Abstract
Background
Posttraumatic stress disorder (PTSD) is a common and debilitating mental disorder that occurs following exposure to a traumatic event. However, most individuals do not develop PTSD following even a severe trauma, leading to a search for new variables—such as genetic and other molecular variation— associated with vulnerability and resilience in the face of trauma exposure.
Method
We examined whether serotonin transporter (SLC6A4) promoter genotype and methylation status modified the association between number of traumatic events experienced and PTSD in a subset of 100 individuals from the Detroit Neighborhood Health Study.
Results
Number of traumatic events was strongly associated with risk of PTSD. Neither SLC6A4 genotype or nor methylation status were associated with PTSD in main effects models. However, SLC6A4 methylation levels modified the effect of number of traumatic events on PTSD after controlling for SLC6A4 genotype. Persons with more traumatic events were at increased risk for PTSD but only at lower methylation levels. At higher methylation levels, individuals with more traumatic events were protected from this disorder. This interaction was observed whether the outcome was PTSD diagnosis, symptom severity, or number of symptoms.
Conclusions
Gene-specific methylation patterns may offer potential molecular signatures of increased risk for and resilience to PTSD.
Keywords: posttraumatic stress disorder, epigenetic, methylation, SLC6A4, trauma
Introduction
Posttraumatic stress disorder (PTSD) occurs following exposure to a traumatic event and is characterized by three symptom clusters: reexperiencing, avoidance and numbing, and hyperarousal. PTSD is distinct from other commonly occurring mental disorders in that exposure to a traumatic stressor is a prerequisite for diagnosis. Although a majority[1] of adults in the United States have been exposed to traumatic events, only a minority go on to develop the disorder: lifetime rates of PTSD are estimated at approximately 6.8%, and 12-month prevalence rates at approximately 3.5%[2, 3]. Even among individuals exposed to objectively high levels of trauma, the conditional risk of PTSD is less than 50%[1]. The contrast between the prevalence of trauma exposure and the prevalence of PTSD has driven the search for risk and protective factors - beyond burden of trauma exposure - that influence vulnerability and resilience to developing the disorder. However, the traditional risk factor models supported by meta-analytic studies explain only about 20% of the interpersonal variance in PTSD[4]. The limitations of established risk factor models has led to a search for new variables - such as genetic and other molecular variation – associated with vulnerability and resilience in the face of trauma exposure.
The serotonin transporter (SLC6A4) locus has been the most commonly studied gene in relation to PTSD. The protein encoded by SLC6A4 serves many functions; however, its action has been particularly well-studied in the brain where it transports serotonin at synaptic terminals and other neuronal areas[5] and serves to regulate emotional aspects of behavior[6]. Previous work on SLC6A4 has examined whether DNA sequence variation in this gene is associated with PTSD diagnosis or symptoms. Of the total of 40 published candidate gene association studies for PTSD, 9 have focused on SLC6A4 and specifically on the commonly occurring VNTR polymorphism found in the promoter region (5-HTTLPR)[7]. The alleles in the promoter region of this gene have traditionally been classified as short (s) and long (l), with the latter conferring higher expression than the former[8] and the former being associated with greater amygdala activity to fearful stimuli[9]. Published findings for this locus and PTSD have been somewhat contradictory with 6 of 9 studies finding the s-allele or s/s genotype associated with elevated risk of PTSD diagnosis[10–14] or symptoms[15], two studies finding association between the high expression variant and PTSD[16, 17], and one study finding no evidence for an association[18]. Importantly, four of the 9 studies involving this locus report evidence for a significant genotype by environment interaction[11, 12, 14, 17] whereby the effect of genotype on risk of PTSD was stronger among individuals under high stress (versus low stress) conditions. Taken together, these studies provide evidence for the role of SLC6A4 in the etiology of PTSD. However, the picture presented is complex in that the association appears to be modified by environmental conditions and other factors.
The complexity of the association between SLC6A4 and PTSD may be related, in part, to emerging evidence suggesting that methylation at CpG sites (downstream of the 5-HTTLPR region) contributes to SLC6A4 expression. DNA methylation is one of the major mechanisms of epigenetic regulation or the regulation of genetic functions mediated through mechanisms that are independent of DNA sequences. DNA methylation occurs in vertebrates predominantly through covalent modification of DNA, whereby methyl groups are coupled to cytosine residues when cytosine and guanine are separated by a phosphate (i.e., at a CpG site)[19]. Methylation involves chemical modifications that regulate DNA accessibility, which in turn alters the transcriptional activity of the surrounding loci. In many cases, increased methylation in specific gene regions (e.g. promoter) is associated with reduced transcriptional activity and, therefore, gene expression.
Work by Philbert et al using samples from the Iowa Adoption Studies cohort provided initial evidence that increased methylation levels in the CpG island overlapping with the transcriptional start site of SLC6A4 was associated with decreased levels of SLC6A4 RNA, and that those with the 5-HTTLPR s allele showed a trend toward higher methylation levels across CpG sites located in this upstream island[20, 21]. This same group showed that child abuse was associated with significantly elevated methylation levels across multiple CpG sites and, among females, at specific CpG sites as well[22]. Taken together, these Iowa Adoption study results are suggestive of a relation between SLC6A4 methylation and stress-related outcomes. Nevertheless, one limitation of these studies is the use of DNA derived from EBV-transformed lymphoblast cell lines, which have been demonstrated to undergo alterations in methylation status with increasing cell passages[23].
Studies of both SLC6A4 genotype and methylation suggest this gene may be salient to the development of stress-related outcomes. This paper examines whether methylation status of the serotonin transporter gene (SLC6A4) modifies the association between number of traumatic events and risk of PTSD. To address this we tested whether SLC6A4 genotype and methylation levels were associated with PTSD and modified the effect of number of traumatic events on risk for PTSD. Samples were drawn from100 individuals in the Detroit Neighborhood Health Study (DNHS) using microarray-derived methylation data for two CpG sites upstream of SLC6A4. The first CpG site (cg05016953) occurs within a 799 bp CpG[24] island that overlaps with the first exon of SLC6A4[25], upstream of the gene's predicted transcription start sites but downstream of the 5-HTTLPR VNTR locus[26]. The second site (cg22584138) occurs within the first intron of SLC6A4, upstream of the gene's start codon but downstream of the gene's predicted start sites, CpG island, and 5-HTTLPR locus.
Materials and Methods
Detroit Neighborhood Health Study
The DNHS is a study of adults, 18 years or older, from the Detroit population. A probability sample of 1,547 households within the city limits of Detroit was initially chosen and one individual per household was then randomly selected for interview. Participants were administered a 40-minute assessment which included questions on exposure to traumatic events, socio-demographic characteristics, and a standardized assessment of PTSD and depression. The DNHS was approved by the Institutional Review Board at the University of Michigan.
Respondents were also asked to provide blood specimen by way of venipuncture; 612 samples were collected from consenting participants. We performed a two-tailed chi-square test to determine if participants who provided a blood sample (n=612) were significantly different from the total sample (n=1,547); results show that the socio-demographic characteristics of the consenting participants of the blood draw were comparable to the complete sample.
The sample for this study consisted of 100 of these 612 consenting participants. Descriptive statistics for the sample are presented in Table 1. We compared the 100 individuals in our final sample to the 612 consenting participants of the blood draw and found that the two samples differ only on age – our final sample consisted of slightly higher proportion of younger individuals.
Table 1.
Descriptive statistics and bivariate comparisons of participants with and without PTSD
| Overall n/mean | Sample %/sd | Trauma exposed/no PTSD (n=77) | PTSD (n=23) | Test p-value | |||
|---|---|---|---|---|---|---|---|
| n/mean | %/sd | n/mean | %/sd | ||||
| SLC6A4 methylation beta-value | 0.35 | 0.13 | 0.35 | 0.13 | 0.33 | 0.12 | 0.50 |
| Number of traumatic events | 6.04 | 3.6 | 5.58 | 3.48 | 7.57 | 3.65 | 0.02 |
| Age | 45.32 | 16.78 | 44.91 | 17.29 | 46.70 | 15.19 | 0.66 |
| Female | 60 | 60 | 45 | 58.4 | 15 | 65.2 | 0.56 |
| African American | 79 | 79 | 62 | 80.5 | 17 | 73.9 | 0.56 |
| Education equal to or greater than high school | 86 | 86 | 67 | 87.01 | 19 | 82.61 | 0.73 |
| Ever smoke | 58 | 58 | 41 | 53.3 | 17 | 73.9 | 0.08 |
| PBMC | 23.34 | 8.05 | 23.35 | 8.31 | 23.29 | 7.25 | 0.97 |
| Depression diagnosis | 33 | 33 | 21 | 27.30 | 12 | 52.20 | 0.03 |
| SLC6A4genotype (functional classification) SS, SLg, LgLg | 21 | 21 | 17 | 22.10 | 4 | 17.40 | 0.70 |
| SLa, LgLa | 46 | 46 | 33 | 42.9 | 13 | 56.5 | |
| LaLa | 29 | 29 | 24 | 31.20 | 5 | 21.70 | |
| LaXL, LgXL | 4 | 4 | 3 | 3.90 | 1 | 4.40 | |
Note: PBMC = peripheral blood mononuclear cells
Number of traumatic events
Participants were initially asked to identify traumatic events that they experienced from a list of 19. These included: military combat, rape, other sexual assault, shot/stabbed, held captive/tortured/kidnapped, mugged/held up/threatened with a weapon, badly beat up, serious car or motor vehicle crash, any other kind of serious accident or injury, fire, flood, earthquake or other natural disaster, diagnosed with a life threatening illness, witnessed someone being killed or seriously injured, unexpectedly discovering a dead body, learning about a loved one being raped, seriously physically attacked, seriously injured in a motor vehicle crash or seriously injured in any other accident, sudden unexpected death of a close friend or relative. Number of traumatic events was a count of the different types of traumat event and ranged from 0–19 for each person.
Assessment of posttraumatic stress disorder
Individual assessment of PTSD symptoms was conducted via telephone interview using a modified version of the PTSD checklist (PCL-C), a 17-item self-report measure of Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) symptoms, and additional questions about duration, timing, and impairment or disability due to the symptoms. PTSD symptoms were then assessed by referencing the event that the participant regarded as the worst. Respondents were considered affected by lifetime PTSD if all six DSM-IV criteria were met in reference to either the worst event. All 100 individuals included in this study were exposed to at least one traumatic event; among these, 23 were PTSD-affected and 77 were unaffected.
Assessment of lifetime depression
Individual assessment of lifetime depression was conducted using a modified version of the Patient Health Questionnaire checklist (PHQ-9), a 9-item self-report measure based on the DSM-IV criteria[27]. Each of the nine symptoms was scored from 0 (not at all) to 3 (nearly every day). Depression was considered present when 2 or more symptoms have been present at least “more than half the days” in a two week period, and one of the symptoms is depressed mood, or anhedonia, and the respondent reported that the symptoms seem to occur together. One symptom, “thoughts that you would be better off dead or of hurting yourself in some way” counts if at all present, regardless of frequency/duration.
Validation of posttraumatic stress disorder and depression
We assessed the validity of our identification of PTSD and depression via in-person clinical interviews among a random subsample of 51 participants[28]. As described previously, a trained counselor conducted one-hour in-person clinical interviews, using the Clinician-Administered PTSD Scale for DSM-IV for PTSD[29] and the Structured Clinical Interview for DSM-IV Disorders[30]. The counselor was blinded to the information obtained from the main study. Comparison of the clinical interviews with the main study showed excellent concordance for both PTSD and depression[28].
Microarray Analyses
DNA was isolated from whole blood using the Qiagen (Valencia, CA) QIAamp® DNA Mini Kit. Bisulfite conversion of whole blood-derived DNA samples was performed using the EZ-96 DNA methylation kit from Zymo Research (Orange, CA). One microgram (μg) of each sample (including controls) was subjected to bisulfite conversion following manufacturer's recommended protocol. Experimental controls included replicates for two samples to assess variation throughout the experimental process (i.e. from initial bisulfite conversion through microarray analysis), as well as one sample of completely unmethylated and completely methylated human DNA, commercially available through Zymo Research, in each of the two 96 well plates used in the bisulfite conversion step. All control replicates were placed on separate microarray chips, and the remaining samples were assigned to microarray chips at random, without regard to PTSD status. Bisulfite converted DNA samples were subjected to methylation profiling via the humanmethylation27 DNA Analysis BeadChip by Illumina following the manufacturer's recommended protocol. Using this platform, methylation levels were determined for 27,578 CpG dinucleotides spanning 14,495 genes in each of the 100 test samples. The resulting data were background normalized using Bead Studio. Correlation coefficients of the two replicated samples were 0.81 and 0.89, respectively. Methylation microarray data were initially validated via pyrosequencing and DNA sequencing of a subset of individuals tested on the original microarray and are reported in detail elsewhere[28].
Methylation of SLC6A4 was assessed at two CpG sites represented on the HM27 beadchip. The first CpG site (cg05016953) occurs within a 799 bp CpG[24] island that overlaps with the first exon of SLC6A4[25], upstream of the gene's predicted transcription start sites but downstream of the 5-HTTLPR VNTR locus[26]. The second site (cg22584138) occurs within the first intron of SLC6A4, upstream of the gene's start codon but downstream of the gene's predicted start sites, CpG island, and 5-HTTLPR locus. Methylation beta values of <0.2 and >0.8 have previously been characterized as unmethylated and methylated, respectively[28].
Pyrosequencing validation
We used locus-specific pyrosequencing to validate methylation data at at cg22584138. Pyrosequencing assays were designed and implemented by EpigenDx (Worcester, MA). We observed a highly significant (r = .45, p < .001) correlation between the methylation value at the same locus in the the pyrosequencing assay as from the Illumina Beadchip based on data from 85 of the original 100 individuals tested in the microarray analysis.
Genotyping
Samples were initially genotyped using the method described for genotyping the serotonin transporter promoter polymorphism in Gelertner et al[31]. Genotyping in this initial phase was performed using Qiagen©'s Taq PCR Core Kit and associated protocols, along with the cycling parameters of 94°C initial at 2 minutes, followed by 33 cycles of: 94°C denature at 15 seconds, 66°C annealing temperature at 15 seconds and 72°C extension at 30 seconds; and a final temperature at 72°C at 5 minutes. PCR products were then visualized as short (S) or long (L) or extra long (XL) alleles through visualization on a 2% agarose gel stained with ethidium bromide. Amplification and visualization were conducted at least twice in each individual in order to accurately call each genotype. In order to further discern the functionally distinct La and Lg alleles[32], all samples from individuals with non-homozygote SS genotypes (n=87) were subjected to a second phase of analysis. Samples in this second phase were amplified using Takara LA Taq™ polymerase with the manufacturers' buffer and dNTPs, again using primers previously reported[31]. Thermocycling conditions included a 94°C initial at 1 minute, followed by 33 cycles of: 94°C denature at 30 seconds, 66°C annealing temp at 30 seconds and 72°C extension at 2 minutes; and a final temperature at 72°C for 5 minutes. Amplified products were then subjected to restriction enzyme digestion with MspI[33]. Digested products were then size fractionated and visualized using a 2% agarose gel. For analyses, genotypes were grouped according to functional classification as 1) SS, SLg, LgLg (low), 2) SLa, LgLa (intermediate), 3) LaLa (high). The function of XL is unknown, therefore, one individual with LaXL was classified as “high” and three individuals as LgXL were classified as low.
Analysis
Data were analyzed using R v2.10.0 and SAS v9.2. DNA methylation beta values are continuous variables between 0 (completely unmethylated) and 1 (completely methylated). Initially, variance in methylation was assessed at each CpG site. Next, bivariate associations were assessed for methylation and PTSD for each of the variables of interest. Main effects and interaction effects models were then fitted to establish the relationships of the predictors to PTSD. In order to ensure our results were robust to quantitative and qualitative assessments of PTSD, we modeled the PTSD outcome in three ways. PTSD diagnosis based on the DSM-IV criteria was modeled using logistic regression. PTSD severity is the aggregate of intrusion, avoidance, and hyper-arousal symptom responses to “how much bothered are you by this?” and was modeled using the general linear model. The severity measure was log-transformed for normality. The number of symptoms is the sum of symptoms that met the criteria of being present and bothered by it at least “moderately” was modeled using negative binomial regression. Continuous variables such as age, PBMC count, number of traumatic events, and methylation beta values were centered to the mean. All predictor variables were maintained in all the models for consistency. Estimated coefficients were evaluated at α = 0.05.
Results
Initial results revealed substantial variation in methylation beta-values at cg22584138 but not cg05016953 (data not shown). Thus, statistical analyses focused on cg22584138. Table 1 presents the descriptive statistics and bivariate results for participants with and without PTSD. Participants with PTSD reported exposure to a significantly greater number of traumatic events and were significantly more likely to have met lifetime criteria for depression. We found no significant association between SLC6A4 genotype or methylation beta values and PTSD. Methylation at cg22584138 was significantly associated with female sex (p=.04), and number of traumatic events (p=.04), but not age, PBMCs, race, SES, smoking, depression or SLC6A4 genotype (data not shown).
Table 2 presents results from the multivariable logistic regression models for PTSD diagnosis. In the main effects model, no significant associations were observed. In the interaction model, the interaction term for methylation X number of traumatic events was significant (p = .036). The interaction model also provided a significantly improved fit over the main effects model (χ2(1) = 5.649, p = .02 ). The interaction for SLC6A4 genotype X number of traumatic events was tested but was not significant (data not shown).
Table 2.
Effect of SLC6A4 methylation and number of traumatic events on risk of lifetime posttraumatic stress disorder diagnosis
| Main Effects Model −2 Log L = 93.268 | Interaction Model −2 Log L = 87.619 | |||||||
|---|---|---|---|---|---|---|---|---|
| b | p | 95% CI of b | b | p | 95% CI of b | |||
| Intercept | −2.7355 | 0.0074 | −4.7366 | 0.7343 | −2.5973 | 0.0113 | −4.6059 | −0.5886 |
| centered SLC6A4 methylation beta-value | −2.9250 | 0.1878 | −7.2777 | 1.4278 | −0.3743 | 0.8845 | −5.4253 | 4.6768 |
| Number of traumatic events | 0.1218 | 0.1159 | −0.0300 | 0.2736 | 0.1638 | 0.0669 | −0.0114 | 0.3391 |
| Age | 0.00329 | 0.8489 | −0.0306 | 0.0372 | 0.00612 | 0.7397 | −0.0300 | 0.0422 |
| sex | 0.5428 | 0.3622 | −0.6247 | 1.7103 | 0.7154 | 0.2500 | −0.5036 | 1.9344 |
| African American | −0.1657 | 0.8044 | −1.4775 | 1.1461 | −0.4456 | 0.5290 | −1.8328 | 0.9416 |
| Ever smoke | 1.0813 | 0.0792 | −0.1262 | 2.2888 | 1.0194 | 0.1136 | −0.2435 | 2.2824 |
| PBMC | −0.0120 | 0.6990 | −0.0726 | 0.0487 | −0.00530 | 0.8644 | −0.0662 | 0.0556 |
| Depression diagnosis | 1.0446 | 0.0597 | −0.0428 | 2.1320 | 1.1343 | 0.0517 | −0.00854 | 2.2772 |
| SLC6A4 sl genotype | 0.3284 | 0.6004 | −0.9005 | 1.5573 | 0.2726 | 0.6752 | −1.0026 | 1.5479 |
| SLC6A4 ss genotype | −0.1116 | 0.8864 | −1.6427 | 1.4194 | 0.1072 | 0.8946 | −1.4781 | 1.6924 |
|
|
||||||||
| SLC6A4 methylation * number of traumatic events | −1.5596 | 0.0360 | −3.0170 | −0.1022 | ||||
Note: PBMC = peripheral blood mononuclear cells
Note: 's' allele includes S and Lg; 'l' allele includes La and XL
Note: continuous variables were centered to the mean
Table 3 presents results from the multivariable linear regression models for PTSD symptom severity. In the main effects model, sex, number of traumatic events, and lifetime depression were associated with greater PTSD symptom severity. In the interaction model, the interaction term for methylation X number of traumatic events was significant (p = .04). Based on the adjusted R2, the interaction model also explained 3% more of the variance in PTSD symptom severity than the main effects model. The interaction for SLC6A4 genotype X number of traumatic events was tested but was not significant (data not shown).
Table 3.
Effect of SLC6A4 methylation and number of traumatic events on risk of lifetime posttraumatic stress disorder symptom severity
| Main Effects Model Adjusted R-square = 0.2791 | Interaction Model Adjustted R-square = 0.3051 | |||||||
|---|---|---|---|---|---|---|---|---|
| b | p | 95% CI of b | b | p | 95% CI of b | |||
| Intercept | 3.17171 | <.0001 | 2.90733 | 3.43608 | 3.19094 | <.0001 | 2.93069 | 3.45120 |
| centered SLC6A4 methylation beta-value | −0.08024 | 0.7899 | −0.67686 | 0.51638 | 0.11063 | 0.7210 | −0.50296 | 0.72422 |
| Number of traumatic events | 0.04389 | 0.0001 | 0.02192 | 0.06585 | 0.04848 | <.0001 | 0.02647 | 0.07050 |
| Age | −0.00186 | 0.4215 | −0.00642 | 0.00271 | −0.00158 | 0.4873 | −0.00607 | 0.00291 |
| sex | 0.16367 | 0.0439 | 0.00457 | 0.32277 | 0.18439 | 0.0223 | 0.02691 | 0.34188 |
| African American | 0.09231 | 0.3226 | −0.09207 | 0.27669 | 0.07555 | 0.4110 | −0.10621 | 0.25732 |
| Ever smoke | 0.11959 | 0.1441 | −0.04165 | 0.28083 | 0.10766 | 0.1812 | −0.05108 | 0.26641 |
| PBMC | 0.00700 | 0.1331 | −0.00217 | 0.01617 | 0.00745 | 0.1041 | −0.00157 | 0.01647 |
| Depression diagnosis | 0.18103 | 0.0262 | 0.02197 | 0.34009 | 0.17302 | 0.0305 | 0.01664 | 0.32940 |
| SLC6A4 sl genotype | 0.13842 | 0.1144 | −0.03408 | 0.31091 | 0.13951 | 0.1053 | −0.02988 | 0.30890 |
| SLC6A4 ss genotype | 0.15616 | 0.1327 | −0.04831 | 0.36063 | 0.18129 | 0.0783 | −0.02093 | 0.38350 |
|
|
||||||||
| SLC6A4 methylation * number of traumatic events | −0.15546 | 0.0404 | −0.30398 | −0.00694 | ||||
Note: PBMC = peripheral blood mononuclear cells
Note: 's' allele includes S and Lg; 'l' allele includes La and XL
Note: continuous variables were centered to the mean
Table 4 presents results from the multivariable binomial regression models for number of PTSD symptoms. In the main effects model, sex and number of traumatic events was associated with more PTSD symptoms. In the interaction model, the interaction term for methylation X number of traumatic events was significant (p = .018). The interaction model also provided a significantly improved fit over the main effects model (χ2(1) = 5.32, p = .02 ). The interaction for SLC6A4 genotype X number of traumatic events was tested but was not significant (data not shown).
Table 4.
Effect of SLC6A4 methylation and number of traumatic events on risk of lifetime number of posttraumatic stress disorder symptoms
| Main Effects Model −2 Log L = 540.0076 | Interaction Model −2 Log L = 543.69 | |||||||
|---|---|---|---|---|---|---|---|---|
| b | p | 95% CI of b | b | p | 95% CI of b | |||
| Intercept | 0.6600 | 0.0406 | 0.0283 | 1.2916 | 0.6755 | 0.0340 | 0.0509 | 1.3001 |
| centered SLC6A4 methylation beta-value | −0.0361 | 0.9590 | −1.4097 | 1.3375 | 0.6208 | 0.4017 | −0.8301 | 2.0718 |
| Number of traumatic events | 0.1012 | 0.0001 | 0.0498 | 0.1526 | 0.1170 | <.0001 | 0.0659 | 0.1682 |
| Age | −0.0027 | 0.6290 | −0.0135 | 0.0082 | −0.0017 | 0.7513 | −0.0123 | 0.0089 |
| sex | 0.3853 | 0.0407 | 0.0164 | 0.7542 | 0.4196 | 0.0230 | 0.0578 | 0.7814 |
| African American | 0.4105 | 0.0764 | −0.0435 | 0.8644 | 0.3844 | 0.0890 | −0.0586 | 0.8273 |
| Ever smoke | 0.1836 | 0.3346 | −0.1894 | 0.5566 | 0.1649 | 0.3707 | −0.1961 | 0.5258 |
| PBMC | 0.0110 | 0.2867 | −0.0093 | 0.0314 | 0.0130 | 0.2035 | −0.0070 | 0.0330 |
| Depression diagnosis | 0.3553 | 0.0576 | −0.0115 | 0.7221 | 0.3737 | 0.0411 | 0.0152 | 0.7321 |
| SLC6A4 sl genotype | 0.4177 | 0.0426 | 0.0140 | 0.8215 | 0.4108 | 0.0419 | 0.0150 | 0.8067 |
| SLC6A4 ss genotype | 0.3265 | 0.1801 | −0.1509 | 0.8039 | 0.3803 | 0.1128 | −0.0897 | 0.8503 |
|
|
||||||||
| SLC6A4 methylation * number of traumatic events | −0.4122 | 0.0184 | −0.7548 | −0.0695 | ||||
Note: PBMC = peripheral blood mononuclear cells
Note: 's' allele includes S and Lg; 'l' allele includes La and XL
Note: continuous variables were centered to the mean
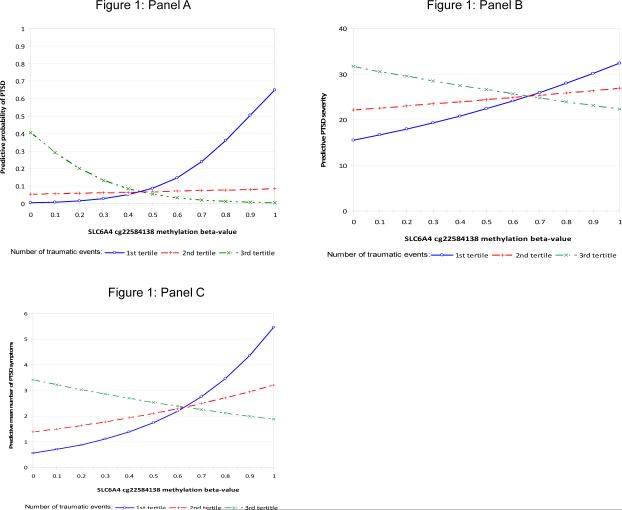
Figure 1 presents the interactions for number of traumatic events by methylation beta-value for the predicted PTSD prevalence (Figure 1A), number of symptoms (Figure 1B), and symptom severity (Figure 1C). Exposure to a greater number of traumatic events was associated with increased risk of PTSD diagnosis, greater number of symptoms, and greater severity of symptoms at low levels of SLC6A4 methylation. In contrast, at high levels of SLC6A4 methylation, exposure to a greater number of traumatic events was associated with resilience to developing PTSD.
Figure 1.
The prevalence, number of symptoms, and symptom severity of posttraumatic stress disorder (PTSD) by number of traumatic events and methylation beta-value. The results show that the association between number of traumatic events and PTSD diagnosis is modified by SLC6A4 methylation. Exposure to a greater number of potentially traumatic events (PTEs) is associated with increased risk of PTSD diagnosis (logistic), greater number of symptoms (linear), and greater severity of symptoms (negative binomial) at low levels of SLC6A4 methylation; in contrast, at high levels of SLC6A4 methylation, exposure to a greater number of traumatic events is associated with resilience to developing PTSD. Methylation beta values of <0.2 and >0.8 have previously been characterized as unmethylated and methylated, respectively[28].
Discussion
We found that number of traumatic events was associated with PTSD diagnosis in this subsample of the DNHS. However, this association was modified by SLC6A4 methylation level at cg22584138, a CpG site within the first intron of SLC6A4 located upstream of the gene's start codon but downstream of transcription start sites and the 5-HTTLPR VNTR locus. Specifically, number of traumatic events was strongly associated with risk of PTSD but only at lower methylation levels: at higher methylation levels, individuals with more traumatic events were protected from this disorder. This interaction was observed whether the outcome was PTSD diagnosis, symptom severity or number of symptoms. These results suggest that methylation levels at SLC6A4 modify the effects of traumatic events in a manner salient to PTSD etiology, offering a potential molecular signature of increased risk for and resilience to this disorder. In contrast to some prior studies, neither the main effect of SLC6A4 genotype nor the interaction of SLC6A4 genotype and number of traumatic events was related to PTSD[7]. Future work in this longitudinal cohort will help to shed light on the extent to which such epigenetic marks modify–or are modified by–traumatic stress.
Notably, variation in methylation levels was observed only for one of the two CpG sites examined (cg22584138). This site differs in location from previous reports of SLC6A4 methylation in relation to other stress-related outcomes[20–22]. Our observations of disease-associated methylation differences in the first intron of the gene are not inconsistent with other reports of methylation-mediated gene expression differences in other diseases[34]. In addition, the role of noncoding, intronic DNA is coming under increasing scrutiny for its role in regulating gene expression[35]. For these reasons, our results offer a potentially novel site for future investigations of stress-related outcomes involving the SCL6A4 locus as well as point to the potential for targeting methylation- mediated gene regulation.
Study Limitations
Our study includes five limitations that should be considered. First, the cross-sectional analyses reported here leave us unable to determine whether the SLC6A4 methylation differences were a consequence of PTSD or whether they are indicative of biologic vulnerabilities that existed among the PTSD-affected prior to the onset of their disorder. Ongoing work using samples from this same longitudinal cohort should help to shed light on this issue. Second, our necessary reliance on peripheral tissues may limit the inferences that can be drawn about SLC6A4 methylation in the brain. Nevertheless, the use of blood-derived tissues for use in assessing psychiatric disorders is increasingly being recognized[36] and a growing body of research is documenting correspondence between brain- and blood derived gene expression signals[37]. Furthermore, recent work in nonhuman primates confirms that SLC6A4 shows concordant methylation levels in the blood and brain [38], suggesting that results presented here may have relevance to the “target organ” of PTSD. Third, data were unavailable to assess the correlation between methylation and gene expression in the same individuals for this locus, limiting our ability to infer the proximal phenotypic impact (i.e. gene expression differences) of the observed interaction. Fourth, PTSD in this sample, as in the general population, is highly comorbid with depression. We attempted to address this issue by adjusting for depression in our models. However, due to our small sample size, we did not have power to test whether our findings were consistent among those with PTSD along versus those with PTSD and depression. Finally, our sample size (n=100) is small. Nonetheless, we did observe statistically significant results across PTSD diagnosis, number of symptoms, and symptom severity suggesting that the findings reported here are robust. Future work in other, independent cohorts, are warranted to confirm these initial findings.
Conclusions
Posttraumatic stress disorder becomes chronic in over half of the individuals who develop the disorder[39]. Despite three decades of research, we still have limited understanding of why, among those exposed to trauma, only some individuals will develop PTSD. Our findings suggest that gene-specific methylation patterns may offer potential molecular signatures of increased risk for and resilience to this disorder. Future work in our longitudinal cohort will help to shed light on the mechanisms underlying our observations and the extent to which such epigenetic marks modify–or are modified by–traumatic stress.
Acknowledgements
We thank Rebecca M. Coulborn for overseeing DNHS specimen collection, Janie Slayden for coordinating the overall DNHS project, and Amy Weckle and Richelo Soliven for handling the DNHS specimen processing and laboratory technical assistance; the many Detroit residents who chose to participate in the DNHS; and Jorge Delva, Larry Gant, Bob Marans, and Trivellore Raghunathan for contributing to the conceptual development of the DNHS. We also thank Dr. Sue Land and staff (Wayne State University, Applied Genomic Technology Center) for running microarrays. This study was supported by National Institutes of Health Grants DA022720, DA022720-S1, MH088283, MH078152, and MH082729 (to S.G. & A.A.) andMH070627 and MH078928 (to K.K.).Additional support was provided by the Robert Wood Johnson Health and Society Scholars Small Grant Program and the University of Michigan Office of the Vice President for Research Faculty Grants and Awards Program (M.U.); and by the Wayne State University Research Excellence Fund (D.W.).
Footnotes
Disclosures: The authors have no disclosures to report.
References
- 1.Kessler RC, Sonnega A, Bromet E, Hughes M, Nelson CB. Posttraumatic stress disorder in the National Comorbidity Survey. Arch Gen Psychiatry. 1995;52:1048–60. doi: 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- 2.Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 3.Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:617–27. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozer EJ, Best SR, Lipsey TL, Weiss DS. Predictors of posttraumatic stress disorder and symptoms in adults: A meta-analysis. Psychol Bull. 2003;129:52–73. doi: 10.1037/0033-2909.129.1.52. [DOI] [PubMed] [Google Scholar]
- 5.Tao-Cheng JH, Zhou FC. Differential polarization of serotonin transporters in axons versus soma-dendrites: an immunogold electron microscopy study. Neuroscience. 1999;94:821–30. doi: 10.1016/s0306-4522(99)00373-5. [DOI] [PubMed] [Google Scholar]
- 6.Meyer-Lindenberg A. Neural connectivity as an intermediate phenotype: brain networks under genetic control. Hum Brain Mapp. 2009;30:1938–46. doi: 10.1002/hbm.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis MC, Nugent NR, Amstadter AB, Koenen KC. Genetics of posttraumatic stress disorder: Review and recommendations for genome-wide association studies. Current Psychiatry Reports. doi: 10.1007/s11920-010-0126-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–31. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 9.Munafo MR, Brown SM, Hariri AR. Serotonin transporter (5-HTTLPR) genotype and amygdala activation: a meta-analysis. Biol Psychiatry. 2008;63:852–7. doi: 10.1016/j.biopsych.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee HJ, Lee MS, Kang RH, Kim H, Kim SD, Kee BS, et al. Influence of the serotonin transporter promoter gene polymorphism on susceptibility to posttraumatic stress disorder. Depress Anxiety. 2005;21:135–9. doi: 10.1002/da.20064. [DOI] [PubMed] [Google Scholar]
- 11.Kilpatrick DG, Koenen KC, Ruggiero KJ, Acierno R, Galea S, Resnick HS, et al. The serotonin transporter genotype and social support and moderation of posttraumatic stress disorder and depression in hurricane-exposed adults. Am J Psychiatry. 2007;164:1693–9. doi: 10.1176/appi.ajp.2007.06122007. [DOI] [PubMed] [Google Scholar]
- 12.Koenen KC, Aiello AE, Bakshis E, Amstadter AB, Ruggiero KJ, Acierno R, et al. Modification of the association between serotonin transporter genotype and risk of posttraumatic stress disorder in adults by county-level social environment. Am J Epidemiol. 2009;169:704–11. doi: 10.1093/aje/kwn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolassa I, Ertl V, Eckart M, Glockner F, Kolassa S, Papassotiropoulos A, et al. Association study of trauma load and SLC6A4 promoter polymorphism in PTSD: evidence from survivors of the Rwandan genocide. J Clin Psychiatry. doi: 10.4088/JCP.08m04787blu. in press. [DOI] [PubMed] [Google Scholar]
- 14.Xie P, Kranzler HR, Poling J, Stein MB, Anton RF, Brady K, et al. Interactive effect of stressful life events and the serotonin transporter 5-HTTLPR genotype on posttraumatic stress disorder diagnosis in 2 independent populations. Arch Gen Psychiatry. 2009;66:1201–9. doi: 10.1001/archgenpsychiatry.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sayin A, Kucukyildirim S, Akar T, Bakkaloglu Z, Demircan A, Kurtoglu G, et al. A prospective study of serotonin transporter gene promoter (5-HTT gene linked polymorphic region) and intron 2 (variable number of tandem repeats) polymorphisms as predictors of trauma response to mild physical injury. DNA Cell Biol. 29:71–7. doi: 10.1089/dna.2009.0936. [DOI] [PubMed] [Google Scholar]
- 16.Thakur GA, Joober R, Brunet A. Development and persistence of posttraumatic stress disorder and the 5-HTTLPR polymorphism. J Trauma Stress. 2009;22:240–3. doi: 10.1002/jts.20405. [DOI] [PubMed] [Google Scholar]
- 17.Grabe HJ, Spitzer C, Schwahn C, Marcinek A, Frahnow A, Barnow S, et al. Serotonin transporter gene (SLC6A4) promoter polymorphisms and the susceptibility to posttraumatic stress disorder in the general population. Am J Psychiatry. 2009;166:926–33. doi: 10.1176/appi.ajp.2009.08101542. [DOI] [PubMed] [Google Scholar]
- 18.Mellman TA, Alim T, Brown DD, Gorodetsky E, Buzas B, Lawson WB, et al. Serotonin polymorphisms and posttraumatic stress disorder in a trauma exposed African American population. Depress Anxiety. 2009;26:993–7. doi: 10.1002/da.20627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 20.Philibert R, Madan A, Andersen A, Cadoret R, Packer H, Sandhu H. Serotonin transporter mRNA levels are associated with the methylation of an upstream CpG island. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:101–5. doi: 10.1002/ajmg.b.30414. [DOI] [PubMed] [Google Scholar]
- 21.Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, Madan A. The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:543–9. doi: 10.1002/ajmg.b.30657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beach SR, Brody GH, Todorov AA, Gunter TD, Philibert RA. Methylation at SLC6A4 is linked to family history of child abuse: an examination of the Iowa Adoptee sample. Am J Med Genet B Neuropsychiatr Genet. 153B:710–3. doi: 10.1002/ajmg.b.31028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grafodatskaya D, Choufani S, Ferreira JC, Butcher DT, Lou Y, Zhao C, et al. EBV transformation and cell culturing destabilizes DNA methylation in human lymphoblastoid cell lines. Genomics. 95:73–83. doi: 10.1016/j.ygeno.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Gardiner-Garden M, Frommer MJ. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 25.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Research 2011. 2010 Oct 18; doi: 10.1093/nar/gkq963. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Down TA, Hubbard TJP. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–61. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brody DS, Hahn SR, Spitzer RL, Kroenke K, Linzer M, deGruy FV, 3rd, et al. Identifying patients with depression in the primary care setting: a more efficient method. Arch Intern Med. 1998;158:2469–75. doi: 10.1001/archinte.158.22.2469. [DOI] [PubMed] [Google Scholar]
- 28.Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de los Santos R, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2010;Early edition:1–6. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Charney DS, Keane TM. Clinician-Administered PTSD Scale for DSM-IV (CAPS-IV): National Center for Posttraumatic. Stress Disorder. 1998 [Google Scholar]
- 30.First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSMIV Axis I Disorders (SCID-I Research. Version 2.0 Biometrics Research; New York: 1996. [Google Scholar]
- 31.Gelernter J, Kranzler H, Cubells JF. Serotonin transporter protein (SLC6A4) allele and haplotype frequencies and linkage disequilibria in African- and European-American and Japanese populations and in alcohol-dependent subjects. Hum Genet. 1997;101:243–6. doi: 10.1007/s004390050624. [DOI] [PubMed] [Google Scholar]
- 32.Hu XZ, Zhu G, Lipsky R, Goldman D. HTTLPR allele expression is codominant, correlating with gene effects on fMRI and SPECT imaging intermediate phenotypes, and behavior. Biol Psychiatry. 2004;55:191S. [Google Scholar]
- 33.Stein MB, Seedat S, Gelernter J. Serotonin transporter gene promoter polymorphism predicts SSRI response in generalized social anxiety disorder. Psychopharmacology (Berl) 2006;187:68–72. doi: 10.1007/s00213-006-0349-8. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Wu M, Xiao H, Lee MT, Levin L, Leung YK, et al. Methylation of a single intronic CpG mediates expression silencing of the PMP24 gene in prostate cancer. Prostate. 70:765–76. doi: 10.1002/pros.21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lomelin D, Jorgenson E, Risch N. Human genetic variation recognizes functional elements in noncoding sequence. Genome Res. 20:311–9. doi: 10.1101/gr.094151.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wrona D. Neural-immune interactions: an integrative view of the bidirectional relationship between the brain and immune systems. J Neuroimmunol. 2006;172:38–58. doi: 10.1016/j.jneuroim.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 37.Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:155–81. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- 38.Suomi SJ. Cultural neuroscience: Bridging natural and social sciences. Institute for Social Research, Center for Culture, Mind and the Brain, University of Michigan; 2010. Risk, resilience and gene-environment interplay in primates. [Google Scholar]
- 39.Kessler RC. Posttraumatic stress disorder: the burden to the individual and to society. J Clin Psychiatry. 2000;61:4–12. [PubMed] [Google Scholar]