Abstract
Copy number variations (CNVs), a major source of human genetic polymorphism, have been suggested to have an important role in genetic susceptibility to common diseases such as cancer, immune diseases and neurological disorders. Nasopharyngeal carcinoma (NPC) is a multifactorial tumor closely associated with genetic background and with a male preponderance over female (3:1). Previous genome-wide association studies have identified single-nucleotide polymorphisms (SNPs) that are associated with NPC susceptibility. Here, we sought to explore the possible association of CNVs with NPC predisposition. Utilizing genome-wide SNP-based arrays and five CNV-prediction algorithms, we identified eight regions with CNV that were significantly overrepresented in NPC patients compared with healthy controls. These CNVs included six deletions (on chromosomes 3, 6, 7, 8 and 19), and two duplications (on chromosomes 7 and 12). Among them, the CNV located at chromosome 6p21.3, with single-copy deletion of the MICA and HCP5 genes, showed the highest association with NPC. Interestingly, it was more specifically associated with an increased NPC risk among males. This gender-specific association was replicated in an independent case–control sample using a self-established deletion-specific polymerase chain reaction strategy. To the best of our knowledge, this is the first study to explore the role of constitutional CNVs in NPC, using a genome-wide platform. Moreover, we identified eight novel candidate regions with CNV that merit future investigation, and our results suggest that similar to neuroblastoma and prostate cancer, genetic structural variations might contribute to NPC predisposition.
INTRODUCTION
Nasopharyngeal carcinoma [NPC (MIM 161550 and 607107)] is a malignancy that originates from the epithelial lining of the nasopharynx. This neoplasm exhibits a distinct geographic and ethnic distribution. The incidence of NPC is low in most parts of the world, with an age-adjusted annual incidence of less than 1/100 000 (1). However, the disease occurs with much greater frequency in southern China (25–50 per 100 000). Intermediate incidence rates among Chinese people who have migrated abroad (2,3) and in populations of admixed Chinese heritage seem to suggest that genetic and environmental factors play important roles in NPC (1,3,4). Within endemic areas, NPC is also known to be closely associated with Epstein–Barr virus (EBV) infection (5). In addition, studies have shown a male preponderance over the female with a ratio of about 3:1 (3,4,6,7). Together, these findings indicate that NPC is likely to have a complex etiology involving genetic, viral and environmental factors.
The previous studies on the genetic factors involved in the development of NPC have largely focused on identifying the genetic determinants associated with NPC susceptibility. Several linkage analyses have suggested an association between the disease and the human leukocyte antigen (HLA) region, with haplotypes such as HLA-A2 being closely associated with NPC development (8,9). In other studies, candidate-gene-based approaches have demonstrated that polymorphisms in DNA repair-, carcinogen metabolism- and detoxification-related genes appear to be associated with NPC susceptibility (10–12). Recently, our laboratory and two other research groups (13–15) conducted genome-wide association studies (GWAS) in unrelated NPC patients and healthy population controls, and identified a number of new susceptibility loci in addition to HLA-A, including GABBR1, ITGA9, TNFRSF19, MDS1-EVI1 and CDKN2A-CDKN2B. However, the estimated odds ratios (ORs) for these putative loci were between 1.49 and 1.88, suggesting that more substantial components of the genetic variance remain to be identified.
Copy number variations (CNVs), including the duplication, insertion or deletion of chromosomal segments that are ≥1 kb, account for a major proportion of human genetic structural variation (16). Compared with single-nucleotide polymorphisms (SNPs), CNVs represent a pool of rarer structural variants that may help explain some of the heritability that is not accounted by common SNPs. Accumulating evidence has demonstrated that certain CNVs are associated with a low to moderate cancer risk in neuroblastoma and prostate cancer (17,18). To date, however, no study has investigated the potential association of CNVs with NPC predisposition. Here, we used a high-resolution genome-wide SNP-based array and five commonly used CNV detection algorithms to detect potential NPC-associated CNVs. Furthermore, our results were successfully validated in an independent case–control group using a deletion-specific PCR (Supplementary Material, Fig. S1).
RESULTS
During the discovery stage, 288 patients initially diagnosed with NPC at the Chang-Gung Memorial Hospital at Lin-kou (Taoyuan County, Taiwan) and 297 healthy ethnically matched local residents of Taoyuan County were enrolled. The genotyping data from our previous study (14), which had been generated on Illumina Hap550v3_A BeadChips, were used for CNV discovery. The demographics of the case and control samples are provided in Supplementary Material, Table S1. A total of 10 NPC patients and 12 healthy individuals were removed because of a low call rate (<99%), gender mismatch and failure of the identity-by-descent (IBD) (pi-hat >0.5) or identity-by-state (IBS) genetic-outlier tests (Supplementary Material, Fig. S2). Thus, 278 cases and 285 healthy subjects passed the quality control check and were used for CNV identification and analysis.
To maximize the detection rate of potential NPC-associated CNVs, we used five algorithms to identify CNVs from intensity data generated on Illumina Hap550v3_A BeadChips. The algorithms included QuantiSNP (19), PennCNV (20), CNV partition, Partek Hidden-Markov model (HMM) and Partek-Segmentation. CNV filtering and quality control procedures were performed on 15 duplicate samples to generate a subset of higher accuracy calls and remove false positive signals. Detailed information is provided in the Materials and Methods section, as well as in Supplementary Material, Figure S3 and Table S2. When overlapping CNVs were identified in cases and controls, they were merged into unique CNV regions (CNVRs; see Supplementary Material, Fig. S4).
In brief, PennCNV generated the most CNV calls (2418 and 2263 from 280 controls and 275 NPC patients, respectively), whereas Partek HMM generated the fewest (115 and 116 from 285 controls and 287 NPC patients, respectively). The CNVs inferred by PennCNV were the smallest (median size = 32.22 and 34.76 kb in controls and NPC patients, respectively), whereas those identified by Partek HMM were the largest (median size = 247.17 and 183.03 kb in controls and NPC patients, respectively). The numbers and sizes of the deleted and duplicated CNVRs inferred by all five methods were found to be similar in the healthy controls and NPC patients (Supplementary Material, Table S3).
To identify potential NPC-associated genomic regions, the frequencies of the deletion and duplications of each CNVR were compared between the case and control groups using the Fisher's exact test. Furthermore, we assessed the significance level of CNVRs by using 10 000 permutations and multiple testing corrections. Eight regions were found to be significantly overrepresented in NPC cases (P < 0.05); they are summarized in Table 1 and Supplementary Material, Table S4. The putative NPC-associated CNVRs included six loci with deletions (Copy number, CN = 1; found on chromosomes 3, 6, 7, 8 and 19) and two loci with duplications (CN = 3; found on chromosomes 7 and 12). Among them, the most significant association with NPC was seen for a deletion (∼96.2 kb) located at chr6p21.3. On the basis of the QuantiSNP results, a significantly higher frequency of this deletion was found in NPC patients (11 of 275 cases, 4%) compared with controls (none in 276 controls, 0%; Fisher's exact test, P = 0.0004). This region contains the protein-encoding genes for major histocompatibility complex Class I chain-related (MIC) A [MICA (MIM 600169)] and HLA Complex P5 [HCP5 (MIM 604676)], as well as the non-protein coding gene for HLA complex group 26 (HCG26).
Table 1.
List of CNVRs significantly over-represented in NPC patients
| Cytoband | Risk allele | Frequency |
P-value | OR (95% CI) | Gene lista | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case (%) | Control (%) | QuantiSNP | PennCNV | CNV partition | Partek segmentation | Partek HMM | ||||
| 3p14.1 | CN = 1 | 25/275 (9.09%)b | 12/280 (4.29%)b | 0.0268 | 0.0974 | 2.23 (1.10–4.54) | None | |||
| 6p21.33 | CN = 1 | 11/275 (4%)c | 0/276 (0%)c | 0.0004* | 0.0004 | 0.0004 | 1.06E–05* | 0.0004* | N.A. | MICA, HCP5, HCG26 |
| 7p22.2 | CN = 3 | 7/275 (2.55%)c | 0/276 (0%)c | 0.0074 | 0.0293 | 0.0288 | 0.0293 | N.A. | CARD11, GNA12 | |
| 7q11.23 | CN = 1 | 7/278 (2.52%)d | 1/283 (0.35%)d | 0.3727 | 0.1731 | 0.0363 | 7.28 (0.89– 59.60) | LOC100133091, POMZP3, PMS2L11, LOC100132832, CCDC146 | ||
| 8p22 | CN = 1 | 36/275 (13.09%)b | 17/280 (6.07%)b | 0.0058 | 0.4946 | 2.33 (1.28–4.26) | TUSC3 | |||
| 12p13.31 | CN = 3 | 11/278 (3.96%)d | 3/283 (1.06%)d | 0.0532 | 0.1102 | 0.0317 | 3.85 (1.06–13.94) | CLEC4C, NANOGNB, NANOG, SLC2A14, SLC2A3 | ||
| 19p13.3 | CN = 1 | 18/278 (6.47%)d | 8/283 (2.83%)d | 0.7608 | 0.8207 | 0.0453 | 2.38 (1.02–5.57) | ZNF253, ZNF93, ZNF682, ZNF90, ZNF486, ZNF826P, MIR1270-1, MIR1270-2 | ||
| 19q13.42 | CN = 1 | 28/275 (10.18%)b | 13/280 (4.64%)b | 0.0146 | 2.33 (1.18–4.60) | LILRA6 | ||||
The regions listed represent the optimal overlap of cases and significance with respect to controls. The statistical significances of the results from each method were determined using Fisher's two-tailed exact test; values of P < 0.05 are shown in bold.
CN, copy number; 95% CI, 95% confidence intervals; N.A., not available.
aFully and partially included RefSeq genes based on methods-inferred boundaries and all genomic analyses used NCBI Build 36.1/hg18.
bGenomic region and frequencies of samples with CNV identified by the PennCNV method.
cGenomic region and frequencies of samples with CNV identified by the QuantiSNP method.
dGenomic region and frequencies of samples with CNV identified by the Partek-Segmentation method.
*Regions with a significant multiple testing corrected P-value (P < 0.05). For detailed information, see Supplementary Material, Table S4.
MICA, which belongs to the highly divergent MIC family, encodes a stress-inducible protein that functions as a ligand for the NKG2D/DAP10 complex on natural killer (NK) cells, γδT cells and CD8+ T cells (21). Engagement of NKG2D can activate the cytolytic responses of γδ T cells and NK cells against MICA-expressing epithelial tumor cells (21,22), and the expression levels of NKG2D ligands (e.g. MICA) on NPC cell lines have been correlated with NK- cell-mediated cytolysis (23). HCP5, which belongs to the P5 gene family, is known to be specifically transcribed in lymphoid cells and tissues (24), but its biological function has not yet been elucidated. Because in endemic areas, NPC is known to be closely associated with EBV infection, we hypothesize that such immune genes could influence susceptibility to NPC. Thus, the deleted region at chr6p21.3, which was also the only locus detected by all five analytical methods, was chosen for further examination.
We first mapped the CNV breakpoints, as this information was needed to design a robust, qualitative assay for our validation experiments. We deduced the putative breakpoint on the basis of the SNP-probe allele intensities on the Illumina HumanHap550K SNP arrays. PCR primers were designed to amplify a fragment containing the breakpoint, and these primers were tested against two samples inferred to carry the deletion (which we designated the MICA/HCP5 deletion) and two samples that were not predicted to carry the deletion (for detailed primer sequences, see Supplementary Material, Table S4). The amplified PCR products were sequenced, and the resulting sequences were basic local alignment search tools-searched against the reference genome sequence (NCBI build 36.1/hg18). Our results confirmed the presence of a deletion in the two suspected subjects, and further indicated that the deleted sequences were identical. According to a minor difference in the flanking sequence and the presence of an additional quad-nucleotide (GAAA), we predicted that the deletion spans ∼96.95 kb and is located between 31464142 and 31561089 on chromosome 6.
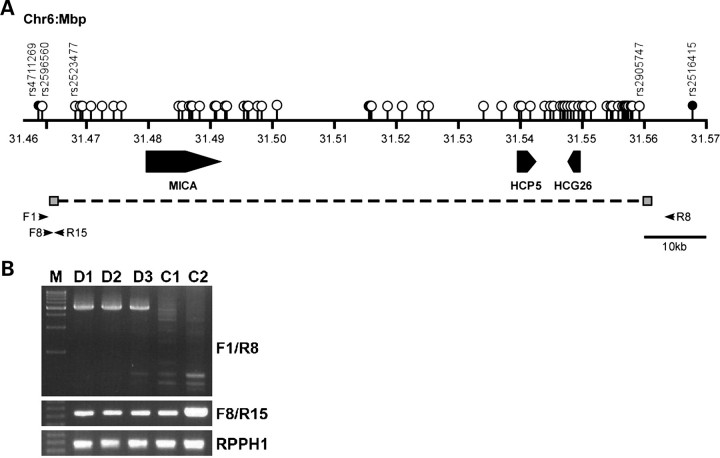
On the basis of these findings, we designed primer pairs F1/R8 and F8/R15, and used them to screen samples for the presence or absence of the deletion. In short, primers F1 and R8 only amplified a PCR product (∼5 kb in length) from samples containing the 90 kb deletion (Fig. 1A), whereas primers F8 and R15, which were located outside and within the deletion, respectively, only generated a PCR product from non-deleted samples. Gene encoding the RNA moiety for the RNase P enzyme [RPPH1 (MIM 608513)], which was found to be single-copy in all our studied samples, was used as reference in this assay. Our deletion-specific PCR assay confirmed our microarray-based identification of the MICA/HCP5 deletion. All samples inferred to carry the MICA/HCP5 deletion (n = 11) yielded a PCR product with primers F1/R8, whereas non-deleted samples did not. Notably, all of the tested samples yielded products with primers F8/R15 and RPPH1. These PCR results confirmed our prediction and demonstrated that all carriers of the MICA/HCP5 deletion were heterozygous for the deletion.
Figure 1.
Genomic organization of the chromosome 6p21.3 region containing the NPC-associated MICA/HCP5 deletion. (A) The three genes are shown with their transcription orientations noted. The white, gray and black circles, respectively indicate the positions of the SNPs located in, on and outside the boundaries of the CNVR inferred by QuantiSNP. The horizontal arrows show the relative positions of the primers used for the CNV-deletion-specific PCR assay. Primers F1 and R8 were used to detect the deleted allele, whereas primers F8 and R15 were used to detect the non-deleted allele. (B) Representative PCR results. Samples predicted to be deleted (D1–D3) and non-deleted (C1–C2) are shown. RPPH1 was amplified from each sample as an internal control.
NPC patients show a male-to-female ratio of approximately 3:1, and a gender-specific association was previously found between the MICA short tandem repeat and NPC in a southern Chinese Han population (25). To examine whether the MICA/HCP5 deletion exhibited a gender-specific association, we stratified samples by gender. As shown in Table 2, the frequency of the MICA/HCP5 deletion was significantly higher in male NPC patients (3.9%) than in male controls (0%; Fisher's exact test, P = 0.0081), whereas a less significant difference was found between the female groups (P = 0.0734). These results indicated a potential gender-specific association between the MICA/HCP5 deletion and NPC in our Taiwanese study population.
Table 2.
Association of MICA/HCP5 deletion and NPC in an independent set of case–control samples
| Characteristica | Discovery phase |
P-valueb | Validation phase |
P-valueb | Combined |
P-valueb | Odds ratio (95% CI) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Controls (n = 276) | NPC patients (n = 275) | Controls (n = 458) | NPC patients (n = 428) | Controls (n = 734) | NPC patients (n = 703) | |||||
| With deletion | 0 (0%) | 11 (4%) | 4.32E–04 | 5 (1.1%) | 13 (3%) | 0.0548 | 5 (0.7%) | 24 (3.4%) | 2.27E–04 | 5.15 (1.96–13.58) |
| Without deletion | 276 (100%) | 264 (96%) | 453 (98.9%) | 415 (97%) | 729 (99.3%) | 679 (96.6%) | ||||
| Male | 183 | 207 | 0.0081 | 316 | 312 | 0.003 | 499 | 519 | 3.65E–05 | 18.92 (2.52–141.91) |
| With deletion | 0 (0%) | 8 (3.9%) | 1 (0.3%) | 11 (3.5%) | 1 (0.2%) | 19 (3.7%) | ||||
| Without deletion | 183 (100%) | 199 (96.1%) | 315 (99.7%) | 301 (96.5%) | 498 (99.8%) | 500 (96.3%) | ||||
| Female | 93 | 68 | 0.0734 | 142 | 116 | 0.694 | 235 | 184 | 0.514 | 1.61 (0.43–6.09) |
| With deletion | 0 (0%) | 3 (4.4%) | 4 (2.8%) | 2 (1.7%) | 4 (1.7%) | 5 (2.7%) | ||||
| Without deletion | 93 (100%) | 65 (95.6%) | 138 (97.2%) | 114 (98.3%) | 231 (98.3%) | 179 (97.3%) | ||||
a‘Deletion’ means the MICA/HCP5 deletion (CN = 1) in this case.
bStatistical significance was determined using Fisher's exact test; values of P < 0.05 are shown in bold.
To validate the association of the MICA/HCP5 deletion with NPC, we used our deletion-specific PCR to assess the frequency of the MICA/HCP5 deletion in an independent group of NPC patients (n = 428) and healthy controls (n = 458). As shown in Table 2, the frequency of the deletion in NPC patients was moderately higher than that in the control subjects (P = 0.0548). Notably, the gender-specific association was also replicated. The frequencies of the MICA/HCP5 deletion in male NPC patients and healthy controls were significantly different at 3.5 and 0.3%, respectively (P = 0.003), whereas no significant difference was detected in the female group (1.7 and 2.8% in female NPC patients and healthy control, respectively; P = 0.694).
DISCUSSION
The MICA gene (11.7 kb) encodes a 383-amino-acid polypeptide that has a predicted mass of 43 kDa (26) and functions as a ligand for NKG2D on γ/δT cells, CD8+ α/β T cells and NK cells. Engagement of NKG2D with the MICA was shown to activate the cytolytic responses of γ/δ T cells and NK cells against MICA-expressing epithelial tumor cells (21,22). MICA polymorphisms have been associated with a number of conditions related to NK activity, including viral infection, cancer, allograft rejection and graft-versus-host disease (27). Moreover, the expression levels of NKG2D ligands (including MICA) in NPC cell lines have been correlated with NK-cell-mediated cytotoxicity (23), with NK cells showing a higher cytotoxicity against parental CNE2 cells (which had a high NKG2D-ligand expression) compared with multidrug-resistant CNE2/DPP cells (which had low NKG2D-ligand expression). In future studies, it is of high significance to examine MICA expression levels in tissues representing different genotypes (e.g. with or without the MICA/HCP5 deletion).
A number of studies have shown associations between certain MICA variants and NPC predisposition. For example, Douik et al. (28) demonstrated that a functionally relevant dimorphism of the MICA gene, MICA-129 val/met, was associated with NPC risk in a Tunisian population. Moreover, Tian et al. (25) reported a gender-specific association between the disease and a short tandem repeat polymorphism in exon 5 of the MICA gene (MICA*A9) in a southern Chinese Han population. Their results also suggested that MICA*A9 could be a genetic susceptibility marker for NPC in males. However, little is known regarding the functional consequences of the various MICA variants, or why there appears to be a gender-specific effect in the risk of NPC. Additional work will be needed to elucidate the biological meanings of these associations.
In addition to the MICA/HCP5 deletion, we identified seven other regions with CNV that were overrepresented in NPC patients versus healthy controls. These regions contain several potential candidates and are worthy of further investigation. For example, the duplication region of chr7p22.2, which was identified by four of the utilized methods (P = 0.0304, QuantiSNP), includes the gene encoding caspase recruitment domain family member 11 (CARD11, MIM 607210). When overexpressed in cells, CARD11 can activate nuclear factor of kappa light chain gene enhancer in B cells (NF-κB) signaling (29). Because NF-κB activation plays important roles in EBV-encoded latent membrane protein 1-mediated tumorigenesis in NPC (30), it seems possible that amplification of this genomic region could promote EBV-mediated tumorigenesis by increasing CARD11 expression and subsequent NF-κB activation. Another of the identified CNV that might logically promote cancer development was the duplication of a region on chr12p13.31, including the genes that encode the homeobox transcription factor, Nanog (NANOG) and C-type lectin domain family 4 member C (CLEC4C). NANOG (MIM 607937) is an important transcription factor that is crucial for maintaining embryonic stem cell self-renewal and pluripotency (31), whereas CLEC4C (MIM 606677) functions in inflammation, immune responses, cell adhesion and cell-cell signaling (32). Additional validations and evaluations will be needed to characterize the potential roles of these CNVRs in the development of NPC.
Currently, the CNV detection method is still immature and several previous reports suggested that the number of CNV inferred by array-based technologies depends on the algorithm used (33–35). Moreover, so far no single method has promised to provide the best results. Therefore, identification of CNVs from array-based technology using multiple algorithms was recommended. In this study, we used five different algorithms for CNV identification and 16 repeated samples for parameter optimization, which would significantly increase the reliability of our discovery. The successful validation using deletion-specific PCR assay in an independent case–control study supports the effectiveness of using multiple algorithms in CNV discovery.
In sum, we herein used the GWAS database and five different algorithms to identify putative CNVs from 288 NPC patients and 297 healthy controls. A deletion of a region on chr6p21.3 including the MICA and HCP5 genes (P = 0.0004) showed the most significant association with NPC and was identified by all five methods. A deletion-specific PCR assay confirmed this finding and revealed that there was a gender-specific association of this deletion with NPC predisposition (P = 0.0081 and 0.0734 in males and females, respectively). Most significantly, we then successfully replicated this finding in an independent group containing 428 NPC patients and 458 controls (P = 0.003 and 0.694 in males and females, respectively). To the best of our knowledge, this is the first report of a GWAS on constitutional CNVs in NPC patients (Table 3). As our findings are based on a single-ethnicity population, it would be important to further validate the results reported here in other Southeast Asian populations having a high prevalence of NPC.
Table 3.
Significant loci identified by previous and current genome-wide association studies
| Locus | Chr. | Risk allele | OR (95% CI)a | P-valuea | Nearest gene | Reference |
|---|---|---|---|---|---|---|
| rs2212020 | 3p22.2 | T | 2.24 (1.59–3.15) | 8.27E−07 | ITGA9 | (13) |
| rs6774494 | 3q26.2 | A | 1.18 (1.11–1.25) | 5.05E−08 | MDS1-EVI1 | (15) |
| rs2267633 | 6p22.1 | A | 1.57 (1.36–1.82) | 1.28E−09 | GABBR1 | (14) |
| rs2076483 | 6p22.1 | A | 1.57 (1.36–1.82) | 1.49E−09 | GABBR1 | (14) |
| rs29230 | 6p22.1 | A | 1.56 (1.34–1.80) | 4.77E−09 | GABBR1 | (14) |
| rs29232 | 6p22.1 | A | 1.67 (1.48–1.88) | 8.97E−17 | GABBR1 | (14) |
| rs3129055 | 6p22.1 | G | 1.51 (1.34–1.71) | 7.36E−11 | HLA-F | (14) |
| rs9258122 | 6p22.1 | A | 1.49 (1.32–1.69) | 3.33E−10 | HLA-F | (14) |
| rs2860580 | 6p22.1 | G | 1.72 (1.61–1.85) | 3.65E−38 | HLA-A | (15) |
| rs2517713 | 6p22.1 | A | 1.88 (1.65–2.15) | 3.90E−20 | HLA-A | (14) |
| rs2975042 | 6p22.1 | A | 1.86 (1.63–2.13) | 1.60E−19 | HLA-A | (14) |
| rs9260734 | 6p22.1 | G | 1.85 (1.61–2.12) | 6.77E−18 | HCG9 | (14) |
| rs3869062 | 6p22.1 | A | 1.78 (1.55–2.05) | 8.68E−16 | HCG9 | (14) |
| rs5009448 | 6p22.1 | G | 1.72 (1.51–1.96) | 1.30E−15 | HCG9 | (14) |
| rs16896923 | 6p22.1 | A | 1.66 (1.42–1.94) | 2.49E−10 | HCG9 | (14) |
| MICA/HCP5 deletion | 6p21.33 | CN = 1 | 5.15 (1.96–13.58) | 2.27E−04 | MICA, HCP5 | Current study |
| rs2894207 | 6p21.33 | A | 1.64 (1.49–1.75) | 1.83E−31 | HLA-B/C | (15) |
| rs28421666 | 6p21.32 | A | 1.52 (1.39–1.67) | 1.40E−18 | HLA-DQ/DR | (15) |
| rs1412829 | 9p21.3 | A | 1.28 (1.16–1.41) | 3.51E−07 | CDNK2A/2B | (15) |
| rs9510787 | 13q12.12 | G | 1.14 (1.06–1.22) | 9.57E−09 | TNFRSF19 | (15) |
| rs1572072 | 13q12.12 | C | 1.16 (1.10–1.25) | 5.50E−08 | TNFRSF19 | (15) |
aOR and P-values were calculated based on combined samples. Ng et al. (13) consisted of 447 NPC patients and 764 healthy controls. Tse et al. (14) consisted of 912 NPC patient and 1925 healthy controls. Bei et al. (15) consisted of 5090 NPC patients and 4957 healthy controls. Our current study consisted of 703 NPC patients and 734 healthy controls.
MATERIALS AND METHODS
Samples
Blood samples of 288 patients initially diagnosed with NPC in Chang-Gung Memorial hospital at Lin-kou (Taoyuan County, Taiwan) were collected. For comparison, 297 healthy ethnically matched local residents of Taoyuan County were recruited through a project designated ‘Integrated Delivery System of Health Screening, Taoyuan, Taiwan’ by Chang-Gung University, CGMH and the Health Bureau of Taoyuan County, Taiwan. Controls samples were randomly selected according to male/female ratio and age distribution. Controls affected by any type of cancer and with a personal family history of NPC were excluded. All cases and controls were of a homogenous Han Chinese origin. This study was reviewed and approved by the institutional review board and ethics committee of CGMH. Informed consent was obtained from all study participants. Demographics of the case and control samples are provided in Supplementary Material, Table S1.
Genotyping and data cleaning
DNA from all individuals was genotyped on the Illumina Hap550v3_A BeadChips, by the Illumina-certificated service provider, Genizon Biosciences (Genizon Biosciences, Canada). A total of 10 NPC patients and 12 healthy individuals were removed because of low call rate (<99%), gender mismatch and failure in IBD (PI-HAT>0.5) or IBS genetic-outlier tests (Supplementary Material, Fig. S1). Two hundred and seventy-eight cases and 285 healthy subjects that passed the sample quality control check were entered into the subsequent CNV identification and analysis procedures.
CNV detection and quality control evaluation
To maximize the finding of potential NPC-associated CNVRs, five algorithms—QuantiSNP(19) (v 2.3 Beta), PennCNV(20) (v 2009 Aug27, hh550 and hg18), CNV partition (v1.0.2 implemented in the Illumina BeadStudio software), Partek HMM and Partek-Segmentation in Partek Genomic Suite version 6.5 (Partek Inc., St. Louis, MO, USA)—were used for identifying CNV from the intensity data of an SNP-based microarray. Among these methods, QuantiSNP, PennCNV, CNV-partition and Partek HMM were developed based on HMM, whereas Partek-Segmentation was developed based on a segmentation-based algorithm. PennCNV combines Log R ratio (LRR) and B-allele frequency (BAF) in each SNP marker to generate a hidden state for copy neutral loss of heterozygosity, and uses each population-based BAF of the SNP to infer CNVs. QuantiSNP uses LRR and BAF independently, and a fixed rate of heterozygosity for each SNP is applied. As for segmentation-based methods that consider intensity (LRR) alone, such as Partek-Segmentation, a reference base-line is generated based on a pool of reference samples.
In order to acquire the subset of higher accuracy calls for further analysis and exclude the potential false positive signals, the raw CNV calls need to be filtered. To explore the best filtering parameters for each method, 15 duplicated samples were tested by comparing the result of CNV calls generated from the same sample. The concordance rate for each pair of duplicates is defined as follows:
The parameters resulting best average concordance rate (≥94%) was applied to each method. The parameters adopted by each method were summarized in Supplementary Material, Table S2.
Several quality control procedures were performed to remove the possible false positive signals in our HumanHap550K array when generating CNV calls. For PennCNV, only samples with standard deviation (SD) of normalized intensity (LRR) <0.20, SD of BAF <0.2, BAF-drifting value <0.01 and the wave factor value between −0.04 and 0.04 were included. The median values of LRR and BAF were adjusted to 0 and 0.5, respectively. Appropriate LRR adjusting based on GC model that was incorporated in PennCNV is also applied. For QuantiSNP, only samples with SD of LRR <0.25, SD of BAF <0.3 and outlier rate <0.01 were included. For CNV-partition, the confidence score threshold was set to 35, and the threshold of probe gap size was defined as 1 m bp. All markers in sex and mitochondria chromosomes were not included in our analysis. Samples with extreme CNV call count, of which standardized Z-score >6, were also excluded. In total, nine controls and three NPC cases in QuantiSNP, five controls and three cases in PennCNV, one control and one case in CNV partition and two controls in Partek Segmentation were removed because of the CNV quality control failure. Detailed procedures for CNV detection and filtering were shown in Supplementary Material, Figure S2. When overlapping CNVs were identified in cases and controls, they were merged into unique CNVRs (see Supplementary Material, Fig. S3). To verify the CNVRs involving deletions, we rechecked the genotyping data to confirm that the SNPs had been called homozygous or ‘no call’ (i.e. for hemizygous or homozygous deletions, respectively). The number of samples used, the characteristics of the CNV calls and the CNVRs identified by each method are shown in Supplementary Material, Table S3.
Deletion-specific polymerase chain reaction
Primers (sequences were shown in Supplementary Material, Table S5) were designed using Primer 3 (http://frodo.wi.mit.edu/primer3/input.htm) adjacent to the maximal deleted region, such that PCR products would only be expected in the presence of the deletion. PCR was carried out using the Long PCR enzyme mix (Fermentas, Thermo Fisher Scientific, Waltham, MA, USA) using the manufacturers’ suggested protocol. PCR products were resolved using agarose gels and visualized with Ethidium Bromide stain and UV illumination.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the Ministry of Education (to Chang Gung University), the National Science Council (NSC 94-2314-B-182A-188, 94-3112-B-182-005, 95-2320-B-182-001 and 97-3112-B-182-008) and Chang Gung Memorial Hospital (CMRPD150961 and CMRPG360221), Taiwan.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the patients who kindly agreed to participate, as well as the physicians who recruited them. We are grateful to Cathy S.J. Fann (Institute of Biomedical Sciences, Academic Sinica, Taipei, Taiwan) for helpful data discussion.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Boyle P., Levin B. International Agency for Research on Cancer and World Health Organization. World Cancer Report 2008. Lyon/Geneva: International Agency for Research on Cancer; Distributed by WHO Press; 2008. [Google Scholar]
- 2.Buell P. The effect of migration on the risk of nasopharyngeal cancer among Chinese. Cancer Res. 1974;34:1189–1191. [PubMed] [Google Scholar]
- 3.Chang E.T., Adami H.O. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol. Biomarkers Prev. 2006;15:1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. doi:10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong R.W., Kannan Kutty M., Dharmalingam S.K., Ponnudurai J.R. Incidence of nasopharyngeal carcinoma in Malaysia, 1968–1977. Br. J. Cancer. 1979;40:557–567. doi: 10.1038/bjc.1979.221. doi:10.1038/bjc.1979.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.zur Hausen H., Schulte-Holthausen H., Klein G., Henle W., Henle G., Clifford P., Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056–1058. doi: 10.1038/2281056a0. doi:10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]
- 6.Devi B.C., Pisani P., Tang T.S., Parkin D.M. High incidence of nasopharyngeal carcinoma in native people of Sarawak, Borneo Island. Cancer Epidemiol. Biomarkers Prev. 2004;13:482–486. [PubMed] [Google Scholar]
- 7.Lee A.W., Foo W., Mang O., Sze W.M., Chappell R., Lau W.H., Ko W.M. Changing epidemiology of nasopharyngeal carcinoma in Hong Kong over a 20-year period (1980–99): an encouraging reduction in both incidence and mortality. Int. J. Cancer. 2003;103:680–685. doi: 10.1002/ijc.10894. doi:10.1002/ijc.10894. [DOI] [PubMed] [Google Scholar]
- 8.Lu S.J., Day N.E., Degos L., Lepage V., Wang P.C., Chan S.H., Simons M., McKnight B., Easton D., Zeng Y., et al. Linkage of a nasopharyngeal carcinoma susceptibility locus to the HLA region. Nature. 1990;346:470–471. doi: 10.1038/346470a0. doi:10.1038/346470a0. [DOI] [PubMed] [Google Scholar]
- 9.Lu C.C., Chen J.C., Tsai S.T., Jin Y.T., Tsai J.C., Chan S.H., Su I.J. Nasopharyngeal carcinoma-susceptibility locus is localized to a 132 kb segment containing HLA-A using high-resolution microsatellite mapping. Int. J. Cancer. 2005;115:742–746. doi: 10.1002/ijc.20946. doi:10.1002/ijc.20946. [DOI] [PubMed] [Google Scholar]
- 10.Hildesheim A., Chen C.J., Caporaso N.E., Cheng Y.J., Hoover R.N., Hsu M.M., Levine P.H., Chen I.H., Chen J.Y., Yang C.S., et al. Cytochrome P4502E1 genetic polymorphisms and risk of nasopharyngeal carcinoma: results from a case-control study conducted in Taiwan. Cancer Epidemiol. Biomarkers Prev. 1995;4:607–610. [PubMed] [Google Scholar]
- 11.Nazar-Stewart V., Vaughan T.L., Burt R.D., Chen C., Berwick M., Swanson G.M. Glutathione S-transferase M1 and susceptibility to nasopharyngeal carcinoma. Cancer Epidemiol. Biomarkers Prev. 1999;8:547–551. [PubMed] [Google Scholar]
- 12.Cho E.Y., Hildesheim A., Chen C.J., Hsu M.M., Chen I.H., Mittl B.F., Levine P.H., Liu M.Y., Chen J.Y., Brinton L.A., et al. Nasopharyngeal carcinoma and genetic polymorphisms of DNA repair enzymes XRCC1 and hOGG1. Cancer Epidemiol. Biomarkers Prev. 2003;12:1100–1104. [PubMed] [Google Scholar]
- 13.Ng C.C., Yew P.Y., Puah S.M., Krishnan G., Yap L.F., Teo S.H., Lim P.V., Govindaraju S., Ratnavelu K., Sam C.K., et al. A genome-wide association study identifies ITGA9 conferring risk of nasopharyngeal carcinoma. J. Hum. Genet. 2009;54:392–397. doi: 10.1038/jhg.2009.49. doi:10.1038/jhg.2009.49. [DOI] [PubMed] [Google Scholar]
- 14.Tse K.P., Su W.H., Chang K.P., Tsang N.M., Yu C.J., Tang P., See L.C., Hsueh C., Yang M.L., Hao S.P., et al. Genome-wide association study reveals multiple nasopharyngeal carcinoma-associated loci within the HLA region at chromosome 6p21.3. Am. J. Hum. Genet. 2009;85:194–203. doi: 10.1016/j.ajhg.2009.07.007. doi:10.1016/j.ajhg.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bei J.X., Li Y., Jia W.H., Feng B.J., Zhou G., Chen L.Z., Feng Q.S., Low H.Q., Zhang H., He F., et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat. Genet. 2010;42:599–603. doi: 10.1038/ng.601. [DOI] [PubMed] [Google Scholar]
- 16.Feuk L., Carson A.R., Scherer S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. doi:10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 17.Diskin S.J., Hou C., Glessner J.T., Attiyeh E.F., Laudenslager M., Bosse K., Cole K., Mosse Y.P., Wood A., Lynch J.E., et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. doi:10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W., Sun J., Li G., Zhu Y., Zhang S., Kim S.T., Wiklund F., Wiley K., Isaacs S.D., Stattin P., et al. Association of a germ-line copy number variation at 2p24.3 and risk for aggressive prostate cancer. Cancer Res. 2009;69:2176–2179. doi: 10.1158/0008-5472.CAN-08-3151. doi:10.1158/0008-5472.CAN-08-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colella S., Yau C., Taylor J.M., Mirza G., Butler H., Clouston P., Bassett A.S., Seller A., Holmes C.C., Ragoussis J. QuantiSNP: an objective Bayes Hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. doi:10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. doi:10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer S., Groh V., Wu J., Steinle A., Phillips J.H., Lanier L.L., Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. doi:10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 22.Wu J., Song Y., Bakker A.B., Bauer S., Spies T., Lanier L.L., Phillips J.H. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. doi:10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 23.Mei J.Z., Guo K.Y., Wei H.M., Song C.Y. [Expression of NKG2D ligands in multidrug-resistant nasopharyngeal carcinoma cell line CNE2/DDP and their effects on cytotoxicity of natural killer cells] Nan Fang Yi Ke Da Xue Xue Bao. 2007;27:887–889. [PubMed] [Google Scholar]
- 24.Vernet C., Ribouchon M.T., Chimini G., Jouanolle A.M., Sidibe I., Pontarotti P. A novel coding sequence belonging to a new multicopy gene family mapping within the human MHC class I region. Immunogenetics. 1993;38:47–53. doi: 10.1007/BF00216390. doi:10.1007/BF00216390. [DOI] [PubMed] [Google Scholar]
- 25.Tian W., Zeng X.M., Li L.X., Jin H.K., Luo Q.Z., Wang F., Guo S.S., Cao Y. Gender-specific associations between MICA-STR and nasopharyngeal carcinoma in a southern Chinese Han population. Immunogenetics. 2006;58:113–121. doi: 10.1007/s00251-006-0093-6. doi:10.1007/s00251-006-0093-6. [DOI] [PubMed] [Google Scholar]
- 26.Bahram S., Bresnahan M., Geraghty D.E., Spies T. A second lineage of mammalian major histocompatibility complex class I genes. Proc. Natl Acad. Sci. USA. 1994;91:6259–6263. doi: 10.1073/pnas.91.14.6259. doi:10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choy M.K., Phipps M.E. MICA polymorphism: biology and importance in immunity and disease. Trends Mol. Med. 2010;16:97–106. doi: 10.1016/j.molmed.2010.01.002. doi:10.1016/j.molmed.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Douik H., Ben Chaaben A., Attia Romdhane N., Romdhane H.B., Mamoghli T., Fortier C., Boukouaci W., Harzallah L., Ghanem A., Gritli S., et al. Association of MICA-129 polymorphism with nasopharyngeal cancer risk in a Tunisian population. Hum. Immunol. 2009;70:45–48. doi: 10.1016/j.humimm.2008.10.008. doi:10.1016/j.humimm.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Bertin J., Wang L., Guo Y., Jacobson M.D., Poyet J.L., Srinivasula S.M., Merriam S., DiStefano P.S., Alnemri E.S. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J. Biol. Chem. 2001;276:11877–11882. doi: 10.1074/jbc.M010512200. doi:10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 30.Morris M.A., Dawson C.W., Young L.S. Role of the Epstein-Barr virus-encoded latent membrane protein-1, LMP1, in the pathogenesis of nasopharyngeal carcinoma. Future Oncol. 2009;5:811–825. doi: 10.2217/fon.09.53. doi:10.2217/fon.09.53. [DOI] [PubMed] [Google Scholar]
- 31.Kashyap V., Rezende N.C., Scotland K.B., Shaffer S.M., Persson J.L., Gudas L.J., Mongan N.P. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–1108. doi: 10.1089/scd.2009.0113. doi:10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanazawa N., Tashiro K., Miyachi Y. Signaling and immune regulatory role of the dendritic cell immunoreceptor (DCIR) family lectins: DCIR, DCAR, dectin-2 and BDCA-2. Immunobiology. 2004;209:179–190. doi: 10.1016/j.imbio.2004.03.004. doi:10.1016/j.imbio.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Winchester L., Yau C., Ragoussis J. Comparing CNV detection methods for SNP arrays. Brief Funct. Genomic Proteomic. 2009;8:353–366. doi: 10.1093/bfgp/elp017. doi:10.1093/bfgp/elp017. [DOI] [PubMed] [Google Scholar]
- 34.Tsuang D.W., Millard S.P., Ely B., Chi P., Wang K., Raskind W.H., Kim S., Brkanac Z., Yu C.E. The effect of algorithms on copy number variant detection. PLoS ONE. 2010;5:e14456. doi: 10.1371/journal.pone.0014456. doi:10.1371/journal.pone.0014456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang D., Qian Y., Akula N., Alliey-Rodriguez N., Tang J., Gershon E.S., Liu C. Accuracy of CNV Detection from GWAS Data. PLoS ONE. 2011;6:e14511. doi: 10.1371/journal.pone.0014511. doi:10.1371/journal.pone.0014511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.