Abstract
Ten Salmonella enterica serovar Typhimurium isolates producing CTX-M-2 extended-spectrum β-lactamase were identified from clinical and poultry sources in two distant cities in Brazil between 2003 and 2004. They included two isolates from pediatric patients and eight isolates from poultry or its environment. All isolates exhibited coresistance to non-β-lactam antimicrobials including tetracycline and trimethoprim/sulfamethoxazole. The CTX-M-2 gene was located on transferable plasmids with sizes between 90 and 170 kb that also carried other resistance determinants in some isolates. By pulsed-field gel electrophoresis, the genetic similarity of the isolates including clinical and poultry ones ranged from 89% to 100%.
Introduction
Salmonella enterica is a common cause of human gastroenteritis worldwide, and a large variety of animals, particularly food animals, have been identified as reservoirs for nontyphoidal Salmonella spp. Among more than 2,500 serotypes of the genus Salmonella described, two of them, Enteritidis and Typhimurium, are the most common causes of human salmonellosis in many countries, including Brazil.7,10
Salmonella infections that cause severe diarrhea as well as systemic infections, such as bacteremia and meningitis, require antimicrobial treatment. Fluoroquinolones and expanded-spectrum cephalosporins are essential drugs that are often used for treating patients with complicated Salmonella infections. Resistance to different β-lactams, primarily caused by the production of acquired extended-spectrum β-lactamases (ESBLs), has emerged worldwide during the last two decades, mostly associated with the Enterobacteriaceae, and in particular Klebsiella pneumoniae and Escherichia coli.16 ESBL production in Salmonella has been considered relatively rare. However, the number of reported cases in various ESBL-producing Salmonella serotypes has been increasing worldwide in recent years, with the CTX-M group being predominant.2,14 ESBL-producing Salmonella enterica serotypes, especially of CTX-M-type, have recently been detected in poultry and poultry products in different countries.1,11,20,23 Growing evidence indicates that ESBL-producing organisms, and CTX-M producers in particular, represent an emerging problem in the community.18 There is a legitimate concern that food-producing animals may serve as a reservoir for ESBL-producing human pathogens and that resistance genes may be transferred to humans through the food supply and ultimately cause treatment failure in patients receiving cephalosporin therapy for serious infections.2,14 In the present study, we report 10 isolates of ESBL-producing S. enterica serotype Typhimurium identified from clinical and poultry sources in Brazil.
Materials and Methods
Bacterial strains
A total of 153 isolates of Salmonella Typhimurium were identified from specimens referred to Instituto Adolfo Lutz in São Paulo, Brazil, between 2003 and 2004. Of the 153 isolates, 73 and 80 were from human and nonhuman sources, respectively. The human isolates were collected from six public hospitals and originated from stool (52), blood (15), cerebrospinal fluid (3), and urine (3). The nonhuman isolates were from poultry and its environment (47), swine and its environment (25), and foodstuffs (8). Of these, 10 displayed an ESBL phenotype, as determined by double-disk diffusion testing. These isolates originated in São Paulo and Porto Alegre, Brazil. The two cities are approximately 1100 km apart. In São Paulo, two isolates were identified, one from the stool of a hospitalized child with gastroenteritis and the other from poultry. In Porto Alegre, one isolate was identified from a blood culture of a hospitalized child and the remaining seven were identified from drag swabs collected from the floors of poultry environment (Table 1).
Table 1.
Antimicrobial Susceptibility, Pulsed-Field Gel Electrophoresis Types and Extended-Spectrum β-Lactamase in Salmonella Typhimurium isolates
| |
|
|
MICs (μg/ml) of: |
|
|
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Origin | City | AMP | CAZ | CPM | CTX | CRO | Additional resistance | PFGE | ESBL |
| 581/03 | Patient | Porto Alegre | ≥ 256 | 24 | ≥ 32 | ≥ 32 | ≥ 32 | AMCa ATM CO SF SFT TT | A | CTX-M2 |
| 390/04 | Patient | São Paulo | ≥ 256 | 8 | ≥ 32 | ≥ 32 | ≥ 32 | AMCa ATM ET SF SFT TT | B | CTX-M2 |
| 804/04 | Poultry | São Paulo | ≥ 256 | 8 | ≥ 32 | ≥ 32 | ≥ 32 | ATM ET SF SFT TT | C | CTX-M2 |
| 1523/04 | Drag swab | Porto Alegre | ≥ 256 | 12 | ≥ 32 | ≥ 32 | ≥ 32 | AMC ATM ET SF SFT TT | C | CTX-M2 |
| 1524/04 | Drag swab | Porto Alegre | ≥ 256 | 8 | ≥ 32 | ≥ 32 | ≥ 32 | ATM ET SF SFT TT | D | CTX-M2 |
| 1526/04 | Drag swab | Porto Alegre | ≥ 256 | 16 | ≥ 32 | ≥ 32 | ≥ 32 | AMC ATM ET GN SF SFT TT | B | CTX-M2 |
| 1527/04 | Drag swab | Porto Alegre | ≥ 256 | 12 | ≥ 32 | ≥ 32 | ≥ 32 | AMC ATM ET SF SFT TT | B | CTX-M2 |
| 1528/04 | Drag swab | Porto Alegre | ≥ 256 | 12 | ≥ 32 | ≥ 32 | ≥ 32 | AMCa ATM ET SF SFT TT | B | CTX-M2 |
| 1612/04 | Drag swab | Porto Alegre | ≥ 256 | 12 | ≥ 32 | ≥ 32 | ≥ 32 | AMCa ATM ET SF SFT TT | B | CTX-M2 |
| 1613/04 | Drag swab | Porto Alegre | ≥ 256 | 12 | ≥ 32 | ≥ 32 | ≥ 32 | AMCa ATM ET SF SFT TT | B | CTX-M2 |
Intermediate resistance.
AMC, amoxicillin/clavulanic acid; AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; CTX, cefotaxime; CRO, ceftriaxone; CPM, cefepime; CO, chloramphenicol; ET, streptomycin; GN, gentamicin; SFT, trimethoprim/sulfamethoxazole; SF, sulfonamide; TT, tetracycline; MIC, minimum inhibitory concentration; PFGE, pulsed-field gel electrophoresis; ESBL, extended-spectrum β-lactamase.
Serotyping
The isolates were serotyped on the basis of somatic O and phase 1 and phase 2 of H flagellar antigens by agglutination tests with antisera (prepared in the Laboratory of Enteric Pathogens, Instituto Adolfo Lutz, São Paulo), as specified in the Kauffmann–White scheme for Salmonella serotyping.19
Susceptibility testing
Antimicrobial susceptibility was determined by the disk diffusion method using Mueller–Hinton agar plates (Merck KGaA, Darmstadt, Germany) according to the guidelines of the Clinical and Laboratory Standards Institute.3,4 The following antimicrobials disks (Oxoid, Hampshire, United Kingdom) were used: nalidixic acid, amoxicillin/clavulanic acid, ampicillin, aztreonam, ceftazidime, ceftazidime/clavulanic acid, cefotaxime, cefotaxime/clavulanic acid, ceftriaxone, cefepime, ciprofloxacin, chloramphenicol, streptomycin, gentamicin, imipenem, kanamycin, trimethoprim/sulfamethoxazole, sulfonamide, and tetracycline. Minimum inhibitory concentrations (MICs) were determined for ampicillin, ceftazidime, cefotaxime, and ceftriaxone by Etest (AB Biodisk, Solna, Sweden) according to the manufacturer's recommendations.
ESBL production was confirmed by double-disk diffusion testing when the key-hole effect was observed between the cephalosporin and amoxicillin/clavulanate disks, indicating partial or total restoration of the activity of ceftazidime or cefotaxime by clavulanic acid.12 E. coli ATCC25922, E. coli ATCC35218, and K. pneumoniae ATCC700603 were used as reference strains for antimicrobial susceptibility testing.
Conjugation assays and plasmid analysis
Conjugal transfer was carried out in mixed broth cultures by using E. coli K-12 (Lac−Thr−Leu−Thi−Strr) as the recipient strain. Transconjugants were selected on Mueller–Hinton agar plates containing cefotaxime (10 μg/ml) and nalidixic acid (50 μg/ml). Plasmid DNA was extracted from E. coli transconjugant cells13 and were analyzed by electrophoresis in 0.7% agarose gel (Tris–acetate buffer), by using plasmids of known sizes as standards.
Pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis (PFGE) analysis was performed according to the Centers for Disease Control and Prevention (CDC) PulseNet protocol (www.cdc.gov/pulsenet/protocols.htm). Briefly, cell lysis was followed by proteinase K treatment and DNA restriction with XbaI (New England Biolabs, Ipswich, MA). PFGE was performed with a CHEF DRII system (Bio-Rad, Hercules, CA) and the following run parameters: a switch time of 2.2 to 63.8 seconds and a run time of 20 hours. S. enterica Braenderup H9812 was used as molecular size marker. The macrorestriction patterns were compared by using the Gel Compar II software (Applied Maths, Sint-Martens-Latem, Belgium). Dice coefficient of 1.5 was used to calculate the similarity by using the unweighted pair group method with arithmetic averages. A difference of at least one restriction fragment in the patterns was considered the criterion for distinguishing between different profiles.
PCR analysis and DNA sequencing
PCR analysis to detect various ESBL genes was carried out as previously described.5 PCR products were resolved on 1% agarose gels, stained with ethidium bromide, and photographed with ultraviolet illumination. DNA sequencing of the PCR products was performed with an ABI 3130 instrument (Applied Biosystems, Foster City, CA). Specifically, the entire coding region of the CTX-M-2 gene was sequenced using external primers.5
Results
Antimicrobial susceptibility
All 10 Salmonella Typhimurium isolates that displayed an ESBL phenotype, as determined by double-disk diffusion testing, were resistant to ampicillin, aztreonam, cefotaxime, ceftriaxone, cefepime, trimethoprim/sulfamethoxazole, sulfonamide, and tetracycline. Nine were resistant to streptomycin, one to chloramphenicol, and another to gentamicin, as shown in Table 1. Six isolates were susceptible to ceftazidime, whereas four showed intermediate resistance. Three isolates were resistant to amoxicillin/clavulanic acid, five intermediately resistant, and two susceptible. All 10 isolates were susceptible to nalidixic acid, ciprofloxacin, and imipenem. The MICs of β-lactams are also shown in Table 1. Most isolates exhibited a higher level of resistance to cefotaxime, cefepime, ceftriaxone (MICs ≥ 32 μg/ml) than to ceftazidime (MICs 8–16 μg/ml).
Conjugation experiments
Cefotaxime-resistant E. coli transconjugants were readily obtained from each Salmonella Typhimurium donor at a frequency of approximately 10−4 transconjugants/recipient. The E. coli transconjugants showed cross resistance to multiple β-lactams. Except for one isolate, all of them also showed additional resistance to sulfonamide, trimethoprim/sulfamethoxazole, and tetracycline (Table 2). The plasmid sizes of E. coli tranconjugants were estimated to be between 90 and 170 kb (Table 2).
Table 2.
Antimicrobial Resistance and Plasmid Characterization of Escherichia coli Transconjugants
| Escherichia coli transconjugants | Origin | City | Cotransferred resistance | Plasmid size (kb) |
|---|---|---|---|---|
| 581/03 | Patient | Porto Alegre | AMP ATM CAZ CPM CTX CRO | 90 |
| 390/04 | Patient | São Paulo | AMP ATM CAZ CPM CTX CRO SF SFT TT | 90 |
| 804/04 | Poultry | São Paulo | AMP ATM CAZ CPM CTX CRO SF SFT TT | 170 |
| 1523/04 | Drag swab | Porto Alegre | AMP ATM CAZ CPM CTX CRO SF SFT TT | 170 |
| 1524/04 | Drag swab | Porto Alegre | AMP ATM CAZ CPM CTX CRO SF SFT TT | 170 |
| 1526/04 | Drag swab | Porto Alegre | AMP ATM CAZ CPM CTX CRO | 90 |
| 1527/04 | Drag swab | Porto Alegre | AMP ATM CAZ CPM CTX CRO | 90 |
| 1528/04 | Drag swab | Porto Alegre | AMP ATM CAZ CPM CTX CRO | 90 |
| 1612/04 | Drag swab | Porto Alegre | AMP ATM CAZ CPM CTX CRO | 90 |
| 1613/04 | Drag swab | Porto Alegre | AMP ATM CAZ CPM CTX CRO | 90 |
AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; CPM, cefepime; CTX, cefotaxime; CRO, ceftriaxone; SF, sulfonamide; SFT, trimethoprim–sulfamethoxazole; TT, tetracycline.
PFGE
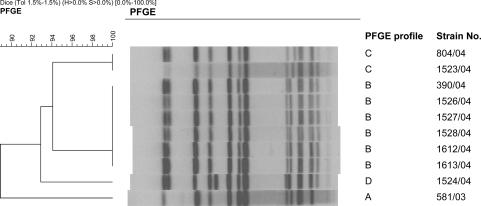
The PFGE patterns of the Salmonella Typhimurium isolates are shown in Fig. 1. Four PFGE patterns (A, B, C, and D) were identified. The genetic relatedness of these strains ranged from 89% to 100%. All PFGE types were identified among the strains isolated in Porto Alegre. The two isolates from São Paulo belonged to groups B and C. The clinical isolate from São Paulo and an avian isolate from Porto Alegre shared an identical pattern in group C.
FIG. 1.
Dendrogram for Salmonella Typhimurium isolates generated by pulsed-field gel electrophoresis (PFGE).
PCR for detection of ESBL resistance genes and sequencing results
All 10 isolates had positive PCR with primers specific for the CTX-M-type β-lactamase genes. DNA sequencing revealed the genes to encode CTX-M-2.
Discussion
In this study, we identified CTX-M-2–producing Salmonella Typhimurium isolates from humans and poultry sources in two distant cities in Brazil. These isolates appeared genetically related based on PFGE using XbaI as the restriction enzyme. While not performed in our study, the use of BlnI as the second enzyme may have further increased the discriminatory power of PFGE.24 CTX-M-2 has commonly been identified in K. pneumoniae and E. coli in Brazil and thus appears to be already widely disseminated in this country.6,9,21 Various ESBLs have been identified in Salmonella spp. in Brazil, including OXA-53, CTX-M-8, and CTX-M-9.8,15,17 Our findings add to the growing list of ESBLs that are produced by Salmonella in Brazil and also suggest possible interspecies transfer of CTX-M-2 into this species from other species in the family Enterobacteriaceae, where this ESBL is already established.
The ESBL-producing Salmonella Typhimurium in our study were resistant not only to β-lactam antimicrobials but also to a variety of antimicrobials including chloramphenicol, streptomycin, gentamicin, trimethoprim/sulfamethoxazole, sulfonamide, and tetracycline. Over the years, an increasing proportion of Salmonella isolates have acquired resistance to various antimicrobials, including Salmonella Typhimurium strains isolated in Brazil.10 Other reports have also highlighted the emergence of ESBL-producing Salmonella strains endowed with an extremely wide spectrum of antimicrobials, as also observed in this study.11,20,22 The ESBL gene responsible for cephalosporin resistance is frequently cotransferred with other resistance determinants, contributing to multidrug resistance phenotype. In the present study, some of the isolates were found to mobilize resistance to tetracycline and trimethoprim/sulfamethoxazole along with the CTX-M-2 gene.
The use of antimicrobial drugs for therapeutic purposes in veterinary medicine and as growth promoters in food-producing animals is speculated to be a major cause of development of resistance in Salmonella, thereby presenting a potential risk to public health from zoonotic infections.2,14 These animals and the products derived from them are then frequently transported regionally as well as internationally, providing ample opportunities for dissemination of multidrug-resistant isolates that possess highly mobile and clinically important resistance mechanisms into the community in a large geographic area.21 Among the isolates studied, two were isolated from hospitalized children, and the others were isolated from poultry and its environment. While the investigation was limited to poultry and swine in our present study, other animals such as cattle are also potential carrier of ESBL-producing Salmonella spp. In summary, our findings of genetically related, ESBL-producing Salmonella isolates identified from patients as well as poultry and its environment underscore the importance of implementing control measures to minimize development of antimicrobial resistance in zoonotic organisms such as Salmonella spp.
Acknowledgments
We thank Claudio Sachi for technical assistance. Part of this study was supported by a Fogarty International Center Global Infectious Diseases Research Training Program grant, National Institutes of Health (D43TW006592; principal investigator, Lee H. Harrison).
Disclosure Statement
No competing financial interests exist.
References
- 1.Bertrand S. Weill F.X. Cloeckaert A. Vrints M. Mairiaux E. Praud K. Dierick K. Wildemauve C. Godard C. Butaye P. Imberechts H. Grimont P.A. Collard J.M. Clonal emergence of extended-spectrum β-lactamase (CTX-M-2)-producing Salmonella enterica serovar Virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003) J. Clin. Microbiol. 2006;44:2897–2903. doi: 10.1128/JCM.02549-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carattoli A. Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 2008;14(Suppl 1):117–123. doi: 10.1111/j.1469-0691.2007.01851.x. [DOI] [PubMed] [Google Scholar]
- 3.Clinical, Laboratory Standards Institute. Performance standards for antimicrobial disk susceptibility testing. Sixteenth informational supplement, document M100-S16 2006.
- 4.Clinical, Laboratory Standards Institute. Performance standards for antimicrobial disk susceptibility tests. Approved standard, ninth edition, document M2-A9 2006.
- 5.de Oliveira Garcia D. Doi Y. Szabo D. Adams-Haduch J.M. Vaz T.M. Leite D. Padoveze M.C. Freire M.P. Silveira F.P. Paterson D.L. Multiclonal outbreak of Klebsiella pneumoniae producing extended-spectrum β-lactamase CTX-M-2 and novel variant CTX-M-59 in a neonatal intensive care unit in Brazil. Antimicrob. Agents Chemother. 2008;52:1790–1793. doi: 10.1128/AAC.01440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Do Carmo Filho J.R. Silva R.M. Castanheira M. Tognim M.C. Gales A.C. Sader H.S. Prevalence and genetic characterization of blaCTX-M among Klebsiella pneumoniae isolates collected in an intensive care unit in Brazil. J. Chemother. 2008;20:600–603. doi: 10.1179/joc.2008.20.5.600. [DOI] [PubMed] [Google Scholar]
- 7.Fernandes S.A. Tavechio A.T. Ghilardi A.C. Dias A.M. Almeida I.A. Melo L.C. Salmonella serovars isolated from humans in Sao Paulo State, Brazil, 1996–2003. Rev. Inst. Med. Trop. Sao Paulo. 2006;48:179–184. doi: 10.1590/s0036-46652006000400001. [DOI] [PubMed] [Google Scholar]
- 8.Fonseca E.L. Mykytczuk O.L. Asensi M.D. Reis E.M. Ferraz L.R. Paula F.L. Ng L.K. Rodrigues D.P. Clonality and antimicrobial resistance gene profiles of multidrug-resistant Salmonella enterica serovar infantis isolates from four public hospitals in Rio de Janeiro, Brazil. J. Clin. Microbiol. 2006;44:2767–2772. doi: 10.1128/JCM.01916-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia Dde O. Doi Y. Szabo D. Adams-Haduch J.M. Vaz T.M. Leite D. Padoveze M.C. Freire M.P. Silveira F.P. Paterson D.L. Multiclonal outbreak of Klebsiella pneumoniae producing extended-spectrum β-lactamase CTX-M-2 and novel variant CTX-M-59 in a neonatal intensive care unit in Brazil. Antimicrob. Agents Chemother. 2008;52:1790–1793. doi: 10.1128/AAC.01440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghilardi A.C. Tavechio A.T. Fernandes S.A. Antimicrobial susceptibility, phage types, and pulse types of Salmonella Typhimurium, in Sao Paulo, Brazil. Mem. Inst. Oswaldo Cruz. 2006;101:281–286. doi: 10.1590/s0074-02762006000300010. [DOI] [PubMed] [Google Scholar]
- 11.Hasman H. Mevius D. Veldman K. Olesen I. Aarestrup F.M. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 2005;56:115–121. doi: 10.1093/jac/dki190. [DOI] [PubMed] [Google Scholar]
- 12.Jarlier V. Nicolas M.H. Fournier G. Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 13.Kado C.I. Liu S.T. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X.Z. Mehrotra M. Ghimire S. Adewoye L. β-Lactam resistance and β-lactamases in bacteria of animal origin. Vet. Microbiol. 2007;121:197–214. doi: 10.1016/j.vetmic.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Mulvey M.R. Boyd D.A. Baker L. Mykytczuk O. Reis E.M. Asensi M.D. Rodrigues D.P. Ng L.K. Characterization of a Salmonella enterica serovar Agona strain harbouring a class 1 integron containing novel OXA-type β-lactamase (blaOXA-53) and 6′-N-aminoglycoside acetyltransferase genes [aac(6′)-I30] J. Antimicrob. Chemother. 2004;54:354–359. doi: 10.1093/jac/dkh347. [DOI] [PubMed] [Google Scholar]
- 16.Paterson D.L. Bonomo R.A. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peirano G. Agerso Y. Aarestrup F.M. dos Reis E.M. dos Prazeres Rodrigues D. Occurrence of integrons and antimicrobial resistance genes among Salmonella enterica from Brazil. J. Antimicrob. Chemother. 2006;58:305–309. doi: 10.1093/jac/dkl248. [DOI] [PubMed] [Google Scholar]
- 18.Pitout J.D. Laupland K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 2008;8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 19.Popoff M.Y. Bockemuhl J. Brenner F.W. Gheesling L.L. Supplement 2000 (no. 44) to the Kauffmann-White scheme. Res. Microbiol. 2001;152:907–909. doi: 10.1016/s0923-2508(01)01274-8. [DOI] [PubMed] [Google Scholar]
- 20.Riano I. Moreno M.A. Teshager T. Saenz Y. Dominguez L. Torres C. Detection and characterization of extended-spectrum β-lactamases in Salmonella enterica strains of healthy food animals in Spain. J. Antimicrob. Chemother. 2006;58:844–847. doi: 10.1093/jac/dkl337. [DOI] [PubMed] [Google Scholar]
- 21.Warren R.E. Ensor V.M. O'Neill P. Butler V. Taylor J. Nye K. Harvey M. Livermore D.M. Woodford N. Hawkey P.M. Imported chicken meat as a potential source of quinolone-resistant Escherichia coli producing extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 2008;61:504–508. doi: 10.1093/jac/dkm517. [DOI] [PubMed] [Google Scholar]
- 22.Weill F.X. Guesnier F. Guibert V. Timinouni M. Demartin M. Polomack L. Grimont P.A. Multidrug resistance in Salmonella enterica serotype Typhimurium from humans in France (1993 to 2003) J. Clin. Microbiol. 2006;44:700–708. doi: 10.1128/JCM.44.3.700-708.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weill F.X. Lailler R. Praud K. Kerouanton A. Fabre L. Brisabois A. Grimont P.A. Cloeckaert A. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multiresistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J. Clin. Microbiol. 2004;42:5767–5773. doi: 10.1128/JCM.42.12.5767-5773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao S. White D.G. Friedman S.L. Glenn A. Blickenstaff K. Ayers S.L. Abbott J.W. Hall-Robinson E. McDermott P.F. Antimicrobial resistance in Salmonella enterica serovar Heidelberg isolates from retail meats, including poultry, from 2002 to 2006. Appl. Environ. Microbiol. 2008;74:6656–6662. doi: 10.1128/AEM.01249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]