Abstract
Chronic Myeloid Leukaemia (CML) is a myeloproliferative disorder characterized by the expression of the oncoprotein, Bcr-Abl kinase. CCN3 normally functions as a negative growth regulator, but it is downregulated in CML, the mechanism of which is not known. MicroRNAs (miRNAs) are small non-coding RNAs, which negatively regulate protein translation by binding to the complimentary sequences of the 3′ UTR of messenger RNAs. Deregulated miRNA expression has emerged as a hallmark of cancer. In CML, BCR-ABL upregulates oncogenic miRNAs and downregulates tumour suppressor miRNAs favouring leukaemic transformation. We report here that the downregulation of CCN3 in CML is mediated by BCR-ABL dependent miRNAs. Using the CML cell line K562, we profiled miRNAs, which are BCR-ABL dependent by transfecting K562 cells with anti-BCR-ABL siRNA. MiRNA expression levels were quantified using the Taqman Low Density miRNA array platform. From the miRNA target prediction databases we identified miRNAs that could potentially bind to CCN3 mRNA and reduce expression. Of these, miR-130a, miR-130b, miR-148a, miR-212 and miR-425-5p were significantly reduced on BCR-ABL knockdown, with both miR-130a and miR-130b decreasing the most within 24 h of siRNA treatment. Transfection of mature sequences of miR-130a and miR-130b individually into BCR-ABL negative HL60 cells resulted in a decrease of both CCN3 mRNA and protein. The reduction in CCN3 was greatest with overexpression of miR-130a whereas miR-130b overexpression resulted only in marginal repression of CCN3. This study shows that miRNAs modulate CCN3 expression. Deregulated miRNA expression initiated by BCR-ABL may be one mechanism of downregulating CCN3 whereby leukaemic cells evade negative growth regulation.
Keywords: BCR-ABL, CCN3, CML, micro RNA, miR-130a, miR-130b
Introduction
Chronic Myeloid Leukaemia (CML) is a haematopoietic stem cell (HSC) disorder. CML is characterized by the presence of the Philadelphia chromosome (Nowell and Hungerford 1960), which encodes the BCR-ABL oncogene. BCR-ABL is a fusion gene resulting from the reciprocal translocation between BCR and ABL genes on the chromosomes 22 and 9 respectively (Rowley 1973). BCR-ABL translates to the constitutively active Bcr-Abl tyrosine kinase protein, which deregulates downstream signalling pathways. This results in deregulated cell proliferation, differentiation, apoptosis and adhesion (Deininger et al. 2000). CML is currently treated using tyrosine kinase inhibitors (TKIs). Imatinib mesylate (Gleevec, STI571) is now the front line treatment for CML patients in all disease stages (Druker and Lydon 2000). Clinical studies have demonstrated that imatinib induces complete cytogenetic response in more than 80% of patients in chronic phase CML (Hochhaus et al. 2009). Resistance to imatinib develops over a period of time and may be attributed to Bcr-Abl kinase domain mutations, additional genetic aberrations or the BCR-ABL independent state of the disease at later stages (Volpe et al. 2009). CML is a stem cell disorder; persistence of the quiescent population of BCR-ABL+ CD34+ stem cells even after treatment with TKIs gives rise to patient relapses. Since CML stem cells do not rely on BCR-ABL for maintaining their quiescent state(Bhatia et al. 2003), alternate targets have to be sought for identifying new treatment strategies.
DNA microarray analysis of a CML murine stem cell model performed to identify the initial effects of Bcr-Abl kinase activity, detected downregulation of CCN3 in CML. Bone marrow cells from CML patients had reduced Ccn3 expression and the expression was restored upon treatment with imatinib. Increased secretion of Ccn3 is also observed in progenitor CD34+ CML cells, indicating loss of CCN3 as an early event in CML (McCallum et al. 2006). In vitro studies using CML cell lines overexpressing Ccn3 or treated with recombinant Ccn3 showed reduced cell proliferation and increased apoptosis (McCallum et al. 2009). In CML, reduced CCN3 expression confers growth advantage and survival means for CML cells and CCN3 deregulation could be a key step in the transformation of hematopoietic cells to CML phenotype (McCallum et al. 2006).
The mechanism by which BCR-ABL downregulates CCN3 is not known. Small regulatory noncoding RNA molecules called MicroRNAs (miRNAs) that negatively regulate protein expression reduce the expression of several tumour suppressor proteins (Schickel et al. 2008). In CML, BCR-ABL upregulates oncogenic miRNAs, which reduces the expression of key tumour suppressor proteins thereby favouring malignant transformation (Faber et al. 2008). In this study, we investigated if BCR-ABL upregulates any miRNA(s) resulting in the downregulation of CCN3 using the CML cell line model K562.
Materials and methods
Cell lines
The K562 CML and HL60 acute myeloid leukaemia (AML) cell lines were obtained from Deutsche Sammlung von Mikrorganismen und Zellkulturen (DSMZ GmbH, Braunschweig, Germany). The cells were maintained in RPMI-1640 supplemented with 10% fetal calf serum (Gibco BRL, Paisley, United Kingdom), and incubated at 37°C in a humidified incubator with 5% CO2. They were passed twice weekly to maintain log phase growth.
BCR-ABL siRNA transfection
K562 cells (1 × 106) were transfected with either siRNA targeting BCR-ABL (Qiagen, Cat. No: 1027020) or scrambled control using cell line nucleofector Solution V, program T-16 (Amaxa GmbH, Cologne, Germany) according to the manufacturer’s instructions. Cells after transfection were incubated at 37°C with 5% CO2 for 24 h and 72 h post transfection.
Real time PCR analysis for BCR-ABL and CCN3 expression
K562 cells transfected with siRNA targeting BCR-ABL or scrambled control were harvested and total RNA was extracted at 24 h and 72 h post transfection using mirVana miRNA isolation kit (Ambion). Samples were run on a denaturing polyacrylamide gel (15%) with 7 M Urea to assess the quality of the RNA. Real time PCR (RT-PCR) using Taqman chemistry was used to detect the levels of BCR-ABL and CCN3 transcripts. The primer-probe sets for BCR-ABL were used according to the Europe Against Cancer protocol as previously described (Gabert et al. 2003). For CCN3 detection, the following primer-probe sets were designed using primer express software (Applied Biosystems) forward primer—5′-TGC GAC CTG CAC CTG TCA-3′, reverse primer- 3′-TCC TGG AGG AAG GCC TCA T-5′, probe-5′-FAM-CCA ACT GTC CTA AGA AC-NFQ MGB-3′. The levels of BCR-ABL and CCN3 were normalized to endogenous 18S rRNA control. The RT-PCR was carried out using an ABI PRISM 7500 Sequence Detector and data analysis was performed using Sequence Detector v1.6.3 software (Applied Biosystems); relative transcript levels were quantified using 2−ΔΔCT method (Ginzinger 2002).
Western blot analysis
K562 cells (1 × 106) transfected with siRNA targeting BCR-ABL were harvested and suspended in RIPA buffer consisting of 1x PBS, 1% Igepal CA-630 (Sigma-Aldrich), 0.1% SDS and protease inhibitor (Complete Mini Cocktail, Roche Diagnostics Ltd, Lewes, UK) and lysed on ice for 10 min. Samples were then sonicated for 15 s and centrifuged at 13,000 rpm at 4°C for 15 min to remove insoluble material. The supernatant was recovered and protein concentration was estimated by the Bradford protein method (BCA protein assay kit, Pierce, Cramlington, United Kingdom).
For Western blotting, 40 μg of the total protein was loaded onto a pre-cast NuPAGE® Novex 3–8% Tris-Acetate Gel (Invitrogen, Paisley, UK) and subsequently transferred to a 0.45-μm pore sized PVDF membrane (Invitrogen). Bcr-Abl expression was determined using a polyclonal rabbit c-Abl antibody (Cell Signalling; dilution 1:1000). For Ccn3 detection, an NH5 antibody raised against the C-terminus of Ccn3 was used at 1:1000 (Subramaniam et al. 2008). All dilutions of the antibodies and blocking steps were done using 5% milk in TBS-Tween. Equivalent protein loading was censured by determining beta-actin levels (Sigma). Chemiluminescence was used to detect the protein bands in the immunoblots (Supersignal; Pierce, Rockford, IL). Optical densitometry was performed using the Autochemi System (Ultra-Violet Products, Cambridge, United Kingdom) and corrected for protein loading via the beta-actin signal.
MiRNA expression profiling using Taqman Low Density Array cards
Total RNA (1 μg) from K562 cells and from anti-BCR-ABL siRNA treated cells at 24 h and 72 h was reverse transcribed using TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems). The profiling of 667 mature miRNAs was performed using Taqman Low Density Array (TLDA) cards on a 7900 HT fast RT PCR using ABI sequence detection software v2.3. The results were analyzed by SDS Relative Quantification (RQ) Manager. The expression levels of miRNAs were normalized to the endogenous RNU48 and the fold changes of individual miRNAs were calculated using the 2−ΔΔCT method.
Database prediction of miRNAs targeting CCN3
The miRNAs potentially targeting CCN3 mRNA were identified using the bioinformatic program miRGEN http://www.diana.pcbi.upenn.edu/miRGen.
MicroRNA transfection
HL60 (BCR-ABL negative) AML cells were transfected with 30 nm pre-miR miRNA precursor molecules of hsa-miR-130a (PM10506) and hsa-miR-130b (PM10777) from Applied Biosystems using program T-19 Amaxa nucleofection. As a control 30 nm of pre-miR miRNA precursor molecule-negative control # 1 (NC1, AM17110, Applied Biosystems) was used. Total RNA and protein were extracted 24 h and 48 h post transfection as described previously.
Quantitative PCR for miRNAs
To determine the expression of candidate miRNAs, 10 ng of total RNA was reverse transcribed using stem loop primers for specific miRNAs and RT-PCR was carried out using individual Taqman miRNA assays (Cat No: 000454 and 000456, Applied Biosystems). The expression levels of miRNAs were normalized to RNU48 (Cat No: 001006, Applied Biosystems) (Wong et al. 2007).
Results
BCR-ABL reduction increases CCN3 expression
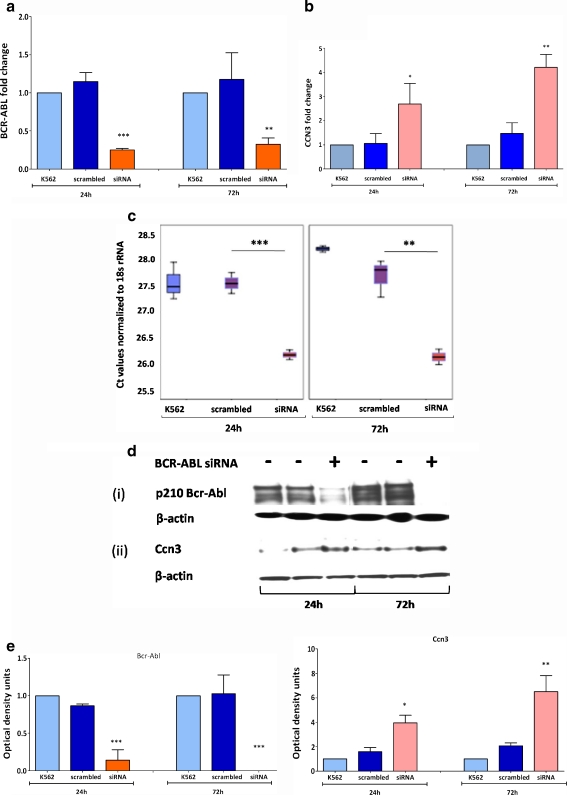
Initially, we established that we were using a robust cell line model. K562 cells were transfected with siRNA directed against BCR-ABL. The levels of BCR-ABL mRNA decreased 4.5 fold at 24 h and 3.6 fold at 72 h of siRNA transfection compared to the scrambled control (Fig. 1a). The reduction in BCR-ABL was associated with a significant increase in CCN3 transcripts (Fig. 1b). CCN3 levels increased 2.5 fold within 24 h of BCR-ABL reduction (p < 0.005). CCN3 levels further increased 2.8 fold at 72 h of BCR-ABL reduction (p < 0.001). The difference in CCN3 transcript levels observed in K562 cells, scrambled control and in anti-BCR-ABL siRNA transfected cells is expressed in terms of Ct (cycle threshold) values normalized to 18SrRNA (Fig. 1c). A lower Ct value indicates higher gene expression. These transcript changes were mirrored at the protein level as demonstrated by western blotting (Fig. 1d) and by the corresponding densitometric plots (Fig. 1e). These findings were also confirmed at 48 h post BCR-ABL silencing (data not shown), but the initial 24 h and later 72 h time points were chosen for the analysis of BCR-ABL dependent miRNA expression.
Fig. 1.
BCR-ABL silencing increases CCN3 expression. siRNA directed to target BCR-ABL was used to investigate CCN3 expression levels. K562 cells transfected with siRNA targeting BCR-ABL, a scrambled sequence and untransfected control cells were compared at 24 h and 72 h post transfection. a Real time PCR was performed to quantify BCR-ABL transcript levels. A significant reduction in BCR-ABL was observed within 24 h of siRNA treatment. b RQ-PCR was performed to detect levels of CCN3 expression as a result of siRNA targeting BCR-ABL. BCR-ABL reduction corresponded with an increase in CCN3. cBox plots indicating the difference in CCN3 gene expression in each set of samples. Data represent normalized Ct values; higher the Ct value, lower the gene expression. d Western blotting was performed on protein lysates to detect expression of Bcr-Abl (i), Ccn3 (ii) and β-actin. e Optical densitometry was performed for the blots in (c) and the data was graphically formatted to compare levels of Bcr-Abl and Ccn3 protein expression normalized to β-actin. Data represent the mean of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001
Candidate miRNAs binding to CCN3 3′ UTR
Four different miRNA target prediction algorithms (Targetscan, miRbase target, PicTar and miRanda) were used to predict miRNAs which could bind to CCN3 3′ UTR. These databases identified 149, 51, 1 and 15 candidate miRNAs respectively. To reduce the number of false positives we used miRGen target that uses the union of the above-mentioned algorithms. This narrowed down the search results to 16 miRNAs: miR-92, miR-92b, miR-132, miR-212, miR-323, miR-539, miR-758, miR-130a, miR-130b, miR-148a, miR-182*, miR-299-5p, miR-302b*, miR-302c, miR-30e-5p and miR-425-5p. Of these, miR-92, miR-92b, miR-132, miR-212, miR-130a and miR-130b were predicted by Targetscan, miRbase target and miRanda.
MiRNAs predicted to target CCN3 are BCR-ABL dependent
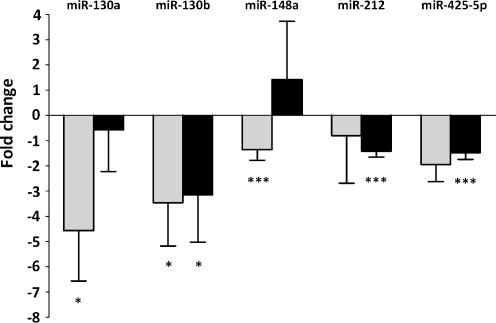
To identify the miRNAs expressed by the K562 cell line model, miRNA expression profiling was carried out using Taqman Low Density Array cards. These arrays detect mature forms of 667 human miRNAs using Taqman chemistry. Of the 16 candidate miRNAs, K562 cells did not express miR-299-5p, miR-302c, miR-363 and miR-539. The remaining 12 miRNAs had varied levels of expression. To detect if the 12 candidate miRNAs are dependent on BCR-ABL, we used siRNA specific to BCR-ABL and analyzed the changes in miRNA signatures at 24 h and 72 h post transfection. Five of the miRNA candidates decreased with BCR-ABL reduction (Fig. 2), of which miR-130a (−4.5 fold), miR-130b (−3.5 fold), miR-148a (−1.3 fold), miR-425-5p (−2 fold) had significant reduction at the initial 24 h. At 72 h, miR-130b (−3.1 fold) and miR-425-5p (−1.5 fold) still had reduced expression. We focused further studies on miR-130a, which showed significant decrease at 24 h and on miR-130b, which decreased at both 24 h and 72 h time points.
Fig. 2.
The expression of miRNAs predicted to target CCN3 decreases with BCR-ABL reduction. Analysis of BCR-ABL dependent miRNA expression of K562 cells was carried out using TLDA miRNA cards. The miRNA expression of K562 cells transfected with siRNA directed against BCR-ABL was compared with the control K562 cells at 24 h and 72 h post transfection. Data represents the fold reduction observed in the expression of 5 miRNAs predicted to target CCN3 with each BCR-ABL silencing experiment (n = 3), *, p < 0.05; ***, p < 0.001. The expression of RNU48 was used as the endogenous control. Light bars represent 24 h and dark bars represent 72 h time points
Validation of the TLDA results
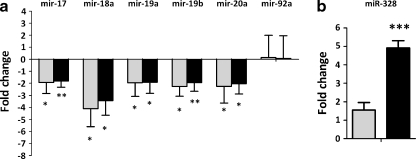
To validate our approach, we compared the expression of BCR-ABL associated miRNAs reported in the literature with the results generated from the TLDA platform. The miR-17-92 cluster is a group of oncogenic miRNAs upregulated by the BCR-ABL–c-Myc pathway (Venturini et al. 2007). We found a decrease of all the miRNAs in this cluster with the exception of miR-92 (Fig. 3a), similar to the findings reported previously by Venturini et al. (Venturini et al. 2007). Bcr-Abl kinase activity results in the loss of miR-328 in the blast crisis of CML that impairs myeloid cell differentiation (Eiring et al. 2010). In our studies, we found miR-328 levels increasing with BCR-ABL reduction, with significant increase at 72 h of BCR-ABL silencing (Fig. 3b).
Fig. 3.
BCR-ABL associated miRNAs change expression with respect to BCR-ABL reduction. The expression of two of the BCR-ABL associated miRNAs was identified using the TLDA platform. a The miR-17-92 cluster upregulated by BCR-ABL showed significant reduction at both 24 h and 72 h post BCR-ABL siRNA transfection in K562 cells. b The tumour suppressor miR-328 was significantly increased at 72 h of BCR-ABL silencing. Data represent the mean fold reductions observed in three independent experiments. *, p < 0.05;; ***, p < 0.001. Light bars represent 24 h and dark bars represent 72 h time points
Imatinib treatment of K562 cells reduces miR-130a and miR-130b expression
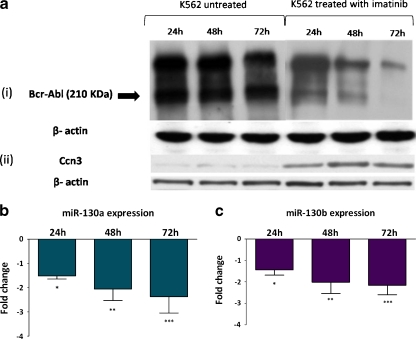
To confirm BCR-ABL dependent expression of miR-130a and miR-130b, K562 cells were treated with 1 μM imatinib for 24, 48 and 72 h to inhibit Bcr-Abl kinase activity. Decrease in Bcr-Abl protein resulted in increased Ccn3 expression (Fig. 4a). The expression of miRNAs was determined by miRNA specific Taqman real time PCR assays. Imatinib treatment resulted in significant decrease of both miRNAs at all time points (Fig. 4b and c).
Fig. 4.
Imatinib treatment of K562 cells reduces miR-130a and miR-130b expression. K562 cells were treated with 1 μM imatinib for 24, 48 and 72 h and the expression levels of miR-130a and miR-130b were compared with untreated K562 cells. Quantitative PCR for individual miRNAs were performed and expression was normalized to the endogenous control RNU48. (a) Western blotting showing the decrease in Bcr-Abl expression (i) and the corresponding increase in Ccn3 (ii) in K562 cells with 24, 48 and 72 h of imatinib treatment (b) Expression levels of miR-130a and (c) miR-130b following 24, 48 and 72 h of imatinib treatment of K562 cells. Data represent the mean fold reduction in miR-130a and miR-130b expression relative to the untreated K562 cells (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001
Overexpression of miR-130a and miR-130b reduces CCN3 levels
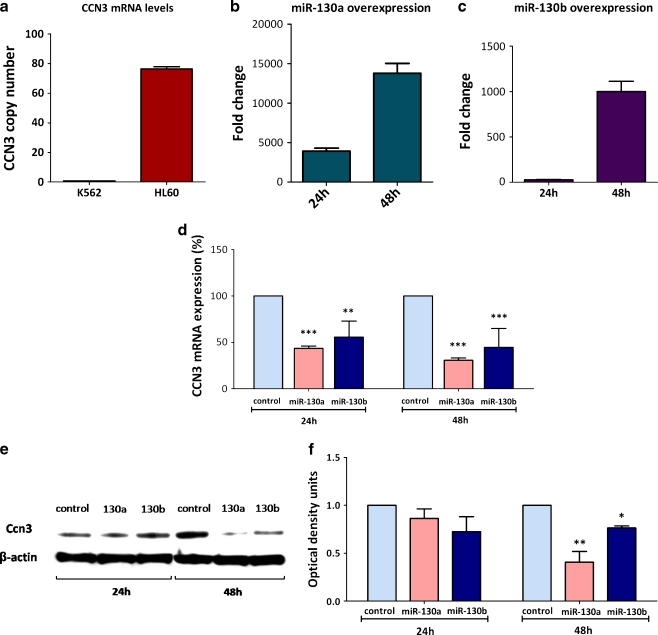
To confirm the effect of miR-130a and miR-130b on CCN3 expression, the AML cell line HL60, which does not express BCR-ABL, was used, as BCR-ABL+ cells do not express CCN3. We compared the expression of CCN3 in HL60 cells with K562 cells using real time PCR. Each amplification reaction contained 50 ng of cDNA equivalents. HL60 had 76.3 copies of CCN3 compared to the 0.8 copies of CCN3 in K562 (Fig. 5a). HL60 cells were transfected with Taqman Pre-miR miRNA precursors, of miR-130a and miR-130b. These precursors are chemically modified double stranded RNA molecules that mimic endogenous mature miRNA molecules. Pre-miR™ Negative control #1, which is a random sequence of Pre-miR molecules having no identified effects on known miRNA function was used as a control. Transfection of both miR-130a and miR-130b mimics resulted in significant increase in the expression of both miRNAs (Fig. 5b and c). HL60 cells do not express miR-130a; therefore the transfection of miR-130a mimic into these cells results in a considerable fold increase in miR-130a transcript levels. The expression of CCN3 was determined by quantitative PCR at 24 h and 48 h post transfection of the miRNA mimics. Both miR-130a and miR-130b overexpression resulted in the decrease of CCN3 mRNA levels. The levels of CCN3 in HL60 decreased by 43 ± 2% at 24 h (p < 0.001) and by 31 ± 2% (p < 0.001) at 48 h when transfected with miR-130a (Fig. 5d). Whereas, miR-130b transfection decreased CCN3 transcripts by 55 ± 17% at 24 h (p < 0.01) and by 44 ± 20% at 48 h (p < 0.001) (Fig. 5d). Significant reduction in Ccn3 protein levels were seen with miR-130a transfection at 48 h where as miR-130b overexpression resulted only in modest decrease of Ccn3 (Fig. 5e and f). Though at 24 h there was clearly a decrease at RNA levels, a significant decrease at the Ccn3 protein levels was not observed with miR-130b overexpression.
Fig. 5.
Overexpression of miR-130a and miR-130b reduces CCN3 levels. (a) Real time PCR analysis of CCN3 copy number in K562 cells and BCR-ABL negative HL60 cells. (b) Amaxa transfection of miRNA mimics of miR-130a showed a significant increase in miR-130a transcripts at 24 h and 48 h following transfection, (c) transfection of miR-130b miRNA mimics show increased expression of miR-130b in HL60 cells at both 24 h and 48 h (d) CCN3 mRNA levels decrease with miR-130a and miR-130b overexpression at 24 h and 48 h post transfection of the miRNA mimics (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001. (e) Corresponding protein level determination, miR-130a overexpression decreases Ccn3 at 48 h where as miR-130b transfection shows a slight decrease at Ccn3 protein levels. (f) Corresponding optical density readings for the blots in (e) comparing the levels of Ccn3 protein normalized to β-actin
Discussion
MiRNAs are non-coding single stranded RNA molecules, typically 21–23 nucleotides in length. They are posttranscriptional gene regulatory molecules, which inhibit protein translation by binding to the target messenger RNAs. MiRNAs control key cellular processes like proliferation, differentiation, apoptosis and aberrant expression of miRNAs have been identified in various pathologies including cancer (Zimmerman and Wu 2011). In CML, BCR-ABL upregulates the expression of oncogenic miRNAs; for example miR-17-92 cluster of oncomirs which facilitate tumour progression (Venturini et al. 2007). Simultaneously, BCR-ABL downregulates the expression of tumour suppressor miRNAs like miR-203 (Bueno et al. 2008) and miR-328 thereby blocking differentiation of the myeloid cells (Eiring et al. 2010). Downstream to this is the imbalance created in their protein targets, whereby oncoproteins are upregulated and tumour supressors are downregulated, a characteristic feature of malignancy. The repression of tumour suppressor mRNAs by miRNAs is emerging as an important deregulatory switch in cancer. We hypothesized that BCR-ABL regulated miRNAs might play a role in downregulating the levels of CCN3 in CML.
Using the CML cell line model K562, which expresses Bcr-Abl kinase, we identified miRNAs that are BCR-ABL dependent by using siRNA against BCR-ABL. BCR-ABL knockdown resulted in the upregulation of 56 miRNAs and the downregulation of 76 miRNAs by >1.5 fold within 24 h of siRNA transfection. Among these, 12 miRNAs predicted to target CCN3 mRNA, showed different levels of expression with respect to BCR-ABL status. We focused only on those miRNAs, which showed a consistent decrease in their expression levels in the triplicate experiments. From the panel of 5 miRNAs, we selected miR-130a and miR-130b to examine their effect on CCN3 expression; miR-130a decreased −4.5 fold and miR-130b decreased −3.5 fold within 24 h of BCR-ABL knockdown.
Both miR-130a and miR-130b are paralogous miRNA sequences located on chromosomes 11 and 22 respectively with their mature sequences differing at two nucleotides at positions 11 and 13. The mature sequence of miR-130a is CAGUGCAAUGUUAAAAGGGCAU and miR-130b is CAGUGCAAUGAUGAAAGGGCAU; miR-130a and miR-130b are predicted to target the same region of CCN3 mRNA from bases 300–321. Transfection of mature sequences of both miRNAs individually into HL60 cells resulted in the repression of CCN3 at varying levels. MiR-130a transfection suppressed Ccn3 protein translation significantly, whereas the downregulation of Ccn3 conferred by miR-130b was only partial. Transfection of miR-130a and miR-130b decreased CCN3 mRNA, but the decrease in Ccn3 protein levels was prominent only with miR-130a overexpression. This suggests that miR-130a might be the key miRNA in reducing Ccn3 expression.
We also examined the expression of miR-130a and miR-130b when K562 cells were treated with imatinib for 24, 48 and 72 h to inhibit Bcr-Abl kinase activity. Imatinib treatment resulted in the decrease of both miR-130a and miR-130b, but the extent of downregulation obtained was different from the siRNA treatment against BCR-ABL. The decrease in expression of both the miRNAs was greatest with siRNA silencing of BCR-ABL, rather than with imatinib treatment. This could reflect the mechanistic differences in the extent of Bcr-Abl reduction obtained by these treatments.
MiR-130a has been implicated in platelet differentiation. MiRNA expression profiling of CD34+ progenitors and platelets showed the downregulation of miR-130a during haematopoietic differentiation. The downregulation of miR-130a increases the expression of its target protein MAFB; a transcription factor required for promoting platelet development (Garzon et al. 2006). A block in cellular differentiation is a hallmark of CML; BCR-ABL mediated miR-130a upregulation might be a means of downregulating proteins associated with myeloid differentiation. A recent study using mouse bone marrow cells showed miR-130a levels enriched in the haematopoietic stem cell fraction (O'Connell et al. 2010). These independent studies indicate miR-130a as a highly expressed miRNA in haematopoietic stem cells and that its expression reduces during differentiation and maturation. In CML, altering the levels of miR-130a by BCR-ABL could be a means of arresting differentiation.
MiR-130b was identified as upregulated in adult T-cell Leukaemia (ATL) cell lines transformed by Human T-cell lymphotrophic virus 1 (HTLV-1). Increased expression of miR-130b was also identified in the peripheral blood mononuclear cells of ATL patients. In ATL, miR-130b downregulates the expression of tumor protein 53–induced nuclear protein 1 (TP53INP1), a key protein required for the induction of cell cycle arrest and apoptosis (Yeung et al. 2008). In hepatocellular carcinoma (HCC), miR-130b was preferentially expressed in the tumour initiating cells. Overexpression of miR-130b in HCC cell lines increased their proliferation, self-renewal capacity and chemoresistance (Ma et al. 2010). A recent study showed that TAp63, an isoform of p63, increases miR-130b expression by enhancing dicer activity. TAp63 upregulation of miR-130b reduces the invasive capacity of mouse embryonic fibroblasts (Su et al. 2010). Increased expression of mir-130b is also reported in primary gastric tumours. Overexpression of miR-130b in different gastric cell lines resulted in the reduction of RUNX3 tumour suppressor facilitating tumour progression (Lai et al. 2010).
Recently, there have been reports on the miRNA regulation of CCN proteins. MiR-155 is found to target CCN1 resulting in reduced placental angiogenesis (Zhang et al. 2010). CCN1 is also regulated by miR-30a-3p; overexpression of this miRNA reduced CCN1 expression in the HepG2, human hepatocellular liver cancer cell line (Nakamoto et al. 2005). In cardiomyocytes and fibroblasts, miR-30 and miR-133 overexpression decreased CCN2 expression resulting in the reduced production of collagen. Decreased expression of these two miRNAs is observed in cardiac pathologies resulting in increased CCN2 expression. This results in increased collagen production leading to cardiac fibrosis (Duisters et al. 2009). In chondrocytes, CCN2 is downregulated by miR-18a, resulting in their decreased differentiation potential (Ohgawara et al. 2009). The miR-17-92 cluster, of which miR-18a is a member is found to downregulate CCN2 expression in gliomas leading to altered differentiation (Ernst et al. 2010). In primary pigmented nodular adrenocortical disease (PPNAD), decreasing levels of miR-449 was correlated with increased CCN5 expression (Iliopoulos et al. 2009).
We report that in CML, the expression of miRNAs, miR-130a and miR-130b is BCR-ABL dependent. The overexpression of mature sequences of either of these miRNAs resulted in the downregulation of CCN3 transcripts in HL60 cells. A significant reduction in Ccn3 protein levels was achieved with miR-130a overexpression. In conclusion, this study for the first time identified the post-transcriptional regulation of CCN3 expression by miRNA activity. Taken together this data suggests that, BCR-ABL downregulation of CCN3 observed in CML might be mediated by the expression of miR-130a/b.
Acknowledgements
This work has been supported by Cancer Research UK graduate studentship.
Abbreviations
- CML
Chronic Myeloid Leukaemia
- MiRNA
MicroRNA
References
- Bhatia R, Holtz M, Niu N, Gray R, Snyder DS, Sawyers CL, Arber DA, Slovak ML, Forman SJ. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 2003;101:4701–4707. doi: 10.1182/blood-2002-09-2780. [DOI] [PubMed] [Google Scholar]
- Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernández-Piqueras J, Malumbres M. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;10:3343–3356. [PubMed] [Google Scholar]
- Druker BJ, Lydon NB. Lessons learned from the development of an abl tyrosine kinase inhibitor for chronic myelogenous leukemia. J Clin Invest. 2000;105:3–7. doi: 10.1172/JCI9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, Made I, Herias V, Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–178. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, Becker H, Chandler JC, Andino R, Cortes J, Hokland P, Huettner CS, Bhatia R, Roy DC, Liebhaber SA, Caligiuri MA, Marcucci G, Garzon R, Croce CM, Calin GA, Perrotti D. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A, Campos B, Meier J, Devens F, Liesenberg F, Wolter M, Reifenberger G, Herold-Mende C, Lichter P, Radlwimmer B. De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene. 2010;29:3411–3422. doi: 10.1038/onc.2010.83. [DOI] [PubMed] [Google Scholar]
- Faber J, Gregory RI, Armstrong SA. Linking miRNA regulation to BCR-ABL expression: the next dimension. Cancer Cell. 2008;13:467–469. doi: 10.1016/j.ccr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Gabert J, Beillard E, Velden VH, Bi W, Grimwade D, Pallisgaard N, Barbany G, Cazzaniga G, Cayuela JM, Cave H, et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—a Europe Against Cancer program. Leukemia. 2003;12:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, Volinia S, Bhatt D, Alder H, Marcucci G, Calin GA, Liu CG, Bloomfield CD, Andreeff M, Croce CM. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci USA. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol. 2002;30:503–512. doi: 10.1016/S0301-472X(02)00806-8. [DOI] [PubMed] [Google Scholar]
- Hochhaus A, O'Brien SG, Guilhot F, Druker BJ, Branford S, Foroni L, et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–1061. doi: 10.1038/leu.2009.38. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Bimpaki EI, Nesterova M, Stratakis CA. MicroRNA signature of primary pigmented nodular adrenocortical disease: clinical correlations and regulation of Wnt signaling. Cancer Res. 2009;69:3278–3282. doi: 10.1158/0008-5472.CAN-09-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KW, Koh KX, Loh M, Tada K, Subramaniam MM, Lim XY, et al. MicroRNA-130b regulates the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer. 2010;46:1456–1463. doi: 10.1016/j.ejca.2010.01.036. [DOI] [PubMed] [Google Scholar]
- Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A, Ng I, Man K, Wong N, To KF, Zheng BJ, Lai PB, Lo CM, Chan KW, Guan XY. miR-130b promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694–707. doi: 10.1016/j.stem.2010.11.010. [DOI] [PubMed] [Google Scholar]
- McCallum L, Price S, Planque N, Perbal B, Pierce A, Whetton AD, Irvine AE. A novel mechanism for BCR-ABL action: stimulated secretion of CCN3 is involved in growth and differentiation regulation. Blood. 2006;5:1716–1723. doi: 10.1182/blood-2006-04-016113. [DOI] [PubMed] [Google Scholar]
- McCallum L, Lu W, Price S, Lazar N, Perbal B, Irvine AE. CCN3: a key growth regulator in chronic myeloid leukaemia. J Cell Commun Signal. 2009;3:115–124. doi: 10.1007/s12079-009-0058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto M, Jin P, O'Donnell WT, Warren ST. Physiological identification of human transcripts translationally regulated by a specific microRNA. Hum Mol Genet. 2005;14:3813–3821. doi: 10.1093/hmg/ddi397. [DOI] [PubMed] [Google Scholar]
- Nowell PC, Hungerford D. A minute chromosome in human chronic granulocytic leukaemia. Science. 1960;132:1497. [Google Scholar]
- O'Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci USA. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgawara T, Kubota S, Kawaki H, Kondo S, Eguchi T, Kurio N, Aoyama E, Sasaki A, Takigawa M. Regulation of chondrocytic phenotype by microRNA 18a: involvement of Ccn2/Ctgf as a major target gene. FEBS Lett. 2009;583:1006–1010. doi: 10.1016/j.febslet.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, El-Naggar A, Creighton CJ, Suraokar MB, Wistuba I, Flores ER. TAp63 suppresses metastasis through coordinate regulation of dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam MM, Lazar N, Navarro S, Perbal B, Llombart-Bosch A. Expression of CCN3 protein in human Wilms’ tumors: immunohistochemical detection of CCN3 variants using domain-specific antibodies. Virchows Arch. 2008;452:33–39. doi: 10.1007/s00428-007-0523-3. [DOI] [PubMed] [Google Scholar]
- Venturini L, Battmer K, Castoldi M, Schultheis B, Hochhaus A, Muckenthaler MU, Ganser A, Eder M, Scherr M. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109:4399–4405. doi: 10.1182/blood-2006-09-045104. [DOI] [PubMed] [Google Scholar]
- Volpe G, Panuzzo C, Ulisciani S, Cilloni D. Imatinib resistance in CML. Cancer Lett. 2009;274:1–9. doi: 10.1016/j.canlet.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Wong L, Kathy L, Russell I (2007) Endogenous controls for real-time quantitation of miRNA using TaqMan® microRNA assays. Application note 127AP11-01, Applied Biosystems
- Yeung ML, Yasunaga J, Bennasser Y, Dusetti N, Harris D, Ahmad N, Matsuoka M, Jeang KT. Roles for microRNAs, miR-93 and miR-130b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res. 2008;68:8976–8985. doi: 10.1158/0008-5472.CAN-08-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Diao Z, Su L, Sun H, Li R, Cui H, Hu Y. MicroRNA-155 contributes to preeclampsia by down-regulating CYR61. Am J Obstet Gynecol. 2010;202:466.e1-7. doi: 10.1016/j.ajog.2010.01.057. [DOI] [PubMed] [Google Scholar]
- Zimmerman AL, Wu S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011;300:10–19. doi: 10.1016/j.canlet.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]