iron depleted media cells were centrifuged at 1500× g, frozen in liquid nitrogen, and disrupted by maceration. Cells were then treated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The manufacturer's protocol was followed to extract total RNA. The RNA was reversibly transcribed using the high capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA, USA). We confirmed the specificity of each primer pair for the target cDNA by the visualization of a single PCR product following agarose gel electrophoresis and melting curve analysis. Primer sequences are listed in Table S4. The cDNA was quantified by qRT-PCR using a SYBR green PCR master mix (Applied Biosystems Step One Plus PCR System). qRT-PCR analysis was performed in triplicate for each cDNA sample as previously described [53]. Data were normalized to the ribosomal L34 gene (GenBank accession number EEH37825). Standard curves were generated by diluting the cDNA solution 1∶5. The standard curve method was used for relative quantification of genes [54]. Statistical comparisons were performed using the student's t test and p-values≤0.05 were considered statistically significant. The data were presented as fold change relation between iron deprivation and control conditions, plotted in a log2 scale of the fold change values (−Fe/Control).
2D-gel electrophoresis
Following P. brasiliensis growth in iron depleted media as described above cells were centrifuged at 1500× g, frozen in liquid nitrogen, and disrupted by maceration. Cells were then resuspended in a solution containing 20 mM Tris-HCl, pH 8.8, 2 mM CaCl2 [55]. Protein concentrations were determined using the Bradford reagent (Sigma-Aldrich) and bovine serum albumin (BSA) was utilized as a standard [56].
For each sample 300 µg of total protein were loaded onto the gel. The 2-D Clean-up Kit (GE Healthcare, Uppsala, Sweden) was used according to the manufacturer's instructions. Proteins samples were treated with 250 µL of buffer containing 7 M urea, 2 M thiourea, 130 mM 3- [(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 0.002% (w/v) dithiothreitol (DTT), ampholyte-containing buffer (IPG buffer, GE Healthcare), and trace amounts of bromophenol blue [57]. Samples were loaded onto a 13 cm Immobiline™ DryStrip gel (GE Healthcare) with a linear separation range of pH 3–11. Isoelectric focusing was conducted on a Multiphor-II electrophoresis system (GE Healthcare) using a step gradient from 500 V for 1 hour, 500–1000 V for 1 hour, 1000–8000 V over 12.5 hours, and 8000 V for 2.5 hours. The isoelectric focusing was preceded by a rehydration step at 30 V for 14 hours.
Following isoelectric focusing strips were transferred to an equilibration buffer [50 mM Tris-HCl pH 8.8, 6 M urea, 30% (v/v) glycerol, 2% (w/v) sodium dodecyl sulfate (SDS), and 0.002% (w/v) bromophenol blue] containing 18 mM of DTT for 40 minutes as previously described [58]. The strips were next transferred to a fresh equilibration buffer containing 135 mM iodoacetamide. Strips were incubated for another 40 minutes. Each IPG strip was placed on top of a polyacrylamide SDS-PAGE gel [59] and covered with agarose [0.5% (w/v) agarose in running buffer, 25 mM Tris-HCl, 192 mM glycine, 0.1% (w/v) SDS]. SDS-PAGE gels were run at 10°C for 1 hour at 150 V and increased to 250 V for 3 hours. Proteins were stained using silver or Coomassie brilliant blue (PlusOne™ Silver Staining Kit or PlusOne Coomassie Tablets PhastGel Blue R-350, GE Healthcare) according to the manufacturer's instructions.
2D-gel image analysis
Gel images were produced using the Image Scanner III (GE Healthcare). 2D gel spot detection, matching, and spot intensity calculations were performed using Image Master 2D Platinum v7.0 (GE Healthcare). Automated spot matching was also checked manually and corrected if needed. Spot intensity values for all protein spot matches within each analysis were calculated and normalized to the intensity of all the spots detected.
2D-gel statistical analysis
ANOVA statistical analysis was used to compare the differences in intensity of each matched protein spot. A comparison with a p-value≤0.05 was considered statistically significant. The p-values were calculated using the Image Master 2D Platinum v7.0 software (GE Healthcare). To check population distribution the Shapiro-Wilk test [60] was applied using the software Statistical Package for Social Science (Version 13.0, SPSS Inc, Cary, NC, USA), considering that the hypothesis of the non-normality of the sample distribution could be rejected at p-value≤0. 05.
In-gel protein digestion
Protein spots were excised from the 2D-gel and diced into small pieces. Spots were next incubated with 250 µL of a solution containing 50 mM sodium thiosulfate and 15 mM potassium ferricyanide for 5 minutes. Gel pieces were washed twice with water to remove reducing agents. Next, the gel pieces were dehydrated in 100 µL of acetonitrile (ACN) and dried in a speed vacuum. The gel pieces were incubated in 50 µL of 10 mM DTT for 1 hour. The DTT solution was removed and 50 µL of 55 mM iodoacetamide was added. The gel pieces were incubated in the dark for 45 minutes. Next, the gel pieces were washed with 100 µL of 25 mM ammonium bicarbonate followed vortexing for 10 minutes. The solution covering the gel pieces was removed and gel pieces were dehydrated in 100 µL of a solution containing 25 mM ammonium bicarbonate/ACN 50% (v/v). The gel pieces were vortexed for 5 minutes and centrifuged. This step was repeated one time. Next, the gel pieces were dried and 25 µL of a 12.5 ng/ml trypsin solution was added (Sequencing Grade Modified Trypsin Promega, Madison, WI, USA). The gel pieces were placed on ice for 10 minutes. The solution covering the gels was removed and 25 µL of 25 mM ammonium bicarbonate was added. The gel pieces were incubated at 37°C for 16 hours. Following digestion the solution covering the gel pieces was transferred to a clean tube. Next, 50 µL of a solution containing 50% (v/v) ACN and 5% (v/v) trifluoroacetic acid acid (TFA) was added to the gel pieces. The gel pieces were then vortexed for 10 minutes and sonicated for 3 minutes. Next, the solution covering the gel pieces was combined with the original aqueous extraction. The samples were dried in a speed vacuum and peptides were dissolved in 10 µL of water for MS analysis [3].
Mass Spectrometry analysis
Two microliters of each peptide sample were deposited on a matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) target plate and dried at room temperature. Next, the peptide mixtures were covered with 2 µL of MALDI matrix solution (10 mg/mL alphacyano-4-hydroxycinammic acid in 50% (v/v) ACN, 5% (v/v) TFA). ZipTips were used to concentrate samples containing low amounts of digested protein (Milipore, Bedford, MA, USA). Dried peptides solutions were analyzed by MS. MALDI quadrupole time-of-flight (Q-TOF) MS, and MALDI-TOF-TOF MS (Synapt, Waters , Manchester, UK and Reflex IV, Bruker Daltonics, Karlsruhe, Germany, respectively). Mass spectra were collected and processed using X-TOF (Bruker Daltonics) and Mass Lynx (Waters). MS data were searched against the NCBI non redundant database and matched with their corresponding proteins sequences using MASCOT (http://www.matrixscience.com). The mass tolerance was set at 100 ppm for each MASCOT search. Each search was restricted to fungi while allowing for one missed trypsin cleavage.
Western blot analysis
P. brasiliensis protein extracts of yeast cells were probed using aconitase, formamidase, isocitrate lyase, and triosephosphate isomerase antibodies. Ten micrograms of protein samples prepared as described above were loaded onto a 12% SDS PAGE gel and separated by electrophoresis. Gels were run at 150 V for ∼2 hours. Protein bands were transferred from gels to nitrocellulose membrane at 30 V for 16 hours in buffer containing 25 mM Tris-HCl, 190 mM glicine, 20% (v/v) methanol. Gels were stained with Ponceau red staining to confirm complete protein transfer. Next, each membrane was submerged in blocking buffer [Phosphate buffered saline solution 1× (PBS; 1.4 mM KH2PO4, 8 mM Na2HPO4, 140 mM NaCl, 2.7 mM KCl; pH 7.3), 5% (w/v) non-fat dried milk, 0.1% (v/v) Tween 20] for 1–2 hours. Membranes were washed with buffer [PBS 1×, 0.1% (v/v) Tween 20] and incubated with primary antibodies for 2 hours at room temperature. Primary antibodies were used at a 1/5000 (v/v) ratio of antibody to buffer. Primary polyclonal antibodies used were anti-aconitase [61], anti-isocitrate lyase (protein molecular weight 60 KDa, unpublished), anti-formamidase [62] and anti-triosephosphate isomerase [63]. This was followed by three 15 minutes washes in blocking buffer. Membranes were incubated with the appropriate conjugated secondary antibody [anti-rabbit or anti-mouse immunoglobulin G coupled to alkaline phosphatase (Sigma-Aldrich)] in a 1/5000 (v/v) ratio. Membranes were developed with 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (BCIP-NBT).
Enzyme activity assays
Formamidase activity was determined by measuring the amount of ammonia formation as previously described [64]. One µg of P. brasiliensis total protein extract prepared as described above were added to 200 µl of a 100 mM formamide substrate solution in 100 mM phosphate buffer containing 10 mM of EDTA, pH 7.4. Samples were incubated at 37°C for 30 minutes. Following the 30 minutes incubation period 400 µl of phenol-nitroprusside and 400 µl of alkaline hypochlorite (Sigma Aldrich, Co.) were added. Next, samples were incubated for 6 minutes at 50°C and the absorbance was read at 625 nm. The amount of ammonia released for each sample was determined by comparing to a standard curve. One unit (U) of formamidase specific activity was defined as the amount of enzyme required to hydrolyze 1 µmol of formamide (corresponding to the formation of 1 µmol of ammonia) per minute per mg of total protein.
Catalases activity was determined by measuring a decrease in the absorbance at 240 nm from the conversion of hydrogen peroxide (H2O2) to oxygen as previously described [65]. Catalases activity for each sample was calculated using a standard curve generated by five different concentrations of H2O2. One unit of catalase activity was defined as the amount of enzyme required to catalyze the consumption of 1 µmol of H2O2 per minute per mg of total protein.
Isocitrate lyase activity was determined by measuring the formation of glyoxylate as its phenylhydrazone derivative [66]. Glyoxylate-phenylhydrazone formation was determined by measuring the absorbance at 324 nm, using an extinction coeficient of 16.8 mM−1 cm−1, in a reaction mixture containing 2 mM threo-D,L-isocitrate (Sigma Aldrich, Co.), 2 mM MgCl2, 10 mM phenylhydrazine HCl (Sigma Aldrich, Co.), 2 mM dithiothreitol, and 50 mM potassium phosphate at pH 7.0. Specific activity was determined as the amount of enzyme required to form 1 µmol of glyoxylate-phenylhydrazone per minute per mg of total protein.
Supporting Information
Iron starvation does not affect the viability of P. brasiliensis yeast cells. Viability of P. brasiliensis yeast cells incubated in MMcM medium containing 3.5 µM iron (circles) and incubation of yeasts cells in MMcM iron depleted media containing 50 µM BPS (triangles). Viability was determined using trypan blue. Error bars represent standard deviation from three biological replicates while * represents p≤0.05.
(TIF)
Categorical representation of regulated P. brasiliensis proteins following iron starvation. Identified proteins were classified according to their respective functional categories determined by MIPS. (A) Categorization of differentially expressed proteins during iron starvation. Classification of proteins with induced (B) and repressed expression (C) in iron limiting condition.
(TIF)
Relative fold induction of iron metabolism related genes in the time course of iron starvation using real time- RT-PCR. 1Values represent the mean of each triplicate sample ± standard deviation.
(DOC)
Additional information about P. brasiliensis identified proteins with increased expression during iron starvation. ** Spots visualized only in iron-depleted condition. 1 Spots numbers refers to Figure 3. 2 p values were accessed by ANOVA statistical test.
(DOC)
Additional information about P. brasiliensis identified proteins with reduced expression during iron starvation. ** Spots visualized only in iron replete condition. 1 Spots numbers refers to Figure 3. 2 p values were accessed by ANOVA statistical test.
(DOC)
Oligonucleotide primers used in quantitative RT-PCR.
(DOC)
Acknowledgments
We thank Mirelle Garcia Silva for the helpful suggestions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work at the Universidade Federal de Goiás was supported by grants from “Financiadora de Estudos e Projetos - FINEP - 0107055200”. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Doherty CP. Host-Pathogen Interactions: The Role of Iron. J Nutr. 2007;137:1341–1344. doi: 10.1093/jn/137.5.1341. [DOI] [PubMed] [Google Scholar]
- 2.Johnson L. Iron and siderophores in fungal–host interactions. Mycol Res. 2008;112:170–183. doi: 10.1016/j.mycres.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Winters MS, Spellman DS, Chan Q, Gomez FJ, Hernandez M, et al. Histoplasma capsulatum proteome response to decreased iron availability. Proteome Sci. 2008;6:36. doi: 10.1186/1477-5956-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lian T, Simmer MI, D'Souza CA, Steen BR, Zuyderduyn SD, et al. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2005;55:1452–1472. doi: 10.1111/j.1365-2958.2004.04474.x. [DOI] [PubMed] [Google Scholar]
- 5.Lan C, Rodarte G, Murillo LA, Jones T, Davis RW, et al. Regulatory networks affected by iron availability in Candida albicans. Mol Microbiol. 2004;53:1451–1469. doi: 10.1111/j.1365-2958.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 1996;15:3377–3384. [PMC free article] [PubMed] [Google Scholar]
- 7.Blaiseau PL, Lesuissse E, Camadro JM. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J Biol Chem. 2001;276:34221–34226. doi: 10.1074/jbc.M104987200. [DOI] [PubMed] [Google Scholar]
- 8.Haas H, Zadra I, Stoffler G, Angermayr K. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J Biol Chem. 1999;274:4613–4619. doi: 10.1074/jbc.274.8.4613. [DOI] [PubMed] [Google Scholar]
- 9.Schrettl M, Bignell E, Kragi C, Joechl C, Rogers T, et al. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med. 2004;200:1213–1219. doi: 10.1084/jem.20041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hortschansky P, Eisendle M, Al-Abdallah Q, Schmidt AD, Bergman S, et al. Interaction of HapX with the CCAAT-binding complex-a novel mechanism of gene regulation by iron. EMBO J. 2007;26:157–3168. doi: 10.1038/sj.emboj.7601752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrettl M, Kim HS, Eisendle M, Kragl C, Nierman WC, et al. Sre-A mediated iron regulation in Aspergillus fumigatus. Mol Microbiol. 2008;70:27–43. doi: 10.1111/j.1365-2958.2008.06376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercier A, Pelletier B, Labbé S. A transcription factor cascade involving Fep 1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2006;5:1866–1881. doi: 10.1128/EC.00199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Gravelat FN, Chiang LY, Chen D, Vanier G, et al. Aspergillus fumigatus AcuM regulates both iron acquisition and gluconeogenesis. Mol Microbiol. 2010;78:1038–1054. doi: 10.1111/j.1365-2958.2010.07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Restrepo A, Tobón A. Paracoccidioides brasiliensis. In: Dismukes WE, Pappas PG, Sobel J, editors. Principles and Practice of Infectious Diseases. Philadelphia: Elsevier; 2005. pp. 3062–3068. [Google Scholar]
- 15.Coutinho ZF, Silva D, Lazera M, Petri V, Oliveira RM, et al. Paracoccidioidomycosis mortality in Brazil (1980–1995). Cad Saúde Pública. 2002;18:1441–1454. doi: 10.1590/s0102-311x2002000500037. [DOI] [PubMed] [Google Scholar]
- 16.Prado M, da Silva MB, Laurenti R, Travassos LR, Taborda CP. Mortality due to systemic mycoses as a primary cause of death or in association with AIDS in Brazil: a review from 1996 to 2006. Mem Inst Oswaldo Cruz. 2009;104:513–521. doi: 10.1590/s0074-02762009000300019. [DOI] [PubMed] [Google Scholar]
- 17.Silva MG, Schrank A, Bailão EFLC, Bailão AM, Borges CL, et al. The homeostasis of iron, copper and zinc in Paracoccidioides brasiliensis, Cryptococcus neoformans var. grubii, and Cryptococcus gattii: a comparative analysis. Frontiers Microbiol. 2011 doi: 10.3389/fmicb.2011.00049. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shakoury-Elizeh M, Tiedeman J, Rashford J, Ferea T, Demeter J, et al. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:1233–1243. doi: 10.1091/mbc.E03-09-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philpott CC, Protchenko O. Response to Iron Deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:20–27. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jo WJ, Kim JH, Oh E, Jaramillo D, Holman P, et al. Novel insight into iron metabolism by integrating deletome and transcriptome analysis in an iron deficiency model of the yeast Saccharomyces cerevisiae. BMC Genomics. 2009;10:130. doi: 10.1186/1471-2164-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo H, Chang Y, Chung Y, Kim K. Proteomic analysis of recombinant Saccharomyces cerevisiae upon iron deficiency induced via human H-Ferritin production. J Microbiol Biotechnol. 2008;18:1368–1376. [PubMed] [Google Scholar]
- 22.Barluzzi R, Saleppico S, Nocentini A, Boelaert JR, Neglia R, et al. Iron overload exacerbates experimental meningloencephalitis by Cryptococcus neoformans. J Neuroimmunol. 2002;132:140–146. doi: 10.1016/s0165-5728(02)00324-7. [DOI] [PubMed] [Google Scholar]
- 23.Kontoyiannis DP, Chamilos G, Lewis RE, Giralt S, Cortes J, et al. Increased bone marrow iron stores is an independent risk factor for invasive aspergillosis in patients with high-risk hematologic malignancies and recipients of allogeneic hematopoietic stem cell transplantation. Cancer. 2007;110:1303–1306. doi: 10.1002/cncr.22909. [DOI] [PubMed] [Google Scholar]
- 24.Kosman DJ. Molecular mechanisms of iron uptake in fungi. Mol Microbiol. 2003;47:1185–1197. doi: 10.1046/j.1365-2958.2003.03368.x. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan J, Ward DM, Crisp RJ, Philpott CC. Iron-dependent metabolic remodeling in S. cerevisiae. Biochim Biophys Acta. 2006;1763:646–651. doi: 10.1016/j.bbamcr.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Foster LA. Utilization and cell-surface binding of hemin by Histoplasma capsulatum. Can J Microbiol. 2002;48:437–442. doi: 10.1139/w02-037. [DOI] [PubMed] [Google Scholar]
- 27.Santos R, Buisson N, Knight S, Dancis A, Camadro JM, et al. Haemin uptake and use as an iron source by Candida albicans: role of CaHMX1-encoded haem oxygenase. Microbiology. 2003;149:579–588. doi: 10.1099/mic.0.26108-0. [DOI] [PubMed] [Google Scholar]
- 28.Weissman Z, Kornitzer D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol. 2004;53:1209–1220. doi: 10.1111/j.1365-2958.2004.04199.x. [DOI] [PubMed] [Google Scholar]
- 29.Weissman Z, Shemer R, Conibear E, Kornitzer D. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol Microbiol. 2008;69:201–217. doi: 10.1111/j.1365-2958.2008.06277.x. [DOI] [PubMed] [Google Scholar]
- 30.Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, et al. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008;4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 32.Rutherford JC, Bird AJ. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot Cell. 2004;3:1–13. doi: 10.1128/EC.3.1.1-13.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006;4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, et al. HapX-mediated adaptation to iron starvation is crucial for virulence of Aspergillus fumigatus. PLos Pathog. 2010;6:e1001124. doi: 10.1371/journal.ppat.1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imsande J. Iron-sulfur clusters: Formation, perturbation, and physiological functions. Plant Physiol Biochem. 1999;37:81–97. [Google Scholar]
- 36.Oexle H, Gnaiger E, Weiss G. Iron-dependent changes in cellular energy metabolism: influence on citric acid cycle and oxidative phosphorylation. Biochim Biophys Acta. 1999;1413:99–107. doi: 10.1016/s0005-2728(99)00088-2. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan J, Ward DM, Crisp RJ, Philpott CC. Iron-dependent metabolic remodeling in S. cerevisiae. Biochim Biophys Acta. 2006;1763:646–651. doi: 10.1016/j.bbamcr.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Passarella S, Marra E, Atlante A, Barile M, Doonan S, et al. Uptake of aspartate aminotransferase into mitochondria in vitro causes efflux of malate dehydrogenase and vice versa. Biochim Biophys Acta. 1990;1022:273–282. doi: 10.1016/0005-2736(90)90274-r. [DOI] [PubMed] [Google Scholar]
- 39.Weiss RG, Stern MD, de Albuquerque CP, Vandegaer K, Chacko VP, et al. Consequences of altered aspartate aminotransferase activity on 13C-glutamate labelling by the tricarboxylic acid cycle in intact rat hearts. Biochim Biophys Acta. 1995;1243:543–548. doi: 10.1016/0304-4165(95)00031-6. [DOI] [PubMed] [Google Scholar]
- 40.Ensign SA. Revisiting the glyoxylate cycle: alternate pathways for microbial acetate assimilation. Mol Microbiol. 2006;61:274–276. doi: 10.1111/j.1365-2958.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- 41.Kornberg HL, Krebs HA. Synthesis of cell constituents from C2 units by a modified tricarboxylic acid cycle. Nature (London) 1957;179:988–991. doi: 10.1038/179988a0. [DOI] [PubMed] [Google Scholar]
- 42.Bramer CO, Silva LF, Gomez JG, Priefert H, Steinbuchel A. Identification of the 2-methylcitrate pathway involved in the catabolism of propionate in the polyhydroxyalkanoate-producing strain Burkholderia sacchari IPT101(T) and analysis of a mutant accumulating a copolyester with higher 3-hydroxyvalerate content. Appl Environ Microbiol. 2002;68:271–279. doi: 10.1128/AEM.68.1.271-279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muñoz-Elías EJ, Upton AM, Cherian J, McKinney JD. Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol Microbiol. 2006;60:1109–1122. doi: 10.1111/j.1365-2958.2006.05155.x. [DOI] [PubMed] [Google Scholar]
- 44.Saraste M. Oxidative phosphorylation at the fin de siècle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- 45.Friedman DB, Stauff DL, Pishchany G, Whitwell CW, Torres VJ, et al. Staphylococcus aureus redirects central metabolism to increase iron availability. PLOS Pathog. 2006;2:777–789. doi: 10.1371/journal.ppat.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalapos MP. Methylglyoxal in living organisms. Chemistry, biochemistry, toxicology and biological implications. Toxicol Lett. 1999;110:145–175. doi: 10.1016/s0378-4274(99)00160-5. [DOI] [PubMed] [Google Scholar]
- 47.Hoque A, Uraji M, Banu MN, Mori IC, Nakamura Y, et al. The effects of methylglyoxal on glutathione S-transferase from Nicotiana tabacum. Biosci Biotechnol Biochem. 2010;74:2124–2126. doi: 10.1271/bbb.100393. [DOI] [PubMed] [Google Scholar]
- 48.Barelle CJ, Priest CL, Maccallum DM, Gow NA, Odds FC, et al. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 2006;8:961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fava Netto C, Vegas VS, Sciannamea JJ, Guarnieri DB. Antígeno polissacarídeo do Paracoccidioides brasiliensis. Estudo do tempo de cultura do P. brasiliensis necessário ao preparo do antígeno. Rev Inst Med Trop. 1969;11:177–181. [PubMed] [Google Scholar]
- 50.Bailão AM, Schrank A, Borges CL, Dutra V, Molinari-Madlum EEWI, et al. Differential gene expression by Paracoccidioides brasiliensis in host interaction conditions: Representational difference analysis identifies candidate genes associated with fungal pathogenesis. Microbes Infect. 2006;8:2686–2697. doi: 10.1016/j.micinf.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 51.Andes D, Lepak A, Pitula A, Marchillo K, Clark J. A simple approach for estimating gene expression in Candida albicans directly from a systemic infection site. J Infect Dis. 2005;192:893–900. doi: 10.1086/432104. [DOI] [PubMed] [Google Scholar]
- 52.Restrepo A, Jiménez BE. Growth of Paracoccidioides brasiliensis yeast phase in a chemically defined culture medium. J Clin Microbiol. 1980;12:279–281. doi: 10.1128/jcm.12.2.279-281.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dantas SF, Vieira de Rezende TC, Bailão AM, Taborda CP, da Silva Santos R, et al. Identification and characterization of antigenic proteins potentially expressed during the infectious process of Paracoccidioides brasiliensis. Microbes Infect. 2009;11:895–903. doi: 10.1016/j.micinf.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF. High-throughput real-time quantitative reverse transcription PCR. Curr Protoc Mol Biol. 2006;15:1–28. doi: 10.1002/0471142727.mb1508s73. [DOI] [PubMed] [Google Scholar]
- 55.Fonseca CA, Jesuino RS, Felipe MS, Cunha DA, Brito WA, et al. Two-dimensional electrophoresis and characterization of antigens from Paracoccidioides brasiliensis. Microbes Infect. 2001;3:535–542. doi: 10.1016/s1286-4579(01)01409-5. [DOI] [PubMed] [Google Scholar]
- 56.Bradford MM. A dye binding assay for protein. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 57.Shaw MM, Riederer BM. Sample preparation for two-dimensional gel electrophoresis. Proteomics. 2003;3:1408–1417. doi: 10.1002/pmic.200300471. [DOI] [PubMed] [Google Scholar]
- 58.Herbert B, Galvani M, Hamdan M, Olivieri E, MacCarthy J, et al. Reduction and alkylation of proteins in preparation of two-dimensional map analysis: why, when, and how? Electrophoresis. 2001;22:2046–2057. doi: 10.1002/1522-2683(200106)22:10<2046::AID-ELPS2046>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 59.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 60.Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples). Biometrika. 1965;52:591–611. [Google Scholar]
- 61.Brito WA, Rezende TC, Parente AF, Ricart CAO, de Sousa MV, et al. Identification, characterization and regulation studies of the aconitase of Paracoccidioides brasiliensis. Fungal Biol. doi: 10.1016/j.funbio.2011.02.011. In press. [DOI] [PubMed] [Google Scholar]
- 62.Borges CL, Parente JA, Barbosa MS, Santana JM, Báo SN, et al. Detection of a homotetrameric structure and protein-protein interactions of formamidase lead to new functional insights. FEMS Yeast Res. 2010;10:104–113. doi: 10.1111/j.1567-1364.2009.00594.x. [DOI] [PubMed] [Google Scholar]
- 63.Pereira LA, Báo SN, Barbosa MS, Da Silva JLM, Felipe MSS, et al. Analysis of the Paracoccidioides brasiliensis triosephosphate isomerase suggests the potential for adhesin function. FEMS Yeast Res. 2007;7:1381–1388. doi: 10.1111/j.1567-1364.2007.00292.x. [DOI] [PubMed] [Google Scholar]
- 64.Skouloubris SA, Labigne A, De Reuse H. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol Microbiol. 1997;25:989–998. doi: 10.1111/j.1365-2958.1997.mmi536.x. [DOI] [PubMed] [Google Scholar]
- 65.Aebi H. Assays of catalase in vitro. In: Parker L, editor. Catalase in vitro. Vol. 105. New York: Academic Press; 1984. pp. 122–126. [Google Scholar]
- 66.Ebel F, Schwienbacher M, Beyer J, Heesemann J, Brakhage AA, et al. Analysis of the regulation, expression, and localization of the isocitrate lyase from Aspergillus fumigatus, a potential target for antifungal drug development. Fungal Genet Biol. 2006;43:476–489. doi: 10.1016/j.fgb.2006.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Iron starvation does not affect the viability of P. brasiliensis yeast cells. Viability of P. brasiliensis yeast cells incubated in MMcM medium containing 3.5 µM iron (circles) and incubation of yeasts cells in MMcM iron depleted media containing 50 µM BPS (triangles). Viability was determined using trypan blue. Error bars represent standard deviation from three biological replicates while * represents p≤0.05.
(TIF)
Categorical representation of regulated P. brasiliensis proteins following iron starvation. Identified proteins were classified according to their respective functional categories determined by MIPS. (A) Categorization of differentially expressed proteins during iron starvation. Classification of proteins with induced (B) and repressed expression (C) in iron limiting condition.
(TIF)
Relative fold induction of iron metabolism related genes in the time course of iron starvation using real time- RT-PCR. 1Values represent the mean of each triplicate sample ± standard deviation.
(DOC)
Additional information about P. brasiliensis identified proteins with increased expression during iron starvation. ** Spots visualized only in iron-depleted condition. 1 Spots numbers refers to Figure 3. 2 p values were accessed by ANOVA statistical test.
(DOC)
Additional information about P. brasiliensis identified proteins with reduced expression during iron starvation. ** Spots visualized only in iron replete condition. 1 Spots numbers refers to Figure 3. 2 p values were accessed by ANOVA statistical test.
(DOC)
Oligonucleotide primers used in quantitative RT-PCR.
(DOC)