Abstract
The ETV6/RUNX1 fusion gene, present in 25% of B-lineage childhood acute lymphoblastic leukemia (ALL), is thought to represent an initiating event, which requires additional genetic changes for leukemia development. To identify additional genetic alterations, 24 ETV6/RUNX1-positive ALLs were analyzed using 500K single nucleotide polymorphism arrays. The results were combined with previously published data sets, allowing us to ascertain genomic copy number aberrations (CNAs) in 164 cases. In total, 45 recurrent CNAs were identified with an average number of 3.5 recurrent changes per case (range 0–13). Twenty-six percent of cases displayed a set of recurrent CNAs identical to that of other cases in the data set. The majority (74%), however, displayed a unique pattern of recurrent CNAs, indicating a large heterogeneity within this ALL subtype. As previously demonstrated, alterations targeting genes involved in B-cell development were common (present in 28% of cases). However, the combined analysis also identified alterations affecting nuclear hormone response (24%) to be a characteristic feature of ETV6/RUNX1-positive ALL. Studying the correlation pattern of the CNAs allowed us to highlight significant positive and negative correlations between specific aberrations. Furthermore, oncogenetic tree models identified ETV6, CDKN2A/B, PAX5, del(6q) and +16 as possible early events in the leukemogenic process.
INTRODUCTION
The t(12;21)(p13;q22) that generates the ETV6/RUNX1 fusion gene (also known as TEL/AML1) is the most common specific genetic abnormality in childhood B-lineage acute lymphoblastic leukemia (ALL), occurring in 25% of the cases (1,2). This translocation is believed to constitute the first step in the transformation of a normal cell, even in utero, into the malignant cells that characterize the disease, but additional changes are most likely necessary for leukemia to develop. This view is supported by several notable features. For example, retrospective studies have shown that the fusion gene can be detected in the blood of children long before presentation of overt leukemia; and studies of monochorionic twins with concordant ETV6/RUNX1-positive ALLs have shown that these have identical genomic breakpoints (3,4). In fact, a recent investigation successfully identified a preleukemic ETV6/RUNX1-positive clone in the healthy twin of a patient diagnosed with ETV6/RUNX1-positive ALL and also demonstrated that expression of ETV6/RUNX1 alone can mimic the preleukemic clone but not induce leukemia in an in vivo model (5). Taken together, these data indicate that ETV6/RUNX1-positive leukemia is generated through a multi-step mechanism, and that accumulation of additional genetic changes is necessary for the development of overt leukemia. Hence, to understand fully the genetic evolution of this disorder, identification of the complete spectrum of genetic changes that accompany the ETV6/RUNX1 fusion gene is necessary. Moreover, critical pathogenetic insights may be gained from studying the correlation pattern of the different copy number changes.
High resolution single nucleotide polymorphism arrays (SNP arrays) and comparative genomic hybridization arrays (CGH arrays) have provided powerful tools for identifying genetic changes in childhood leukemia. Recent studies of childhood ALL using such genome-wide techniques have revealed the presence of several submicroscopic genetic changes (6–12). Importantly, these studies have shown that genes involved in the regulation of B-cell development, such as PAX5 and EBF1, are targeted by deletions in around 40% of B-lineage ALL.
In the present study, we first profiled 24 ETV6/RUNX1-positive cases using 500K SNP array analysis, providing a very high resolution view of copy number changes over the whole genome. To enable more reliable frequency estimates of recurrent copy number changes and to study their correlation patterns, we next combined our local data set with data from two recently published series, acquired using 250K and 50K SNP arrays, respectively (6,7). This yielded a total of 164 ETV6/RUNX1-positive ALLs that could be conclusively investigated. By applying newly developed algorithms in addition to different previously described ones, we provide a comprehensive analysis of the interdependencies among the recurrently gained copy number changes in this leukemia subtype.
RESULTS
High resolution genomic profiling of local cases
Twenty-three ETV6/RUNX1 positive ALLs and the ETV6/RUNX1 positive cell line REH were studied using 500K SNP array analysis. Seventeen of these have previously been analyzed using CGH arrays of lower resolution (8). All described lesions were confirmed in the present study using the 500K SNP array platform. Moreover, the increased resolution enabled us to identify additional deletions less than 300 kb in size (referred to as focal deletions) in all 24 cases, typically affecting only one or a small number of genes. In fact, the three cases where no copy number aberrations (CNAs) were seen in the previous study (8) all displayed focal deletions when analyzed on the higher resolution arrays. In total, 29 recurrent CNAs were found in the 24 cases, of which 16 were recurrent focal deletions (Table 1). The most common recurrent focal deletions involved PAX5 (9p13.2; 25%), the adjacent genes RAG1 and RAG2 (11p12, 21%), TBL1XR1 (3q26.32, 21%) and CD200/BTLA (3q13.2, 21%). Notably, apart from TBL1XR1, which encodes a transcriptional regulator implicated in nuclear hormone response, all these genes encode proteins with established functions in the immune system or during B-cell differentiation.
Table 1.
Recurrent copy number changes in 164 ETV6/RUNX1-positive ALLs
| Gain/loss | Chr | Smallest affected regiona (Mb) | Size (Mb) | Affected gene(s) | Labelb | Local cases (n = 24) | External Data Set 1 (n = 47) | External Data Set 2 (n = 93) | Total (n = 164) |
|---|---|---|---|---|---|---|---|---|---|
| L | 1 | 66.5–66.6 | 0.1 | PDE4B | PDE4B | 2 | 0 | 4 | 6 (4%) |
| L | 1 | 93.5–95.1 | 1.6 | >4 genes | del(1p21.3p22.1) | 0 | 2 | 0 | 2 (1%) |
| L | 1 | 187.5–187.6 | 0.1 | No gene | del(1q31.2) | 4 | 2 | NA | NA |
| L | 1 | 191.4–191.4 | 0.0 | No gene | del(1q31.3) | 3 | 4 | 9 | 16 (10%) |
| L | 2 | 3.6–3.8 | 0.2 | No gene | del(2p25.3) | 0 | 4 | 1 | 5 (3%) |
| L | 2 | 9.7–9.7 | 0.0 | YWHAQ | YWHAQ | 1 | 1 | NA | NA |
| L | 2 | 127.1–127.1 | 0.0 | GYPC | GYPC | 2 | 0 | NA | NA |
| L | 2 | 237.9–242.7 | 4.8 | >4 genes | del(2q37.1q37.3) | 0 | 2 | 2 | 4 (2%) |
| L | 3 | 35.3–35.6 | 0.3 | ARPP-21 | ARPP-21 | 0 | 2 | 1 | 3 (2%) |
| L | 3 | 41.7–41.7 | 0.0 | ULK4 | ULK4 | 1 | 2 | 1 | 4 (2%) |
| L | 3 | 48.2–49.3 | 1.1 | >4 genes | del(3p21.31) | 1 | 4 | 6 | 11 (6%) |
| L | 3 | Various | FHIT | FHIT | 5 | 5 | 5 | 15 (9%) | |
| L | 3 | 113.5–113.6 | 0.1 | CD200, BTLA | CD200/BTLA | 5 | 6 | 10 | 21 (13%) |
| L | 3 | 128.4–128.4 | 0.0 | No gene | del(3q21.3) | 0 | 2 | NA | NA |
| L | 3 | Various | TBL1XR1 | TBL1XR1 | 5 | 7 | 7 | 19 (12%) | |
| L | 4 | 44.5–44.5 | 0.0 | YIPF7 | YIPF7 | 1 | 1 | NA | NA |
| L | 4 | 150.1–150.3 | 0.2 | NR3C2 | NR3C2 | 2 | 5 | 7 | 14 (9%) |
| L | 5 | 142.6–142.7 | 0.1 | NR3C1 | NR3C1 | 3 | 5 | 2 | 10 (6%) |
| L | 5 | 158.4–158.5 | 0.1 | EBF1 | EBF1 | 1 | 5 | 1 | 7 (4%) |
| L | 6 | 26.3–26.4 | 0.1 | HIST cluster | HIST cluster | 4 | 2 | NA | NA |
| L | 6 | Various | >4 genes | del(6q) | 6 | 8 | 18 | 32 (20%) | |
| L | 7 | 106.0–106.1 | 0.1 | PIK3CG | PIK3CG | 0 | 2 | 0 | 2 (1%) |
| L | 8 | 0–36.3 | 36.3 | >4 genes | del(8p) | 0 | 2 | 2 | 4 (2%) |
| L | 8 | Various | TOX | TOX | 1 | 4 | 1 | 6 (4%) | |
| G | 8 | 83.7–145.9 | 62.1 | >4 genes | dup(8q) | 1 | 1 | 4 | 6 (4%) |
| L | 9 | 21.9–22.0 | 0.1 | CDKN2A, CDKN2B | CDKN2A/B | 4 | 10 | 22 | 36 (22%) |
| L | 9 | Various | PAX5 | PAX5 | 6 | 13 | 14 | 33 (20%) | |
| L | 9 | 96.1–96.2 | 0.1 | HSD17B3, SLC35D2 | HSD17B3/SLC35D2 | 0 | 3 | 3 | 6 (4%) |
| G | 10 | 0.0–20.8 | 20.8 | >4 genes | dup(10p) | 1 | 5 | 1 | 7 (4%) |
| G | 10 | 0.0–135.3 | 135.3 | >4 genes | +10 | 0 | 3 | 11 | 14 (9%) |
| L | 10 | 111.8–111.9 | 0.1 | ADD3 | ADD3 | 2 | 2 | 0 | 4 (2%) |
| L | 11 | 36.6–36.6 | 0.0 | RAG1, RAG2 | RAG1/2 | 5 | 5 | NA | NA |
| L | 11 | 62.2–62.3 | 0.1 | BSCL2, GNG3, HNRPUL2, TTC9C | del(11q12.3) | 2 | 2 | 3 | 7 (4%) |
| L | 11 | Various | >4 genes | del(11q) | 3 | 3 | 6 | 12 (7%) | |
| L | 12 | 11.7–11.8 | 0.1 | ETV6 | ETV6 | 10 | 33 | 54 | 97 (59%) |
| L | 12 | 14.4–14.5 | 0.1 | ATF7IP | ATF7IP | 9 | 23 | NA | NA |
| L | 12 | 90.8–90.9 | 0.1 | BTG1 | BTG1 | 1 | 6 | 7 | 14 (10%) |
| L | 13 | 27.2–27.2 | 0.0 | No gene | del1(3q12.2) | 1 | 2 | NA | NA |
| L | 13 | Various | >4 genes | del(13q) | 3 | 3 | 2 | 8 (5%) | |
| L | 14 | 72.3–72.4 | 0.1 | DPF3 | DPF3 | 1 | 2 | 2 | 5 (3%) |
| L | 15 | 39–39.8 | 0.8 | >4 genes | del(15q15.1) | 2 | 3 | 5 | 10 (6%) |
| L | 15 | 87.8–88.3 | 0.5 | >4 genes | del(15q26.1) | 1 | 1 | 3 | 5 (3%) |
| G | 16 | 0.0–88.7 | 88.7 | >4 genes | +16 | 3 | 1 | 10 | 14 (9%) |
| L | 17 | 25.9–26.3 | 0.4 | CRLF3, C17orf41, C17orf42 | del(17q11.2) | 0 | 2 | 1 | 3 (2%) |
| L | 17 | 57.0–61.5 | 4.5 | >4 genes | del(17q23.2q24.1) | 0 | 2 | 0 | 2 (1%) |
| L | 18 | Various | TCF4 | TCF4 | 2 | 4 | 5 | 11 (7%) | |
| L | 19 | 4.9–4.9 | 0.0 | UHRF1 | UHRF1 | 2 | 0 | NA | NA |
| L | 19 | 39.4–39.6 | 0.2 | >4 genes | del(19q13.11) | 0 | 3 | 1 | 4 (2%) |
| L | 19 | 52.1–52.3 | 0.2 | GRLF1, NPAS1, TMEM160 | del(19q13.32) | 2 | 2 | 2 | 6 (4%) |
| L | 20 | 10.4–10.4 | 0.0 | C20orf94 | C20orf94 | 1 | 4 | 9 | 14 (9%) |
| L | 21 | Various | RUNX1 | RUNX1 | 1 | 3 | 2 | 6 (4%) | |
| G | 21 | 0.0–46.9 | 46.9 | >4 genes | +21 | 2 | 4 | 13 | 19 (12%) |
| G | 21 | Various | >4 genes | dup(21q) | 2 | 3 | 3 | 8 (5%) | |
| G | 12, 21 | 21.5 | >4 genes | +der(21)t (12;21)c | 6 | 2 | 11 | 19 (12%) | |
| G | X | 130.3–154.5 | 24.2 | >4 genes | dup(Xq) | 6 | 10 | 11 | 27 (16%) |
Chr, chromosome; L, loss; G, gain, NA, not available.
aGenomic positions according to the hg17 genome assembly (NCBI Build 35).
bThe labels are used to describe copy number changes in text and figures.
cInferred from coinciding gains of 12p and 21q, the size includes both 12p and 21q material.
Combined analysis of local and external data
To obtain a more reliable frequency estimate of the genetic changes occurring in ETV6/RUNX1-positive ALL and to enable analysis of the correlation pattern of these changes, the locally produced data set was combined with two external data sets: (i) Mullighan et al. (6). (External Data Set 1, abbreviated EDS1; n = 47 cases) and (ii) Kawamata et al. (7) (External Data Set 2, abbreviated EDS2; n = 93 cases), yielding a combined data set of 164 ETV6/RUNX1-positive cases. In total, 55 recurrent genetic lesions were identified when the local data set was combined with EDS1 (71 cases analyzed; Table 1). Since EDS2 was of lower resolution, only 45 of the 55 recurrent changes could be conclusively identified in this data set (Table 1; Supplementary Material, Fig. S1). Of the 10 recurrent changes that could not be studied in EDS2, only 5 were present in more than 2 cases with high resolution data (Table 1), namely ATF7IP (this gene, located on 12p13.1, was co-deleted with ETV6 in 28 cases and removed by focal deletions in 4 cases), RAG1/2 (10 cases), del(1q31.2) (6 cases), histone cluster 1 deletions on 6p21–p22 (6 cases) and del(13q12.2) (3 cases). All further analyses were done on the 45 recurrent genetic lesions that could be analyzed in all three data sets (Fig. 1A). The average number of recurrent aberrations in each case was 3.5 (range 0–13, median 3). The majority of the recurrent lesions resulted in loss of genetic material (37 aberrations, 82%), with only eight (18%) recurrent gains being identified. The most common aberrations were deletions of ETV6 (12p13.2; 59%), CDKN2A/B (9p21.3; 22%), focal deletions of PAX5 (20%), deletions of a large region on 6q (20%) and gain of Xq [referred to as dup(Xq), 16%]. Twenty-six (55%) of the deletions covered regions where recurrent focal deletions identified one or two genes as potential targets (Table 1). Several of the genes targeted by focal deletions are involved in B-cell development or play an important role in normal hematopoiesis, e.g. ETV6 (97 cases; 59%), PAX5 (33 cases; 20%), TCF4 (18q21.2; 11 cases; 7%) and EBF1 (5q33.3, 7 cases; 4%).
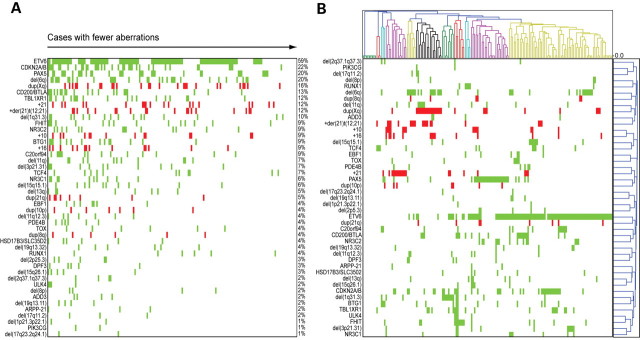
Figure 1.
Recurrent copy number changes in ETV6/RUNX1-positive pediatric ALLs. (A) The 45 recurrent copy number aberrations (CNAs) and 164 ETV6/RUNX1-positive ALLs sorted by frequency and number of recurrent aberrations, respectively. The cases are sorted with decreasing number of recurrent CNAs from left to right, and the CNAs are sorted by frequency from top to bottom. A green box indicates loss of material and a red box indicates gain of material. (B) Hierarchical clustering using 1-Pearson correlation and average linkage of the 164 ETV6/RUNX1-positive ALLs (top graph) and the 45 recurrent CNAs (right graph). Patients are indicated above the graph and CNAs on the left. A green box indicates loss of material and a red box indicates gain of material. Eleven distinct clusters of patients are detected; these are indicated in alternating colors of green, red, blue, purple, yellow and black.
Of note, the aberrations detected in the ETV6/RUNX1-positive cell line REH showed high similarity to the aberrations found in the patient-derived samples. Although this cell line had the highest number of recurrent CNAs, 13 CNAs, the number of changes was unexpectedly low, suggesting a rather stable genetic constitution of this cell line despite serial passaging in vitro.
Given the large number of recurrent chromosomal changes that were identified in the combined data set, we next used various methods and algorithms to explore the correlation pattern among the different changes.
Hierarchical clustering analysis reveals cases with identical patterns of recurrent CNAs
We first applied hierarchical clustering analysis (HCA) on the 164 cases with regard to their pattern of CNAs using Pearson correlation and average linkage (Fig. 1B). This analysis revealed the formation of 10 clusters (indicated by different colors in Fig. 1B) that were defined mainly by the presence of the following abnormalities: TCF4 (present in 3/3 cases in the cluster), BTG1 (3/3 cases), +21 (11/14 cases), del(6q) (6/6 cases), dup(Xq) (17/17 cases), del(1q31.3) (8/8 cases), FHIT (7/7 cases), RUNX1 (4/4 cases), PAX5 (24/25 cases) or ETV6 (67/68 cases). Also, a cluster of nine cases lacked recurrent abnormalities.
Cases with identical patterns of CNAs will form tight subclusters in the analysis (clusters with zero distance between cases in Fig. 1B). We identified 13 such clusters, with 42 cases (26%) having an identical pattern of recurrent CNAs to that of another case in the data set. The cases with identical patterns included, apart from the nine cases without recurrent aberrations, 16 cases with a single recurrent abnormality (6 with ETV6, 4 with PAX5, 2 with +21, 2 with del(1q31.3) and 2 with TCF4), and 15 cases with two recurrent abnormalities; 2 of these had del(6q) together with dup(Xq); the remaining cases had ETV6 in combination with either del(6q) (4 cases), CDKN2A/B (3 cases), dup(Xq) (2 cases), PAX5 (2 cases) or C20orf94 (2 cases). Also, two cases had an identical pattern of recurrent CNAs with the three aberrations ETV6, CDKN2A/B and TBL1XR1. The majority of cases (74%), however, displayed a unique pattern of CNAs, suggesting that the process of acquiring CNAs is unique for each case (Fig. 1B).
Connected pair analysis reveals co-occurring CNAs
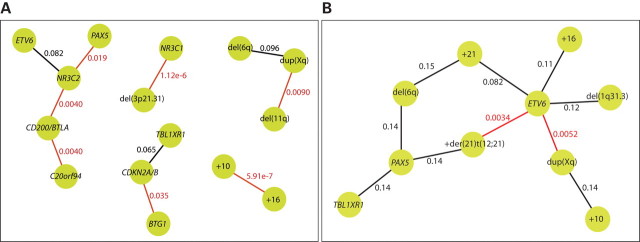
A problem when studying the relationship between CNAs by hierarchical cluster analysis using common dissimilarity measures is that rare CNAs often appear to be closely related simply because both CNAs are absent in a large number of cases. In order to highlight aberration pairs based on their co-occurrence rather than their absence, we performed connected pair analysis (CPA) with a dissimilarity measure that is based on how the number of co-occurrences of two CNAs differs from what is expected if they are independent. We plotted the pairs corresponding to the 1% smallest and 1% largest dissimilarities, as determined by this dissimilarity measure (Fig. 2). This analysis showed that the presence of trisomy 10 and 16 is strongly associated. A number of significant dependencies (q < 0.05, based on chi-square tests), not visible in the cluster analysis, were also seen. These include a strong association between NR3C1 and del(3p21.31) and an association between del(11q) and dup(Xq). The CD200/BTLA deletion was associated both with deletion of C20orf94 and deletion of NR3C2, with the deletion of NR3C2 also being associated with deletion of PAX5. There was also an association between deletions of BTG1 and the CDKN2A/B locus. Among the node pairs with the largest dissimilarities, only two significant negative connections were detected; the presence of an additional der(21)t(12;21) and duplication of Xq material were both negatively associated with deletion of ETV6.
Figure 2.
Positive and negative correlations between copy number abnormalities. Dissimilarity scores were calculated for all possible pairs of copy number abnormalities, and the 1% largest and the 1% smallest dissimilarities are illustrated here as nodes with a connection. The nodes represent copy number abnormalities, and the connections represent either positive or negative associations between the copy number changes. The q-value, based on chi-square tests, for each association is indicated at the connection. Statistically significant associations (q < 0.05) are indicated in red. (A) Illustration of positive associations between copy number abnormalities. In this data set, the strongest association is seen between gains of chromosomes 10 and 16 (q = 0.00000059). (B) Illustration of negative associations between copy number abnormalities. Deletion of ETV6 is significantly negatively associated with both dup(Xq) (q = 0.0052) and an additional copy of the der(21)t(12;21) (q = 0.0034).
Oncogenetic trees suggest pathways for leukemia development
Oncogenetic trees enable studying the relationship among CNAs and have successfully been used to study mutations and CNAs involved in several different tumor types (13–15). Branching oncogenetic trees were developed as a method to infer a sequential order in which abnormalities most likely have occurred (16). This model has since been revised with distance-based oncogenetic trees which do not imply the same rigid sequential dependency between aberrations (17).
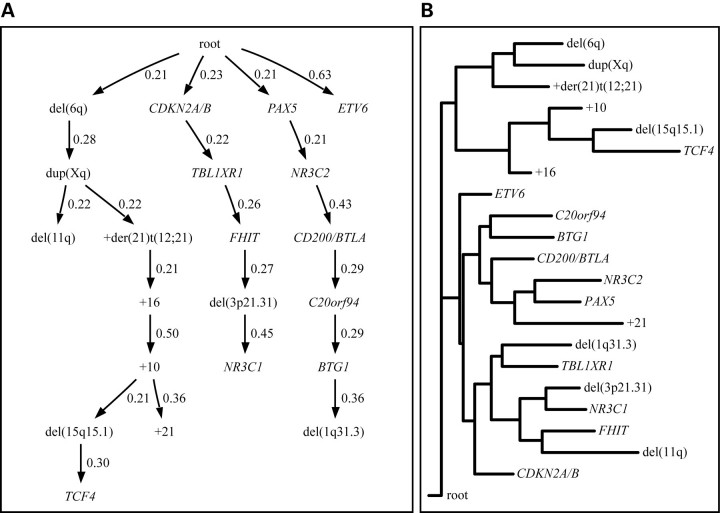
According to a branching tree model of the data (Fig. 3A), deletion of PAX5, deletion of CDKN2A/B and del(6q) are important early events as they are close to the root and also the root of a new subtree. Deletion of ETV6 is also close to the root, but is not a necessary precursor for other changes, making its role more uncertain. Interestingly, the subtree of abnormalities rooted in del(6q) mainly includes abnormalities affecting large chromosomal regions and whole chromosomal gains. In contrast, the subtrees rooted in CDKN2A/B and PAX5 mostly include small focal deletions, possibly suggesting different genetic pathways of leukemia development with different underlying mechanisms (large chromosomal changes versus small focal deletions).
Figure 3.
Oncogenetic tree analysis of recurrent copy number changes. The distribution of 21 copy number changes, determined to be non-random by the method described by Brodeur et al. (29), in 164 ETV6/RUNX1-positive cases was used to create oncogenetic trees. The root indicates a cell that has already acquired the t(12;21). (A) In the branching oncogenetic tree, a probable sequential order for the acquisition of copy number abnormalities is given, as indicated by arrows. The probability of acquiring a copy number abnormality for a cell that has acquired the preceding copy number abnormality is indicated above each arrow. The aberrations del(6q), CDKN2A/B and PAX5 are potentially early events as these are close to the root and also the root of a new subtree. (B) In the distance-based oncogenetic tree, the copy number changes are represented as leaves while the internal nodes represent hidden or unknown events. The horizontal leaf-to-leaf distance depends on the association between copy number changes meaning that copy number changes forming a cluster have a large probability of co-occurrence. Three distinct clusters can be seen, with one cluster containing most of the larger aberrations such as +10, +16 and +der(21)t(12;21). The two remaining clusters are preceded by ETV6 and mainly contain smaller aberrations.
Distance-based oncogenetic trees give a model that does not infer a specific sequential path for alterations; instead alterations occurring together are clustered, with alterations close to the root being expected to constitute early events. Application of this model revealed that the data are divided into three main clusters, showing a very high agreement with the three main branches of the branching tree model. The CNAs ETV6, +16 and CDKN2A/B are closest to the root, indicating that these represent early genetic events (Fig. 3B).
As also revealed by CPA, the association between +10 and +16 is clearly visible also in the two oncogenetic tree models. Since chromosomes 10 and 16 are two of only four recurrently gained chromosomes in this data set (the other two being der(21)t(12;21) and chromosome 21), we speculated that they occur together because a subgroup of cases has a tendency to acquire additional whole chromosomes. We therefore examined if cases with gain of either chromosome 10 or 16 had a higher proportion of additional chromosomal gains than cases without. We found that the 21 cases with an additional chromosome 10 or 16 harbored 39 whole chromosomal gains affecting a chromosome other than 10 or 16 (range 0–7 gained chromosomes, median 1), while the remaining 142 cases had 37 additional chromosomes in total (range 0–5, median 0, P = 0.000013, Wilcoxon rank-sum test). This supports the assumption that a subgroup of cases had a tendency to acquire whole chromosomal gains.
DISCUSSION
In the present study, we have investigated copy number changes in 164 cases of ETV6/RUNX1-positive ALL, providing the largest genome-wide copy number study of childhood ALL harboring this abnormality. This allowed us to identify a total set of 45 recurrent CNAs with an average number of 3.5 recurrent changes per case (range 0–13). The recurrent nature of these CNAs suggests that they are important ‘driver mutations’ for the establishment and/or progression of ETV6/RUNX1-positive leukemia; although a few may represent particularly break prone regions and, thus, constitute ‘passenger mutations’ that does not provide any selective advantage to the leukemic clone. Both the high number of recurrent aberrations and the fact that a majority of cases (74%) display a unique pattern of CNAs indicate that a large genetic heterogeneity exists within the ETV6/RUNX1-positive subgroup of ALL. To characterize this heterogeneity further, we have used HCA, CPA and oncogenetic tree models to identify recurrent CNAs that co-occur more or less often than expected by chance and to highlight probable sequential orders for the acquisition of these CNAs.
The most common types of abnormalities, present in 46% of the recurrently altered regions, were small focal deletions encompassing only one or two genes. The focal deletions that were most common in the combined data set affected ETV6 (59%), CDKN2A/B (22%), genes involved in B-cell development such as PAX5 (20%), TCF4 (7%) and EBF1 (4%), or other genes with an established function in the immune system such as CD200 and BTLA (13%), well in agreement with previous studies (6,9,10). Genes governing B-cell development were deleted in 28% of the cases. Indeed, these findings, together with studies in murine models demonstrating that loss of PAX5, EBF1 and TCF4 all lead to impaired production of mature B-cells (18–20), suggest that deletions in any one of these genes are likely to contribute to arresting the cells at an immature state. Interestingly, deletions in this pathway were not mutually exclusive, as 11% of the cases with deletions of either PAX5, EBF1 or TCF4 also had a deletion affecting one of the two other genes. Previous studies describing deletions of genes governing B-cell development in B-ALLs have focused on PAX5 and EBF1 (6). In the current large data set, however, deletions of TCF4 (7%) were more common than deletions of EBF1 (4%). Hence, the importance of this deletion might previously not have been fully appreciated.
The fifth most common focal deletion, found in 12% of the cases, occurred within or directly 5′ of the gene TBL1XR1. This gene encodes a protein that is required for nuclear hormone receptor transcriptional repression via the SMRT/N-CoR complex (21). Deletion of TBL1XR1 in ETV6/RUNX1-positive ALL is associated with overexpression of genetic targets of the retinoic acid and thyroid hormone receptors, presumably due to loss of repression (10). In this context, it is noteworthy that deletions in or directly 5′ of NR3C1 and NR3C2, both encoding nuclear hormone receptors binding glucocorticoids, were detected in the present data set in 6 and 9% of the cases, respectively. In the data set of Mullighan et al. (6), these three deletions were found to be recurrent only in the ETV6/RUNX1-positive subgroup of ALL. Hence, perturbed nuclear hormone receptor function, occurring at both the level of the receptors and the level of co-factors, appears to be a specific feature, present in 24% of ETV6/RUNX1-positive ALL. This frequency is well on par with that of deletions affecting genes involved in B-cell development.
Of the larger lesions, loss of 12p and 9p chromosomal material and gain of Xq were the most frequent changes. We have previously reported that dup(Xq) is much more prevalent in males than in females with ETV6/RUNX1-positive ALL (8), a finding clearly confirmed in the present data set in which 25/27 cases with dup(Xq) were males.
Previous studies of childhood ALL using high resolution SNP arrays have mainly focused on the frequencies with which genetic changes occur (6,7,9,11). In the present study, taking advantage of the relatively large combined data set, we also applied three different algorithms, found to offer three unique ways of describing the distribution of recurrent CNAs in the data set.
Using cluster analysis (Fig. 1B) we found that, while there is a large number of recurrent CNAs, most cases (74%) have acquired a unique subset of these CNAs. Only cases with relatively few recurrent CNAs (i.e. three, two, one or zero recurrent CNAs) were found to have non-unique patterns of CNAs. Most likely, this illustrates that the acquisition of CNAs is a stochastic process where the probability of having identical patterns of CNAs decreases with the number of CNAs. CPA was used to highlight CNAs that co-occur more or less often than expected by chance. This analysis, visualized by the connected nodes in Figure 2, identified a number of significant deviations from independence in the co-occurrences of specific CNAs. For example, a strong connection between trisomies 10 and 16 was observed. Since cases harboring gain of either chromosomes 10 or 16 (or both) were more likely to have additional whole chromosomal gains (P = 0.000013), we conclude that gain of chromosomes 10 and 16 occur together in cases that have acquired random chromosome gains, either sequentially or through a single abnormal mitosis. The latter event is believed to be the most common origin of the characteristic chromosomal gains in high hyperdiploid ALL (22). Another connection identified by CPA where the CNAs seem to have a shared origin is the connection between dup(Xq) and del(11q). In this case, it is likely that their co-occurrence can be explained by the fact that duplicated material from Xq often represent an unbalanced translocation between chromosomes 10 and 11, resulting in concurrent dup(Xq) and del(11q) (8).
CPA also identified a significant negative correlation between deletion of ETV6 and duplication of der(21)t(12;21), most likely due to a functional relation between deletion of ETV6 and duplication of the fusion gene. This agrees well with previous data suggesting that heterodimerization between ETV6 and ETV6/RUNX1 hampers the function of the fusion protein, and that increasing the ratio of ETV6/RUNX1 over normal ETV6 makes the fusion protein more potent (23). Both deletion of ETV6 and duplication of the fusion gene can increase this ratio, indicating that the presence of both these aberrations would be redundant, which could explain their negative association. Interestingly, deletion of ETV6 was also negatively linked to dup(Xq), possibly indicating that the currently unknown functional outcome of dup(Xq) is related to ETV6 function or expression.
The relationship between CNAs and their putative temporal order of appearance was also studied using branching and distance-based oncogenetic trees (16,17). Overall, good agreement occurs between the two tree models, separating three groups of aberrations. Both trees identified CDKN2A/B and ETV6 as early aberrations while del(6q), PAX5 and +16 were identified as early events in only one of the trees. Hence, the oncogenetic tree models imply that these genetic changes occur early in the leukemogenic process, whereas the other recurrent changes might well represent events that occur later during evolution of the leukemic clone. This, however, needs to be confirmed by studies where the sequential order of acquisition for different CNAs can be observed directly. Most of the aberrations affecting larger chromosome regions were confined to one of the three groups identified in the oncogenetic trees, while the two other groups consisted of small deletions, illustrating that large and small genetic changes commonly occur together with similar-sized CNAs. The larger sized aberrations include the whole chromosome gains that are likely to co-occur in a subset of cases with random whole chromosome gains, as discussed earlier. As for smaller aberrations, some of the focal deletions have been reported to be the result of aberrant RAG activity (24,25). Hence, a predisposing mutation affecting RAG specificity could be responsible for the clusters of small deletions.
In conclusion, we have investigated the presence and distribution of genetic changes in the largest set of ETV6/RUNX1-positive childhood ALL described to date. A majority of the cases had a unique set of recurrent CNAs, illustrating a previously unrecognized heterogeneity in this ALL subtype. We could confirm earlier studies showing that genes in the B-cell development pathway are common targets of focal deletions in B-cell ALL, but we also identified that the nuclear hormone receptor response pathway is targeted equally often as the B-cell development pathway in ETV6/RUNX1-positive ALL. By studying the correlation pattern of the recurrent CNAs, we suggest a probable sequential order for the aberrations and highlight significant positive and negative correlations between CNAs.
MATERIALS AND METHODS
Patient material and high resolution profiling
Between 1997 and 2006, 132 childhood B-cell precursor ALLs were genetically analyzed at the Department of Clinical Genetics at Lund University Hospital, Sweden. Of these, 35 (27%) were found to harbor the ETV6/RUNX1 fusion gene by reverse transcription PCR and/or fluorescent in situ hybridization. All patients were treated at Lund or Linköping University Hospitals, Sweden. DNA from bone marrow (BM) was available from 23 of the ETV6/RUNX1 positive children (9 girls, 14 boys, median age 5 years, range 2–14 years). In addition, DNA from the ETV6/RUNX1-positive cell line REH (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) was also used. Seventeen of the 23 ALLs from local patients have been described earlier in a study using CGH arrays of lower resolution (8). DNA from patient material was extracted from BM at diagnosis (20 cases) or relapse (3 cases) using standard methods. This study was reviewed and approved by the Research Ethics Committees of Lund and Linköping Universities. All cases were hybridized to the Affymetrix mapping 500K array set (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's instructions. Detailed information is available in Supplementary Methods. The raw and processed data from these cases have been deposited in the Gene Expression Omnibus database (26) and are accessible through accession number GSE19996.
External data sets
To identify recurrent genetic aberrations in greater detail and to study their correlation pattern, we also analyzed our data together with two recently published data sets. The first data set was produced by Mullighan et al. (6) (EDS1) and the second by Kawamata et al. (7) (EDS2). EDS1 consisted of 250K and 100K SNP array analyses of 242 cases of childhood ALL, 47 (19%) of which harbored the ETV6/RUNX1 fusion gene (6). Only the 250K Sty analyses were used in the present study, since the additional resolution provided by the two 50K arrays was found to be counteracted by noise introduced by adding data from two additional platforms. The CEL and genotyping files for all ETV6/RUNX1-positive cases were downloaded from http://www.stjuderesearch.org/data/ALL-SNP1/ (the data have since been moved to http://hospital.stjude.org/forms/genome-download/request/). EDS2 consisted of 50K SNP array analyses of 399 cases of childhood ALL, of which 96 (24%) harbored the ETV6/RUNX1 fusion gene (7). Reliable copy number profiles could not be generated for 3 of the 96 arrays. Hence, 93 were of sufficient quality to be included in our analysis. The external data together with the local data comprised 164 cases (23 local patients, 47 patients from EDS1, 93 patients from EDS2 and 1 ETV6/RUNX1-positive cell line).
Identification of copy number aberrations
The initial data analysis was performed in dChip (27), with all three data sets being analyzed separately. Circular binary segmentation was performed on the copy number data using DNAcopy (28). This analysis produced three lists of CNAs, one for each data set. The lists from the two data sets of highest resolution (the local data set and EDS1) were analyzed together and regions found to harbor CNAs in two or more cases in these two data sets were considered to be recurrent. Only regions found to be recurrent in the two high resolution studies were analyzed in EDS2. This data set was produced using a platform of lower resolution and for regions where EDS2 only contained between three and five probes, changes were identified by studying the copy number and genotype of individual SNPs in the affected region. A more detailed description of the analysis is available in Supplementary Methods.
Data analysis
To visualize the structure of the data set and infer subsets of patients or aberrations showing similarities, a binary matrix describing the presence of each of the detected CNAs in every case was constructed and different methods were applied: (i) HCA was performed using 1-Pearson correlation as dissimilarity measure and average linkage, (ii) oncogenetic branching trees and (iii) distance-based oncogenetic trees were created as described (16,17). Only the subset of CNAs determined to be non-random using the method described by Brodeur et al. (29) was used for creating the oncogenetic trees.
The data were also visualized using a dissimilarity measure, which was developed by us to highlight pairs of recurrent changes that co-occur either less often or more often than expected by chance. The pairs of aberrations showing the smallest and largest dissimilarities, respectively, were connected to form networks of similarly or dissimilarly occurring aberrations. This analysis is referred to as CPA. The dissimilarity measure D was defined as
where N is the total number of samples in the data set, NAB is the number of co-occurrences of the CNAs A and B, NA is the total number of occurrences of A and NB is the total number of occurrences of B. Finally, chi-square tests were performed to test the independence between the occurrence patterns for each pair of aberrations. The R package ‘qvalue’ was used to estimate the q-value (the minimum false discovery rate at which the test can be considered significant) for each pairwise comparison (30). The data analysis is described in more detail in the Supplementary Methods.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Swedish Cancer Society, the Swedish Childhood Cancer Foundation, the Swedish Research Council, the Inga-Britt and Arne Lundberg Foundation, the Gunnar Nilsson Cancer Foundation and the Medical Faculty of Lund University.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr C.G. Mullighan and Dr J.R. Downing for making External Data Set 1 publicly available. We also acknowledge the help of Dr M. Kato and Dr M. Schrappe for help with annotations of External Data Set 2.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Shurtleff S., Buijs A., Behm F., Rubnitz J., Raimondi S., Hancock M., Chan G., Pui C., Grosveld G., Downing J. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9:1985–1989. [PubMed] [Google Scholar]
- 2.Forestier E., Heyman M., Andersen M.K., Autio K., Blennow E., Borgström G., Golovleva I., Heim S., Heinonen K., Hovland R., et al. Outcome of ETV6/RUNX1-positive childhood acute lymphoblastic leukaemia in the NOPHO-ALL-1992 protocol: frequent late relapses but good overall survival. Br. J. Haematol. 2008;140:665–672. doi: 10.1111/j.1365-2141.2008.06980.x. doi:10.1111/j.1365-2141.2008.06980.x. [DOI] [PubMed] [Google Scholar]
- 3.Wiemels J., Cazzaniga G., Daniotti M., Eden O., Addison G., Masera G., Saha V., Biondi A., Greaves M. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/s0140-6736(99)09403-9. doi:10.1016/S0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 4.Horsley S.W., Colman S., McKinley M., Bateman C.M., Jenney M., Chaplin T., Young B.D., Greaves M., Kearney L. Genetic lesions in a preleukemic aplasia phase in a child with acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2008;47:333–340. doi: 10.1002/gcc.20537. doi:10.1002/gcc.20537. [DOI] [PubMed] [Google Scholar]
- 5.Hong D., Gupta R., Ancliff P., Atzberger A., Brown J., Soneji S., Green J., Colman S., Piacibello W., Buckle V., et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–339. doi: 10.1126/science.1150648. doi:10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 6.Mullighan C.G., Goorha S., Radtke I., Miller C.B., Coustan-Smith E., Dalton J.D., Girtman K., Mathew S., Ma J., Pounds S.B., et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. doi:10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 7.Kawamata N., Ogawa S., Zimmermann M., Kato M., Sanada M., Hemminki K., Yamatomo G., Nannya Y., Koehler R., Flohr T., et al. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111:776–784. doi: 10.1182/blood-2007-05-088310. doi:10.1182/blood-2007-05-088310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lilljebjörn H., Heidenblad M., Nilsson B., Lassen C., Horvat A., Heldrup J., Behrendtz M., Johansson B., Andersson A., Fioretos T. Combined high-resolution array-based comparative genomic hybridization and expression profiling of ETV6/RUNX1-positive acute lymphoblastic leukemias reveal a high incidence of cryptic Xq duplications and identify several putative target genes within the commonly gained region. Leukemia. 2007;21:2137–2144. doi: 10.1038/sj.leu.2404879. doi:10.1038/sj.leu.2404879. [DOI] [PubMed] [Google Scholar]
- 9.Kuiper R.P., Schoenmakers E.F.P.M., van Reijmersdal S.V., Hehir-Kwa J.Y., van Kessel A.G., van Leeuwen F.N., Hoogerbrugge P.M. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–1266. doi: 10.1038/sj.leu.2404691. doi:10.1038/sj.leu.2404691. [DOI] [PubMed] [Google Scholar]
- 10.Parker H., An Q., Barber K., Case M., Davies T., Konn Z., Stewart A., Wright S., Griffiths M., Ross F.M., et al. The complex genomic profile of ETV6-RUNX1 positive acute lymphoblastic leukemia highlights a recurrent deletion of TBL1XR1. Genes Chromosomes Cancer. 2008;47:1118–1125. doi: 10.1002/gcc.20613. doi:10.1002/gcc.20613. [DOI] [PubMed] [Google Scholar]
- 11.Mullighan C.G., Miller C.B., Radtke I., Phillips L.A., Dalton J., Ma J., White D., Hughes T.P., Le Beau M.M., Pui C., et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. doi:10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 12.Tsuzuki S., Karnan S., Horibe K., Matsumoto K., Kato K., Inukai T., Goi K., Sugita K., Nakazawa S., Kasugai Y., et al. Genetic abnormalities involved in t(12;21) TEL-AML1 acute lymphoblastic leukemia: analysis by means of array-based comparative genomic hybridization. Cancer Sci. 2007;98:698–706. doi: 10.1111/j.1349-7006.2007.00443.x. doi:10.1111/j.1349-7006.2007.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang F., Desper R., Papadimitriou C.H., Schäffer A.A., Kallioniemi O.P., Richter J., Schraml P., Sauter G., Mihatsch M.J., Moch H. Construction of evolutionary tree models for renal cell carcinoma from comparative genomic hybridization data. Cancer Res. 2000;60:6503–6509. [PubMed] [Google Scholar]
- 14.Sweeney C., Boucher K.M., Samowitz W.S., Wolff R.K., Albertsen H., Curtin K., Caan B.J., Slattery M.L. Oncogenetic tree model of somatic mutations and DNA methylation in colon tumors. Genes Chromosomes Cancer. 2009;48:1–9. doi: 10.1002/gcc.20614. doi:10.1002/gcc.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pathare S., Schäffer A.A., Beerenwinkel N., Mahimkar M. Construction of oncogenetic tree models reveals multiple pathways of oral cancer progression. Int. J. Cancer. 2009;124:2864–2871. doi: 10.1002/ijc.24267. doi:10.1002/ijc.24267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desper R., Jiang F., Kallioniemi O.P., Moch H., Papadimitriou C.H., Schäffer A.A. Inferring tree models for oncogenesis from comparative genome hybridization data. J. Comput. Biol. 1999;6:37–51. doi: 10.1089/cmb.1999.6.37. doi:10.1089/cmb.1999.6.37. [DOI] [PubMed] [Google Scholar]
- 17.Desper R., Jiang F., Kallioniemi O.P., Moch H., Papadimitriou C.H., Schäffer A.A. Distance-based reconstruction of tree models for oncogenesis. J. Comput. Biol. 2000;7:789–803. doi: 10.1089/10665270050514936. doi:10.1089/10665270050514936. [DOI] [PubMed] [Google Scholar]
- 18.Nutt S.L., Heavey B., Rolink A.G., Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 19.Lin H., Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. doi:10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 20.Zhuang Y., Cheng P., Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol. Cell. Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon H., Choi Y., Cole P.A., Wong J. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 2005;25:324–335. doi: 10.1128/MCB.25.1.324-335.2005. doi:10.1128/MCB.25.1.324-335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulsson K., Johansson B. High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2009;48:637–660. doi: 10.1002/gcc.20671. doi:10.1002/gcc.20671. [DOI] [PubMed] [Google Scholar]
- 23.McLean T., Ringold S., Neuberg D., Stegmaier K., Tantravahi R., Ritz J., Koeffler H., Takeuchi S., Janssen J., Seriu T., et al. TEL/AML-1 dimerizes and is associated with a favorable outcome in childhood acute lymphoblastic leukemia. Blood. 1996;88:4252–4258. [PubMed] [Google Scholar]
- 24.Mullighan C.G., Downing J.R. Genome-wide profiling of genetic alterations in acute lymphoblastic leukemia: recent insights and future directions. Leukemia. 2009;23:1209–1218. doi: 10.1038/leu.2009.18. doi:10.1038/leu.2009.18. [DOI] [PubMed] [Google Scholar]
- 25.Novara F., Beri S., Bernardo M.E., Bellazzi R., Malovini A., Ciccone R., Cometa A.M., Locatelli F., Giorda R., Zuffardi O. Different molecular mechanisms causing 9p21 deletions in acute lymphoblastic leukemia of childhood. Hum. Genet. 2009;126:511–520. doi: 10.1007/s00439-009-0689-7. doi:10.1007/s00439-009-0689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. doi:10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin M., Wei L., Sellers W.R., Lieberfarb M., Wong W.H., Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. doi:10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 28.Venkatraman E.S., Olshen A.B. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. doi:10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 29.Brodeur G.M., Tsiatis A.A., Williams D.L., Luthardt F.W., Green A.A. Statistical analysis of cytogenetic abnormalities in human cancer cells. Cancer Genet. Cytogenet. 1982;7:137–152. doi: 10.1016/0165-4608(82)90010-3. doi:10.1016/0165-4608(82)90010-3. [DOI] [PubMed] [Google Scholar]
- 30.Storey J.D. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B Stat. Methodol. 2002;64:479–498. doi:10.1111/1467-9868.00346. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.