Abstract
We carried out the first study of Enterococcus faecalis clinical isolates in Cuba by multilocus sequence typing linking the molecular typing data with the presence of virulence determinants and the antibiotic resistance genes. A total of 23 E. faecalis isolates recovered from several clinic sources and geographic areas of Cuba during a period between 2000 and 2005 were typed by multilocus sequence typing. Thirteen sequence types (STs) including five novel STs were identified, and the ST 64 (clonal complex [CC] 8), ST 6 (CC2), ST 21(CC21), and ST 16 (CC58) were found in more than one strain. Sixty-seven percent of STs corresponded to STs reported previously in Spain, Poland, and The Netherlands, and other STs (ST115, ST64, ST6, and ST40) were genetically close to those detected in the United States. Prevalence of both antimicrobial resistance genes [aac(6′)-aph(2″), aph(3′), ant(6), ant(3″)(9), aph(2″)-Id, aph(2″)-Ic, erm(B), erm(A), erm(C), mef(A), tet(M), and tet(L)] and virulence genes (agg, gelE, cylA, esp, ccf, and efaAfs) were examined by polymerase chain reaction. Aminoglycoside resistance genes aac(6′)-Ie-aph(2″)-Ia, aph(3′), ant(6), ant(3″)(9) were more frequently detected in ST6, ST16, ST23, ST64, and ST115. The multidrug resistance was distributed to all STs detected, except for ST117 and singleton ST225. The presence of cyl gene was specifically linked to the ST64 and ST16. Presence of the esp, gel, and agg genes was not specific to any particular ST. This research provided the first insight into the population structure of E. faecalis in Cuba, that is, most Cuban strains were related to European strains, whereas others to U.S. strains. The CC2, CC21, and CC8, three of the biggest CCs in the world, were evidently circulating in Cuba, associated with multidrug resistance and virulence traits.
Introduction
Enterococci have been documented in recent years as important nosocomial pathogens worldwide. Enterococcus faecalis is the most prevalent species associated with human infections in both community and hospital settings.21 The biggest concern of enterococci is that they become multiresistant pathogens due to acquisition of resistance via incorporation of mobile genetic elements, in addition to intrinsic resistances.16 In addition, the virulence factors of enterococci have been recognized as important for pathogenicity of this organism and survival in the host.26
Numerous genetic typing methods have been applied to the epidemiological investigations of enterococci, including multilocus enzyme electrophoresis, insertion sequence element-based typing, pulsed-field gel electrophoresis (PFGE), restriction fragment length polymorphism analysis, ribotyping, repetitive sequence-based polymerase chain reaction (PCR), arbitrary primed PCR, and random amplification of polymorphic DNA.15
Recently, multilocus sequence typing (MLST), a molecular typing method based on identification of alleles from sequences of housekeeping genes, was developed for determination of genetic lineages within E. faecalis and Enterococcus faecium and has successfully differentiated population structure of these species distributed globally.11,21 Nevertheless, a few studies have employed MLST for molecular epidemiology of enterococcus so far, therefore evolutionary history and distribution of various lineages in the world is still poorly understood.
The importance of Enterococcus in Cuban hospitals, and intra- and interhospital dissemination of some E. faecalis clones have been reported in previous studies by the use of PFGE.19,20 However, PFGE is not adequate to characterize the genetic evolution of bacterial clones.15 Therefore, in the present study, we carried out MLST of E. faecalis isolates from Cuba as a first study and analyzed the relatedness of the sequence type (ST) to the presence of virulence determinants and the antibiotic resistance genes.
Materials and Methods
Bacterial strains, identification
Twenty-three E. faecalis isolates from different geographic locations in Cuba isolated between 2001 and 2005 were chosen for this study (Table 1). E. faecalis isolates were selected proportionally according to the number of reports of isolations per year, per province, and hospital. The species identification of E. faecalis was performed using API 20 Strep (bioMerieux, Marcy, L'Etoile, France), and it was confirmed by PCR as described by Dutka-Malen et al.6
Table 1.
Allelic Profile of Enterococcus faecalis Sequence Types and Epidemiological Data
| |
MLST allele |
|
|
|
|
|
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | gdh | gyd | pstS | gki | aroE | xpt | yqiL | ST | CC | Source | Year | Place | Province/city |
| DQ195 | 12 | 7 | 3 | 7 | 6 | 1 | 5 | 6 | 2 | Endot. sec | 2003 | Hospital | Havana City |
| DQ203 | 12 | 7 | 3 | 7 | 6 | 1 | 5 | 6 | 2 | Blood | 2003 | Hospital | Havana City |
| DQ210 | 12 | 7 | 3 | 7 | 6 | 1 | 5 | 6 | 2 | Surgical wound | 2003 | Hospital | Santiago |
| DQ132 | 10 | 1 | 11 | 6 | 5 | 1 | 4 | 64 | 8 | Catheter | 2001 | Hospital | Holguin |
| DQ152 | 10 | 1 | 11 | 6 | 5 | 1 | 4 | 64 | 8 | Blood | 2001 | Hospital | Holguin |
| DQ199 | 10 | 1 | 11 | 6 | 5 | 1 | 4 | 64 | 8 | Blood | 2003 | Hospital | Havana City |
| DQ320 | 10 | 1 | 11 | 6 | 5 | 1 | 4 | 64 | 8 | Blood | 2005 | Hospital | Havana City |
| DQ118 | 1 | 7 | 9 | 1 | 1 | 1 | 1 | 21 | 21 | Surgical wound | 2002 | Hospital | Santiago |
| DQ120 | 1 | 7 | 9 | 1 | 1 | 1 | 1 | 21 | 21 | Catheter | 2002 | Hospital | Santiago |
| DQ295 | 1 | 7 | 9 | 1 | 1 | 1 | 1 | 21 | 21 | Surgical wound | 2005 | Hospital | Holguin |
| DQ256 | 5 | 1 | 1 | 3 | 7 | 7 | 6 | 16 | 58 | Vagina | 2004 | Community | Guantanamo |
| DQ269 | 5 | 1 | 1 | 3 | 7 | 7 | 6 | 16 | 58 | CSF | 2004 | Hospital | Santiago |
| DQ297 | 5 | 1 | 1 | 3 | 7 | 7 | 6 | 16 | 58 | Ammniotic fluid | 2005 | Hospital | Holguin |
| DQ314 | 5 | 1 | 1 | 3 | 7 | 7 | 6 | 16 | 58 | Vagina | 2005 | Community | Holguin |
| DQ138 | 14 | 2 | 18 | 10 | 16 | 2 | 12 | 59 | N | Perianal fistula | 2001 | Hospital | Holguin |
| DQ213 | 3 | 6 | 23 | 12 | 9 | 10 | 7 | 40 | 40 | Blood | 2003 | Hospital | Havana City |
| DQ165 | 27 | 2 | 16 | 28 | 26 | 2 | 1 | 81 | N | CSF | 2002 | Hospital | Holguin |
| DQ243 | 2 | 3 | 13 | 11 | 3 | 2 | 2 | 23 | 25 | Peritoneal fluid | 2004 | Hospital | Holguin |
| DQ190 | 1 | 1 | 11 | 1 | 21 | 1 | 2 | 115 | N | CSF | 2003 | Hospital | C. de Ávila |
| DQ287 | 17 | 2 | 22 | 1 | 14 | 14 | 1 | 116 | 116 | Catheter | 2004 | Hospital | Havana City |
| DQ298 | 1 | 1 | 9 | 6 | 1 | 1 | 1 | 117 | 21 | Blood | 2005 | Hospital | Havana City |
| DQ261 | 15 | 2 | 33 | 16 | 17 | 15 | 35 | 118 | N | Vagina | 2004 | Community | Guantanamo |
| DQ317 | 39 | 2 | 49 | 45 | 47 | 2 | 48 | 225 | N | Surgical wound | 2005 | Hospital | Havana City |
CC, clonal complex; N, not assigned; Endot. Sec., endotracheal secretion; CSF, cerebrospinal fluid.
Antimicrobial susceptibility test
The minimal inhibitory concentrations (MICs) for gentamicin, streptomycin, amikacin, ampicillim, vancomycin, teicoplanin, ciprofloxacin, norfloxacin, levofloxacin, rifampin, fosfomycin, and nitrofurantoin, erythromycin, chloramphenicol, and tetracycline were determined by the agar dilution method according to the Clinical Laboratory Standards Institute guidelines (2007).3 MIC was defined as the lowest concentration of the antimicrobial agent that inhibited visible growth at 35°C after overnight incubation. To interpret MICs as susceptible or resistant, the breakpoints suggested by the Clinical Laboratory Standards Institute3 were applied.
Detection of antibiotic resistance genes by PCR
Presence of different antibiotic resistance genes was investigated by PCR using specific primers, as previously described [erm(B), erm(A), erm(C), and mef(A),24 tet(M) and tet(L)17 and aac(6′)-aph(2″), aph(3′), ant(6), ant(3″)(9), aph(2″)-id, and aph(2″)-ic].1 Strains for positive and negative control were included in all the PCRs.
Detection of virulence genes
Virulence factor genes of E. faecalis were detected by PCR assay using previously reported primers.2,7,13,25,26 Six genes coding aggregation substance (agg), gelatinase (gelE), cytolysin (cylA), enterococcal surface protein (esp), adhesion protein (efaAfs), and sex pheromones (ccf) were targeted.
MLST
MLST was carried out according to the scheme described previously.21 The seven E. faecalis housekeeping genes, gdh (glucose-6-phosphate dehydrogenase), gyd (glyceraldehyde-3-phosphate dehydrogenase), pstS (phosphate ATP-binding cassette transporter), gki (glucokinase, putative), aroE (shikimate 5-dehydrogenase), xpt (xanthine phosphoribosyltransferase), and yqil (acetyl-CoA acetyltransferase) were amplified by PCR. Information of the E. faecalis MLST scheme, sequences of PCR primers, and the PCR conditions are available at MLST web site (http://efaecalis.mlst.net). The ST was further analyzed to determine the clonal complex (CC) by the eBURST version.3 software. The genetic relationship among strains was analyzed by constructing a dendogram using MEGA4 program.
Results
Clonal lineages identified by MLST
Twenty-three E. faecalis strains were assigned to 13 different STs of which five STs were novel types (ST115, ST116, ST117, ST118, and ST225). The most frequent types found were ST64 (four isolates) and ST16 (four isolates), followed by ST21 (three isolates) and ST6 (three isolates), whereas other nine STs were represented by a single isolate (ST23, ST40, ST59, ST81, and the five novel STs detected) (Table 1).
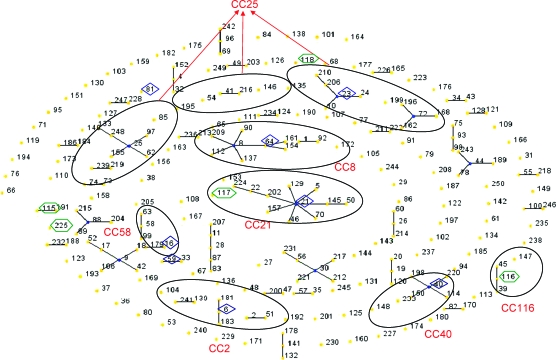
Clustering of these types with the eBURST revealed the presence of seven CCs, that is, CC2 (ST6), CC8 (ST64), CC21 (ST21 and ST117), CC25 (ST23), CC40 (ST40), CC58 (ST16), CC116 (ST116), and three singletons (ST81, ST118, and ST225) as shown in Fig. 1. Two STs, ST115 and ST59, represented a single-locus variant (SLV) of ST191 and ST33, respectively. ST6 is a double-locus variant of ST2 belonging to the CC2 described in Spain. The ST64 represented SLV of ST8 belonging to the CC8. The ST116, a new ST detected in Cuba, was SLV of ST45 and ST39 that were isolated from hospitalized patients in Spain. ST117 is a double-locus variant of ST21 in CC21 (one of the biggest CCs detected in Spain), which includes strains from animals, hospitalized patients, and healthy volunteers from the community (P. Ruiz-Garbajosa, personal communication).
FIG. 1.
eBURST diagram of Enterococcus faecalis STs including 23 STs identified in Cuba in this study. Each number represents ST, and oval indicates CC of STs. Numbers in hexagons indicate newly identified STs in Cuban strains, whereas other STs previously described worldwide and found in Cuba in the present study are shown in lozenges. ST, sequence type; CC, clonal complex. Color images available online at www.liebertonline.com/mdr.
Epidemiology of CC
E. faecalis isolates characterized by MLST were grouped into CCs independent of their epidemiologic and geographical origin. The ST16 was found in both the hospital and community and was responsible for either invasive disease or simple colonization. The singleton ST118 was detected only in the community. The rest of STs and CCs was detected in hospitals only (Table 1).
ST64 (CC8), one of the most predominant STs detected in Cuba, has persisted for several years in the hospitals (2001–2005) and dispersed in the western and the eastern region of Cuba, similar to ST6 (CC2) but it was detected only in one year (2003). ST16 (CC58) was detected between 2004 and 2005 in the hospitals and community from eastern region of Cuba (Table 1).
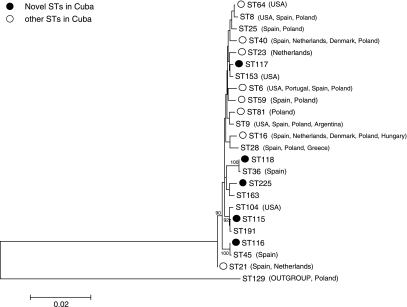
The dendogram revealed the genetic relationship of STs among the E. faecalis isolates from Cuba (n = 23) and representative STs (ST8, ST9, ST25, ST28, and ST104) belonging to dominant CCs from various countries (Fig. 2).
FIG. 2.
Dendogram of sequences of housekeeping genes used for multilocus sequence typing showing genetic relationship among Enterococcus faecalis isolates from Cuba and from various countries.
Most STs detected in Cuba have also been reported in European countries (ST6, ST16, ST21, ST23, ST40, ST59, ST81) and 67% of the isolates corresponded to isolates from Spain, Poland, and The Netherlands. Some of the novel STs (ST116, ST117, ST118) were close to those detected in Europe, whereas ST64, ST6, ST40, and the other novel ST (ST115) were close to those detected in the United States (Fig. 2).
Distribution of resistance and virulence genes among the clonal lineages detected
No isolate was resistant to ampicillim, vancomycin, teicoplanin, and moxifloxacin among the strains characterized by MLST (Table 2). Nevertheless, high rates of high-level resistance were found to aminoglycosides, that is, gentamicin (48%), streptomycin (52%), and amikacin (69.5%). Similar results were obtained for resistance to erythromycin (69.5%), chloramphenicol (43.4%), tetracycline (91%), and rifampicin (56.5%). The low-resistance rates were found for ciprofloxacin (22%), norfloxacin (13%), levofloxacin (22%), fosfomycin (4.3%), and nitrofurantoin (8.7%).
Table 2.
Virulence Genes, Drug Resistance Genes, and Antimicrobial Susceptibility in Enterococcus faecalis Strains of Individual Sequence Types
| |
|
Virulence genes |
Aminoglycoside resistance gene |
Tetracycline resistance gene |
Macrolide resistance gene |
MIC (μg/mL) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | ST | agg | gelE | cylA | esp | acc | efa Afs | aac(6′)-aph(2″) | aph(3′)- Illa | ant(6)- Ia | ant (3″) (9) | tet(M) | tet(L) | erm(B) | GM | AMK | SM | TC | EM | CP | CIP | RIF |
| DQ195 | 6 | − | − | − | + | − | + | + | − | + | − | + | − | − | ≥4096 | 2048 | ≥4096 | 32 | 2 | 64 | 16 | ≤0.5 |
| DQ203 | 6 | − | − | − | + | − | + | + | − | + | − | + | − | + | ≥4096 | 2048 | 2048 | 128 | 16 | 64 | 32 | 16 |
| DQ210 | 6 | − | − | − | + | + | + | − | + | + | − | + | − | − | 128 | ≥4096 | 2048 | 64 | 4 | 64 | 32 | ≤0.5 |
| DQ132 | 64 | − | + | + | + | − | + | + | + | − | − | + | − | + | ≥4096 | ≥4096 | 64 | ≥256 | 32 | 2 | 2 | 16 |
| DQ152 | 64 | − | + | + | + | − | + | − | − | − | − | + | − | + | 64 | 128 | 128 | ≥256 | 64 | ≥128 | ≤0.5 | 8 |
| DQ199 | 64 | − | − | + | + | − | + | + | − | − | + | + | − | + | ≥4096 | ≥4096 | 2048 | ≥256 | 128 | ≥128 | ≤0.5 | 8 |
| DQ320 | 64 | − | + | − | + | − | + | − | − | + | − | + | − | + | 32 | 64 | ≥4096 | 128 | 32 | ≥128 | 4 | 16 |
| DQ118 | 21 | + | + | − | + | + | + | − | + | − | − | + | − | − | 32 | 2048 | 64 | 64 | 2 | 4 | 1 | ≤0.5 |
| DQ120 | 21 | + | + | − | + | + | + | − | + | − | − | + | − | − | 32 | ≥4096 | 32 | 32 | ≤1 | 2 | 1 | 1 |
| DQ295 | 21 | − | − | − | + | + | + | − | − | − | − | + | − | − | 16 | 64 | 128 | 64 | 16 | 2 | 2 | ≤0.5 |
| DQ256 | 16 | + | − | + | + | − | + | + | + | − | + | + | − | + | 2048 | ≥4096 | 2048 | 128 | 16 | 8 | ≤00.5 | 8 |
| DQ269 | 16 | + | − | + | + | + | + | + | + | + | − | + | − | − | ≥4096 | ≥4096 | ≥4096 | 128 | 2 | 2 | 1 | 1 |
| DQ297 | 16 | + | − | + | + | + | + | + | + | + | − | + | − | − | ≥4096 | ≥4096 | ≥4096 | 64 | 8 | 4 | ≤0.5 | 1 |
| DQ314 | 16 | + | − | + | + | + | + | + | + | − | − | + | − | − | ≥4096 | ≥4096 | 512 | 32 | 8 | 8 | 1 | ≤0.5 |
| DQ138 | 59 | − | + | − | + | + | + | + | − | − | − | − | + | − | 2048 | 2048 | 2048 | 128 | 16 | 2 | 16 | ≤0.5 |
| DQ213 | 40 | − | + | − | + | + | + | − | + | − | − | + | − | + | 32 | 2048 | ≥4096 | ≥256 | 32 | 4 | 2 | ≤0.5 |
| DQ165 | 81 | − | + | − | + | + | + | − | + | − | − | + | − | + | 8 | 2048 | 128 | 64 | 64 | ≥128 | 2 | 16 |
| DQ243 | 23 | − | − | + | − | + | + | + | + | + | + | − | − | 2048 | 2048 | 2048 | 64 | 2 | 2 | 1 | ≤0.5 | |
| DQ190 | 115 | − | − | − | − | + | + | + | + | − | + | + | + | ≥4096 | ≥4096 | ≥4096 | 128 | 64 | 32 | 1 | ≤0.5 | |
| DQ287 | 116 | − | − | − | − | + | + | − | − | − | − | − | − | + | 16 | 64 | 64 | ≤2 | 32 | 64 | 1 | 1 |
| DQ298 | 117 | − | − | − | + | + | + | − | − | − | − | − | − | − | 32 | 64 | 32 | ≤2 | ≤1 | 2 | 2 | 8 |
| DQ261 | 118 | + | + | − | + | + | + | − | + | + | − | − | + | + | 128 | 2048 | 2048 | 32 | 128 | 4 | 1 | 1 |
| DQ317 | 225 | + | − | − | − | + | + | − | − | − | − | − | + | + | 32 | 16 | 128 | 16 | 128 | 2 | ≤0.5 | ≤0.5 |
GM, gentamicin; AMK, amikacin; SM, streptomycin; TC, tetracycline; EM, erythromycin; CP, chlormaphenicol; CIP, ciprofloxacin; RIF, rifarmpin.
Ninety-one percent of the isolates were multiresistant (resistance to three or more different families of antimicrobial agents). Multidrug resistance (MDR) was distributed among all STs detected, except for ST117 and singleton ST225.
High-level resistance to gentamicin was significantly more frequently detected among isolates belonging to ST6 (CC2), ST16 (CC58), and ST64 (CC8) (relative risk >1.5, p < 0.05), whereas none of the isolates belonging to ST21 (CC21) was resistant to this aminoglycoside. Aminoglycoside resistance genes aac(6′)-Ie-aph(2″)-Ia, aph(3′), ant(6), ant(3″)(9) were more frequently detected in ST6, ST16, ST64, ST23, ST115, and ST118. The fluoroquinolone resistance was linked to ST6 (CC2), ST59, and ST116 (CC116).
All E. faecalis strains examined in this study in Cuba carried the ccf and the efa genes, whereas 61.5%, 46%, 38.5%, and 23% of the strains carried the esp, gel, agg, and cyl genes, respectively. The presence of cyl gene was specifically linked to the ST64 (CC8) and the ST16 (CC58) that accounted for 35% of all the isolates studied (relative risk >1.5, p < 0.05).
Presence of the esp, gel, and agg genes was not specific to any particular ST. Two STs, ST6 and ST16, contained both esp-positive and esp-negative isolates, whereas all isolates belonging to ST64 were esp negative, and all the ST21 isolates were esp positive.
Discussion
MLST in E. faecalis worldwide has not yet been well characterized so far, and this fact implies the need for careful interpretation of the data obtained through the molecular epidemiology of this species in different countries. The characterization of 23 strains of E. faecalis isolated in Cuba using MLST provided the first data for the knowledge on the genetic population structure for this species in Cuba, which allowed us to compare with those obtained worldwide, and to know the spread of some E. faecalis clones to Cuba.
In the present study, no correlation was found between the clinical source of isolation and the clonal structure of the E. faecalis. Similarly, it was reported by other authors that no host-specific lineage of E. faecalis was found.22
Isolates from the community were recovered from vagina, and these isolates were multiresistant and carried some virulence genes. It could represent a risk to the production of endogenous infection in the patients from this primary site of colonization, overalls, if the patient suffering a urinary tract instrumentation, bladder cauterization, urinary stones, or other forms of urinary obstruction.9 This finding is an evidence of the spread of multiresistant and virulent enterococci in the community in Cuba and highlights the need of monitoring of enterococci and further research on the impact of the horizontal transfer of these genes to enterococci in patients.
ST64, the most persistent ST detected in Cuba during the period of study, showed MDR, harboring aac(6′)-aph(2″), ant(3)(9), aph(3′), tet(M), erm(B), and possessed virulence genes ccf, efa, cyl, gelE, and agg. It is suggested that such characteristics contributed to the persistence of ST64 in the hospital environment and further increased the transmission and spread of the antimicrobial resistance in this species.22
The results of this study demonstrated a wide dissemination of E. faecalis STs in Cuba because some lineages were dispersed in the western and eastern regions of Cuba during long periods of time (ST6, ST16, ST21, and ST64). The antimicrobial drug use might have favored the selection of an enterococcal subpopulation with enhanced antibacterial resistance, virulence, and ability to spread.
Some significant genetic relationships between isolates recovered from Cuba and other countries were found. ST16, ST6, ST21, and ST40 are an example of spread of some genetic lineage of E. faecalis in different parts of the world as they have been detected in several countries, that is, Spain, Poland, The Netherlands, Hungary, Denmark, and the United States.12,21 Isolates belonging to ST16 had been previously isolated from hospitalized patient, nonhospitalized humans, and from animals, whereas isolates belonging to ST40 had been previously isolated from a hospitalized patient, a food handler from Spain, and a seal from The Netherlands.23
E. faecalis isolates belonging to CC40 were detected in Europe, and presumably originated in Poland or its neighboring countries, according to the hypothesis of Kawalec et al.12 The results of the present work indicated the spread of this CC40 in the American continent. It also reported the circulation of CC21 lacking the MDR phenotype in Poland.12 Similarly, Cuban strains belonging to CC21 were not multiresistant or resistant to a few antimicrobials.
Ruiz-Garbajosa et al. reported four major CCs in Spain: the CC9, CC2, CC21, and CC10. CC2 and CC9 were considered to represent hospital-adapted clonal groups, with vancomycin-resistant strains and β-lactamase production.21 One year later, Kawalec et al. reported in Poland the circulation of CC2 and they found that only one-third of the isolates belonging to this CC were vancomycin resistant.12 In our study, we detected the circulation of ST6 that belongs to CC2, but none of the Cuban isolates described in this study was vancomycin resistant or β-lactamase–producing isolates. This finding supports the hypothesis that this clone has originally spread as vancomycin susceptible and later subsets of this clone acquired vancomycin-resistant gene through horizontal gene transfer, as it has been previously reported in Cuba, Spain, Portugal, and Poland.4,12,18,20
Among STs reported in this study, ST23, ST59, and ST81 had also been found previously in a few countries (Spain, Poland, The Netherlands),12,21 whereas five STs (ST115, ST116, ST117, ST118, and ST225) were novel STs. These STs have not been reported in other countries so far, and the high diversity in their allelic profiles (Table 1) suggests that they are genetically unrelated. The lack of information on the population structure of E. faecalis in neighboring countries makes it difficult to speculate about the regional spread of these novel STs detected in Cuba.
The finding of all STs detected in this study, carrying ccf gene, is very important because this gene codes for sexual pheromone that activates the conjugation of the plasmid pCF10. This plasmid plays a significant role in the dissemination of virulence factors and of resistance genes among enterococcus.5 Previously, we detected the ccf gene in 81% of the enterococci isolates from HIV-AIDS patients admitted to the hospital of the “Pedro Kouri” Institute in Cuba and it was found in several enterococcus species (E. faecalis, E. faecium, Enterococcus durans, and Enterococcus avium).10 Our results suggest that ccf gene may be widely distributed in E. faecalis strains that circulate in Cuba.
High prevalence of esp gene was detected also among STs described in this study. Recently, it has been suggested that esp may serve as a marker for the presence of the pathogenicity island harboring multiple virulence factors in E. faecalis and E. faecium.23 Shankar et al. reported a clonal expansion of esp-positive E. faecalis isolates of porcine origin in Denmark which belong to ST16 and ST40.23 This finding is similar to our result because three strains related to ST16 and the ST40 possessed this virulence factor gene.
In addition, it was notable in the present study that cylA gene encoding cytolysin was linked to ST16 and ST64, and three of the four ST64 strains were recovered from blood samples. This finding shows the distribution of cylA in specific lineages of E. faecalis and suggests the association of cytolysin with severity of enterococcal infections as reported previously.8,14,26
In summary, this research provided the first insight into the population structure of E. faecalis in Cuba. Most Cuban strains are related to European strains, and some strains are related to U.S. strains. CC2, CC8, CC21 (three of the biggest CCs in the world), and CC58 were evidently circulating in Cuba, associated with MDR and virulence traits. Specific STs of E. faecalis were identified in isolates from hospital and community, suggesting a wide spread of these STs in this country.
Acknowledgments
We thank Julio Vazquez for helpful discussions and critical reading of the manuscript. This study was supported in part by a Grant-in-Aid for Scientific Research (No. 20590608) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Disclosure Statement
The authors of this paper have no commercial associations that might create a conflict of interest in connection with the submitted manuscript.
References
- 1.Alam M.M. Kobayashi N. Ishino M. Sumi A. Kobayashi K. Uehara N. Watanabe N. Detection of a novel aph(2″) allele (aph[2″]-Ie) conferring high-level gentamicin resistance and a spectinomycin resistance gene ant(9)-Ia (aad 9) in clinical isolates of enterococci. Microb. Drug Resist. 2005;11:239–247. doi: 10.1089/mdr.2005.11.239. [DOI] [PubMed] [Google Scholar]
- 2.Camargo I.L. Gilmore M. Darini A. Multilocus sequence typing and analysis of putative virulence factors in vancomycin-resistant and vancomycin-sensitive Enterococcus faecium isolates from Brazil. Clin. Microbiol. Infect. 2006;12:1123–1130. doi: 10.1111/j.1469-0691.2006.01496.x. [DOI] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI document M100-S17. Clinical and Laboratory Standards Institute; Wayne, PA: 2007. [Google Scholar]
- 4.Del Campo R. Tenorio C. Zarazaga M. Gomez-Lus R. Baquero F. Torres C. Detection of a single vanA-containing Enterococcus faecalis clone in hospitals in different regions in Spain. J. Antimicrob. Chemother. 2001;48:746–747. doi: 10.1093/jac/48.5.746. [DOI] [PubMed] [Google Scholar]
- 5.Dunny G.M. Genetic functions and cell-cell interactions in the pheromone-inducible plasmid transfer system of Enterococcus faecalis. Plasmid. 1990;4:689–696. doi: 10.1111/j.1365-2958.1990.tb00639.x. [DOI] [PubMed] [Google Scholar]
- 6.Dutka-Malen S. Evers S. Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton T.J. Gasson M. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 2001;67:1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsner H.A. Sobottka I. Mack D. Claussen M. Laufs R. Wirth R. Virulence factors of Enterococcus faecalis and Enterococcus faecium blood culture isolates. Eur. J. Clin. Microb. Infect. Dis. 2000;19:39–42. doi: 10.1007/s100960050007. [DOI] [PubMed] [Google Scholar]
- 9.Felmingham D. Wilson A. Quintana A. Grüneberg R. Enterococcus species in urinary tract infection. Clin. Infect. Dis. 1992;15:295–301. doi: 10.1093/clinids/15.2.295. [DOI] [PubMed] [Google Scholar]
- 10.Herrera C. Determinación de genes de virulencia y susceptibilidad antimicrobiana en Enterococcus spp. procedentes de pacientes VIH/SIDA [Tesis] Ciudad de La Habana, Cuba: Instituto Pedro Kourí; 2005. (In Spanish) [Google Scholar]
- 11.Homan W.L. Tribe D. Poznanski S. Li M. Hogg G. Spalburg E. Van Embden J.D. Willems R.J. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 2002;40:1963–1971. doi: 10.1128/JCM.40.6.1963-1971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawalec M. Pietras Z. Danilowicz E. Jakubczak A. Gniadkowski M. Hryniewicz W. Willems R. Clonal structure of Enterococcus faecalis isolated from Polish hospitals: the characterization of epidemic clones. J. Clin. Microbiol. 2007;1:147–153. doi: 10.1128/JCM.01704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin B. Garriga M. Hugas M. Aymerich T. Genetic diversity and safety aspects of enterococci from slightly fermented sausages. J. Appl. Microbiol. 2005;98:1177–1190. doi: 10.1111/j.1365-2672.2005.02555.x. [DOI] [PubMed] [Google Scholar]
- 14.Mundy L.M. Sahm D. Gilmore M. Relationship between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev. 2000;13:513–522. doi: 10.1128/cmr.13.4.513-522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nallaparedy S.R. Duh R.W. Singh K.V. Murray B.E. Molecular typing of selected Enterococcus faecalis Isolates: pilot study using multilocus sequence typing and pulsed-field gel electrophoresis. Microbiology. 2002;40:868–876. doi: 10.1128/jcm.40.3.868-876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nallaparedy S.R. Wenxiang H. Weinstock G.M. Murray B.E. Molecular characterizations of a widespread, pathogenic, and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. J. Bacteriol. 2005;187:5709–5718. doi: 10.1128/JB.187.16.5709-5718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishimoto Y. Kobayashi N. Alam M.M. Ishino M. Uehara N. Watanabe N. Analysis of the prevalence of tetracycline resistance genes in clinical isolates of Enterococcus faecalis and Enterococcus faecium in a Japanese hospital. Microb. Drug Resist. 2005;11:146–153. doi: 10.1089/mdr.2005.11.146. [DOI] [PubMed] [Google Scholar]
- 18.Novais C. Coque T.M. Sousa J.C. Baquero F. Peixe L. Local genetic patterns within a vancomycin-resistant Enterococcus faecalis clone isolated in three hospitals in Portugal. Antimicrob. Agents Chemother. 2004;48:3613–3617. doi: 10.1128/AAC.48.9.3613-3617.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quiñones D. Goñi P. Rubio M.C. Baquero F. Gomez-Lus R. Del Campo R. Genetic relatedness and antimicrobial resistance determinants among clinical isolates of enterococci from Cuba. Clin. Microbiol. Infect. 2006;12:793–797. doi: 10.1111/j.1469-0691.2006.01421.x. [DOI] [PubMed] [Google Scholar]
- 20.Quiñones D. Goñi P. Rubio M.C. Duran E. Gomez-Lus R. Enterococci spp. isolated from Cuba: species frequency of occurrence and antimicrobial susceptibility profile. Diagn. Microbiol. Infect. Dis. 2005;51:63–67. doi: 10.1016/j.diagmicrobio.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Garbajosa P. Bonten M.J. Robinson D.A. Top J. Nallapareddy S.R. Torres C. Coque T.M. Canton R. Baquero F. Murray B.E. del Campo R. Willems R. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a back-ground of high rates of recombination. J. Clin. Microbiol. 2006;44:2220–2228. doi: 10.1128/JCM.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz-Garbajosa P. Coque T.M. Cantón R. Willems R. Baquero F. del Campo R. Los complejos clonales de alto riesgo CC2 y CC9 están ampliamente representados en cepas hospitalarias de Enterococcus faecalis. Enferm. Infecc. Microbiol. Clin. 2007;25:513–518. doi: 10.1157/13109988. (In Spanish) [DOI] [PubMed] [Google Scholar]
- 23.Shankar N. Baghdayan A.S. Willems R. Hammerum A.M. Jensen L.B. Presence of pathogenicity island genes in Enterococcus faecalis isolates from pigs in denmark. J.Clin. Microbiol. 2006;44:4200–4203. doi: 10.1128/JCM.01218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutcliffe J. Grebe T. Tait-Kamradt A. Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vankerckhoven V. Van Autgaerden T. Vael C. Lammens C. Chapelle S. Rossi R. Jabes D. Goossens H. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J. Clin. Microbiol. 2004;42:4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vergis E.N. Shankar N. Chow J.W. Hayden M.K. Snydman D.R. Zervos M.J. Linden P.K. Wagener M.M. Muder R.R. Association between the presence of enterococcal virulence factors gelatinase, hemolysin, and enterococcal surface protein and mortality among patients with bacteremia due to Enterococcus faecalis. Clin. Infect. Dis. 2002;35:570–575. doi: 10.1086/341977. [DOI] [PubMed] [Google Scholar]