SUMMARY
Native glutamate receptor ion channels are tetrameric assemblies containing two or more different subunits. NMDA receptors are obligate heteromers formed by coassembly of two or three divergent gene families. While some AMPA and kainate receptors can form functional homomeric ion channels, the KA1 and KA2 subunits are obligate heteromers which function only in combination with GluR5–7. The mechanisms controlling glutamate receptor assembly involve an initial step in which the amino terminal domains (ATD) assemble as dimers. Here we establish by sedimentation velocity that the ATDs of GluR6 and KA2 coassemble as a heterodimer of Kd 11 nM, 32000-fold lower than the Kd for homodimer formation by KA2; we solve crystal structures for the GluR6/KA2 ATD heterodimer and heterotetramer assemblies. Using these structures as a guide we perform a mutant cycle analysis to probe the energetics of assembly, and show that high affinity ATD interactions are required for biosynthesis of functional heteromeric receptors.
INTRODUCTION
During biogenesis most ion channels and neurotransmitter receptors undergo regulated assembly prior to insertion into the plasma membrane. In prokaryotes many ion channels function as homo-oligomers, which for individual subtypes range in size from dimers to hexamers, while in eukaryotes, as a consequence of gene duplication, appropriate members of a diverse subunit pool must be selected to form hetero-oligomeric assemblies of restricted stoichiometry and composition. The glutamate receptor ion channels (iGluRs) which mediate excitatory synaptic transmission are important examples of the biological diversity which arose from gene duplication, and these receptors play key roles in brain development, synaptic plasticity, motor function, information processing and memory formation. In mammals, the diverse functional roles of iGluRs are mediated by a family of 18 genes, several of which undergo alternative splicing and mRNA editing (Traynelis et al., 2010). Genetic, biochemical and functional studies have established that individual iGluR subunits will coassemble with members of the same functional family, but not with other subtypes, to generate the large and diverse receptor population required for normal brain activity (Ayalon et al., 2005; Ayalon and Stern-Bach, 2001; Brose et al., 1994; Burnashev et al., 1992; Leuschner and Hoch, 1999; Partin et al., 1993). A fundamental problem in biology is to understand the mechanisms controlling this selective assembly.
The major families of iGluRs were identified by classical pre-genetic techniques, using selective ligands and functional assays, leading to identification of AMPA, kainate and NMDA receptor subtypes (Watkins and Evans, 1981). For kainate and NMDA receptors, the native receptor assemblies in vivo contain subunits encoded by two or three different gene families, several of which do not generate functional ion channels when expressed as homomeric proteins. For example, GluR5, GluR6 and GluR7 (also called GluK1, GluK2 and GluK3) can form functional homomeric ion channels in heterologous expression systems (Egebjerg et al., 1991; Schiffer et al., 1997), but in vivo they coassemble with the KA1 and KA2 subunits from a second gene family (also called GluK4 and GluK5), which also bind glutamate, but which are functionally inactive when expressed as homomeric proteins (Herb et al., 1992; Werner et al., 1991). For NMDA receptors the principles are similar, with the GluN1subunit, which encodes the binding site for glycine or D-serine, forming obligate heteromeric assemblies with GluN2 subunits which bind glutamate (Furukawa et al., 2005; Monyer et al., 1992). By contrast, AMPA receptors are formed by coassembly of the GluA1–GluA4 subunits, each of which can form functional homomeric receptors (Hollmann et al., 1989; Keinänen et al., 1990), although in vivo most AMPA receptors contain both the GluA2 subunit, and either GluA1, GluA3 or GluA4 (Geiger et al., 1995; Rossmann et al., 2011).
A large number of studies have revealed that control of trafficking plays a key role in regulating iGluR transport to the plasma membrane and synapse (Greger and Esteban, 2007; Mah et al., 2005; Penn et al., 2008; Ren et al., 2003; Shi et al., 2010; Valluru et al., 2005). However, the mechanisms which control the assembly of heteromeric glutamate receptors assembled from two or three different gene families are largely unknown, but likely to involve multiple stages of regulation before transport comes into play. In particular, dimer assembly by the 380 residue amino terminal domain, which emerges from the ribosome before the ligand binding domain and its associated membrane embedded ion channel segments plays a key role in determining how subunits coassemble during the early stages of biogenesis. Recent studies on AMPA and NMDA receptors provide compelling evidence for such a role and highlight the complex mechanisms regulating iGluR assembly (Farina et al., 2011; Rossmann et al., 2011; Shanks et al., 2010). In this study we examine the role of the ATD in assembly of heteromeric kainate receptors assembled from the GluR6 and KA2 subunits, which form the most abundant kainate receptor subtype in the brain (Petralia et al., 1994). We address the issue of whether there exists a unique assembly pattern; define the mechanisms which underlie its formation and which exclude alternative assemblies; and probe the energetics of assembly for heteromeric glutamate receptors.
RESULTS AND DISCUSSION
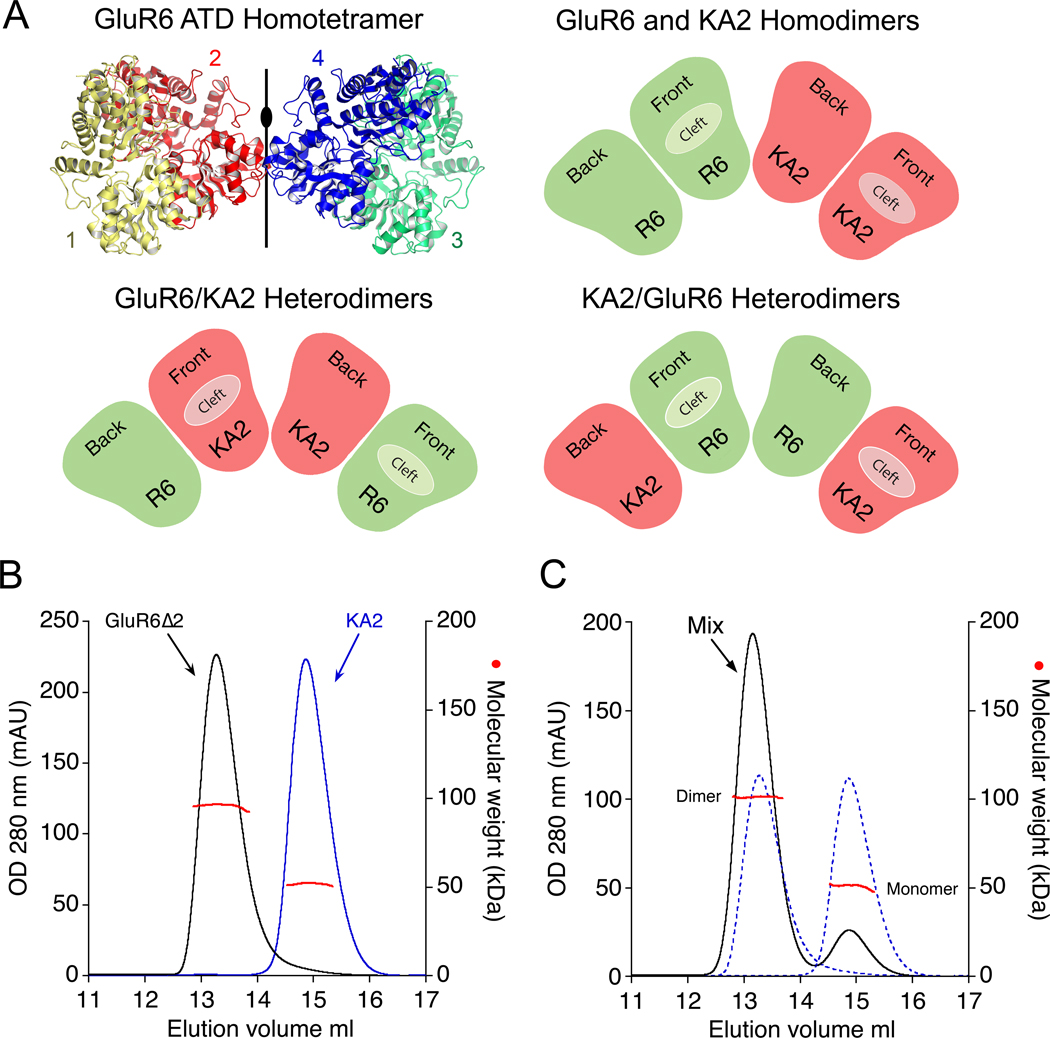
Glutamate receptor ion channels are tetrameric assemblies in which both the ATD and ligand binding domains assemble as a dimer of dimers (Sobolevsky et al., 2009). Because the iGluR ATD dimer of dimers assembly lacks 4-fold rotational symmetry, a receptor generated by coassembly of GluR6 and KA2 subunits could be formed by pairs of homodimers or pairs of heterodimers, and in the latter case, the dimer of dimers interface could be formed by either the GluR6 or KA2 subunits (Figure 1A). To address the issue of whether the initial assembly is formed by homodimers or heterodimers, we used analytical size exclusion chromatography coupled with refractive index and multiangle light scattering detectors (SEC-UV/RI/MALS), combined with analytical ultracentrifugation (AUC), to measure interactions between the GluR6 and KA2 subunit ATDs; we then solved crystal structures of the dimeric and tetrameric ATD assemblies; performed a mutant cycle analysis of intersubunit contacts in the dimer assemblies; we used chemical cross linking to establish the assembly pattern in full length receptors; and determined that high affinity interactions between the GluR6 and KA2 ATDs are necessary for boisynthesis on heteromeric kainate receptors in vivo.
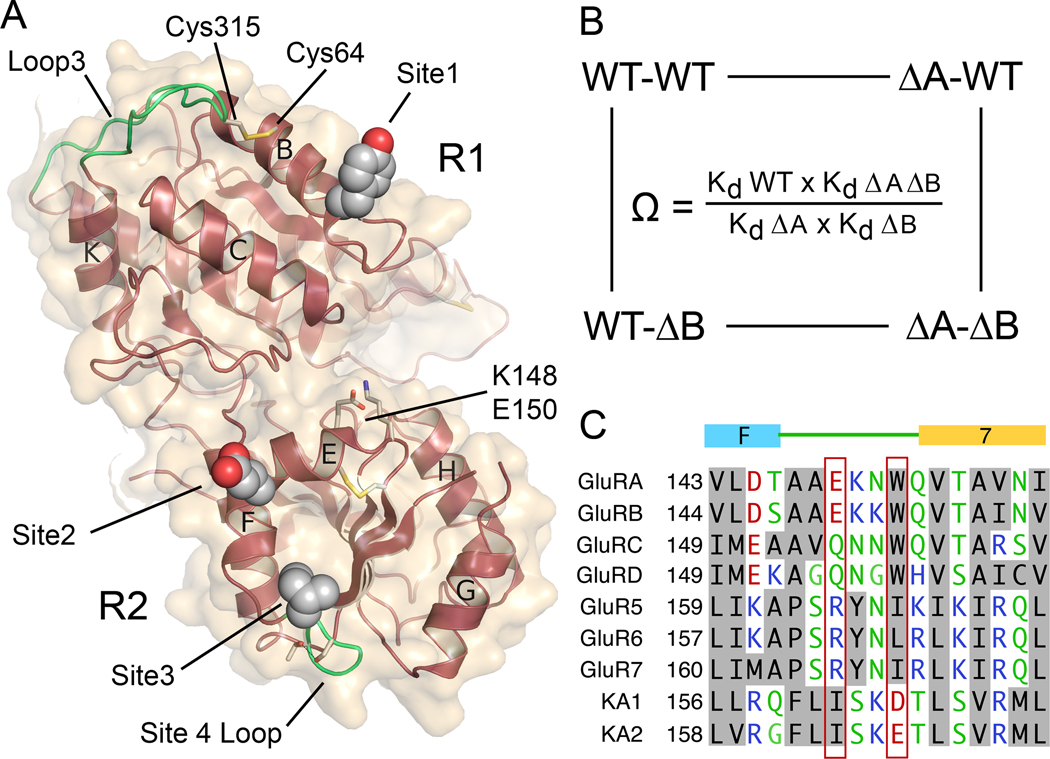
Figure 1. Assembly principles for heteromeric glutamate receptor ATDs.
(A) Crystal structure of the GluR6 ATD homotetramer (PDB 3H6H) with the four subunits individually colored, illustrating the global 2-fold axis of dimer symmetry; the cartoons show three possible subunit arrangements for an ATD heterotetramer assembled from GluR6 (green) and KA2 subunits (red). (B) Superimposed gel permeation chromatography profiles for the GluR6Δ2 and KA2 ATDs when the proteins were injected separately; analysis by SEC-UV/RI/MALS (red data points) revealed elution with mass values corresponding to dimers and monomers respectively. (C) When the two proteins were mixed at approximately equal concentrations prior to injection, the amplitude of the dimer peak increased, with a corresponding decrease in the monomer peak, indicating formation of GluR6/KA2 heterodimers; dashed lines show data from B scaled by 50% to account for the dilution factor when the samples were mixed.
Analysis of GluR6 & KA2 amino terminal domain oligomerization
We expressed the GluR6 and KA2 ATDs as soluble glycoproteins in HEK293 cells and purified them to homogeneity by affinity and ion-exchange chromatography. Analytical size exclusion chromatography revealed a broad and asymmetric elution profile for the GluR6 ATD at physiological pH, with a peak mass of 192 kDa that we initially interpreted as resulting from a monomer-dimer-tetramer equilibrium (Figure S1A). However, sedimentation velocity (SV) experiments at loading concentrations of 2–33 µM revealed a reversible, concentration dependent formation of much larger oligomeric species (Figure S1B). In prior work we found that this behavior was suppressed at pH 5 (Kumar et al., 2009), which was an acceptable compromise for our initial structural studies on iGluR ATDs, but inappropriate for an analysis of assembly mechanisms, since the pH in the cytoplasm, endoplasmic reticulum, and Golgi apparatus is close to neutral. In order to circumvent GluR6 aggregation at physiological pH we resorted to protein engineering, capitalizing on prior structural knowledge of iGluR ATD assembly (Clayton et al., 2009; Jin et al., 2009; Kumar et al., 2009).
The ATDs of iGluRs have a clam-shell like structure for which the upper and lower lobes have been named domain R1 and R2 (Karakas et al., 2009; Kumar et al., 2009). In prior work we noted that in GluR6 ATD crystal structures the dimer assemblies pack via the lateral edges of domain R2 to generate spiral arrays of tetramers (Figure S1C), suggesting a possible mechanism involving domain R2 in the aggregation observed in SV experiments (Figure S1D). An N-linked glycan introduced into this interface would be expected to abolish aggregation of the GluR6 ATD in solution (Figure S1E), without interfering with dimer assembly. We verified this by making two glycan wedge mutants, GluR6Δ1 (A213N/G215S) and GluR6Δ2 (G215N/M217T), both of which showed chromatographic and sedimentation behavior consistent with formation of high affinity homodimers in the complete absence of higher MW species (Figure 1B, Figure S2A and Figure 3C. The X-ray crystal structure of the GluR6Δ1 mutant revealed an essentially identical dimer assembly as found for wild type GluR6 (RMSD 0.53 Å for 649 Cα atoms), but packed in a different space group with the glycan wedge facing solvent channels in the crystal lattice (Figure S1F). Although insertion of a glycan at the ATD dimer of dimers interface would likely disrupt assembly of an intact GluR6 tetramer, this modification allowed us to quantitatively analyze GluR6 ATD dimer assembly in isolation of higher order oligomers. For the KA2 ATD, SEC-UV/RI/MALS analysis revealed essentially monomeric behaviour at a loading concentration of 2.0 mg/ml in striking contrast to dimer formation for the GluR6 ATD (Figure 1B). Notably, when approximately equal concentrations of the GluR6Δ1 or GluR6Δ2 ATDs were mixed with the KA2 ATD at a 1:1 ratio prior to injection, the amplitude of the monomer peak decreased by 77% while that for the dimer increased by 71%, indicating preferential formation of GluR6/KA2 heterodimers (Figure 1C and Figure S2B). Because the GluR6Δ1 and GluR6Δ2 glycan wedge mutants had indistinguishable behavior assayed by SEC-UV/RI/MALS, in the majority of subsequent biochemical experiments we used GluR6Δ2, while for crystallization of heteromeric assemblies we continued to work with GluR6Δ1.
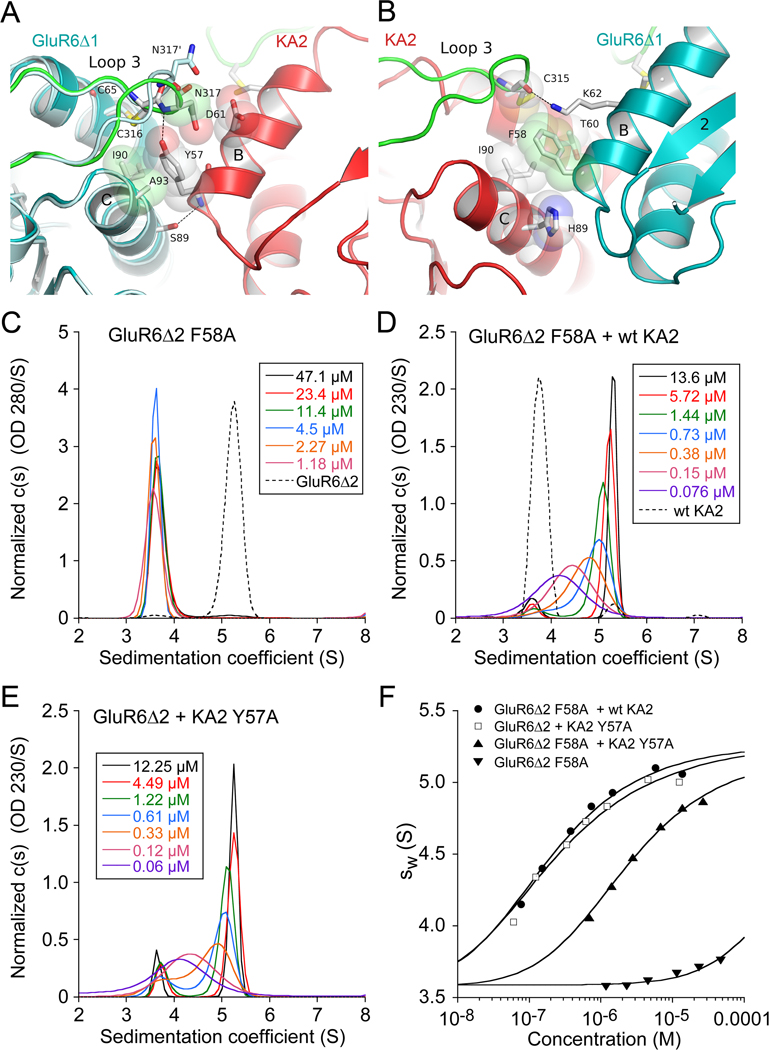
Figure 3. Role of conserved domain R1 aromatic side chains in heterodimer assembly.
(A) Crystal structure of the GluR6Δ1/KA2 heterodimer domain R1 interface showing the interaction of KA2 Tyr57 with residues in α-helices B and C and loop three of the GluR6 subunit. A single subunit from the GluR6 homodimer crystal structure, superimposed on α-helices B and C, is colored light blue and shows the change in conformation of loop 3 and Asn 317 in the heterodimer assembly. (B) The view after rotation by 180° shows the interaction of GluR6 Phe58 with residues in α-helices B and C and loop three of the KA2 subunit. (C) Sedimentation velocity analysis for self assembly of the GluR6Δ2 F58A mutant ATD reveals a shift in monomer-dimer equilibrium due to a 2000-fold increase in Kd for homodimer formation; the profile for 36 µM GluR6Δ2 which sediments as a dimer is shown as a dashed line. (D) When the GluR6Δ2 F58A mutant is mixed with wt KA2 the sedimentation profile shifts to the right due to formation of ATD heterodimers; the profile for 14 µM KA2 which sediments as a monomer is shown as a dashed line. (E) A similar sedimentation profile was obtained for an equimolar mixture of the GluR6Δ2 and KA2 Y57A mutant ATDs. (F) Isotherms of weighted-average sedimentation coefficients fit with a monomer-dimer model reveals that compared to mutations in single subunits, mutation to alanine of both GluR6 F58 and KA2Y57 produces a 13-fold increase in Kd for heterodimer formation.
Preferential assembly of GluR6 and KA2 heterodimers
For mixtures of self associating systems with components of similar molecular weight, like the GluR6 and KA2 ATDs, measurement of the Kds for monomer, dimer and tetramer equilibria by sedimentation analysis is technically challenging. The present study was greatly facilitated by the large difference in Kd for self association of the GluR6 and KA2 ATDs, and, as shown later, by mutants which preferentially disrupt homodimer versus heterodimer assemblies. To quantify the strength of the association between the GluR6 and KA2 ATDs we carried out sedimentation equilibrium (SE) experiments in an analytical ultracentrifuge at 10°C using multiple protein concentrations and rotor speeds. Experiments were performed for GluR6Δ2, KA2, and an approximately equimolar mix of the two proteins. In each case the data was best fit to a reversible monomer–dimer equilibrium model (Figure 2A). The GluR6Δ2 ATD formed homodimers with a Kd of 0.35 µM (95% confidence interval; 0.30 µM – 0.41 µM), compared to a Kd of 11 µM at pH 5 (Kumar et al., 2009), indicating that the ATD dimer assembly is a potential site of proton modulation. On the other hand, the KA2 ATD showed very weak association, with a best-fit binding constant of Kd 410 µM (95% confidence interval 380 µM – 440 µM). The Kd for heterodimer formation was 0.076 µM (95% confidence interval; 0.02 µM – 0.141 µM), with the heterodimer forming the major species when KA2 was in slight excess. Comparable Kd values of 0.25 µM (0.20 – 0.30 µM) for GluR6Δ2, 350 µM (380 – 650 µM) for KA2, and 0.011 µM (0.006 – 0.017 µM) for the heterodimer were obtained from sedimentation velocity (SV) experiments at 20°C, which in addition established the absence of any species of size larger than a dimer. The Kd value for GluR6Δ2/KA2 heterodimer formation from SV analysis is 32000-fold lower than that for homodimer formation by KA2 and 23-fold lower than the Kd for homodimer formation by GluR6Δ2, establishing that the GluR6Δ2 and KA2 ATDs preferentially assemble as heterodimers.
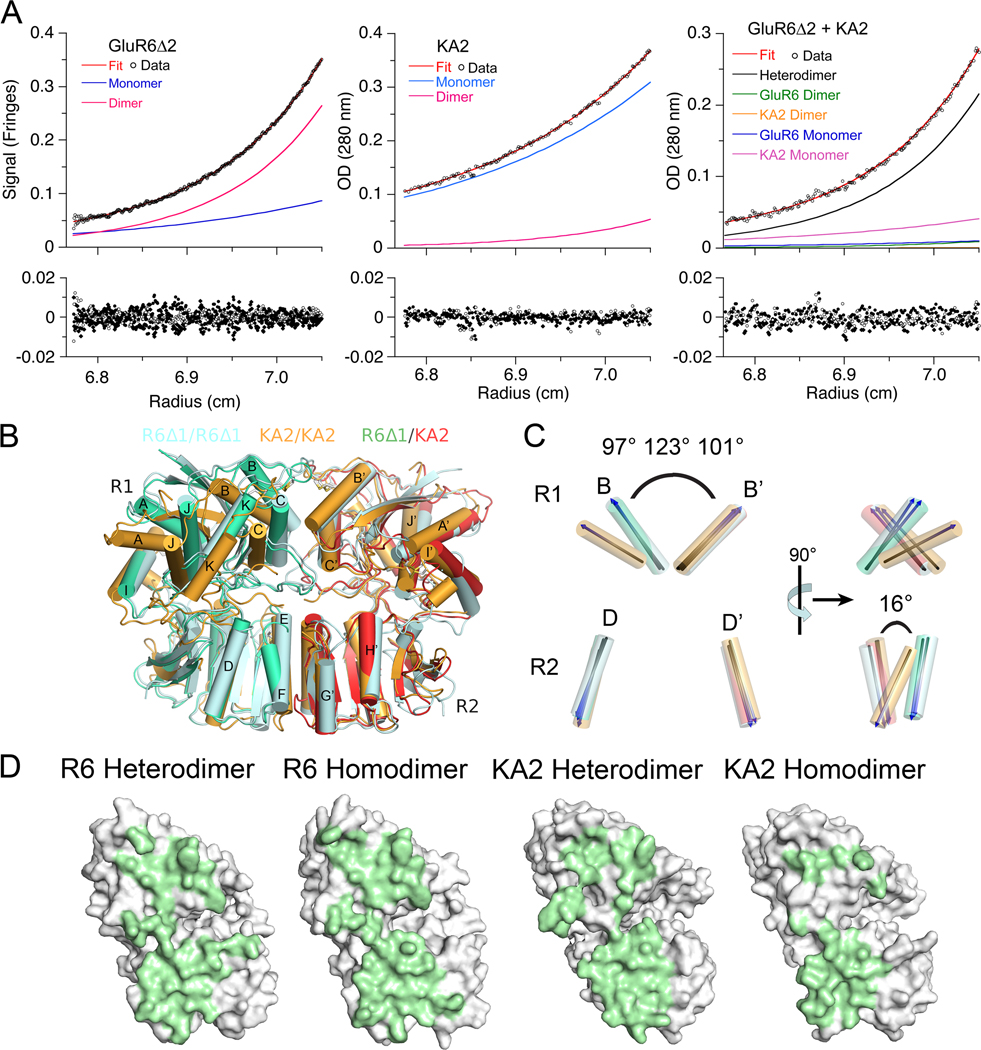
Figure 2. The GluR6 and KA2 ATDs coassemble to form high affinity heterodimers.
(A) Sedimentation equilibrium analysis for the GluR6Δ2 ATD, homodimer Kd 350 nM (left); the KA2 ATD, homodimer Kd 410 µM (middle); and an approximately equimolar mixture of the GluR6Δ2 and KA2 ATDs, heterodimer Kd 77 nM (right); fits to a monomer-dimer model (red line), with the calculated monomer and dimer populations shown as blue and pink lines, are shown for the three experiments (upper panels); the lower panels show residuals for single cells for a global fit to data for 3 loading concentrations each run at three speeds (6,500, 10,000 and 16,000 r.p.m). (B) The GluR6/KA2 heterodimer crystal structure superimposed using domain R2 coordinates on crystal structures for GluR6 (PDB 3G3H) and KA2 (PDB 3OM0) homodimers (C). Superimposed vectors drawn through helix B in domain 1 and helix D in domain 2, for the same structures, colored as in panel B, with the angle between helix B and its dimer partner indicated for the GluR6 homodimer, the KA2 homodimer, and the GluR6/KA2 heterodimer; rotation by 90° (right panel) reveals a 16° change in angle between helix D and helix D’ in domain R2 of the KA2 homodimer assembly. (D) Green shading illustrates buried molecular surfaces for the GluR6 and KA2 subunits for the same heterodimer and homodimer assemblies shown in B.
We also carried out SEC, SV and SE analysis for a mixture of the wild type GluR6 and KA2 ATDs at pH 7.4. The SEC elution profile shows a pronounced rightward shift compared to that obtained for GluR6 in the absence of KA2, but a left shift compared to the profile for GluR6Δ2 mixed with KA2 (Figure S3A). In the SV analysis, the c(s) distributions show a significantly extended reaction boundary compared to the GluR6Δ2 profile, consistent with a system in rapid equilibrium consisting of monomers, dimers, and higher order oligomers, most likely tetramers (Figure S3B). Strikingly, there was no formation of larger complexes like that observed for wt GluR6 at a similar concentration (Figure S1B), suggesting that GluR6/KA2 heterodimer formation is competitive with the assembly-pathway for high order GluR6 oligomers, and that the GluR6/KA2 heterodimer does not aggregate. With a small excess of the KA2 ATD, SE analysis for the wt GluR6/KA2 mixture could be well fit with a model for monomer-homodimer and monomer-heterodimer-heterotetramer equilibria, in which the monomer-homodimer and monomer-heterodimer Kds were constrained to values estimated in independent experiments, as described above; a global fit to nine data sets from multiple rotor speeds and loading concentrations gave an apparent Kd of 3.5 µM for tetramer formation by assembly of heterodimers (Figure S3C). A similar apparent Kd of 6.2 µM for tetramer formation was obtained from SV analysis. However, because the sedimentation mixture contains multiple species, including free GluR6, we cannot exclude other models in which the tetramer species is a mixture of both GluR6/KA2 tetramer assemblies and high order GluR6 oligomers. Thus, although the Kd for tetramer formation by kainate receptor ATDs remains uncertain, the interaction is several orders of magnitude weaker than for dimer formation.
Crystal structure of the GluR6/KA2 heterodimer assembly
To define the molecular mechanisms controlling ATD assembly we solved crystal structures for both the GluR6Δ1/KA2 heterodimer (Figure 2B), and as a control the GluR6Δ1 homodimer (Figure S1F), both at 2.9 Å resolution (Table 1). The conformation and arrangement of subunits closely resembles that observed previously for wild type GluR6 and GluR7 homodimers, with RMSDs of 1.55 Å (563 Cα atoms) and 0.53 Å (649 Cα atoms) for superposition on the wt GluR6 ATD dimer (PDB 3H6H). By contrast, there is a substantial difference in packing for the GluR6Δ1/KA2 heterodimer assembly compared the KA2 ATD homodimer assembly solved previously, RMSD 3.56 Å for 428 Cα atoms (PDB 3OM0, Kumar and Mayer, 2010). The most substantial difference from the KA2 homodimer structure is due to a change in orientation in the upper lobes of the two protomers in the GluR6Δ1/KA2 heterodimer assembly (Figure 2B and C). After superposition using domain R2 coordinates, measurement of the angles between vectors drawn through alpha helix B and its dimer partner, gave values of 97° and 101° for the GluR6 homodimer and heterodimer assemblies, while for the KA2 homodimer assembly the angle increases to 123° reflecting a large separation of the upper lobes (Figure 2C). Rotation by 90° parallel to the plane of quasi 2-fold symmetry between the subunits in the dimer assemblies reveals that in the KA2 homodimer assembly domain R2 has also rotated by 16° relative to the heterodimer assembly (Figure 2C). In combination, these movements bring the R1 domains of the GluR6 and KA2 subunits close together in the heterodimer assembly, and as a result the distance between the Cα positions of Ile90 in α-helix C, and Phe58 or Tyr 57 in α-helix B of GluR6 and KA2 respectively, decreases to 6.7 Å and 5.5 Å (Figure 3A); this close apposition is similar to that found in homomeric dimer assemblies for the GluR6, GluR6Δ1 and GluR7 ATDs, for which the corresponding Cα positions are separated by 5.5, 5.8 and 5.6 Å, while for the KA2 homodimer assembly these residues are separated by 10.7 Å. In addition to this movement, analysis of the extent of domain closure indicates that the KA2 clam shell in the heterodimer is closed by 4.5° – 7.5° compared to KA2 subunits in homodimer crystal structures, while the GluR6 subunit is closed by 4.2° – 6.5° when compared to GluR6 homodimer structures. Solvent accessible surface analysis of the GluR6/KA2 heterodimer interface reveals a total buried area of 2953 Å2 with the KA2 protomer contributing 1496 Å2, a gain of 536 Å2 compared to the KA2 homodimer assembly (Figure 2D). For the GluR6 subunit, although there is little change in buried surface area in the homodimer and heterodimer assemblies, local rearrangements produce key changes in inter-subunit contacts.
Table 1.
Crystallographic Data Collection and Refinement Statistics
| Data Set | GluR6Δ1 | GluR6Δ1/KA2 | GluR6wt/KA2 |
|---|---|---|---|
| Data Collection | |||
| Space group | I4 | P212121 | C2 |
| Unit cell a, b, c (Å) | 191.8, 191.8, 47.0 | 65.6, 139.5, 195.4 | 365.9, 109.0, 155.0 |
| Resolution range (Å)a | 50.00 – 3.0 (3.1) | 30.00 – 2.90 (3.0) | 50.00 – 3.95 (4.1) |
| Observations | 65,539 | 164,845 | 719,425 |
| Unique reflections | 17,783 | 40,232 | 54,061 |
| Completeness (%) b | 99.9 (99.4) | 99.1 (97.2) | 100 (100) |
| Rmergebc | 0.078 (0.706) | 0.114 (0.450) | 0.139 (0.895) |
| I/σ(I)b | 16.0 (2.0) | 11.6 (2.9) | 18.6 (2.8) |
| Structure Refinement | |||
| Resolution range (Å) | 47.95 − 2.98 | 29.97 − 2.90 | 49.0 − 3.9 |
| Reflections used | 16,775 | 39,761 | 50,913 |
| Protein / glycan atoms | 5,886 / 84 | 11,771 / 224 | 29,645 / 0 |
| Rwork / Rfree (%) | 24.2 / 29.3 | 19.7 / 25.6 | 26.4 / 28.0 |
| Rms Deviations | |||
| Bond lengths (Å) | 0.003 | 0.006 | 0.005 |
| Bond angles (°) | 0.608 | 0.918 | 0.0593 |
Values in parenthesis indicate the low resolution limit for the highest-resolution shell of data.
Values in parenthesis indicate statistics for the highest-resolution shell of data.
Rmerge = = (Σ| II – <II>|) / ΣI |II|, where <II> is the mean II over symmetry–equivalent reflections.
Domain R1 interface and loop 3 interactions in heterodimer assembly
The R1 interface in both the GluR6/KA2 heterodimer and in the GluR6 homodimer is formed primarily by a close apposition of α-helices B and C from each protomer. For both subunits, loop 3, which has been proposed to be a major determinant of subtype specific assembly mediated by iGluR ATDs (Jin et al., 2009), projects into the heterodimer interface and is anchored by intramolecular disulfide bonds between Cys65 or Cys64 on α-helix B, and Cys316 or Cys315, for GluR6 and KA2 respectively (Figure 3 and Figure S4A and B). Of note, we observe novel intersubunit interactions in the heterodimer assembly, which are absent in GluR6 and KA2 homodimer structures, and which involve loop 3. Due to formation of a hydrogen bond between the KA2 Tyr57 OH group and the GluR6 main chain nitrogen of Asn317, the tip of loop 3 in the GluR6 subunit undergoes a conformational change in the heterodimer assembly (Figure 3A). This results in a 5Å movement of the Asn317 side chain, which dips down into the heterodimer interface and becomes trapped between the Asp61 and Tyr57 side chains near the base of α-helix B in the KA2 subunit. Due to replacement of Tyr57 by Phe58, in the GluR6 homodimer this hydrogen bond is absent. Additional interactions made by the KA2 protomer at the R1 interface, which are unique to the heterodimer structure, result from movement of α-helices B and C towards the central axis of dimer formation, generating a series of contacts with the GluR6 protomer that are absent in KA2 homodimers. In particular, Tyr57 is inserted into a hydrophobic pocket formed by residues Ile90 and Ala93 from α-helix C of the GluR6 subunit (Figure 3A), while in KA2 homodimer ATD crystal structures the Tyr57 side chain is solvent exposed and adopts multiple conformations. The conformation of the Tyr57 side chain in the heterodimer assembly is also stabilized by van der Waals contacts with the Cys65-Cys316 disulfide bond in loop 3 of the interacting GluR6 protomer, and by a hydrogen bond between the main chain amide of Tyr57 and the hydroxyl group of Ser89 on α-helix C of the GluR6 protomer (Figure 3A). A hydrogen bond between GluR6 Lys62 in α-helix B and the main chain carbonyl of Cys315 in loop 3 of the KA2 protomer further stabilizes the heterodimer interface. On the 2-fold related side of the heterodimer assembly, the side chain of Phe58 at the base of α-helix B in the GluR6 subunit makes hydrophobic contacts with His89, Ile90 and the loop 3 Cys64-Cys315 disulfide bond of the KA2 protomer (Figure 3B), but as noted above cannot form a hydrogen bond contact with loop 3 of the KA2 subunit. Supplementary Movie 1 shows details of these contacts.
Aromatic side chain interactions at the R1 interface stabilize ATD dimer assemblies
To test the importance of intersubunit interactions made by the GluR6 Phe58 and KA2 Tyr57 side chains, which occupy similar positions in the heterodimer and GluR6 homodimer assemblies, we made the GluR6Δ2 F58A and KA2 Y57A mutants, and used sedimentation velocity experiments to measure changes in Kd for assembly of ATD homodimers and heterodimers. Strikingly, for SV runs at loading concentrations of 1.2 µM to 47 µM the c(s) peak distribution for the GluR6Δ2 F58A ATD mutant was largely monomeric (Figure 3C). Analysis of weighted-average sedimentation coefficient isotherms (Figure 3F) yielded a Kd value for homodimer formation of 490 µM (95% confidence interval; 380 µM – 650 µM), 2000-fold higher than for GluR6Δ2. However, when mixed with the KA2 subunit ATD, the sedimentation profile for the GluR6Δ2 F58A mutant shifted to higher S values and showed the characteristic pattern for a reversible monomer-dimer system in rapid equilibrium (Figure 3D). Analysis of sw(S) isotherms gave a Kd for heterodimer formation of 0.109 µM (95% confidence interval; 0.096 µM – 0.121 µM) tenfold weaker than the value measured by SV for wildtype (Kd 11 nM). Likewise, SV analysis for a mixture of the GluR6Δ2 and KA2 Y57A mutant ATDs (Figure 3E) gave a similar Kd for heterodimer assembly of 0.14 µM (95% confidence interval; 0.11 µM – 0.18 µM). However, when the aromatic side chains were mutated to alanine in both subunits (Figure 3F), the Kd for heterodimer assembly by the GluR6Δ2 F58A and KA2 Y57A mutant mix increased 150-fold to 1.63 µM (95% confidence interval; 1.57 µM – 1.70 µM). The fact that the GluR6 Δ2 F58A mutant still forms high affinity heterodimers with KA2, even though its ability to assemble as homodimers is essentially abolished, suggests that while the interaction of Phe58 is very important for GluR6 homodimer formation, other regions, most probably the R2 domain, must make a substantial contribution to heterodimer formation with KA2.
Conformational changes in the KA2 subunit contributing to heterodimer assembly
The domain R2 dimer interface is formed by a hydrophobic core flanked by polar residues mediated by a 2-fold related assembly of α-helices E and F and β-strand 7 from each protomer (Figure S4C and S4D). GluR6 residues Leu151, Ile152, Gln155, Ile158, Lys159 and Ser162 on α-helices E and F and Leu168, Ile170 on β strand 7 form one face of the domain R2 interface. Similarly, Ala149, Leu152, Leu153, Glu156, Val159, Leu163, Ile164 on α-helices E and F, and Val171 and Met173 on β-strand 7, form the complimentary surface of the KA2 subunit in the heterodimer (Figure 4A). For the GluR6 domain R2 surface, comparisons between homodimer and heterodimer assemblies reveals only subtle changes in rotamer conformations. However, similar comparisons for KA2 revealed a striking conformational change in residues Leu163-Thr168 (Figure 4A). In the KA2 homodimer structure α-helix F extends from residues 156– 163. In the heterodimer assembly α-helix F partially unwinds, producing a 10 Å movement of Lys166 at the tip of the loop, that requires a 180° turn over a 4 residue stretch in the loop connecting α-helix F with β-strand 7. This unusual conformation is stabilized by a 2.7 Å intramolecular hydrogen bond between the hydroxyl groups of Ser165 and Thr168 in the KA2 subunit, and by intermolecular hydrogen bonds between main chain atoms of Ile164 in KA2 and Leu168 from the GluR6 protomer (Figure 4A). This movement repositions the side chain of Ile 164 which stacks against Ile158, Pro161 and Ile170 in the center of the hydrophobic patch of the GluR6 protomer, while L163 is repositioned at the periphery. At the top of the domain R2 interface, the side chain of Glu156 forms hydrogen bonds with the hydroxyl group of Tyr145 and the side chain amide of Gln172 in the GluR6 subunit, while in the KA2 homodimer Glu156 forms an intramolecular salt bridge with Arg 160 in α-helix E of the same subunit. Supplementary Movie 2 shows details of these contacts.
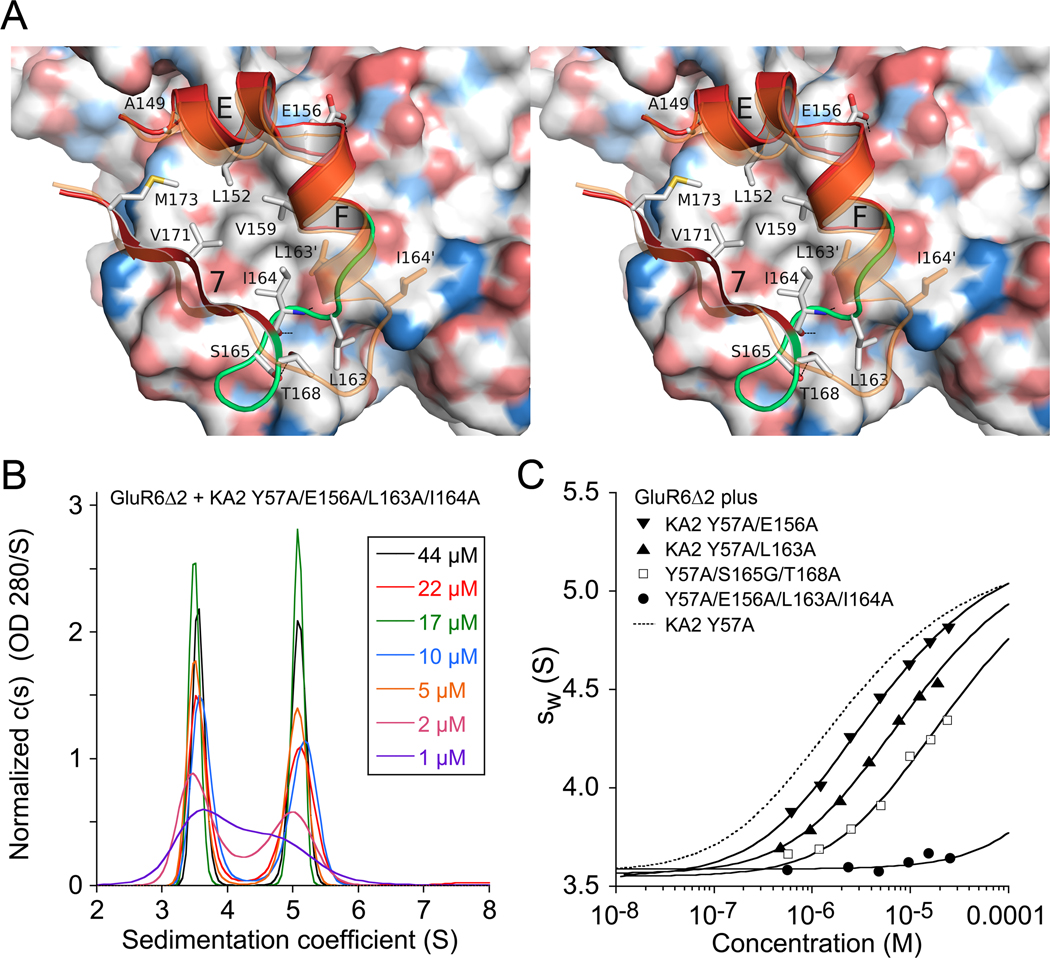
Figure 4. Domain R2 plays a key role in heterodimer assembly.
(A) Stereo diagram of the GluR6/KA2 heterodimer domain R2 interface; the GluR6 subunit is shown as a molecular surface colored by atom type. The red ribbon diagram shows the KA2 subunit in the heterodimer assembly, with the loop connecting α-helix F with β-strand 7 shaded green; key residues forming intermolecular contacts are drawn as sticks; the ribbon diagram in transparent orange shows a subunit from a KA2 homodimer assembly. (B) Sedimentation velocity analysis for an equimolar mix of the GluR6Δ2 and KA2 E156A/L163A/I164A mutant ATDs reveals disruption of heterodimer assembly, with essentially no concentration dependence of the c(s) peak positions corresponding to monomers and dimers formed by the KA2 and GluR6 subunits respectively. (C) Isotherms of weighted-average sedimentation coefficients determined from peak integration of the c(s) data for individual KA2 Y57A domain R2 mutants mixed with GluR6Δ2 F58A; solid lines shows fits of a monomer-dimer model; the dashed line shows the isotherm for KA2 with wild type domain R2 residues.
In view of conformational rearrangements observed in domain R2 of the heterodimer interface, and the persistence of heterodimer formation at µM ATD concentrations when Phe58 and Tyr57 in domain R1 were mutated to alanine, we examined the role of domain R2 in heterodimer assembly, and made alanine mutations in the KA2 subunit for Glu156, Leu163, and I164, all in the background of KA2 Y57A. To destabilize the tight turn in the loop connecting α-helix F to β-strand 7 we made the S165G/T168A double mutant. Sedimentation velocity experiments were used to measure the Kd for heterodimer formation with GluR6Δ2. These experiments established that domain R2 plays a key role since for the KA2 Y57A/E156A/L163A/I164A quad mutant heterodimer assembly was essentially abolished, and in SV experiments we observed two non interacting species corresponding to GluR6Δ2 homodimers and KA2 mutant monomers (Figure 4B). To quantify the effects of individual KA2 domain R2 mutations we performed additional SV experiments in the KA2 Y57A mutant background with GluR6Δ2 F58A as the dimer partner; because neither of these mutants forms homodimers, this facilitated analysis of heterodimer formation, giving easy to interpret isotherms of weighted-average sedimentation coefficients (Figure 4C). The results gave Kd values with 95% confidence intervals of 2.0 µM (0.18 – 2.27 µM) for E156A, 6.7 µM (6.4 – 7.1 µM) for L163A, and 13.0 µM (10 – 15 µM) for the S165G/T168A double mutant. The I164A mutation produced a much greater disruption, such that we can set only a lower limit on the Kd of 200 µM or larger. We also screened two additional sets of KA2 mutants in regions of the structure which might be anticipated to affect heterodimer formation. The 1st set tested, KA2 C64S/C315S, targeted the disulfide bond which holds loop three in place (Figure 5A). The second set targeted Lys148 and Glu150 at the N-terminus of α-helix E, which were candidates for mediating contacts with His105, Ser108 and Asp109 in domain R1 of the GluR6 protomer. However, both the C64S/C315S and K148A/E150A KA2 double mutants produced no change in oligomerization when mixed with either GluR6Δ2 or GluR6Δ2 F58A and analyzed by UV/RI/MALS-SEC (Figure S5). The lack of effect of loop 3 disulfide bond disruption likely occurs because, in a heterodimer assembly with GluR6, loop 3 of the KA2 subunit is held in place by other contacts such as the hydrogen bond between the main chain carbonyl oxygen of Cys315 and the side chain of Lys62 in the GluR6 subunit. We were unable to test the effect of the GluR6 C65S/C316S mutant, because this construct could not be expressed at levels sufficient for biochemical analysis, possibly due to misfolding. The interactions made by Lys148 and Glu150 with the GluR6 subunit are formed in solvent exposed loops with weak electron density, and it is likely that this region is quite mobile, since our results reveal that it does not contribute to dimer stability.
Figure 5. Mutant cycle analysis for interactions between intermolecular contacts in the heterodimer assembly.
(A) The KA2 subunit ATD viewed face on to the heterodimer surface, indicating the position of Tyr57 in site 1; Glu156 in site 2; Ile164 in site 3; Ser165 and Thr168 in site 4 that stabilizes the loop which makes site 3 contacts; the Cys64-Cys315 disulfide bond which holds loop 3 in place; and the location of Lys148 and Glu150. (B) Illustration of a mutant cycle for coupling between two sites indicated as A and B. (C) Amino acid alignment for AMPA and kainate receptors reveals a unique conservation of Ile164 in the KA2 subunits, and exchange of conserved residues in the loop connecting alpha helix F with beta strand 7.
Mutant cycle analysis of interactions between sites in domains R1 and R2
To estimate the strength of the interactions underlying dimer formation we purified a series of 15 mutant ATD proteins and measured their Kd for homodimer and heterodimer formation using SV experiments. To select mutant combinations suitable for analysis by SV we performed SEC-UV/RI/MALS experiments to assay for either depletion of the monomer KA2 peak when mixed with GluR6Δ2, or an increase in dimer peak when mixed with GluR6Δ2F58A (Figure 1C, Figure S5A and Table S1). Out of 30 combinations tested, 13 pairs were selected for analysis by SV; examples of isotherms for weighted-average sedimentation coefficients for KA2 mutants mixed with GluR6Δ2F58A and GluR6Δ2 are shown in Figure 4C and Figure S5C respectively. To calculate ΔΔG values we used SV measurements for the GluR6Δ2 homodimer Kd (250 nM), the KA2 homodimer Kd (350 µM), and the GluR6Δ2/KA2 heterodimer Kd (11 nM), as reference values (Table S1). The formation of GluR6Δ2/KA2 heterodimers is favored by 6.04 kcal/mol compared to the KA2 subunit homodimer Kd. Based on analysis of the heterodimer crystal structure there are four sites which likely contribute to this large free energy change (Figure 5A), which we label site 1 (Tyr57), site 2 (Glu156), site 3 (Leu163 and Leu164), and site 4 (Ser165 and Thr 168 in the loop connecting α-helix F to β-strand 7 in the KA2 subunit). Mutation of site 1 produced a −4.5 kcal/mol loss of binding energy for the GluR6Δ2F58A homodimer, but only a −2.9 kcal/mol loss for the GluR6Δ2F58A/KA2Y57A heterodimer, with equal contributions by the F58A and Y57A mutants, of −1.4 and −1.5 kcal/mol respectively (Table S1). The excess total dimerization energy, totaling 3.1 kcal/mol, must come from other sites in the heterodimer interface. The mutation E156A produced a loss of only −0.43 kcal/mol; for L163A the loss was −1.31 kcal/mol. The S165G/T168A double mutant produced a loss of binding energy of −1.74 kcal/mol. Strikingly, Ser165 and Thr168 do not make contacts with the GluR6 subunit, and instead merely serve to stabilize the loop conformation which positions L163 and I164 in the dimer interface.
To examine whether the binding mechanism for heterodimer formation was an additive or cooperative process we performed a mutant cycle analysis looking at interactions between sites in domains R1 and R2 with both the GluR6Δ2 and GluR6Δ2F58A mutant used as heterodimer partners. Mutant cycles were calculated as shown in Figure 5B, where coupling coefficients (Ω) greater than one indicate positive cooperativity (Carter et al., 1984; Hidalgo and MacKinnon, 1995). The analysis yielded coupling coefficients (Ω values) of only 0.8–1.7 and reveals clearly that the process of heterodimer assembly is an additive process with little cooperativity between domains R1 and R2 (Extended Experimental Procedures). The much larger disruption of heterodimer assembly observed for the I164A mutant likely reflects conformational changes resulting from destabilization of the hydrophobic patch formed by the loop rearrangement in the KA2 subunit. Of note, amino acid sequence alignments (Figure 5C) reveal that in other iGluR subunits Ile164 is replaced by charged or polar residues, consistent with a unique role for Ile164 in mediating heterodimer assembly for KA1 and KA2. This alignment also reveals exchange of Glu167 (Asp165 in KA1) by Trp/Ile/Leu in other iGluR subunits (Figure 5C). The residues exchanged are in a flexible loop region connecting helix F and strand 7, the conformation of which differs in individual iGluR families. In GluR1–4 and GluR5–7 the Trp/Ile/Leu residues form part of the hydrophobic core of domain 2, while in the KA2 subunit the polar residues are surface exposed, and make intersubunit contacts in the heterodimer assembly. In the KA1 and KA2 subunits Phe160/162 fills the space in the hydrophobic core which in other iGluR subunits is occupied by the Trp/Ile/Leu residues which align with Asp165/Glu167 in KA1 and KA2. At the corresponding position in the AMPA receptors and GluR5–7 the Phe residue is replaced by smaller Ala or Pro side chains.
GluR6 forms the dimer of dimers interface in the GluR6/KA2 ATD tetramer
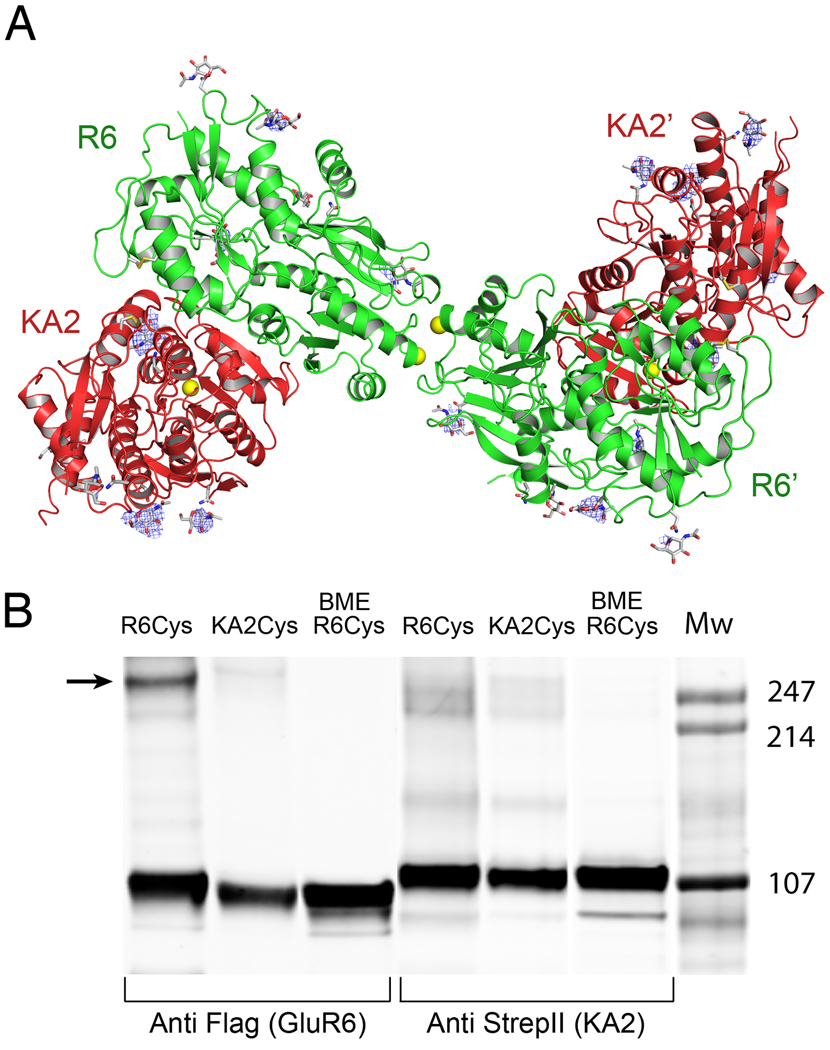
In order to elucidate the structure of the GluR6/KA2 ATD tetramer we crystallized a complex of wt GluR6 and KA2. We first demonstrated by size exclusion chromatography and AUC experiments that wt GluR6 and KA2 likely interact to form tetramers in solution (Figure S3). The wt GluR6 and KA2 complex purified by gel filtration crystallized in a large unit cell with 10 protomers in the asymmetric unit, which assemble as 5 identical heterodimers. Four of the heterodimers assemble to generate two pairs of tetramers, and a third identical tetramer is generated by crystallographic symmetry operations for the remaining dimer (Figure S6A). Although this crystal form diffracted only to 3.9Å resolution (Table 1) the availability of a higher resolution refined heterodimer crystal structure allowed us to use molecular replacement to position the heterodimers in the symmetric unit and to refine the structure with good statistics using deformable elastic network restraints (Schroder et al., 2010). The RMSD of 0.66 Å for least squares superposition of 714 Cα atoms of the GluR6Δ1/KA2 dimer indicates that GluR6/KA2tetramers are formed by rigid body assembly of heterodimer pairs. In each of the tetramer assemblies, the GluR6/KA2 heterodimers are arranged in such a way that the dimer of dimers interface is mediated by the two GluR6 subunits (Figure 6A). Helices G and H of the GluR6 subunit form the 2-fold symmetric interface as found previously for GluR6 ATD homodimer structures (Das et al., 2010; Kumar et al., 2009). Electron density (Fo-Fc) difference maps, which revealed the positions of glycan residues not used in model building or refinement, allowed us to use the unique N-linked glycosylation patterns for the GluR6 and KA2 ATDs as an additional check for subunit identity in the tetramer assemblies (Figure 6A); particularly prominent is the excess density at KA2 Asn200, a site resistant to digestion by Endo H (Kumar and Mayer, 2010).
Figure 6. Tetramer assembly is mediated by the GluR6 subunit.
(A) Crystal structure of the GluR6/KA2 heterotetramer ATD assembly for wt GluR6 and KA2, colored green and red respectively; the ribbon diagram shows one of three identical tetramers for the 10 protomers in the asymmetric unit, with one tetramer formed by non crystallographic symmetry operations; side chains which support N-linked glycosylation are drawn as sticks; yellow spheres indicate the positions in the GluR6 and KA2 subunits at which cysteine mutations were introduced to test for formation of disulfide cross links in full length receptors. Electron density maps (Fo-Fc contoured at 3 σ blue mesh) illustrate features corresponding to glycan residues, which were not included in the model or used for refinement. (B) Western blots run under non-reducing conditions for detergent solubilized affinity-purified (StrepII tag) full-length heteromeric GluR6/KA2, in which Cys mutants were introduced at equivalent sites in the domain R2 lateral surface of either GluR6 or KA2 and probed against Flag (GluR6) and StrepII (KA2) epitopes; lanes 3 and 6 contain the same samples loaded in 1 and 4 but with the addition of 10 mM BME.
To validate that the same ATD tetramer assembly occurs in full-length heteromeric kainate receptors we performed cysteine mutant cross-linking experiments. For these we used the GluR6 G215C 5× cysteine (–) mutant, which we had shown previously to form spontaneous cross links in full-length GluR6 homotetramers (Das et al., 2010), and introduced a cysteine mutation at the equivalent Gly215 position in the KA2 subunit. We tested mutants for oligomer formation by Western blot analysis under non-reducing conditions. Unique FLAG and STREPII tags were also inserted in the GluR6 and KA2 full-length subunits respectively for purification by affinity chromatography and for Western blot analysis. Dimers formed spontaneously when GluR6 G215C and KA2 were expressed together but not when GluR6 wt and KA2 G215C were coexpressed (Figure 6B). This indicates that GluR6 mediates the dimer of dimers interface in a GluR6/KA2 heterotetramer consistent with the ATD heterotetrameric crystal structure.
Assembly of heteromeric kainate receptors in vivo requires high affinity ATD interactions
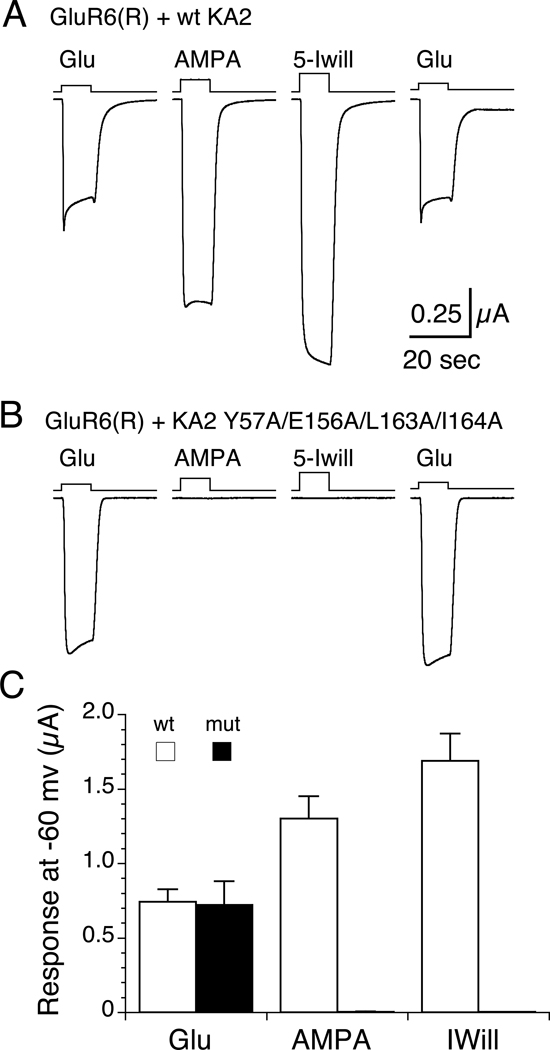
To test whether high affinity interactions between the GluR6 and KA2 ATDs underlies assembly of heteromeric kainate receptors in vivo, we used functional assays in Xenopus oocytes which utilized the different ligand binding selectivity of the GluR6 and KA2 subunits (Herb, et al., 1992; Swanson et al., 1998). When GluR6(R) and KA2 were coexpressed at a ratio of 1:2, following incubation with 0.3 mg/ml concanavalin-A to attenuate desensitization, we recorded robust responses to 60 µM glutamate (0.74 ± 0.08 µA, n=5), 500 µM AMPA (1.30 ± 0.15 µA, n=5) and 500 µM 5-iodowillardiine (1.69 ± 0.18 µA, n=5). For the KA2 Y57A/E156A/L163A/I164A mutant responses to glutamate (0.72 ± 0.16 µA, n=6) were of similar amplitude, while for AMPA and 5-Iodowillardiine responses were too small to record reliably (< 5 nA), indicating that high affinity interactions of the GluR6 and KA2 ATDs is required for assembly of heteromeric kainate receptors in vivo (Figure 7). We also established that interactions between the ATDs play a key role in the selective assembly of iGluRs by performing SEC-UV/RI/MALS for the AMPA receptor GluA2 ATD injected alone, or mixed with the KA2 ATD. In contrast to the decrease in KA2 monomer peak amplitude observed for the GluR6 and KA2 mixture the GluA2 and KA2 ATDs do not interact even at protein concentrations of greater than 10 µM (data not shown).
Figure 7. High affinity ATD interactions underlie assembly of heteromeric kainate receptors in vivo.
(A) Responses to 60 µM glutamate, 500 µA AMPA, and 500 µM 5-iodowillardiine recorded at −60 mV from a Xenopus oocyte injected with 1.5 ng GluR6(R) and 3 ng wt KA2 cRNAs. (B) Responses for the same protocol but with the KA2 Y57A/E156A/L163A/I164A mutant. (C) Bar plot showing the mean ± sem for responses for wt KA2 (n=5) and the KA2 mutant (n=6).
Conclusions
The results of our experiments reveal that in heteromeric kainate receptors the ATDs of the GluR6 and KA2 subunits assemble as pairs of heterodimers, in which the KA2 subunits lie at the lateral edges of the tetramer, while the GluR6 subunits mediate the dimer of dimers assembly on the 2-fold axis of molecular symmetry. The high affinity of the KA2 subunit for GluR6 ensures that ATD heterodimers will form early during the process of biogenesis, before trafficking comes into play, and in addition provides a mechanism which suppresses formation of functional GluR6 homotetramers which lack the KA2 subunit, while ensuring a 2:2 stoichiometry of assembly. The binding mechanism generating the kainate receptor heterodimer assembly involves residues present in both the R1 and R2 lobes of KA2 protomers. By contrast, the emerging picture of AMPA receptor assembly indicates that domain R1 plays a major role in heterodimer assembly (Rossmann et al., 2011), similar to what we find for GluR6 homodimers. For NMDA receptors, quantitative analysis of ATD assembly using sedimentation experiments has not yet been reported.
Despite a major role in iGluR assembly, it is striking that in prior work genetic deletion of the ATD for AMPA, kainate, and NMDA receptors does not abolish the formation of functional ion channels in heterologous expression systems, indicating that its role in assembly is not obligatory (Gielen et al., 2009; Horning and Mayer, 2004; Pasternack et al., 2002; Plested and Mayer, 2007; Yuan et al., 2009). It is notable that prokaryotic iGluRs which entirely lack the ATD domain assemble, activate and desensitize similar to eukaryotic iGluRs (Chen et al., 1999), suggesting that the ATD most likely plays a role in facilitating the efficient assembly of heteromeric iGluR assemblies. Consistent with this, in unpublished work we have found a > 10-fold reduction in cell surface expression of functional homomeric GluA2 AMPA receptors in Xenopus oocytes injected with equal amounts of GluA2ATD(−) cRNA compared to wild type GluA2.
Because the GluR6 and KA2 ATD heterodimer is formed with very high affinity it is unlikely that this assembly undergoes large conformational changes in an intact receptor. Because the dimer is stabilized by contacts mediated by both the R1 and R2 domains, it is also unlikely that the individual subunits in a dimer could undergo substantial changes in domain closure in response to small ligands. By contrast, although we can set only a lower limit of 3–5 µM on the Kd for formation of the tetrameric ATD dimer of dimers assembly, it is very likely that the tetramer is a dynamic assembly in which the two arms formed by ATD dimers can move relative to each other. It is thus tempting to speculate that if the ATD interacts with other proteins in the synaptic cleft, this could affect receptor clustering and mobility, and in addition regulate ion channel activity via conformational changes propagated to the ligand binding domain. The AMPA receptor ATD has been reported to bind to N-cadehrins (Saglietti et al., 2007) and neuronal pentraxins thereby contributing to excitatory synapotogenesis (O'Brien et al., 1999; Ripley et al., 2011; Sia et al., 2007; Xu et al., 2003). Likewise, the GluN1 ATD and the extracellular domain of EphB mediate EphrinB and NMDA receptor interaction(Dalva et al., 2000; Takasu et al., 2002). More recently, the Delta2 receptor ATD has been shown to form trans-synaptic interactions via cerebelin-1 precursor protein and neurexin (Matsuda et al., 2010; Uemura et al., 2010). The exact nature and stoichiometry of these interactions is not known but will be influenced by the different stabilities of the high-affinity dimer and low-affinity dimer of dimers interfaces in individual iGluR subtypes.
EXPERIMENTAL PROCEDURES
Protein Purification and crystallization
The GluR6 and KA2 ATDs were expressed in adherent and suspension cultures of wild type HEK 293T cells for SEC-UV/RI/MALS and AUC studies and purified as described previously (Kumar and Mayer, 2010; Kumar et al., 2009). For crystallization the proteins were expressed in N-acetyl glucosaminyltransferase I–deficient GnTI− HEK293 cells and digested with Endo H (Reeves et al., 2002). Complete descriptions are given in Extended Experimental Procedures.
Data collection, Structure Determination and Model Refinement
X-ray diffraction data sets were collected using synchrotron radiation at the Advanced Photon Source (GM/CA CAT; beamline 23-ID-B) and were indexed, integrated and scaled using HKL2000 (Otwinowski and Minor, 1997). The GluR6Δ1 homodimer and GluR6Δ1/KA2 heterodimer structures were solved by molecular replacement using the program PHASER (McCoy et al., 2007) and search probes composed of monomers of rat GluR6 (PDB ID: 3H6H) and KA2 (PDB ID: 3OM0) ATDs. The structures were iteratively built and refined with riding hydrogens using Coot (Emsley and Cowtan, 2004) and Phenix (Adams et al., 2010). The refined GluR6Δ1/KA2 heterodimer was used as a search probe in PHASER to solve the GluR6wt/KA2 heterotetramer structure, which was initially refined using a deformable elastic network (DEN) model implemented in CNS 1.3 (Schroder et al., 2010) before switching to Phenix. Data collection and refinement statistics are given in Table 1.
Analytical size exclusion chromatography
SEC-UV/RI/MALS was performed using a Superdex 200 HR 10/30 size exclusion column equilibrated with 20 mM HEPES, 200 mM NaCl, 1 mM EDTA, pH 7.4. The protein loading concentration was 2mg/ml unless stated otherwise. Detection was performed using a triple-angle light scattering detector (Mini-DAWN™ TREOS, Wyatt Technology), and a differential refractometer (Optilab® rEX, Wyatt Technology). Molecular weight and hydrodynamic radius determination was performed using ASTRA (Wyatt Technology).
Analytical ultracentrifugation
For AUC proteins were dialyzed against a buffer containing 200 mM NaCl, 1 mM EDTA and 20 mM Na Phosphate, pH 7.5. Sedimentation velocity (SV) experiments were carried out in ProteomeLab XL-I analytical ultracentrifuges (Beckman Coulter, Palo Alto, CA) at 20°C at a rotor speed of 50,000 rpm, following standard protocols (Brown et al., 2008). For the study of GluR6 and KA2 mixtures, parallel dilution series were conducted for each component alone, as well as a stock mixture at an ≈ 1:1 molar ratio, spanning a total loading concentration range from 0.005 to ~2.0 mg/ml. Data were analyzed with SEDFIT applying sedimentation coefficient distributions c(s) (Schuck, 2000) followed by integration to determine the weighted-average sedimentation coefficients sw, which were fitted in SEDPHAT with models for monomers, homodimers and heterodimers in chemical equilibrium (Schuck, 2003). The s-values for monomer and dimer species were fixed to best-fit estimates derived from analysis of GluR6 and KA2 mutants with very low and very high affinity, respectively.
Sedimentation equilibrium (SE) experiments were conducted with ~ 4.5 mm sample columns at a 5–10 fold range of loading concentrations at sequential rotor speeds of 6,500 rpm, 10,000 rpm and 16,000 rpm at 10°C. The radial signal profiles were acquired using both absorbance optics at 230, 250 and 280 nm and interference optics. Data were globally fitted in SEDPHAT with equilibrium models using multi-signal analysis and soft mass conservation constraints (Vistica et al., 2004). Further details of the SV and SE analyses are described in Extended Experimental Procedures.
Cysteine crosslinking and Western blots
Unique FLAG and StrepII tags were inserted into the full length GluR6 G215C 5× cysteine (–) mutant, which we had shown previously to form spontaneous cross links in full-length GluR6 homotetramers (Das et al., 2010), and into the KA2 G215C mutant subunit at their N and C-termini respectively for affinity purification and Western blot analysis. Total cell lysates from HEK293T suspension cultures were prepared on the 4th/5th day post transfection. A StrepII affinity column was used to purify receptor complexes which were than resolved in duplicate on SDS-PAGE under non-reducing conditions. SDS gels were electroblotted onto PVDF membranes and probed by either anti-FLAG or anti-StrepII antibody for detection of GluR6 and KA2 receptor subunits respectively.
Highlights.
Heteromeric kainate receptor ATD tetramer structures solved by Xray crystallography
GluR6/KA2 heterodimer assembles with Kd of 11 nM
Interaction surface involved residues in both upper and lower lobes
Heteromeric ion channel assembly disrupted by ATD alanine mutants
Supplementary Material
ACKNOWLEDGEMENTS
We thank Carla Glasser and Andrea Balbo for technical assistance and Drs. P. Kwong and Yongping Yang for advice with suspension cell cultures. Nucleic acid sequencing was performed by the NINDS DNA sequencing facility. Synchrotron diffraction data was collected at the GM/CA CAT 23 ID-B beamline. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. This work was supported by the intramural research programs of NICHD and NIBIB, NIH, DHHS (MLM and PS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes. Protein Data Bank: Coordinates and structure factors have been deposited with accession codes 3QLT, 3QLU and 3QLV for the GluR6Δ1 homodimer, GluR6Δ1/KA2 heterodimer, and GluR6/KA2 tetramer.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalon G, Segev E, Elgavish S, Stern-Bach Y. Two regions in the N-terminal domain of ionotropic glutamate receptor 3 form the subunit oligomerization interfaces that control subtype-specific receptor assembly. J Biol Chem. 2005;280:15053–15060. doi: 10.1074/jbc.M408413200. [DOI] [PubMed] [Google Scholar]
- Ayalon G, Stern-Bach Y. Functional assembly of AMPA and kainite receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31:103–113. doi: 10.1016/s0896-6273(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Brose N, Huntley GW, Stern-Bach Y, Sharma G, Morrison JH, Heinemann SF. Differential assembly of coexpressed glutamate receptor subunits in neurons of rat cerebral cortex. J Biol Chem. 1994;269:16780–16784. [PubMed] [Google Scholar]
- Brown PH, Balbo A, Schuck P. Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation. Curr Protoc Immunol Chapter. 2008;18(Unit 18):15. doi: 10.1002/0471142735.im1815s81. [DOI] [PubMed] [Google Scholar]
- Burnashev N, Monyer H, Seeburg PH, Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8:189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38:835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- Chen GQ, Cui C, Mayer ML, Gouaux E. Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature. 1999;402:817–821. doi: 10.1038/45568. [DOI] [PubMed] [Google Scholar]
- Clayton A, Siebold C, Gilbert RJ, Sutton GC, Harlos K, McIlhinney RA, Jones EY, Aricescu AR. Crystal structure of the GluR2 amino-terminal domain provides insights into the architecture and assembly of ionotropic glutamate receptors. J Mol Biol. 2009;392:1125–1132. doi: 10.1016/j.jmb.2009.07.082. [DOI] [PubMed] [Google Scholar]
- Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- Das U, Kumar J, Mayer ML, Plested AJ. Domain organization and function in GluK2 subtype kainate receptors. Proc Natl Acad Sci USA. 2010;107:8463–8468. doi: 10.1073/pnas.1000838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egebjerg J, Bettler B, Hermans-Borgmeyer I, Heinemann S. Cloning of a cDNA for a glutamate receptor subunit activated by kainate but not AMPA. Nature. 1991;351:745–748. doi: 10.1038/351745a0. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Farina AN, Blain KY, Maruo T, Kwiatkowski W, Choe S, Nakagawa T. Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. J Neurosci. 2011;31:3565–3579. doi: 10.1523/JNEUROSCI.6041-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- Geiger JRP, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principle neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Esteban JA. AMPA receptor biogenesis and trafficking. Curr Opin Neurobiol. 2007 doi: 10.1016/j.conb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Herb A, Burnashev N, Werner P, Sakmann B, Wisden W, Seeburg PH. The KA-2 subunit of excitatory amino acid receptors shows widespread expression in brain and forms ion channels with distantly related subunits. Neuron. 1992;8:775–785. doi: 10.1016/0896-6273(92)90098-x. [DOI] [PubMed] [Google Scholar]
- Hidalgo P, MacKinnon R. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science. 1995;268:307–310. doi: 10.1126/science.7716527. [DOI] [PubMed] [Google Scholar]
- Hollmann M, O'Shea-Greenfield A, Rogers SW, Heinemann S. Cloning by functional expression of a member of the glutamate receptor family. Nature. 1989;342:643–648. doi: 10.1038/342643a0. [DOI] [PubMed] [Google Scholar]
- Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horning MS, Mayer ML. Regulation of AMPA receptor gating by ligand binding core dimers. Neuron. 2004;41:379–848. doi: 10.1016/s0896-6273(04)00018-2. [DOI] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinänen K, Wisden W, Sommer B, Werner P, Herb A, Verdoorn TA, Sakmann B, Seeburg PH. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- Kumar J, Mayer ML. Crystal Structures of the Glutamate Receptor Ion Channel GluK3 and GluK5 Amino-Terminal Domains. J Mol Biol. 2010;404:680–696. doi: 10.1016/j.jmb.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner WD, Hoch W. Subtype-specific assembly of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits is mediated by their n-terminal domains. J Biol Chem. 1999;274:16907–16916. doi: 10.1074/jbc.274.24.16907. [DOI] [PubMed] [Google Scholar]
- Mah SJ, Cornell E, Mitchell NA, Fleck MW. Glutamate receptor trafficking: endoplasmic reticulum quality control involves ligand binding and receptor function. J Neurosci. 2005;25:2215–2225. doi: 10.1523/JNEUROSCI.4573-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Miura E, Miyazaki T, Kakegawa W, Emi K, Narumi S, Fukazawa Y, Ito-Ishida A, Kondo T, Shigemoto R, et al. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–368. doi: 10.1126/science.1185152. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- O'Brien RJ, Xu D, Petralia RS, Steward O, Huganir RL, Worley P. Synaptic clustering of AMPA receptors by the extracellular immediate-early gene product Narp. Neuron. 1999;23:309–323. doi: 10.1016/s0896-6273(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;277:307–344. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Partin KM, Patneau DK, Winters CA, Mayer ML, Buonanno A. Selective modulation of desensitization at AMPA versus kainate receptors by cyclothiazide and concanavlin A. Neuron. 1993;11:1069–1082. doi: 10.1016/0896-6273(93)90220-l. [DOI] [PubMed] [Google Scholar]
- Pasternack A, Coleman SK, Jouppila A, Mottershead DG, Lindfors M, Pasternack M, Keinanen K. Alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor channels lacking the N-terminal domain. J Biol Chem. 2002;277:49662–49667. doi: 10.1074/jbc.M208349200. [DOI] [PubMed] [Google Scholar]
- Penn AC, Williams SR, Greger IH. Gating motions underlie AMPA receptor secretion from the endoplasmic reticulum. EMBO J. 2008;27:3056–3068. doi: 10.1038/emboj.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Wenthold RJ. Histological and ultrastructural localization of the kainate receptor subunits, KA2 and GluR6/7, in the rat nervous system using selective antipeptide antibodies. J Comp Neurol. 1994;349:85–110. doi: 10.1002/cne.903490107. [DOI] [PubMed] [Google Scholar]
- Plested AJ, Mayer ML. Structure and mechanism of kainate receptor modulation by anions. Neuron. 2007;53:829–841. doi: 10.1016/j.neuron.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Riley NJ, Garcia EP, Sanders JM, Swanson GT, Marshall J. Multiple trafficking signals regulate kainate receptor KA2 subunit surface expression. J Neurosci. 2003;23:6608–6616. doi: 10.1523/JNEUROSCI.23-16-06608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley B, Otto S, Tiglio K, Williams ME, Ghosh A. Regulation of synaptic stability by AMPA receptor reverse signaling. Proc Natl Acad Sci U S A. 2011;108:367–372. doi: 10.1073/pnas.1015163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M, Sukumaran M, Penn AC, Veprintsev DB, Babu MM, Greger IH. Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers. EMBO J. 2011;30:959–971. doi: 10.1038/emboj.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saglietti L, Dequidt C, Kamieniarz K, Rousset MC, Valnegri P, Thoumine O, Beretta F, Fagni L, Choquet D, Sala C, et al. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron. 2007;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Schiffer HH, Swanson GT, Heinemann SF. Rat GluR7 and a carboxy-terminal splice variant, GluR7b, are functional kainate receptor subunits with a low sensitivity to glutamate. Neuron. 1997;19:1141–1146. doi: 10.1016/s0896-6273(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Schroder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- Shanks NF, Maruo T, Farina AN, Ellisman MH, Nakagawa T. Contribution of the global subunit structure and stargazin on the maturation of AMPA receptors. J Neurosci. 2010;30:2728–2740. doi: 10.1523/JNEUROSCI.5146-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Suh YH, Milstein AD, Isozaki K, Schmid SM, Roche KW, Nicoll RA. Functional comparison of the effects of TARPs and cornichons on AMPA receptor trafficking and gating. Proc Natl Acad Sci U S A. 2010;107:16315–16319. doi: 10.1073/pnas.1011706107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia GM, Beique JC, Rumbaugh G, Cho R, Worley PF, Huganir RL. Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron. 2007;55:87–102. doi: 10.1016/j.neuron.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson GT, Green T, Heinemann SF. Kainate receptors exhibit differential sensitivities to (S)-5-Iodowillardiine. Mol Pharmacol. 1998;53:942–949. [PubMed] [Google Scholar]
- Takasu MA, Dalva MB, Zigmond RE, Greenberg ME. Modulation of NMDA receptor-dependent calcium influx and gene expression through EphB receptors. Science. 2002;295:491–495. doi: 10.1126/science.1065983. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- Valluru L, Xu J, Zhu Y, Yan S, Contractor A, Swanson GT. Ligand binding is a critical requirement for plasma membrane expression of heteromeric kainate receptors. J Biol Chem. 2005;280:6085–6093. doi: 10.1074/jbc.M411549200. [DOI] [PubMed] [Google Scholar]
- Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Watkins JC, Evans RH. Excitatory amino acid transmitters. Ann Rev Pharmacol Toxicol. 1981;21:165–204. doi: 10.1146/annurev.pa.21.040181.001121. [DOI] [PubMed] [Google Scholar]
- Werner P, Voigt M, Keinänen K, Wisden W, Seeburg PH. Cloning of a putative high-affinity kainate receptor expressed predominantly in hippocampal CA3 cells. Nature. 1991;351:742–744. doi: 10.1038/351742a0. [DOI] [PubMed] [Google Scholar]
- Xu D, Hopf C, Reddy R, Cho RW, Guo L, Lanahan A, Petralia RS, Wenthold RJ, O'Brien RJ, Worley P. Narp and NP1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron. 2003;39:513–528. doi: 10.1016/s0896-6273(03)00463-x. [DOI] [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.