Abstract
The inactivation of BRCA2, a suppressor of breast, ovarian and other epithelial cancers, triggers instability in chromosome structure and number, which are thought to arise from defects in DNA recombination and mitotic cell division, respectively. Human BRCA2 controls DNA recombination via eight BRC repeats, evolutionarily conserved motifs of ∼35 residues, that interact directly with the recombinase RAD51. How BRCA2 controls mitotic cell division is debated. Several studies by different groups report that BRCA2 deficiency affects cytokinesis. Moreover, its interaction with HMG20b, a protein of uncertain function containing a promiscuous DNA-binding domain and kinesin-like coiled coils, has been implicated in the G2–M transition. We show here that HMG20b depletion by RNA interference disturbs the completion of cell division, suggesting a novel function for HMG20b. In vitro, HMG20b binds directly to the BRC repeats of BRCA2, and exhibits the highest affinity for BRC5, a motif that binds poorly to RAD51. Conversely, the BRC4 repeat binds strongly to RAD51, but not to HMG20b. In vivo, BRC5 overexpression inhibits the BRCA2–HMG20b interaction, recapitulating defects in the completion of cell division provoked by HMG20b depletion. In contrast, BRC4 inhibits the BRCA2–RAD51 interaction and the assembly of RAD51 at sites of DNA damage, but not the completion of cell division. Our findings suggest that a novel function for HMG20b in cytokinesis is regulated by its interaction with the BRC repeats of BRCA2, and separate this unexpected function for the BRC repeats from their known activity in DNA recombination. We propose that divergent tumor-suppressive pathways regulating chromosome segregation as well as chromosome structure may be governed by the conserved BRC motifs in BRCA2.
Keywords: HMG20b, mitosis, BRCA2, BRC repeats, cytokinesis
Introduction
Germline mutations in the BRCA2 tumor suppressor predispose with high penetrance to cancers of the breast, ovary, pancreas and other organs (Rahman and Stratton, 1998; Breast Cancer Linkage Consortium, 1999; Nathanson et al., 2001; Welcsh and King, 2001; Antoniou et al., 2003; King et al., 2007). BRCA2 inactivation triggers instability in the structure and number of chromosomes during cell division (Patel et al., 1998; Yu et al., 2000). There is mounting evidence that BRCA2's tumor-suppressive activity in preserving chromosome structure is mediated through its function in regulating RAD51, an enzyme essential in eukaryotic cells for the repair of DNA breaks by homologous DNA recombination (Chen et al., 1999; Yuan et al., 1999; Pellegrini et al., 2002; Venkitaraman, 2002; Carreira et al., 2009; Shivji et al., 2009). BRCA2 binds directly to RAD51 via the BRC repeats, evolutionarily conserved motifs of ∼35 residues, eight copies of which occur in mammalian BRCA2 orthologues. The eight mammalian BRC repeats are conserved not only in their protein sequence but also in their spacing within the protein, suggestive of conserved function(s) (Bork et al., 1996). Each of the eight BRC repeats (BRC1–BRC8) found in human BRCA2 exhibits a varying capacity to bind RAD51 in vitro (Wong et al., 1997). Peptides encoding individual BRC repeats, or the ∼1123 residue domain encoding the region of BRCA2 containing all eight repeats, regulate the ordered assembly of RAD51 on DNA substrates (Shivji et al., 2006). Thus, they enhance the loading of RAD51 onto single-stranded DNA substrates, while suppressing double-strand DNA binding, and these opposing activities promote the reactions that lead to homologous recombination (Carreira et al., 2009; Shivji et al., 2009).
How BRCA2 may work to maintain chromosome number is less clearly understood, although roles have been proposed in the transition between the G2 and M phases of the cell cycle (Marmorstein et al., 2001), and in the completion of cell division by cytokinesis (Daniels et al., 2004; Jonsdottir et al., 2009; Vinciguerra et al., 2010; Rowley et al., 2011), which have recently been questioned (Lekomtsev et al., 2010). A protein previously reported to bind to BRCA2–HMG20b or BRAF35 (Marmorstein et al., 2001)–has been implicated in the G2–M transition. HMG20b was first isolated (Sumoy et al., 2000) as a novel member of the high-mobility group (HMG) of non-sequence-specific DNA-binding proteins, which at its C terminus also contains two kinesin-like coiled coil regions, a distinctive structural feature shared with the HMG protein BAF57 (Wang et al., 1998). HMG20b can be copurified with BRCA2 in a high-molecular-weight complex of >2 Mda from asynchronous cell cultures, and interacts directly with a region spanning residues 1648–2190 in the human BRCA2 sequence. HMG20b was reported to decorate condensing chromosomes; and injection of an anti-HMG20b antibody was found to delay the transition from G2 into mitosis (Marmorstein et al., 2001). However, subsequent studies on HMG20b reveal that it interacts with KIF4 (Lee and Kim, 2003), a kinesin protein localized predominantly to mitotic microtubules rather than interphase chromatin (Zhu and Jiang, 2005).
Here, we have investigated the function of HMG20b and the significance of its interaction with BRCA2. We report that HMG20b depletion by RNA interference provokes abnormalities in the completion of cell division, delaying mitosis and triggering the formation of binucleate daughters. In vitro, HMG20b interacts directly with BRC5, a repeat that interacts poorly with RAD51, whereas, conversely, BRC4 interacts strongly with RAD51 but not HMG20b. In vivo, BRC4 overexpression inhibits the BRCA2–RAD51 interaction and the ability of RAD51 to assemble at sites of DNA breakage, but has no discernible effect on the completion of cell division. In contrast, BRC5 disrupts the BRCA2–HMG20b interaction, and phenocopies the mitotic effects of BRCA2 deficiency or HMG20b depletion. These observations suggest that HMG20b has a novel function in the completion of cell division by cytokinesis that is mediated via its interaction with the BRC repeats of BRCA2, delineating two dichotomous pathways for the tumor-suppressive activity of BRCA2 controlled by the BRC-repeat motifs.
Results
Depletion of HMG20b delays and disrupts the completion of cell division
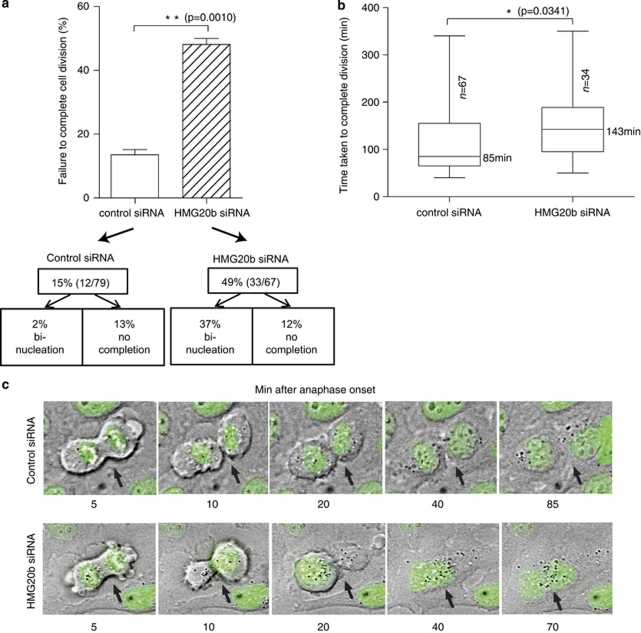
To better define the function of HMG20b during mitosis, we depleted the protein from HeLa cells by small interfering RNA (siRNA) transfection and examined mitotic progression using time-lapse microscopy (Daniels et al., 2004). The extent of HMG20b protein depletion after the exposure of cells to specific siRNAs, or control oliognucleotides, was determined by western blotting (Supplementary Information, Supplementary Figure S1a). We find that HMG20b depletion significantly delays the time taken from anaphase onset to the completion of cell division, a phenotype we have previously detected in BRCA2-deficient cells (Daniels et al., 2004). Typically, HMG20b-depleted cells initiate cleavage furrowing and undergo significant furrow ingression, but frequently fail to complete cell division (49% of HMG20b-depleted mitotic cells (n=67) compared with 15% of controls (n=79). Many exit mitosis to form binucleate cells (37% of mitotic cells). Less frequently, HMG20b-deficient cells remain with advanced furrow ingression without undergoing abscission during a 6-h observation period (12% of mitotic cells; Figure 1a).
Figure 1.
HMG20b is required for the completion of cell division. (a) Frequency of failure to complete cell division after depletion of HMG20b. Cells which either became binucleate or did not complete cell division within a 6-h observation period were counted from time-lapse images. The mean±s.e.m. is shown. (b) Box-and-whisker plot showing time taken from anaphase onset to completion of cell division measured from time-lapse images. The median value is shown; the bottom and top of the box represent the 25 and 75th percentiles respectively. (c) Representative frames from a time-lapse experiment with HeLa cells stably expressing green fluorescent protein-H2B. Cells of interest are marked with black arrows. See also Supplementary Movies 1 and 2.
About half (51%) of HMG20b-depleted cells do ultimately complete cell division, but the majority is significantly delayed in progression. Thus, the time taken from anaphase onset to the completion of division increases markedly from a median value of 85 min (control siRNA, n=67) to 143 min (HMG20b siRNA, n=34) (Figure 1b). Representative frames from time-lapse experiments with HeLa cells expressing green fluorescent protein-tagged histone H2B are shown in Figure 1c, supplemented by movies demonstrating the defects induced by HMG20b depletion (Supplementary Movies 1 and 2). Similar defects are observed when using multiple individual siRNAs targeting HMG20b, further substantiating that it is specifically due to loss of HMG20b function (Supplementary Information, Supplementary Figures S1b–d). Collectively, these data demonstrate that HMG20b depletion delays the completion of mitotic cell division, and triggers abnormalities in the process.
The BRC5 repeat of BRCA2 binds directly to HMG20b but not RAD51, in contrast to BRC4
Human BRCA2 and HMG20b can be reciprocally coimmunoprecipitated with one another from HeLa nuclear extracts (Marmorstein et al., 2001). We confirmed this using extracts of asynchronously dividing 293T cells and rabbit polyclonal antibodies against BRCA2 or HMG20b, coupled to protein A beads. We found that only a small fraction (<5%) of each protein was present in the complex (Figure 2a). Interestingly, complex formation was increased (Supplementary Information, Supplementary Figure S2) when cells were enriched in prometaphase (by nocodazole exposure, lanes 2 and 6) or in cytokinesis (by purvalanol A treatment after nocodazole exposure, lanes 3 and 7). These findings are consistent with a role for the HMG20b–BRCA2 complex in mitotic cell division.
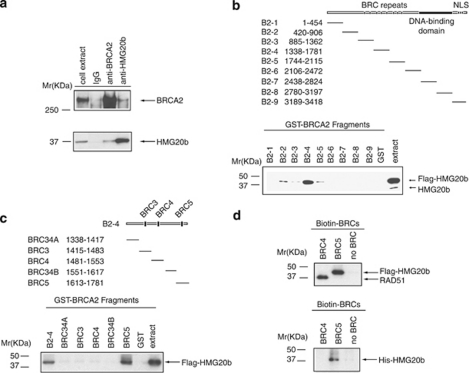
Figure 2.
BRC5 repeat of BRCA2 binds directly to HMG20b. (a) BRCA2 and HMG20b complexes were pulled down from 293T extracts using protein A-coupled anti-BRCA2 antibody (αB2-9) and anti-HMG20b antibody (M10), respectively. Coimmunoprecipitated BRCA2 and HMG20b were detected by western blotting. (b) GST–BRCA2 fragments (whose span is shown in the schematic) were incubated with 293T extracts expressing Flag-HMG20b, and the bound proteins were analyzed by western blotting (‘GST pull-down assay'). See also Supplementary Figures S3a and b. (c) The B2-4 fragment was subcloned into five smaller fragments as shown in the schematic, and the GST pull-down assay was performed as shown in (b). Supplementary Figure S3c shows the controls for sample loading. Note that the B2-4 fragment is distinct from the BRC4 repeat. (d) Biotin-labelled BRC4 (1517–1551) and BRC5 (1661–1695) peptides were incubated with 293T extracts expressing Flag-HMG20b (upper panel) and purified HMG20b protein (His-HMG20b, lower panel) in a streptavidin pull-down assay.
To identify the region of BRCA2 that mediates the interaction, we split its coding sequence into nine overlapping GST-fusion proteins, B2-1 to B2-9, and incubated them with extracts from 293T cells overexpressing FLAG-tagged HMG20b (Figure 2b and Supplementary Information, Supplementary Figures S3a and b). The B2-4 fragment, spanning BRCA2 residues 1338–1781, pulls down both endogenous HMG20b and the overexpressed FLAG-tagged form (bands marked by arrows). Whereas the overexpressed form of HMG20b also binds weakly to other fragments (B2-2, -3 and -5, see Supplementary Information, Supplementary Figure S3a), pull down of the endogenous form is undetectable with these fragments. Collectively, these results suggest that the major site of interaction between endogenous HMG20b and BRCA2 lies within the B2-4 region.
Further dissection of the B2-4 region reveals (Figure 2c and Supplementary Information, Supplementary Figure S3c) that the portion responsible for HMG20b binding lies between BRCA2 residues 1613–1781, which span the BRC5 repeat. Indeed, incubation of cell extracts with biotinylated peptides encoding the BRC4 (1517–1551) or BRC5 (1661–1695) repeats shows that BRC5 binds strongly to HMG20b, whereas binding to RAD51 is negligible. By contrast, BRC4 binds strongly to RAD51 as previously reported (Wong et al., 1997), but binding to HMG20b is undetectable. The BRC5–HMG20b interaction is direct, as indicated by the ability of recombinant His-tagged HMG20b to bind to a synthetic peptide encoding BRC5 (Figure 2d). Thus, our findings provide the first evidence that the BRC repeat motifs of BRCA2 bind to a protein other than the RAD51 recombinase, and in particular, suggest a functional dichotomy between BRC4 and BRC5 in their ability to bind RAD51 and HMG20b.
BRC5 inhibits the interaction in cells between BRCA2 and HMG20b but not RAD51, in contrast to BRC4
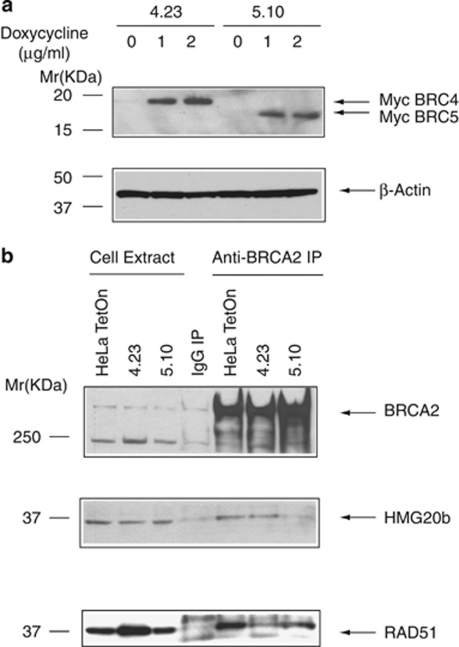
To analyze the functional significance of the BRCA2–HMG20b interaction, we established stable cell lines expressing tetracycline-inducible, myc-tagged fragments encoding BRC4 (spanning BRCA2 residues 1481–1553) or BRC5 (residues 1636–1715), respectively, in the cell line HeLa TetOn. Two clones expressing similar levels of myc-tagged BRC4 or BRC5 (clones 4.23 and 5.10, respectively) upon treatment with doxycycline were chosen for further study (Figure 3a).
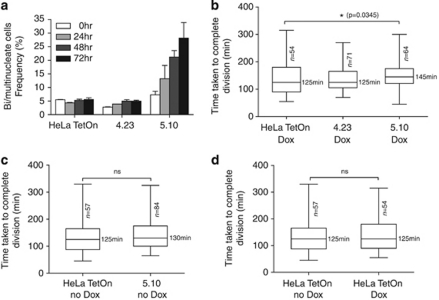
Figure 3.
BRC5 overexpression in cells inhibits the interaction between BRCA2 and HMG20b, whereas BRC4 does not. (a) HeLa TetOn cell clones stably expressing either myc-tagged BRC4 (clone 4.23) or BRC5 (clones 5.10) were induced by doxycycline treatment, and the expression of myc-BRC4 or -BRC5 was analyzed by western blotting with anti-myc antibody. β-Actin was used as a loading control. (b) BRCA2-associated protein complexes were immunoprecipitated from myc-BRC4- or -BRC5-expressing cells using protein A-coupled anti-BRCA2 antibody (αB2-9), and western blotted with anti-HMG20b or anti-RAD51 antibodies, as shown.
Next, we determined whether or not the expression of myc-BRC4 or myc-BRC5 affects the interaction between BRCA2 and HMG20b. We pulled down BRCA2 complexes using an anti-BRCA2 antibody from cells induced to express either myc-BRC4 or myc-BRC5, and probed the immunoprecipitates for HMG20b or RAD51 (Figure 3b). In myc-BRC5-expressing cells (5.10), the amount of HMG20b coimmunoprecipitated with BRCA2 was less than in cells expressing myc-BRC4 (4.23) or control HeLa TetOn cells (compare lanes 5, 6 and 7). In contrast, the amount of RAD51 coimmunoprecipitated with BRCA2 was not affected by myc-BRC5 expression, but was decreased by myc-BRC4 (lanes 5, 7 and 6). These results show that the expression of BRC4 or BRC5 can specifically inhibit the endogenous interaction of BRCA2 with RAD51 or HMG20b, respectively.
BRC5 inhibits the completion of cell division but not damage-induced RAD51 assembly, in contrast to BRC4
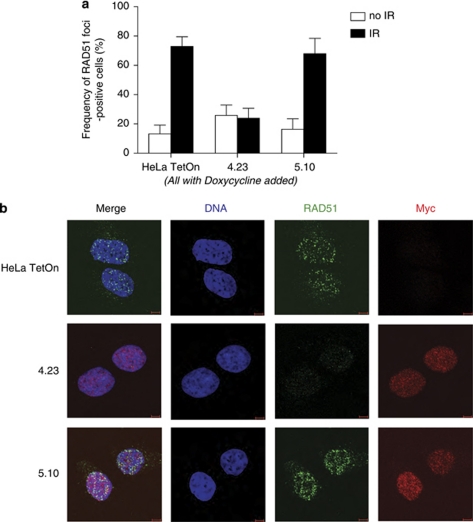
We therefore examined the effect of myc-BRC4 or myc-BRC5 expression on the assembly of RAD51 at sites of DNA damage, a function of BRCA2 mediated through its RAD51-binding BRC repeats including BRC4 (Chen et al., 1999; Yuan et al., 1999; Pellegrini et al., 2002). Cells induced to express either myc-BRC4 or myc-BRC5 were exposed to 5 Gy ionizing radiation, and stained 4 h later with an anti-RAD51 antibody to enumerate the formation of damage-induced RAD51 foci (Figures 4a and b). Consistent with the lack of biotin-BRC5 binding to RAD51 (Figure 2d) as well as the failure of myc-BRC5 expression to inhibit the BRCA2-RAD51 interaction in cells (Figure 3b), cells induced to express myc-BRC5 were still capable of forming damage-induced RAD51 foci (Figures 4a and b). In contrast, myc-BRC4 expression suppressed the induction of RAD51 foci, commensurate with its ability to bind RAD51.
Figure 4.
BRC4 overexpression in cells inhibits ionizing radiation (IR)-induced RAD51 foci formation, whereas BRC5 does not. (a) Doxycycline-induced, myc-BRC4- or -BRC5-expressing cells were treated with IR (5 Gy) and fixed 4 h later for immunostaining with anti-RAD51 and anti-myc antibodies. Cells containing more than 15 foci were counted as positive for RAD51 foci formation. The mean±s.e.m. from three independent experiments is shown. The number of cells examined in each sample was ⩾100. (b) Representative micrographs showing RAD51 foci formation in BRC-repeat-expressing cells. DNA in blue, RAD51 in green and myc in red are shown. Scale bar is 5 μm.
These results prompted us to determine whether cells induced to express myc-BRC5 would exhibit defects in the completion of cell division similar to those induced by HMG20b depletion. We induced myc-BRC4 or myc-BRC5 expression in the stable clones and quantified binucleate cells for a 72 h period (Figure 5a, n>500 for each sample). In myc-BRC5-expressing cells, the binucleate cell population increases up to 28.2% after 72 h, whereas there was no significant increase in HeLa TetOn cells or myc-BRC4-expressing cells (5.6 and 4.9%, respectively) treated in a similar manner.
Figure 5.
BRC5 overexpression inhibits the completion of cell division. (a) BRC4 (4.23) and BRC5 (5.10) clones were treated with doxycycline, and bi- or multinucleated cells were counted from phase-contrast images taken at the indicated time points. The mean±s.e.m. from three independent experiments is shown. The number of cells enumerated in each experiment was ⩾500 cells. Similar results were obtained with two additional, independently derived myc-BRC5-expressing clones (Supplementary Figure S4). (b) Myc-BRC4- or -BRC5-expressing clones were followed by time-lapse imaging. The time taken from anaphase onset to the completion of cell division was measured and plotted in a box-and-whisker graph. The median value is shown; the bottom and top of the box represent the 25 and 75th percentiles, respectively. Statistical comparisons in this and the subsequent panels were by one-way analysis of variance using the Kruskal–Wallis test. See also Supplementary Movies 3 and 4. (c) Clone 5.10 is compared with HeLa TetOn cells before they were treated with doxycycline. (d) HeLa TetOn cells are compared before/after doxycycline treatment.
We further analyzed progression through mitosis using serial time-lapse microscopy in cells induced to express the myc-BRC repeats (Figure 5b, see also Supplementary Movies 3 and 4). The myc-BRC5-expressing cells (5.10) initiate cleavage furrowing and undergo furrow ingression, but the time taken from anaphase onset to the completion of division is delayed from the median value of 125 min (HeLa TetOn, n=54) to 145 min (5.10, n=64). By contrast, expression of myc-BRC4 has no significant effect on the time taken to complete mitosis (median=125 min, n=71). Several further controls also suggest that defects in the completion of cell division are triggered specifically by myc-BRC5 expression. First, 5.10 cells did not exhibit any delay in the completion of mitosis when myc-BRC5 expression was not induced (Figure 5c, median=130 min, n=84) compared with untreated HeLa TetOn cells (median=125 min, n=57). Moreover, doxycycline treatment itself did not affect the completion of mitosis in HeLa TetOn cells (Figure 5d, median=125 min, n=57 or 54). Similar results were obtained using two additional, independently derived cell clones inducibly expressing BRC5 under TetOn control (Supplementary Information, Supplementary Figures S4 a–c). Taken together, these results show that myc-BRC5 expression perturbs the completion of cell division without preventing furrow ingression, recapitulating the abnormalities provoked by HMG20b depletion. Thus, our findings provide evidence that the novel role we have identified for HMG20b in the completion of cell division is regulated by its interaction with the BRC5 repeat of BRCA2.
Discussion
Our work suggests a novel biological function for the poorly characterized HMG20b protein. We observe using serial time-lapse imaging that HMG20b depletion delays the completion of cell division by cytokinesis. Most HMG20b-depleted cells that show a cytokinesis defect can initiate cleavage furrow formation and ingression as observed by phase-contrast microscopy, but are delayed in completing cell division, and frequently fail to separate. The mechanism by which HMG20b regulates cytokinesis warrants further investigation. Interestingly, in a fraction of the HMG20b-depleted cells that become binucleate, we occasionally observe rapid oscillation of the daughter nuclei through the contractile ring (Supplementary Movie 2). This is reminiscent of anillin function in the stabilization of the cleavage furrow (Straight et al., 2005; Zhao and Fang, 2005), raising the possibility that HMG20b may perform a similar role. However, the HMG20b binding-partner KIF4 has been implicated in spindle mid-zone formation and the progression of cytokinesis (Lee and Kim, 2003; Zhu and Jiang, 2005), and so perturbations in KIF4 function could also contribute to the defects that we observe in HMG20b-depleted cells.
Our work also provides fresh insight into the mechanism of tumor suppression by BRCA2. BRCA2 inactivation in mouse models or in human cancers causes the accumulation of aneuploid cells (Patel et al., 1998; Tutt et al., 1999), suggesting that BRCA2 directly or indirectly regulates chromosome segregation. Indeed, multiple studies by different groups have shown that BRCA2-deficient cells exhibit defects in the G2–M transition (Marmorstein et al., 2001; Ayoub et al., 2009) or in the completion of cytokinesis (Daniels et al., 2004; Jonsdottir et al., 2009; Vinciguerra et al., 2010). By contrast, one recent study reported no evidence of mitotic delays or cytokinetic defects in human cells with compromized BRCA2 function, and suggested that BRCA2 might not be involved in the control of cell division at all (Lekomtsev et al., 2010). Notably however, Lekomtsev et al. found that a small percentage of HeLa (Kyoto) cells (which we estimate from their data to approximate ∼5–10%) exhibited bi- or multinucleation after exposure to two out of three individual siRNAs against BRCA2; this effect was low in comparison with their positive control, an siRNA against the well-known cytokinetic mediator, MgcRacGAP (Lekomtsev et al., 2010). Nevertheless, a similarly low percentage of multinucleation recently reported in HeLa cells depleted of FANCD1/BRCA2 was found to be statistically different from controls (Vinciguerra et al., 2010). Moreover, multi-nucleate cell divisions also occur in tissues and cultures of cells obtained from transgenic mice carrying mutant Brca2 (Daniels et al., 2004; Rowley et al., 2011). Consistent with these findings, we too observe a modest but statistically significant increase in failure to complete cell division, and in the time taken for the process, when BRCA2 is depleted from HeLa (Kyoto) cells using multiple distinct siRNAs created with the algorithms developed by different manufacturers (Supplementary Information, Supplementary Figures S5a–c). Moreover, we find that a murine monoclonal antibody against BRCA2 stains the cytokinetic mid-body in HeLa (Kyoto) cells, and that this staining is reduced but not entirely ablated after BRCA2 depletion by siRNA (Supplementary Figures S6a-c).
Collectively, these results support the possibility that BRCA2 participates in mitotic cell division. That the division defects induced by its depletion affect only a fraction of cells suggests that BRCA2 is not essential for cytokinesis, but instead, may directly or indirectly affect the efficiency of its completion, as previously proposed (Daniels et al., 2004). One possible explanation is that BRCA2 depletion exerts its effects on cytokinesis by modulating the function of other proteins with stronger roles in cell division. Moreover, these functions may indirectly affect cytokinesis, for instance, by perturbing other steps in mitosis upon which efficient cell division is contingent. For example, the BRCA2-associated protein BCCIP has been reported to regulate mitotic spindle formation and centrosome number, as well as cytokinesis (Meng et al., 2007).
The findings we report here indicate that BRCA2 may also regulate cell division through its interaction with HMG20b. We show for the first time that the BRC repeats of BRCA2, previously implicated in the control of DNA recombination via their interaction with RAD51, also bind to HMG20b. Interestingly, the BRC5 repeat binds strongly to HMG20b but not RAD51, whereas the BRC4 repeat exhibits the converse properties. Moreover, BRC5 overexpression in cells provokes defects in cell division but not RAD51 assembly, whereas BRC4 elicits the converse effects. The dichotomous roles of BRC4 and BRC5 argue that BRCA2 has functions in the completion of cell division mediated via HMG20b that are separable from its known functions in DNA recombination mediated by RAD51 (Figure 6).
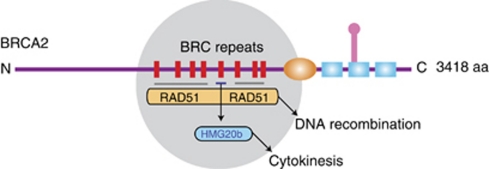
Figure 6.
A model for the divergent tumor-suppressive functions of the BRC repeats of BRCA2. In a schematic depiction of human BRCA2, the gray circle highlights the evolutionarily conserved region that contains eight BRC repeats whose sequence and spacing are preserved among mammalian orthologues. Most BRC repeats, including BRC4, are known to bind and control the RAD51 recombinase in reactions that lead to homologous DNA recombination. BRC5 (but not BRC4) binds directly to HMG20b, and this interaction regulates an unrecognized function for HMG20b in the efficient completion of cell division by cytokinesis. Thus, divergent tumor-suppressive functions maintaining chromosome segregation as well as chromosome structure may be mediated by the conserved BRC repeats of BRCA2.
Proteins other than RAD51 that bind to the BRC repeats of BRCA2 have not hitherto been identified. However, the human BRC5 repeat exhibits only a limited capability to bind RAD51; indeed, one of the two tetrameric clusters of hydrophobic residues essential for RAD51 interaction is poorly functional in BRC5 because of the occurrence of a polar Ser residue (Rajendra and Venkitaraman, 2010). The results we report here provide evidence that the human BRC5 repeat may, instead, be specialized for binding HMG20b. Given the poor affinity of human BRC5 for RAD51 in comparison with HMG20b in vitro, the BRC5–HMG20b interaction may be preferred under physiological conditions. Indeed, BRC5 overexpression in cells decreases the interaction of BRCA2 with HMG20b but not RAD51.
We estimate that only a small fraction of the total cellular pools of HMG20b and BRCA2 (<5%) is involved in complex formation. Nonetheless, disruption of the HMG20b–BRCA2 interaction by overexpression of BRC5 but not BRC4 suffices to induce defective cytokinesis. From this, we suggest that it is the BRCA2-associated fraction of HMG20b that most likely participates in cell division.
In conclusion, our findings suggest a novel biological function for HMG20b in the completion of cell division, and provide evidence that this function is regulated by its interaction with BRCA2. By showing that HMG20b binds to the BRC5 repeat of human BRCA2, and that the inhibition of the BRCA2-HMG20b interaction via BRC5 overexpression can phenocopy the effects of HMG20b depletion on cell division, our work suggests a model (Figure 6) in which the conserved BRC repeats of BRCA2 may control divergent tumor-suppressive pathways that preserve chromosome number as well as chromosome structure.
Materials and methods
Protein expression and purification
Glutathione S-transferase (GST)–BRCA2 fragments were prepared as described before (Lee et al., 2004). Human HMG20b complementary DNA was cloned into pET21a vector (Merck4Biosciences, Darmstadt, Germany). His-HMG20b was expressed in BL21 cells and purified using Ni-NTA agarose beads (Qiagen, Crawley, UK).
GST pull-down assay
GST–BRCA2 fragments bound to Glutathione Sepharose Beads (GE Healthcare, Chalfont St Giles, UK) were incubated with 293T cell extracts expressing Flag-HMG20b in the binding buffer (50 m Tris–HCl (pH 7.4), 150 m NaCl, 1% NP-40, 5 m EDTA, 1 m PMSF, 10 m NaF, Complete protease inhibitor cocktail (Roche, Burgess Hill, UK)) for 30 min at room temperature. The beads were washed extensively with the binding buffer and analyzed by western blotting.
Streptavidin pull-down assay
Biotinylated BRC peptides were synthesized by Cancer Research UK, Protein and Peptide Chemistry Service. BRC4 peptide corresponds to human BRCA2 residues 1517–1551, and BRC5 corresponds to residues 1661–1695. For the pull-down assay, 1 nmol of biotinylated BRC4 and BRC5 peptides were preincubated with Dynabeads M-280 Streptavidin (Invitrogen, Paisley, UK) in the binding buffer (50 m Tris–HCl (pH 7.4), 150 m NaCl, 1% NP-40, 5 m EDTA, 1 m PMSF, 10 m NaF, 0.1% bovine serum albumin, Complete protease inhibitor cocktail (Roche)). In all, 300 μg of 293T cell extracts expressing Flag-HMG20b or 1 μg of His-HMG20b were incubated with the beads for 30 min at room temperature and washed with the binding buffer using a magnetic separation unit. Bound proteins were eluted in sample buffer and analyzed by western blotting.
Affinity pull-down of BRCA2 and HMG20b complexes
Rabbit IgG, rabbit polyclonal anti-BRCA2 Ab (raised against human BRCA2 3189–3418, anti-B2-9) and rabbit polyclonal anti-HMG20b Ab (M10, raised against human HMG20b 138–245) were coupled to protein A beads using DMP (Sigma, St. Louis, MO, USA). The 293T cells were extracted with 1 volume of buffer A (50 m HEPES (pH 7.4), 420 m NaCl, 0.2% NP-40, 1 m EDTA, 25% glycerol, 1 m DTT and Complete protease inhibitor cocktail (Roche)) and the extracts were diluted with 2 volumes of buffer B (50 m HEPES (pH 7.4), 0.2% NP-40, and 1 m EDTA). In all, 2 mg of diluted extracts were incubated with 20 μg of protein A-coupled antibodies. The complexes were washed with the 1:2 mixture of buffers A and B, and eluted with sample buffer for analysis with 3–8% Tris-Acetate gel (Invitrogen). For BRCA2 pull-down from mitotic cell extracts, HeLa (Kyoto) cells were treated with 40 ng/ml of nocodazole overnight to enrich the cells in prometaphase. To enrich the cells undergoing cytokinesis, cells arrested overnight in nocodazole were treated with 22.5 μ Purvalanol A (Merck4Biosciences) for 40 min.
Plasmids and transfection
Human HMG20b complementary DNA was cloned into p3xFLAG-CMV-10 vector (Sigma) and used for transfection of 293T cells using Lipefectamine2000 (Invitrogen) transfection reagent.
Western blotting
Whole-cell extracts were made in the NP-40 lysis buffer (50 m HEPES (pH 7.4), 100 m NaCl, 0.5% NP-40, 10 m EDTA, 20 m β-glycerophosphate, 1 m DTT, 1 m sodium orthovanadate, 1 m PMSF and Complete protease inhibitor cocktail (Roche)). BRCA2 was resolved by 3–8% Tris-Acetate gels (Invitrogen) and detected with anti-BRCA2 mouse mAb Ab-1 (Merck4Biosciences). Other proteins were resolved by 10 or 12% SDS polyacrylamide gel and probed with anti-HMG20b Ab (rabbit polyclonal M10 or mouse monoclonal clone 4.21 (anti-BRAF35, Millipore, Billerica, MA, USA)), anti-Flag mouse mAb (M2, Sigma), anti-6xHis mouse mAb (Clontech, Saint-Germain-en-Laye, France), anti-RAD51 Ab (rabbit polyclonal Ab-1, Merck4Biosciences or mouse mAb14B4 (GeneTex, Irvine, CA, USA) and 9E10 mouse mAb (anti-Myc, Santa cruz Biotechnology, Santa cruz, CA, USA). Anti-β-Actin Ab (mouse mAb AC-15, Sigma) and GAPDH Ab (mouse mAb A-3, Santa cruz Biotechnology) were used to confirm equal loading of samples.
Immunofluorescence
HeLa cells grown overnight on glass coverslips were fixed in 4% paraformaldehyde (PFA) for 5 min, permeabilized with TBS-T (Tris-buffered saline containing 0.1% Triton-X100) and incubated in blocking solution (2% bovine serum albumin in TBS-T). Primary and secondary antibodies were diluted in blocking solution and incubated for1 h and 30 min, respectively. RAD51 was stained with rabbit polyclonal antibody (Ab-1, Merck4Biosciences, 1:1000). Mouse monoclonal antibody (9E10, Santa cruz Biotechnology 1:500) was used to stain myc-BRC repeats. Secondary antibodies used were Alexa 488 or 568-conjugated goat IgGs from Invitrogen. Images were acquired with Zeiss (Welwyn Garden City, UK) LSM510 META confocal microscope.
For immunostaining of BRCA2, HeLa (Kyoto) cells transfected with either control or BRCA2 siRNA (non-targeting siRNA no. 2 and siGENOME SMART pool M-003462-01, respectively, Thermo Scientific, Lafayette, CO, USA) were fixed in 4% PFA and incubated with anti-BRCA2 mouse mAb 5.23 (Millipore, 1:50) and anti-Aurora B rabbit polyclonal Ab (ab2254, 1:1,000, Abcam, Cambridge, UK). DNA was stained with Hoechst 33342 dye (10 μg/ml) and coverslips were mounted with Mowiol (Merck4Biosciences). Mean intensity was measured using Image J software (NIH, Bethesda, MD, USA) from more than 100 cells for nuclear staining and from 40 late-anaphase cells for mid-body staining in each experiment. The results from two independent experiments were analyzed using GraphPad Prism software (La Jolla, CA, USA; unpaired t-test).
siRNA-mediated depletion of HMG20b and BRCA2
Control (mock) or HMG20b siRNAs (siGENOME SMART pool, catalog no. M-020146-01-0005, Thermo Scientific) were transfected into HeLa cells seeded on a 6-well plate using Lipofectamine 2000 reagent (Invitrogen). After 24 h of transfection, cells were plated into a 2-well Lab-Tek chamber slide (Nunc, Roskilde, Denmark) and followed by time-lapse imaging from 30 h after transfection.
Similar methods were used to analyze cells exposed to the following individual HMG20b siRNA sequences (Thermo Scientific):
#2: D-020146-02 5′-GAGAGAAGCAGCAGUACAU-3′ and
#4: D-020146-04 5′-GGACACAGGGCAGACGAAA-3′.
Individual BRCA2 siRNA sequences were used at 20 n to transfect HeLa (Kyoto) cells for serial time-lapse imaging. The sequences are:
Luciferase siRNA (control): 5′-CGUACGCGGAAUACUUCGA-3′
Qiagen #6: SI02653434 (Qiagen) (Lekomtsev et al., 2010)
Invitrogen #7: BRCA2HSS101097 (Invitrogen) (Lekomtsev et al., 2010)
Dharmacon #1: D-003462-01 5′-GAAACGGACUUGCUAUUUA-3′
Dharmacon #2: D-003462-02 5′-GUAAAGAAAUGCAGAAUUC-3′
Dharmacon #3: D-003462-03 5′-GGUAUCAGAUGCUUCAUUA-3′
Dharmacon #4: D-003462-04 5′-GAAGAAUGCAGGUUUAAUA-3′ (all from Thermo Scientific).
Serial time-lapse imaging
Cells were changed into Leibovitzs L15 medium before taking bright-field images using Zeiss Axiovert 200 M microscope equipped with a humidified heated enclosure. Time-lapse images were collected every 5 min using Volocity software (Improvision, Perkin Elmer, Waltham, MA, USA). Cells were followed from anaphase onset, when chromosome separation first becomes visible, until the completion of cell division, for as long as required up to 6 h. Cells that did not complete cytokinesis by the end of observation period, or formed bi- or multinucleated cells, were classified as having failed to complete cell division. For representative time-lapse images, HeLa cells expressing green fluorescent protein-H2B were transfected with HMG20b siRNA before differential interference contrast (DIC), and fluorescence images were captured with an Olympus (Southend-on-Sea, UK) IX81 microscope and Cell software. The unpaired t-test was performed using GraphPad Prism software (La Jolla, CA, USA), except for Figure 5b and Supplementary Figures S1d, S4b, and S5c where one- way analysis of variance was used.
Inducible expression of BRC repeats
Tet-On Inducible Gene Expression System (Clontech) was used to express BRC repeats conditionally. BRCA2 fragments containing BRC repeats (BRC4: residues 1481–1553, BRC5: residues 1636–1715) were cloned into pTRE2pur vector together with 3 × NLS and Myc epitope from pEF/myc/nuc vector (Invitrogen). The constructs were used to transfect HeLa TetOn advanced cell line, and stable clones were selected with puromycin (1 μg/ml). Selected clones were screened for similar levels of expression upon addition of doxycycline (1 μg/ml, Sigma). For pull-downs with anti-BRCA2 antibody, each clone was treated with doxycycline (1 μg/ml) overnight before harvesting, and 6 mg of cell extracts and 100 μg of coupled antibody were used for each pull-down. For time-lapse imaging, doxycycline was added 16 h before the start of imaging and kept for a further 24 h during imaging.
Bi/multinucleation of BRC-expressing cells
HeLa TetOn, clone 4.23 and clone 5.10 cells were plated in 35 mm dishes and treated with doxycycline (1 μg/ml) for 24, 48 and 72 h, respectively. Phase-contrast images were taken at each time point, and over 500 cells were counted for each sample. Results from three independent experiments were analyzed using GraphPad Prism software.
Ionizing radiation-induced RAD51 foci formation
BRC-repeat-expressing clones were grown overnight on coverslips in the presence of doxycycline (1 μg/ml ), treated with 5 Gy of ionizing radiation and fixed with 4% PFA 4 h later. Cells showing more than 15 RAD51 foci were counted as positive. The results are from three independent experiments, and in each experiment, more than 100 cells were counted for each sample.
Acknowledgments
ML performed the experiments reported here and conceived them with ARV. MJD and MJG generated preliminary data, suggested experiments and assisted with data interpretation. We thank Dr Mark Petronczki (Clare Hall Labs, London) for sharing protocols and the siRNA sequences Q#6 and I#7 (Lekomtsev et al., 2010), and the members of our lab for helpful discussions. MJG was supported in part by a fellowship from the Canadian Institute for Health Research, and ARV was supported by a grant from the Medical Research Council, which also funded ML and MJD.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Breast Cancer Linkage Consortium Cancer risks in BRCA2 mutation carriers. The breast cancer linkage consortium. J Natl Cancer Inst. 1999;91:1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Rajendra E, Su X, Jeyasekharan AD, Mahen R, Venkitaraman AR. The carboxyl terminus of Brca2 links the disassembly of Rad51 complexes to mitotic entry. Curr Biol. 2009;19:1075–1085. doi: 10.1016/j.cub.2009.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Blomberg N, Nilges M. Internal repeats in the BRCA2 protein sequence. Nat Genet. 1996;13:22–23. doi: 10.1038/ng0596-22. [DOI] [PubMed] [Google Scholar]
- Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, et al. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J Biol Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–879. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- Jonsdottir AB, Vreeswijk MP, Wolterbeek R, Devilee P, Tanke HJ, Eyfjord JE, et al. BRCA2 heterozygosity delays cytokinesis in primary human fibroblasts. Cell Oncol. 2009;31:191–201. doi: 10.3233/CLO-2009-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TA, Li W, Brogi E, Yee CJ, Gemignani ML, Olvera N, et al. Heterogenic loss of the wild-type BRCA allele in human breast tumorigenesis. Ann Surg Oncol. 2007;14:2510–2518. doi: 10.1245/s10434-007-9372-1. [DOI] [PubMed] [Google Scholar]
- Lee M, Daniels MJ, Venkitaraman AR. Phosphorylation of BRCA2 by the Polo-like kinase Plk1 is regulated by DNA damage and mitotic progression. Oncogene. 2004;23:865–872. doi: 10.1038/sj.onc.1207223. [DOI] [PubMed] [Google Scholar]
- Lee YM, Kim W. Association of human kinesin superfamily protein member 4 with BRCA2-associated factor 35. Biochem J. 2003;374:497–503. doi: 10.1042/BJ20030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekomtsev S, Guizetti J, Pozniakovsky A, Gerlich DW, Petronczki M. Evidence that the tumor-suppressor protein BRCA2 does not regulate cytokinesis in human cells. J Cell Sci. 2010;123:1395–1400. doi: 10.1242/jcs.068015. [DOI] [PubMed] [Google Scholar]
- Marmorstein LY, Kinev AV, Chan GK, Bochar DA, Beniya H, Epstein JA, et al. A human BRCA2 complex containing a structural DNA binding component influences cell cycle progression. Cell. 2001;104:247–257. doi: 10.1016/s0092-8674(01)00209-4. [DOI] [PubMed] [Google Scholar]
- Meng X, Fan J, Shen Z. Roles of BCCIP in chromosome stability and cytokinesis. Oncogene. 2007;26:6253–6260. doi: 10.1038/sj.onc.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson KL, Wooster R, Weber BL. Breast cancer genetics: what we know and what we need. Nat Med. 2001;7:552–556. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, et al. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- Rahman N, Stratton MR. The genetics of breast cancer susceptibility. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- Rajendra E, Venkitaraman AR. Two modules in the BRC repeats of BRCA2 mediate structural and functional interactions with the RAD51 recombinase. Nucleic Acids Res. 2010;38:82–96. doi: 10.1093/nar/gkp873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley M, Ohashi A, Mondal G, Mills L, Yang L, Zhang L, et al. 2011Inactivation of Brca2 promotes Trp53-associated but inhibits KrasG12D-dependent pancreatic cancer development in mice Gastroenterology(e-pub ahead of print; doi: 10.1053/j.gastro.2010.12.039 [DOI] [PMC free article] [PubMed]
- Shivji MK, Davies OR, Savill JM, Bates DL, Pellegrini L, Venkitaraman AR. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivji MK, Mukund SR, Rajendra E, Chen S, Short JM, Savill J, et al. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc Natl Acad Sci USA. 2009;106:13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Field CM, Mitchison TJ. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol Biol Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumoy L, Carim L, Escarceller M, Nadal M, Gratacos M, Pujana MA, et al. HMG20A and HMG20B map to human chromosomes 15q24 and 19p13.3 and constitute a distinct class of HMG-box genes with ubiquitous expression. Cytogenet Cell Genet. 2000;88:62–67. doi: 10.1159/000015486. [DOI] [PubMed] [Google Scholar]
- Tutt A, Gabriel A, Bertwistle D, Connor F, Paterson H, Peacock J, et al. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9:1107–1110. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Vinciguerra P, Godinho SA, Parmar K, Pellman D, D'Andrea AD. Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells. J Clin Invest. 2010;120:3834–3842. doi: 10.1172/JCI43391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Chi T, Xue Y, Zhou S, Kuo A, Crabtree GR. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, et al. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- Zhao WM, Fang G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J Biol Chem. 2005;280:33516–33524. doi: 10.1074/jbc.M504657200. [DOI] [PubMed] [Google Scholar]
- Zhu C, Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc Natl Acad Sci USA. 2005;102:343–348. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.