Abstract
Magnetic resonance spectroscopy (MRS) and spectroscopic imaging (MRSI) are valuable tools to detect metabolic changes in tumors. The currently emerging era of molecular medicine, which is shaped by molecularly targeted anticancer therapies combined with molecular imaging of the effects of such therapies, requires powerful imaging technologies that are able to detect molecular information. MRS and MRSI (MRS/I) are such technologies that are able to detect metabolites arising from glucose and choline metabolism in noninvasive in vivo settings and at higher resolution in tissue samples. The roles that MRS/I plays in diagnosing different types of cancer as well as in early monitoring of tumor response to traditional chemotherapies are reviewed. Emerging roles of MRS/I in the development and detection of novel targeted anticancer therapies that target oncogenic signaling pathways or targets in choline or glucose metabolism are discussed.
Keywords: Magnetic resonance spectroscopy, MRS, metabolism, glucose, choline
1. Introduction
Although a lot of progress has been made in diagnosing cancers early and improving available treatment options, cancer still is a devastating disease that claims one in four death in the United States (1). A unique problem with anticancer therapies is that cancer cells can adapt to these therapies and survive them, leading to treatment resistance. This problem arises due to the genomic instability of cancer cells, which is additionally triggered by their cooperative interaction with stromal cells and the tumor microenvironment (TME). Together, cancer cells, stromal cells, and the TME progress to form a primary tumor, invade surrounding normal tissue, and give rise to metastatic nodules at distant sites. To overcome the problem of resistance to treatment, and to render anticancer chemotherapies less toxic, tremendous research efforts have been invested in the development of molecularly targeted anticancer therapies. The investigation of strategic molecular targets has uncovered successful targets in oncogenic signaling pathways, such as receptor tyrosine kinases and their downstream effectors, as well as targets in key pathways affecting proliferation, apoptosis, angiogenesis, invasion and metastasis, previously defined as the hallmarks of cancer (2). Recent preclinical and clinical cancer research findings have clearly shown that personalized molecular targeting is necessary for the successful treatment of cancer, in which more than one critical target is being hit at once.
One important biological outcome that results from the interplay between genetically altered tumor cells and the physiological TME is the altered tumor and cancer cell metabolism. Glucose metabolism and choline metabolism are significantly different in tumors. This is a result of changes in oncogenic signaling pathways that lead to changes in expression and/or activity of enzymes involved in glucose and choline metabolism. The altered tumor metabolism can be detected with noninvasive imaging modalities such magnetic resonance (MR), positron emission tomography (PET), and single photon emission tomography (SPECT), which are well suited to detect metabolic biomarkers or surrogate markers in cancer diagnosis and anticancer treatment. In this review we have focused on noninvasive magnetic resonance spectroscopy (MRS) and spectroscopic imaging (MRSI) and their roles in guiding molecular medicine by playing important roles in molecular target discovery, molecular targeting, diagnosis, and noninvasive monitoring of choline and glucose metabolism in cancer to assess treatment success or failure.
MRS is very helpful in studying metabolism because the signals of different metabolites occur at different chemical shifts. The aberrations in the genome and proteome of cancer cells result in their unique metabolic phenotype. The cancer metabolic phenotype is characterized by high glucose uptake, increased glycolytic activity and lactate production, decreased mitochondrial activity, low bioenergetic status, and aberrant phospholipid metabolism (3-5). Tissue-specific metabolites such as N-acetyl aspartate in the brain, and citrate in the prostate, show an MRS-detectable decrease as a result of an expanding cancer cell population combined with a reduction of the normal tissue fraction (5-7). Proton or 31P MRS or MRSI detection of these endogenous metabolites can help diagnose cancer (8-15), and monitor anticancer therapy in some cases, as outlined in this review. The fact that MRS/I is noninvasive makes it such a powerful technique because this opens up the possibility to perform dynamic monitoring of tumor metabolism in patients. However, the spectral resolutions achieved in vivo are relatively low at currently approved clinical magnetic field strengths, making it necessary in some cases to observe combined overlapping signals in vivo, such as the total choline containing signal at 3.2 ppm. Ex vivo applications of MRS provide a much higher spectral resolution, and novel techniques based on magic angle spinning that can avoid extraction of the tissue have evolved over the past decade. Such high-resolution (HR) magic angle spinning (MAS) MRS techniques are currently being implemented in the diagnostic clinical workflow following biopsy or surgical resection of tumors, and will be further explained in this review.
MRS studies on the aberrant cancer cell metabolism have resulted in the identification relevant enzymes as novel anticancer targets (4, 16-19). The total choline (tCho) signal is already being explored in the clinical setting as MR biomarker for assisting in tumor diagnosis and early monitoring of tumor response to anticancer therapy (20-22). Molecular targeting of targets in choline phospholipid metabolism can be noninvasively detected and imaged by MRS and MRSI (MRS/I). Pharmacological and molecular approaches to target choline kinase, a key enzyme in choline metabolism, are currently being developed (16, 23), and are discussed in detail below. In this review, we have described some recent biomedical MRS applications of 1H, 31P, and 13C MRS in preclinical models of cancer, and have outlined new developments, such as 13C hyperpolarization to improve the MR detection sensitivity of 13C-labeled substrates.
2. Key molecules driving the altered choline metabolism in cancer
MRS is especially useful in studying metabolism in heterogeneous tissues such as tumors with its ability to identify different compounds by their chemical shifts. MR detects radiofrequency (RF) signals emitted after MR active nuclei such as 1H, 31P, 13C, and 19F are excited by external alternating magnetic fields. MR spectra can be acquired with or without spatial localization. In the absence of spatial information, the signal is acquired from the entire sensitive region of the coil that detects the RF signal. Localized MRS can either be acquired from a single volume element (single-voxel), or from multiple voxels (multi-voxel) to generate images of the spatial distribution of metabolites, referred to as MRSI. When measuring such localized MR spectra, phase-encoding gradients are added to the MRS or MRSI (MRS/I) sequence to generate images of signals with spatially encoded chemical shift information. This phase encoding can be applied in one, two, or three spatial dimensions, and its application is useful in deciphering metabolic distributions in heterogeneous tissues such as tumors.
Elevated metabolites in choline phospholipid metabolism in cancer, which can be noninvasively detected and imaged by MRS/I (16, 23), present unique targets to exploit for molecular targeting. Malignant transformation has been observed to alter the profile of the choline compounds glycerophosphocholine (GPC) and phosphocholine (PC) in breast (24) and ovarian (25) cancer cells, which was detected by using MRS/I. GPC is the predominant signal in nonmalignant breast and ovarian epithelial cells, whereas PC displays the highest concentration in ovarian and breast cancer cells (24, 25). Prostate cancer cells contain elevated PC and GPC levels (26). The choline phospholipid metabolites typically detected in high-resolution 1H MR spectra are free choline (Cho), PC, and GPC. Similarly, in high-resolution 31P MR spectra, signals from PC and phosphoethanolamine (PE), and GPC and glycerophosphoethanolamine (GPE) are detected. In clinical settings, 1H or 31P MRS or MRSI detection of these choline metabolites has been proven useful in the diagnosis of cancer (8-15), and in monitoring the response of tumors to anticancer therapy (20-22), as detailed in subsequent sections.
2.1 Magnetic resonance spectroscopy techniques for detecting and differentiating cancer, and monitoring anticancer therapy, based on choline metabolites
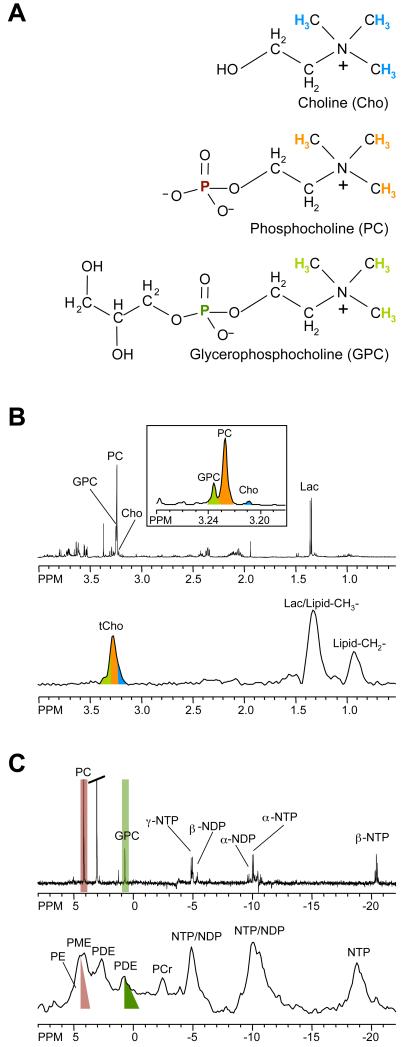
Proton MRS signals from water-soluble choline metabolites arise between 3.2 – 3.3 ppm from the nine chemically equivalent protons in the choline-N(CH3)3 groups. In Figure 1, color-coded 1H nuclei are displayed in the chemical structure formula that give rise to the MR signals that are shown with the same color-code in representative MR spectra. Because nine protons contribute to this signal, it displays a higher signal intensity than the 1H signals in the methylene groups of the same choline metabolites, which only contain two equivalent protons each (see Figure 1). Using 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid (TSP) as a chemical shift reference at pH 7.4, free choline is detected as a peak at 3.21 ppm, PC at 3.23 ppm, and GPC at 3.24 ppm, in high-resolution (HR) 1H MR spectra of cell or tissue extracts, or in HR magic angle spinning (MAS) MR spectra. The chemical structures of Cho, PC, and GPC are shown in Figure 1. It is not possible to resolve Cho, PC, and GPC in vivo, even at higher magnetic field strengths, because of the broader line widths that result from restricted molecular motion and magnetic field inhomogeneities. As an alternative, a single unresolved peak arising from total choline containing compounds (tCho), comprising of these three signals is detected, as shown in Figure 1. Multi-voxel in vivo 1H MRSI of one or multiple slice(s) through a region of interest provides multiple spectra from a slice or volume of tissue that can be processed to obtain the spatial distribution of tCho or other metabolites.
Figure 1.
(A) Chemical structures of the choline phospholipid metabolites free choline (Cho), phosphocholine (PC), and glycerophosphocholine (GPC). (B) High-resolution ex vivo 1H MR spectra of triple-negative human MDA-MB-231 breast cancer cell extracts (top) and in vivo 1H MR spectra of the same cell line grown as orthotopic tumor (bottom). (C) High-resolution ex vivo 31P MR spectra of triple-negative human MDA-MB-231 breast cancer cell extracts (top) and in vivo 31P MR spectra of the same cell line grown as orthotopic tumor (bottom). Assignments: Cho, free choline; GPC, glycerophosphocholine; GPE, glycerophosphoethanolamine; DPDE, diphosphodiester; NDP, nucleoside diphosphate; NTP, nucleoside triphosphate; Lac, lactate; Lipid-CH2-, methylene groups of mobile lipids; Lipid-CH3-, methyl groups of mobile lipids; PC, phosphocholine; PE, phosphoethanolamine; PCr, phosphocreatine; Pi, inorganic phosphate; tCho, total choline-containing compounds (Cho+PC+GPC). The 1H and 31P nuclei in Cho, PC, and GPC and their respective 1H and 31P signals in the MR spectra are color-coded to identify the MR signals that arise from the corresponding nuclei.
2.1.1 1H MRS/MRSI in cancer detection and treatment based on choline metabolites
Clinical multi-center trials are currently underway to establish single-voxel 1H MRS or multi-voxel 1H MRSI as a tool for cancer detection. Numerous of studies investigated the feasibility and diagnostic value of combined MRI and MRS/I in the detection of cancer (9, 13, 14, 27-31). Several research groups investigated the feasibility of combined dynamic contrast enhanced (DCE) MRI and MRS in evaluating breast lesions (14, 30). In a study by Jacobs et al, five patients with breast carcinoma and four with benign lesions were evaluated (30). Persistent enhancement of lesion time intensity, which describes how the inspected tissues absorb and release the contrast agent over the period of time that coincides with the MRI examination, was found in two benign cases, plateau enhancement in two of four benign and four of five malignant lesions, and one malignant case exhibited a washout pattern (30). The tCho signal-to-noise ratio (SNR) was significantly different (P < 0.003) between benign and malignant lesions (30). A combination of DCE MRI and MRS proved feasible, and improved specificity compared to either modality alone (30). Throughout the past few years, several studies with hundreds of breast cancer patients have demonstrated that an increased MRS-detected tCho signal that overlaps with the contrast-enhancing breast lesion in DCE MRI can be used to identify malignant breast tumors and distinguish them from benign breast lesions (14, 31, 32). Recent studies of prostate cancer (28, 29) compared the area under the receiver operating curve (ROC) of combined MRI and MRS, MRI alone, and MRS alone. These studies found that the combination of MRI and MRS yielded superior diagnostic results than either modality alone, in terms of sensitivity (72.3%), specificity (92.6%) and accuracy (80.1%) of combined MRI and MRS for detection of prostate cancer. Combining whole prostate qualitative MRI with MR spectral estimation was shown in previous research studies to be feasible in routine clinical practice (28, 29).
Many studies use peak area ratios as a criterion to describe the changes in metabolic profiles in cancer detection. The advantage of using ratios is that the effects of cell density variations that can affect the calculation of metabolite concentrations are canceled against each other when calculating a ratio. MRS of the prostate provides information of the cellular metabolites of the gland. Normal prostate tissue contains high levels of citrate, occurring at chemical shifts of 2.50 ppm and 2.72 ppm {van der Graaf, 1996 #412}. In prostate cancer (33), the citrate level is diminished or undetectable and the tCho level is elevated (Figure 2). The total creatine (tCr) peak, usually around 3.0 ppm, is very close to the tCho peak (3.2 ppm) in these in vivo MR spectra and may not always be separable. The tCr signal is assumed to remain unaltered between healthy and cancerous tissue. For this reason and practical purposes, the ratio of tCho plus tCr over citrate [choline + creatine]/citrate (CC/Ci) is used for spectral analysis to make it feasible for clinical practice (33). 1H MRS of brain tumors compared to normal brain detected decreased N-acetylaspartate (NAA) and frequently increased tCho. An increase in the ratio of tCho to NAA (tCho/NAA) was highly correlated with increased tumor malignancy and can be used as a noninvasive index of tumor grade (9, 34-36).
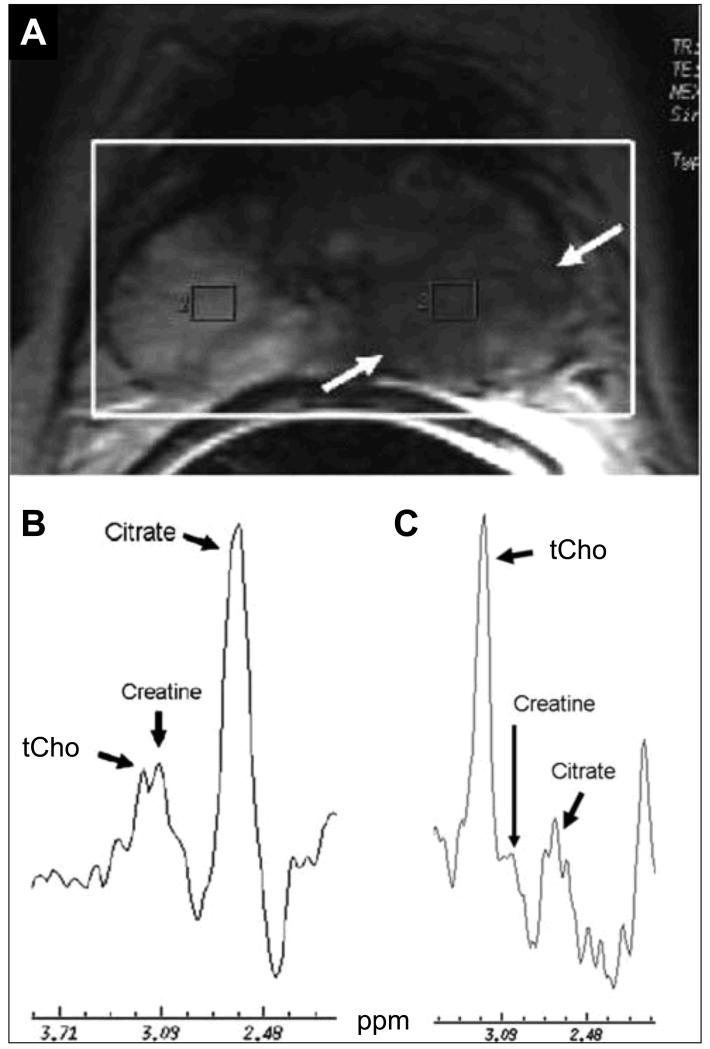
Figure 2.
Axial T2-weighted MR image and MRSI of the normal prostate gland and prostate cancer obtained at 1.5 T. (A) The MRSI was acquired from the whole gland (heavy white box) showing a large low signal intensity lesion (arrows) in the left peripheral gland. (B) The right voxel from the peripheral gland shows a high T2-weighted MRI signal intensity with the normal metabolic profile of prostatic gland, characterized by a high level of citrate and lower levels of choline and creatine. (C) The corresponding spectrum from the left voxel shows increased choline and reduced citrate, indicative of the metabolic profile of prostatic tumor combined with low signal intensity on T2-weighted MRI. Adapted from (33).
Results of numerous ex vivo (37-39) and in vivo (40, 41) studies have shown that neoplastic breast tissue contains elevated levels of tCho. Since the sensitivity of in vivo MRS is limited, adding a quantitative analysis of 1H MRS, such as calculating the tCho concentration of the lesions (14), to the breast imaging examination could help to improve the radiologist’s ability to characterize malignant breast lesions. Two referencing strategies have been used for converting measured signal amplitudes into concentrations. The first method is to use external referencing, in which the in vivo measurement is compared with a measurement from an external phantom of known concentration (42, 43). The second referencing strategy proposed for breast MRS is to use the water peak from a non-suppressed spectrum from the same voxel as an internal reference. The tCho concentration is calculated by comparing the tCho and water signal intensities and correcting for the differences in relaxation (40, 44).
MRS/I cannot only help with the diagnosis of cancers in the clinic, but also monitor clinically approved anticancer treatments. Multiple studies have identified changes in tCho as having a high likelihood of detecting early response to treatment in breast cancer (21, 45, 46). Neoadjuvant chemotherapy (NACT) is the standard treatment for patients with inoperable locally advanced breast cancer (LABC). A recent study (22) compared changes in tCho concentration and in tumor size at follow-up after NACT between patients, who achieved pathologic complete response (pCR) and those who did not, by using MRI and quantitative 1H MRS (22). Figure 3 shows the maximum intensity projection MR images (left) and corresponding MR spectra (right) from a 40-year-old patient who achieved pCR (22). In this study, patients who had a greater reduction in tCho compared with changes in tumor size were more likely to achieve pCR (22). Another recent study (47) was designed to investigate the changes in the signal-to-noise ratio (SNR) of the tCho resonance (ChoSNR) using MRSI and an anatomical parameter, such as the volume of the tumor, in patients with LABC, and then determined the potential clinical utility of these parameters in the assessment of tumor response during the various stages of NACT. This study concluded that ChoSNR might serve as a useful parameter in predicting tumor response in the absence of the absolute tCho concentration (47). ChoSNR gave a sensitivity of 85.7% with a specificity of 91% (47). An early response of prostate cancer to hormone-deprivation therapy (48, 49) and cryosurgery (50) was detected by a decrease in [tCho/citrate] ratios quantified from 1H MR spectra. Anticancer treatments leading to apoptosis or necrosis induced characteristic changes in choline phospholipid metabolites (51). An early response to therapy in lymphomas was also associated with a reduction of tCho (52). Some cancers, e.g. cervical cancer, did not exhibit a change in the tCho signal following neoadjuvant therapy, followed by surgical resection of the tumors (53). However, these cervical cancers displayed a reduced tumor volume along with decreased triglyceride signal and no survival advantage associated with the reduced tumor volume or decreased triglycerides (53).
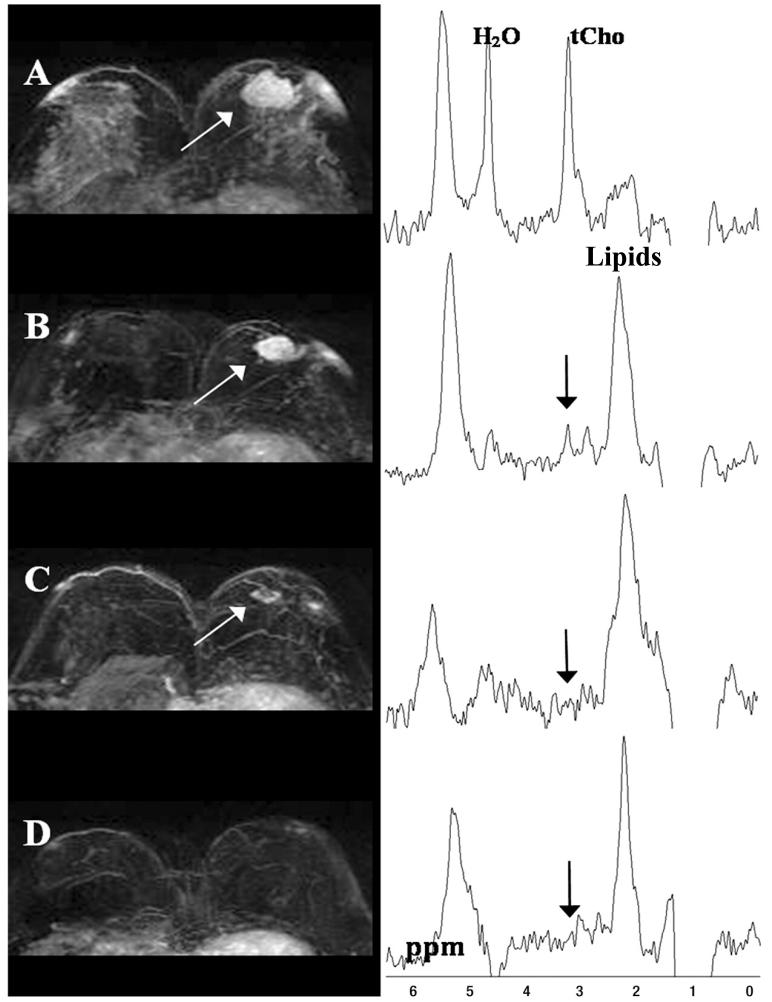
Figure 3.
Maximum intensity projection MR images (left) and corresponding MR spectra (right) from a 40-year-old patient breast cancer patient who achieved pathological complete response. (A) Before treatment, a 3.4-cm lesion (arrow) shows a heterogeneous enhancing pattern. An elevated tCho peak (right) is visible at 3.23 ppm in the water-fat suppressed MR spectrum. Gaussian model fitting of the tCho peak gave 2.33 ± 0.54 mmol/kg tCho. (B) Enhancing lesion (white arrow) and water-fat suppressed spectrum (black arrow) acquired after one neoadjuvant chemotherapy (NACT) cyclophosphamide (AC) cycle. Lesion size was reduced to 2.6 cm, the tCho peak was visible at 3.22 ppm, and tCho = 1.15 ± 0.25 mmol/kg. The tCho level was reduced by 51%, while the tumor size decreased by 23%. (C) At second follow-up after two AC cycles and one taxane regimen, the lesion (white arrow) further shrank to 1.3 cm, indicating good response. (D) After completion of all neoadjuvant chemotherapy cycles, the breast lesion completely regressed. The tCho peak (arrow) was no longer detectable (100% reduction) in the water-fat suppressed spectrum. Adapted from (22).
2.1.2 HR MAS 1H MRS in cancer detection and treatment based on choline metabolites
The relatively new applications of ex vivo high-resolution MR technique HR MAS 1H MRS to fresh tissues, which can be implemented in the clinical workflow, is currently gaining importance in diagnosis and treatment monitoring of cancer. HR MAS 1H MRS is well suited to examine intact biological tissue ex vivo at high spectral resolution (7, 54-56). In the clinic, HR MAS 1H MRS can be useful for analyzing the metabolome of biopsy specimens prior to pathological classification (7, 54-56). In HR MAS 1H MRS, the solid sample is spun at a frequency of up to 12 kHz at the magic angle of θm, which is 54.74° with respect to the direction of the magnetic field. At this magic angle, where (3cos2θm-1)/2 approaches zero, residual dipolar interactions average out, and therefore the normally broad lines become narrower and the spectral resolution increases for better identification and analysis of the spectrum (7, 54-56). One major advantage of this technique is that, unlike in HR MRS of tissue extracts, the HR MAS 1H MRS-analyzed tissue can be used for subsequent histologic, biochemical, and genetic analyses (7, 54-57). The high spectral resolution of HR MAS 1H MR spectra makes their data analysis more challenging. A combination of different multivariate data analysis methods such as principal component analyses or neural network applications is typically used to process these rich data sets (57). A recent study demonstrated that HR MAS 1H MRS was able to detect the metabolic phenotype of tumors, demonstrating a marked metabolic difference between healthy breast and breast tumor tissue evident by clearly separated score plots of three principal components (57). Absolute metabolite quantification can also be performed from HR MAS 1H MR spectra either alone or in combination with multivariate data analysis (58). Knowledge of absolute tumor metabolite levels can help understand the disease process on a biochemical and molecular level. A known amount of TSP can be added to the samples for quantification, or a pre-calibrated Electronic REference To access In vivo Concentrations (ERETIC) can be applied to HR MAS 1H MRS to quantify metabolites in intact cancer samples (58). HR MAS 1H MRS has a great potential to be used clinically as a complement for histopathological analysis, for detecting and characterizing cancers (57, 58).
2.1.3 31P MRS/MRSI in cancer detection and treatment based on choline metabolites
Phosphorus-containing choline metabolites can be detected with 31P MRS, as demonstrated in Figure 1. Following chemical shift calibration to a reference compound such as methylene diphosphonic acid at 18 ppm, a signal from PC at 3.9 ppm and GPC at 0.5 ppm are observed in HR 31P MR spectra of cell or tumor extract, as shown by color-coded signals and their corresponding 31P nuclei with the same color-code in the chemical structures. In the in vivo setting, a mixed phosphomonoester (PME) signal containing unresolved PC and phosphoethanolamine (PE) resonances, and a mixed phosphodiester (PDE) signal containing unresolved GPC and GPE resonances are observed in 31P MR spectra (see color-coding in Figure 1).
Since 1H MRS has a higher sensitivity and a higher availability of a scanning standard compared with 31P MRS, it is much more popular on clinical scanners. However, a multi-institutional group has been created to investigate the utility of in vivo 31P MRS in studying human cancers in vivo (59). The standardization of the acquisition protocol for in vivo-localized 31P MRS across different institutions has resulted in comparable in vivo data, decreasing the possible problems related to a research study carried out under a multi-institutional setting. A recent clinical study demonstrated the feasibility of partially resolving PC from PE, and GPE from GPC by using 1H-decoupled 31P MRS at 1.5 T (60) and at the higher field strength of 3T that is also clinically available. A large-scale multi-center trial across nine institution using 31P MRS for non-invasive monitoring of chemotherapy of solid non-Hodgkin’s lymphoma (NHL) pre- and post-treatment, was performed on either General Electric (GE) or Siemens clinical 1.5T MR scanners (61). In this multi-center 31P MRS trial, the PME/NTP ratio significantly decreased following treatment in complete (P<0.001) and partial (P<0.05) responders, but not in non-responders (P>0.1) (61). The PME/NTP ratio in the pre-treatment 31P MR spectra also correlated with the treatment outcome, indicating that the PME/NTP ratio can serve as a significant predictor of long-term clinical response and time-to-treatment failure in NHL.
Two recent studies reported a novel 1H to 31P polarization transfer method by applying chemical shift selective refocusing pulses at 3T, which can achieve a more than twofold increase of SNR compared to direct 31P MRS methods (62, 63). This selective refocused insensitive nuclei-enhanced polarization transfer (sRINEPT) method provided baseline-resolved PE, PC, GPE, and GPC peaks in human brains (62, 63). The sRINEPT method allowed to generate a three-dimensional 31P MRSI data set with good SNR from voxel sizes of 17 cm3 within a clinically acceptable measurement time of 17 minutes (63). Evaluation of intact tissue samples from experimental tumors has been performed using 1H-decoupled 31P HR MAS MRS (64, 65). The concentrations of the well-resolved tissue metabolites showed good agreement with concentrations measured from extracts of the same pieces of tissue (64, 65).
2.1.4 13C MRS/ MRSI in cancer detection and treatment based on choline metabolites
For studies of choline metabolites, 13C MRS can be useful to detect 13C-labeled metabolites following administration of 13C-labeled free choline or methionine as substrates in cancer cells and solid tumors (23, 66-70). The 13C label in either the −CH2 or the NCH3 groups of free choline can be followed with 13C MRS to detect the incorporation of a 13C label in the downstream metabolites and products, such as PC and GPC. Quantitative flux rates can be calculated by metabolic modeling. Carbon-13 MRS methods that attempt to detect the 13C directly suffer from relatively low sensitivity. However, magnetization transfer techniques can improve the 13C MRS sensitivity. Examples of such magnetization transfer techniques that can drastically improve the detection sensitivity of 13C nuclei are nuclear Overhauser effect (NOE) methods, heteronuclear cross-polarization (HCP) experiments (71), and indirect inverse detection methods (72). In NOE techniques, the 13C signals are enhanced because magnetization is transferred from neighboring protons through space using dipole-dipole spin coupling, or through chemical bonds using the J-coupling between 13C and 1H spins. Direct 13C detection methods using magnetization transfer to enhance 13C signals are NOE, distortionless enhancement by polarization transfer (DEPT), insensitive nuclei enhanced by polarization transfer (INEPT), and HCP. Indirect detection schemes, in which the magnetization is transferred from 1H to 13C and then back to 1H, are heteronuclear multiple quantum coherence (HMQC) and heteronuclear single quantum coherence (HSQC) methods. Carbon-13 signal detection in HMQC and HSQC is done indirectly as 1H frequencies, which significantly enhances the sensitivity due to the higher gyromagnetic ratio γ of 1H (72).
2.2 Altered enzymes in choline metabolism of cancer
1H MRS consistently detects significant differences in choline phospholipid metabolites of malignant versus benign breast lesions. It is critically important to understand the molecular causes underlying these metabolic differences, because this may identify novel targets for anticancer therapy selective for cancer cells. The major molecular causes for the increased PC and tCho levels in cancer cells and tumors include an increased expression and activity of choline kinase alpha (CHK-α) (16, 18, 23, 73-76), a higher rate of choline transport (77, 78), and increased activities of phosphatidylcholine (PtdCho)-specific phospholipase C (PtdCho-PLC) (25, 79) and D (PtdCho-PLD) (25, 80-82), and several phospholipases A2 (83, 84). Other enzymes in the choline phospholipid metabolic pathways are in the process of investigation (85). All these enzymes participate in the biosynthetic and breakdown pathways of the major membrane phospholipid phosphatidylcholine (PtdCho) (23) as shown in the schematic in Figure 4.
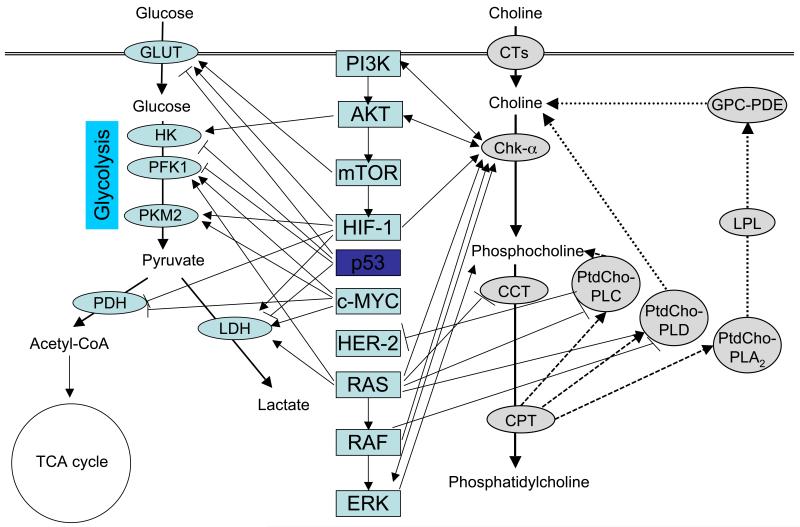
Figure 4.
Schematic showing the most important cancer-related cell signaling pathways (center) that impact upon enzymes in glucose metabolism (left) and enzymes in choline phospholipid metabolism (right). Arrows depict an activation in the direction of the arrow, and short vertical lines at the end of a line symbolize inhibition in the direction of vertical line. Abbreviations: acetyl-CoA, acetyl coenzyme A; AKT, v-akt murine thymoma viral oncogene homolog 1; CCT, CTP:phosphocholine cytidylyltransferase; Chk-α, choline kinase alpha; c-Myc, v-myc myelocytomatosis viral oncogene homolog; CPT, diacylglycerol; CT, choline transporter; CTP, cytidine-5′-triphosphate; Erk, extracellular signal-regulated kinases; GLUT, glucose transporter; GPC-PDE, glycerophosphocholine phosphodiesterase; HIF-1, hypoxia inducible factor 1; HK, hexokinase; LDH, lactate dehydrogenase; LPL, lysophospholipase; mTOR, mammalian target of rapamycin; PDH, pyruvate dehydrogenase; PFK1, phosphofructokinase 1; PI3K, phosphatidylinositol 3-kinase; PKM2, pyruvate kinase muscle isozyme 2; PtdCho-PLA2, phosphatidylcholine-specific phospholipase A2; PtdCho-PLC, phosphatidylcholine-specific phospholipase C; PtdCho-PLD, phosphatidylcholine-specific phospholipase D; TCA, tricarboxylic acid.
CHK-α is responsible for the generation of PC from its precursor Cho in the biosynthesis pathway of PtdCho. Both CHK-α (16, 18) and its product PC (24) have been recently reported as essential molecules in cell proliferation and transformation. One study correlated CHK-α expression and activity with clinical features in breast cancer patients, demonstrating that CHK-α is overexpressed in 17% of human mammary carcinomas, and its activity is increased in 38.5% of the analyzed breast tumors (18). The levels of CHK-α expression were also explored in both normal and tumoral cell lines derived from lung, colon, and prostate samples (86). In keeping with the role of CHK-α and PC in malignant transformation, CHK-α expression was found up-regulated at a high percentage in human tumor-derived cell lines as well as in different human tumoral tissues (86). CTP:phosphocholine cytidylyltransferase (CCT) is the rate-limiting enzyme in the biosynthetic pathway of PtdCho and its downregulation may be contributing to the elevated PC and tCho levels in cancer cells as well (87, 88). Within the breakdown pathways of PtdCho catabolism, PtdCho-PLC activation has been implicated in contributing to the aberrant choline phospholipid metabolism in cancer (25, 79, 89-91). The overall activities of PtdCho-PLC and PtdCho-PLD were higher in human ovarian carcinoma cell lines as compared with normal or immortalized ovary epithelial cells (25, 79). CHK-α (4, 16-19, 92-94) and PtdCho-PLD (81) and PtdCho-PLC (25) have been explored as molecular targets for anticancer therapy as outlined in section 2.4. Additional insight into the enzymes that are altered in choline metabolism in cancer can be found in recent reviews (4, 95, 96).
2.3 Oncogenes and signaling pathways arriving at cancer-phenotypic endpoints of choline metabolism
Multiple growth factors, cytokines, oncogenes, and chemical carcinogens have the ability to regulate and impact upon the enzymes in choline phospholipid metabolism (4, 97, 98). Cancer cells display distinct phenotypic endpoints in choline metabolism characterized by high PC, relatively high GPC, and high tCho as a result of deregulated oncogenic signaling pathways that impact upon the enzymes involved in choline metabolism, some of which are outlined below. While GPC is the dominating signal in normal breast and ovarian epithelium, PC is higher than GPC in ovarian and breast cancer cells (24, 25). Carcinogenesis of prostate epithelium results in elevated PC and GPC levels (26).
Some of the most important players in deregulated oncogenic signaling cascades are mutant ras oncogenes. Transformation with the H-ras oncogene resulted in an inhibitory effect on CCT (87, 88) as shown in Figure 4. Ras oncogene transformation of NIH 3T3 fibroblasts decreased PtdCho-PLC activity, and correlated with PtdCho-PLC translocation from the cytosol to the plasma membrane (90). Gain-of-function mutations in Ras can activate PtdCho-PLD, mediated by positive regulatory pathways through RAL-GDS and phosphatidylinositol 3-kinase (PI3K), and negative feedback signaling through RAF-1 and RAL-GDS (99). Mutant ras oncogene transformation caused an increase in CHK-α activity, which correlated with increased PC levels (100). Ras-mediated regulation of CHK-α also involved RAL-GDS and PI3K (101). Taken together, the drastic increase that can be observed in cellular PC levels following mutant ras-oncogene transformation (88, 102) is most likely due to the combined effects of oncogenic RAS protein regulation of CHK-α (101, 103), CCT (87, 88), PtdCho-PLD (99), and PtdCho-PLC (90) activity.
Mitogen activated protein kinases (MAPK) such as extracellular signal-regulated kinases (ERK), which are activated by phosphorylation, are frequently deregulated in several types of cancer, and are currently being explored as drug targets (104). Inhibition of ERK1/2 phosphorylation resulted in a significant drop in the PC levels of cancer cells (104), indicative of a positive correlation between ERK activation and high PC levels as shown in Figure 4. CHK-α silencing significantly reduced phosphorylated ERK levels, and was able to attenuate MAPK signaling (105). PC may serve as a noninvasive pharmacodynamic MRS-detected marker for monitoring MAPK signaling blockade (104).
The PI3K pathway is deregulated in most cancer, and is a major target for cancer drug development (106, 107). PI3K inhibition resulted in a concentration- and time-dependent decrease in PC and tCho, most likely due to a decreased in CHK-α protein expression levels (106, 107), as indicated in Figure 4. MRS monitoring of the effects of PI3K inhibitors may provide a noninvasive pharmacodynamic biomarker for PI3K inhibition (106, 107). The reversal of this pathway was demonstrated when CHK-α silencing decreased cellular PC, PtdCho, and phosphatidic acid levels as well as signaling through the PI3K/AKT pathway, with reduced levels of phosphorylated AKT detected following CHK-α silencing (105).
A recent study has demonstrated that the transcription factor hypoxia-inducible factor (HIF)-1 was able to bind to the putative Chk-α promoter regions and increase CHK-α expression along with cellular PC and tumor tCho levels detected by MRS/I (108). HIF-1 alpha is stabilized under hypoxic conditions, which can frequently arise within the tumor microenvironment, and enables cancer cells to adapt to hypoxic conditions by inducing the expression of a multitude of genes. Additional HIF-1 responses in cancer cells are discussed in section 3.
Expression of the Her-2 oncogene in nonmalignant human mammary epithelial cells significantly increased PC and tCho levels (24). An interesting link was recently revealed between PtdCho-PLC and Her-2, where inhibition of PtdCho-PLC was able downregulate HER-2 overexpression on the plasma membrane of breast cancer cells (109). Additional oncogenic signaling pathways affecting enzymes in choline metabolism are currently under intense investigation and were recently reviewed in (4, 95, 96).
The connection between oncogenic signaling pathways and choline metabolism makes it possible to use the tCho and PC signals as surrogate markers for molecular therapies that target these oncogenic signaling pathways. For example, a decrease of tCho and PC was observed upon targeting mitogen activated protein kinase (MAPK) (104), fatty acid synthase (110), and BCR-ABL tyrosine kinase (111). However, PC levels increased following histone deacetylase (HDAC) inhibition (112), demonstrating that the treatment effects on choline metabolism when targeting signaling pathways must be carefully examined before designating noninvasive surrogate markers such as PC and tCho for these novel molecular therapies. An overview of the key oncogenic pathways affecting enzymes in choline metabolism is shown in the right part of Figure 4. Several oncogenic signaling pathways that affect choline phospholipid metabolic enzymes can also modulate the expression and/or activity of enzymes in glucose metabolism as outline in section 3.4.
2.4 Targeting of choline metabolism for cancer therapy
CHK-α (4, 16-19, 92-94) and PtdCho-PLD (81) and PtdCho-PLC (25) have been explored as molecular targets for anticancer therapy, and have been silenced or inhibited in studies of MRS-monitored, targeted anticancer therapies. Downregulation or inhibition of CHK(-α) or PtdCho-PLD can be detected noninvasively with 31P or 1H MRS (25, 79, 92, 93). Preclinical anticancer treatments with CHK inhibitors caused tumor growth arrest and apoptosis (19, 92, 94), and silencing CHK-α with siRNA significantly reduced breast cancer cell proliferation and increased differentiation (16). A recent study showed that longitudinal noninvasive single-voxel 31P MRS monitoring of tumoral PC and PME levels was feasible to assess successful gene therapy with an intravenously injected lentiviral vector, which was able to deliver CHK-α-specific short-hairpin RNA (shRNA) in a preclinical breast cancer model (93). A more extensive review of targeting choline metabolism for cancer therapy can be found in recent reviews on his topic (4, 95, 96).
3. Key molecules in the altered glucose metabolism in cancer
A common feature across most, if not all, cancer types is abnormal glucose metabolism. Cancer cells typically exhibit a high rate of glucose uptake and utilization. In contrast to normal cells, glucose is converted to lactate also under aerobic conditions. This phenomenon was first studied by Otto Warburg in the 1920’s and is therefore known as the Warburg effect (113, 114). As the connections between malignant transformation and cellular metabolism on the molecular level gradually have been uncovered, this field of cancer research has enjoyed increased attention in recent years. This research has demonstrated upstream regulation of several metabolic pathways by major oncogenes such as Myc and PI3K/Akt, and that abnormalities in these metabolic systems therefore are interconnected (115, 116). These metabolic features of cancer cells both facilitate cell survival in hypoxic environments and provide a basis for increased cell proliferation both with respect to energy demand and cellular building blocks. Another factor that enables the cancer cells to adapt to hypoxic conditions is the hypoxia-inducible transcription factor (HIF-1). Stabilization of this factor induces the expression of several genes involved in glucose metabolism, angiogenesis and other mechanisms.
The high uptake of glucose and the extensive production of lactate make it feasible to use MRS in the assessment of glucose metabolism. Both glucose and lactate can be observed in proton spectra. In 13C MRS, assessment of glucose and lactate is typically facilitated through the use of isotopically enriched substrate. The most widely used substrate is [1-13C] glucose, which give rise to two separate resonances from the anomers α-glucose (94.8 ppm) and β-glucose (98.7 ppm). The isotopical enrichment is retained throughout glycolysis, resulting in the formation of [3-13C] lactate (22.8 ppm) and [3-13C] alanine (19.0 ppm). In addition, [2-13C] acetyl-CoA enters the TCA cycle giving rise to [4-13C] glutamate and other enriched TCA intermediates.
MRS provides unique possibilities for noninvasive studies of the flux through metabolic pathways in tumor cells. The microenvironment of solid tumors and the regulation of metabolic pathways are closely associated. MRS has an advantage compared to classical biochemical methods in that both spatial and temporal variation in metabolic activity can be studied in in vivo. This has led to clinical exploitation of the altered glucose metabolism in cancer management (117, 118), but MRS is also a versatile research tool. Glucose metabolism in cancer cells has been studied for more than two decades using 13C-labeled substrates (119, 120), allowing detailed description of the metabolic fate of glucose in various experimental systems.
3.1 Magnetic resonance spectroscopy techniques for detecting and differentiating cancer based on glucose metabolism
Glucose metabolism in cancer can be studied using various MRS techniques. High resolution MRS can be applied to cell cultures, tissue samples and extracts. Proton spectroscopy is routinely used in cancer diagnosis and management, and assessment of lactate production is an important parameter in brain cancer. Other metabolites in the glycolysis are present in low concentrations and are seldom considered as biomarkers in proton spectroscopy. Most studies of the glycolytic pathway use 13C MRS for tracing the metabolic fate of [1-13C] glucose or other isotopically enriched substrates. 13C MRS has also been used to study glucose metabolism in vivo (121, 122). Following administration of 13C-enriched substrates to tumor-bearing animals, the fate of the substrates can be studied taking tumor microenvironmental factor into account. However, 13C MRS is not yet routinely employed in clinical diagnosis or management of cancer. This may change as new spectroscopic techniques, such as hyperpolarised 13C MRSI, are further developed.
3.1.1 Clinical 1H MRS/I of glucose metabolism
Clinical 1H MRS has proven valuable in assessment of several cancer types due to tissue-specific changes in metabolite pattern (123). However, assessment of glycolysis using 1H MRS is difficult in several cancer types, as concentrations of most glycolytic metabolites cannot be measured. The exception is lactate, which may be present in measurable concentrations in cancer. Therefore, 1H MRS is widely used in diagnosis, staging and treatment monitoring of brain cancer. Lactate is present in minute concentrations in normal brain tissue, but reaches high concentrations in cancer due to abnormal glycolytic activity. Together with decrease in N-acetyl-aspartate (NAA) due to displacement of healthy neurons, lactate is a functional biomarker for brain cancer.
Typically, 1H MRS/I is used in combination with MR imaging for diagnostic purposes. The relative intensities of lactate, choline and NAA can be used to distinguish between cancer, benign lesions and post-surgery lesions in the brain. High levels of lactate are associated with high-grade tumors, allowing grading of brain tumors based on metabolite concentrations (124). High resolution MRSI may also aid in tumor delineation, which is a key factor in brain tumor surgery planning (125, 126).
In situations where in vivo does not allow evaluation of lactate concentration, ex vivo analysis of biopsy tissue may be an alternative approach. Great care must be taken in sample collection and handling to avoid anaerobic glucose metabolism (127). However, provided that samples are collected, stored and prepared for analysis under controlled conditions, the concentration of lactate can be accurately determined ex vivo using HR MAS MRS (7). In prostate cancer, significant differences in lactate and alanine concentration between benign lesions and prostate cancer biopsies has been reported (128).
3.1.2 Regulation of glycolytic activity studied by 13C MRS
13C MR spectroscopy is a powerful tool for studies of metabolic pathways. The natural abundance of 13C is low (1.1%), which gives this technique low inherent sensitivity. However, this can be turned into an advantage by use of isotopically enriched substrates. The metabolism of 13C-labeled substrates can be traced in both in cultured cells and in vivo, as the fate of the incorporated 13C can be monitored over time with little interference from background signals. The large chemical shift range for carbon allows resolution and identification of metabolites throughout entire metabolic pathways.
The abnormal glucose metabolism in cancer has been studied by 13C MRS for more than two decades (120). For studies of glucose uptake, glycolytic flux and lactate formation, [1-13C] glucose or [1,6-13C]glucose is typically used. These isomers allow evaluation of the uptake and degradation of glucose, as well as the formation of metabolic intermediates and ultimately the formation and elimination of [3-13C] lactate. The rate of glycolysis can be quantitatively estimated through compartmental modeling, which is advantageous in studies of drug efficacy (129).
Initial studies in this field verified high flux through glycolysis in cancer cells (120, 130-133). In vivo investigation of the effect of tumor hemodynamics, hypoxia and substrate supply, has confirmed that high lactate concentrations principally are associated with aerobic glycolysis, not inadequate perfusion (134). It has also repeatedly been demonstrated that the rate of glycolysis can be modulated through cytotoxic drugs or, in the case of breast cancer, endocrine treatment (135-137). The effect of direct glycolytic inhibitors has also been investigated using 13C MRS. Through dynamic studies of both cultured cells and culture medium, the kinetics of glucose metabolism can be studied, including the rates of glucose uptake and lactate secretion (138). 13C MRS of tumors has the advantage of allowing longitudinal studies of glucose metabolism in vivo. In some applications, however, limitations in sensitivity or quantitative performance suggest the use of other methods. Glucose metabolism in tumors may in such cases be evaluated ex vivo using 13C HR MAS MRS. After administration of 13C-enriched tracers to the tumor-bearing animals and subsequent harvesting of tumor tissue, samples are available for immunohistochemistry or gene expression analyses after acquiring the 13C spectra (139).
Overall, 13C MRS is a versatile technique for evaluating the effect of drugs on glucose metabolism in cancer. Targeting the metabolic abnormalities of cancer with new drug entities is likely to benefit from 13C MR spectroscopy as it allows noninvasive monitoring of surrogate biomarkers for therapeutic effect.
The low sensitivity constitutes a problem for clinical use of 13C MRS. However, through optimized MRS protocols and injection of [1-13C] glucose, it has been shown that the kinetics of lactate formation in brain cancer can be investigated (140). Compared to 1H MRS of brain cancer, this approach may be advantageous as it allows assessment of lactate formation rather than just presence of lactate in tumor tissue. The rate of lactate formation reflects the glycolytic capacity of the cancer cells, and are therefore suggested to be a better biomarker for grading brain tumors (134). In order to minimize the dose of [1-13C] glucose needed, polarization transfer pulse sequences and high field strengths (3T and above) are preferential. Although the method described by Wijnen et al suffers from relatively low spatial (50 cm3) and temporal (2.3 min) resolution, it can be used to demonstrate that the rate of lactate formation in tumor tissue is higher than in normal brain tissue.
The potential clinical value of 13C MRS has also been demonstrated ex vivo in biopsies from prostate, lung and colon cancer. In natural abundance spectra, cancer-specific changes in various metabolites were found, allowing differentiation of tumor tissue from that of adjacent non-malignant tissue (141). Although similar data may be obtained more conveniently using proton MRS, the amount of information in 13C spectra could theoretically contain information that is not available in proton spectra. When applying multivariate analysis methods to metabolite patterns, 13C MRS data could potentially add statistical power and be used either alone or in combination with proton MRS data to resolve subtle differences between tissues.
3.1.3 Hyperpolarized MRS
Through the introduction of dynamic nuclear polarization (DNP), the interest in studies of glucose metabolism using MRS has been greatly increased. This technique can increase the signal-to-noise ratio by more than 50,000-fold, using hyperpolarized 13C-labeled substrates (142, 143). At 1.5T, the polarization level of 13C at thermal equilibrium is as low as 1×10−6. Using a relatively rapid hyperpolarization procedure, polarization levels of >2×10−1 can be achieved (143). The substrate to be hyperpolarized is present in the solid state and doped with a single-electron containing compound. At low temperatures (1K), the electrons can be highly polarized due to their high gyromagnetic ratio. In DNP, this polarization is transferred to coupled nuclear spins by microwave irradiation near the resonance frequency of the electrons. The hyperpolarization is retained when the material is rapidly heated and dissolved, producing an injectable liquid within few seconds.
Due to time constraints and specific properties of hyperpolarized material, dynamic MRSI of hyperpolarized substrates offers challenges, which typically are absent in spectroscopic imaging at thermal equilibrium. Firstly, the time window for imaging is restricted to two to three times the T1 relaxation time. This means that hyperpolarized 13C MRSI must be completed within a few minutes after preparation of the tracer. Secondly, the longitudinal magnetization that has been created in the hyperpolarization procedure is not recovering during imaging. Thus, in addition to the inevitable loss due to T1 relaxation, the RF pulses applied in imaging convert longitudinal magnetization to transverse magnetization. In order to minimize magnetization loss, two imaging strategies are employed. Through the use of rapid low flip-angle RF pulses, only a small fraction of longitudinal magnetization is destroyed by each pulse. In addition, as much signal as possible is acquired per excitation through the use of rephasing pulses (144). Another issue is that imaging metabolic conversion is associated with rapid changes in signal. In sequences that use multiple excitations, these changes may convolve into k-space and cause image artifacts (145). Due to the very low interference from background and the more than 50,000-fold increase in signal after DNP, the signal-to-noise ratio is very high in the initial phase of hyperpolarized 13C MRSI. As hyperpolarized 13C imaging primarily is restricted by time and not by signal strength, EPI-based readout has often been found suitable (146). By using metabolite-specific low flip-angle excitation pulses, adiabatic refocusing pulses and echo planar readout, hyperpolarized 13C MRSI is performed in small animals with voxel sizes in the range of 0.135 cm3 with a temporal resolution of around 5 seconds, as shown in an example of a preclinical prostate cancer model in Figure 5 (146, 147).
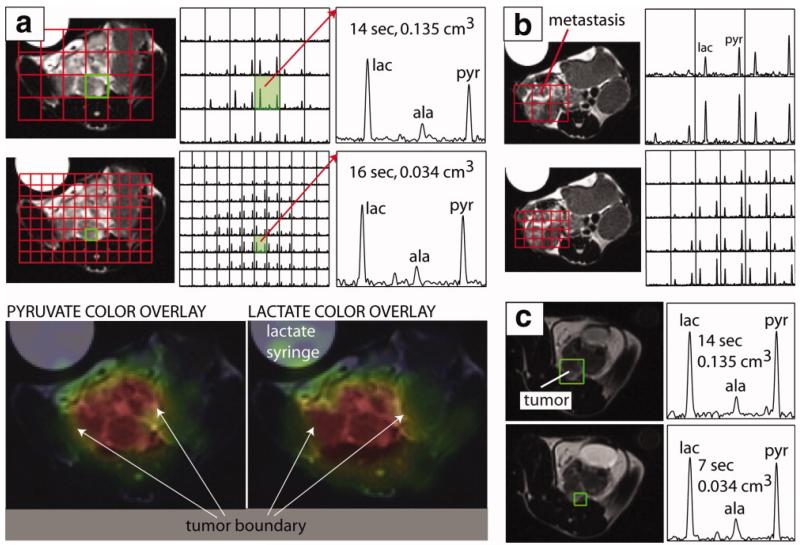
Figure 5.
In vivo MRSI of a transgenic murine mouse model of prostate cancer showing the effect of a compressed sensing imaging protocol. Pyruvate, lactate and alanine were detected in voxels down to 0.034 cm3 with TR=7s. The color overlay maps generated from the accelerated spectra show high intensity regions as brightly colored and highlight the spatial localization of metabolites according to tissue type. Tumor and non-tumor regions showed marked differences in metabolic profile. (a), (b) Comparison of spectra from un-accelerated (top panel) and accelerated (bottom panel) acquisitions with similar scan time demonstrating how increased spatial resolution can characterize metabolic heterogeneity and define tumor boundaries. (c) Comparison of spectra from un-accelerated and accelerated acquisitions demonstrating a 4-fold increase in spatial resolution and a 2-fold decrease in acquisition time. Figure adapted from (146) with permission.
The first substrate to be proven useful in characterization of cancer was [1-13C] pyruvate (148). For both physiochemical and biochemical reasons, this is an attractive substrate for 13C enrichment. Biologically, pyruvate is the end product of glycolysis and a starting point for the TCA cycle, lactate production and alanine production. Among its desirable physiochemical properties is its compatibility with the DNP hyperpolarization method. Pyruvate forms an amorphous solid at low temperatures, which, after hyperpolarization can be readily dissolved in a suitable heated buffer (144). In addition, the signal from the C-1 carbon of both pyruvate, lactate and alanine has a long T1 (approximately 45 seconds in blood) (149). Thus, longitudinal magnetization of [1-13C] pyruvate is maintained for a clinically relevant period of time, allowing spectroscopic imaging of pyruvate uptake and metabolism in distal tissues. In humans, intravenously injected substances reach all major organs within approximately 15-40 seconds (144). Pyruvate is rapidly taken up and metabolized by most organs. After uptake, the pyruvate metabolite pattern is dependent on the nature of the tissue. In tissues where [1-13C] pyruvate predominantly is feeding into the TCA cycle, it is decarboxylated, producing acetyl-CoA. The resulting generation of H13CO 3− can be assessed in real-time using MRS (150). In muscle tissue, transamination of [1-13C] pyruvate leads to formation of [1-13C] alanine, whereas its predominant metabolic fate in cancer is reduction to [1-13C] lactate (148, 151). The rate of [1-13] lactate formation depends on LDH expression and the endogenous pyruvate and lactate concentrations, and application of dynamic modeling has suggested that the rate of conversion may be associated with tumor grade (152, 153). The conversion rate is dose-dependent, which must be taken into account during interpretation of MRSI data following [1-13C] pyruvate injection (153). The blood-brain barrier (BBB) may be a limiting factor for pyruvate uptake in healthy brain tissue within the time frame of hyperpolarized MRSI. However, in xenografted gliomas, it has been shown that uptake and lactate production differs significantly from normal tissue (154). This is assumed to be an effect of disrupted BBB in tumor tissue.
Primarily, [1-13C] pyruvate has been shown to be useful in distinguishing tumor tissue from healthy surrounding tissue, both in prostate cancer, brain cancer and sarcoma. However, as treatment with cytotoxic drugs may reduce the glycolytic rate in cancer, hyperpolarized [1-13C] pyruvate could also potentially be used in therapy monitoring. This was first shown in a solid murine lymphoma model, where the rate of [1-13C] lactate formation and the LDH activity was significantly decreased after treatment with etoposide (155). The potential value of hyperpolarised 13C MRS in monitoring the effect of drugs targeting metabolic pathway has recently been demonstrated by Ward et al, who showed that inhibition of PI3K by LY294002 or everolimus caused a decrease in [1-13C] lactate formation both in glioblastoma (in vitro and in vivo) and breast cancer (in vitro) (156). This reduction was associated with decreased HIF-1 expression and LDH activity, in accordance with downstream effects of PI3K inhibition.
Although [1-13C] pyruvate is well suited for studies of glucose metabolism in cancer, several other substrates have also been suggested. One example is [2-13C] fructose, which can reach sufficient polarization levels through DNP and has a T1 which allows hyperpolarised MRSI studies. Fructose has an early entry into glycolysis through the action of hexokinase, yielding fructose-6-phosphate. In a transgenic mouse model of prostate cancer, it was shown that hyperpolarized [2-13C] fructose was taken up by both benign and malignant tissue, but that significant conversion to [2-13C] fructose-6-phosphate only occurred in voxels containing malignant tissue (157). Another potential marker of tumor response to therapy is [1,4-13C] fumarate. In a murine lymphoma model, the production of [1,4-13C] malate was significantly increased 24 hr after administration of etoposide (158). This change correlated with loss of plasma membrane integrity, suggesting the use of [1,4-13C] fumarate for early detection of cell death following chemotherapy.
For concurrent assessment of different metabolic pathways and tumor microenvironment, several substrates may be hyperpolarized, injected and imaged at the same time. Simultaneous hyperpolarization of [1-13C] pyruvate, H13CO3− (allowing tumor pH measurements), [1,4-13C] fumarate and 13C urea (allowing evaluation of tumor perfusion) has been reported, providing a multimodal approach to evaluation of tumor aggressivity and response to therapy (159).
In summary, labeling and hyperpolarizing endogenous or exogenous substrates with 13C in specified locations allow downstream tracing of metabolic within clinically relevant timeframes. The polarization is retained through serial metabolic conversions, and the ability to image metabolic fluxes is in principle limited only by the longitudinal relaxation (160). This technology is rapidly developing and holds the potential to revolutionize the use of MRSI in molecular imaging of cancer. The method has been applied to a wide range of preclinical applications, and clinical studies have recently been initiated. Due to the combination of endogenous tracers, high sensitivity, non-invasiveness and absence of ionizing radiation, it is expected that hyperpolarized MRSI will be a valuable supplement to existing imaging modalities in the management of cancer.
3.2 Altered enzymes in glycolysis and tricarboxylic acid cycle (TCA) of cancers
The altered glucose metabolism in tumors involves several different processes. Firstly, cancer cells exhibit enhanced glucose uptake through upregulation of facilitative glucose transporters, primarily GLUT1. Overexpression of glucose transporters has been shown in most cancers (161). It has also been demonstrated that glucose transport rates are higher in cancer cell lines than normal cell lines even at the same rates of proliferation, suggesting an association between malignant transformation and increased glucose consumption (162).
Several enzymes in glycolysis have also been shown to be generally upregulated in cancer (163). Both the rate-limiting enzymes 6-phosphofructokinase (PFK1) and pyruvate kinase (PK) as well as concentration-dependant enzymes such as hexokinase 2 (HK2), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), triose-phosphate isomerase (TPI), phosphoglycerate kinase 1 (PGK1) and enolase 1 (ENO1) are upregulated, allowing a sustainable high glycolytic flux.
Increased lactate production is a prerequisite for sustaining a high glycolytic flux. Conversion of pyruvate regenerates NAD+ from NADH, avoiding the depletion that otherwise would occur as a result of glycolytic NAD+ consumption. This is facilitated through upregulation of lactate dehydrogenase A (LDHA). Thus, the net effect of aerobic glycolysis in cancer cells is the formation of large amounts of lactate, which is present inside the cells in abnormally high concentrations and secreted through the action of MCT4, a monocarboxylic acid transporter, which also is upregulated in cancer (164, 165).
3.2.1 Role of the TCA cycle in cancer
Although cancer cells exhibit a significant increase in lactate production from pyruvate, their switch to aerobic glycolysis is not complete. A small fraction of their pyruvate is metabolized via the mitochondrial tricarboxylic acid (TCA) cycle (166). This generates some ATP, but more importantly also supplies the cancer cells with precursor molecules for pathways that consume TCA cycle intermediates. Cancer cells also typically use glutamine as a carbon source for anaplerosis. Using 13C MRS, it has been shown that glutamine catabolism exceeds the nitrogen requirements for nucleotide production. Instead, glutamine provides a supplementary carbon source allowing tumor cells to use glucose-derived carbon and TCA cycle intermediates for biosynthesis of fatty acids and other macromolecules (166). Although the flux trough the TCA cycle is relatively smaller than the flux through glycolysis in many cancers, their TCA cycle is functionally intact and the enzymes of the TCA cycle are not generally dysregulated in cancer. Interesting exceptions are succinate dehydrogenase and fumarate hydratase, which have been established as tumor suppressors (167). Mutations in these genes have been shown to cause certain cancers such as paraganglioma, phaeocytochromas and renal cell carcinoma (168-170). Intriguingly, these cancers conform to Otto Warburg’s original hypothesis that mitochondrial defects are a root cause of cancer.
3.3 Oncogenes and signaling pathways arriving at cancer-phenotypic endpoints of glucose metabolism
The increased glycolysis in cancer is associated with different regulation mechanisms, which are displayed in Figure 4. Firstly, glycolytic enzymes are induced by abnormal activity in several different groups of oncogenes. Secondly, stabilization of HIF-1 may also contribute to this enzyme induction. The relative role of these two systems are not completely clear, but it should be noted that cancer cells which do not experience hypoxia, such as leukemic cells which reside in the blood stream under high oxygen tension, may be highly glycolytic (115). Thus, HIF-1 may not itself contribute to the switch from normal metabolism to extensive aerobic glycolysis, but is rather a mechanism for sustaining high glycolytic flux and cellular proliferation rate under hypoxic conditions.
3.3.1 PI3K/AKT/mTOR, c-Myc, and p53 as regulators of glycolytic activity
In normal cells, glycolysis is tightly controlled through feedback mechanisms such as inhibition of PFK-1 and PK by ATP. In cancer cells, a coordinated regulation of proteins controlling glycolysis is frequently brought about through the action of upstream signaling pathways. Most of the regulatory mechanisms that increase glucose metabolism in cancer cells involve mutations in the PI3K/Akt pathway, Myc or p53, all frequently occuring tumorigenic mutations in cancer.
Mutations that enhance PI3K signaling is a common feature of many cancer types, and amplification of its downstream effectors AKT and mammalian target of rapamycin (mTOR) regulates several cellular responses to growth factor stimulation, which in itself may be sufficient to switch the metabolism from mitochondrial respiration to aerobic glycolysis (115). AKT signaling induces expression of GLUT1 trough the action of mammalian target of rapamycin (mTOR), and also activates HK2 association with mithochondria promoting phosphorylation of glucose (171, 172).
Another oncogenic transcription factor that has been associated with increased glycolytic flux is c-Myc. Increased c-Myc expression upregulates lactate dehydrogenase A (LDHA), which in turn converts pyruvate to lactate. It has also been shown that c-Myc directly regulates other genes in the glycolysis such as GLUT1, HK2, PFK1 and enolase 1 (ENO1) (173, 174).
A major tumor suppressor gene, p53, has also recently been shown to be involved in cancer cell glucose metabolism. Although mechanisms are not completely elucidated, mutations in this gene modulate the balance between oxidative phosphorylation and glycolysis through various transcription factors (175).
3.3.2 HIF-1 as regulator of glycolytic activity
Cancer cells are frequently exposed to decreased oxygen availability. The HIF-1 transcription factor complex is activated in hypoxia and coordinates several adaptive mechanisms. In order to reduce oxygen consumption requirements by reducing oxidative phosphorylation and increasing lactate production, HIF-1 stimulates LDHA activity and reduces pyruvate dehydrogenase-kinase 1 (PDK1) activity. In addition, HIF-1 stimulates expression of both glucose transporters and glycolytic enzymes (176). Several enzymes in the glycolysis are positively regulated by both HIF-1 and c-MYC, potentially making them attractive targets for therapeutic drugs (116).
In summary, mutations in major oncogenes and tumor suppressor genes stimulate glucose transport, increases flux through glycolysis and suppress the entry of pyruvate into the TCA cycle. This abnormal regulation of glucose metabolism is specific for cancer cells, as they acquire a self-sustainable anabolic phenotype that is not depending on extrinsic growth factors.
3.4 Targeting of glucose metabolism for cancer therapy
It may seem contradictory for tumor cells to generate energy through lactate production when oxygen levels would allow oxidative phosphorylation. However, the above-mentioned changes in glycolytic activity all contribute to a significantly increased glycolytic flux in cancer cells. This means that aerobic glycolysis may provide adequate amounts of energy to cancer cells while also providing essential precursors for biosynthesis (115, 177). In addition, the cancer cells gain an increased probability of surviving periods of limited oxygen supply. Secretion of lactate may also play a role in blocking immune responses and aid in invasion of neighbouring tissue (178).
As the high level of aerobic glycolysis appears to be beneficial for the survival and proliferation of tumor cells, metabolic pathways have recently been recognized as attractive drug targets. One possible approach in this respect is the direct targeting of enzymes involved in glucose metabolism. Inhibition of glucose transport has negative effects on tumor growth in vivo, but has currently no clinical utility (179). Several hexokinase inhibitors have also entered clinical trials based on encouraging findings in vitro and in vivo (180-183). Glycolytic inhibitors have been shown to sensitize tumors to other chemotherapeutic agents, providing a new possible approach to treatment of chemoresistant cancers (184-186). Currently, their role in cancer management is unclear. Ongoing clinical studies will hopefully provide new insight in the role of this drug class.
It has become increasingly evident that multiple genetic and epigenetic alterations are a major mechanism in cancer development, and that these alterations in many cases contribute to altered glucose metabolism. This has resulted in development of drugs that target upstream regulation of metabolic pathways rather than effector enzymes. An example is imatinib, which inhibits the chronic myeloid leukemia-specific oncogene product BCR-ABL. Among other effects, imatinib decreases HK and G6PD activity, thereby suppresseing aerobic glycolysis and depriving cells of metabolic intermediates (187). Currently, drugs targeting PI3K, mTOR, AKT and HIF-1 are in clinical trials, suggesting that inhibition of oncogenic signalling pathways may be an important approach to treatment of cancer in the near future (188).
Development of drugs that directly or indirectly inhibit the increased glucose metabolism is likely to benefit from noninvasive techniques, such as MRS, for assessment of drug efficacy. In preclinical drug development, MRS has been frequently used for studies of glucose metabolism kinetics in cell cultures and in vivo model systems (138, 156, 189, 190). An increasing number of studies suggest that MRS also may play a role in clinical cancer management through direct assessment of glucose metabolism (128, 140, 191). The current interest in targeting of oncogenic signalling pathways regulating cancer glycose metabolism combined with the developments in hyperpolarized 13C MRSI for molecular imaging suggest an important role for use of MRS in cancer management in the future.
4. Conclusions
Versatile MRS techniques are currently already available to detect a multitude of metabolic information noninvasively in vivo, and in HR MRS applications ex vivo. The metabolic information provided by MRS can give important information on the molecular make-up of tumors, which is crucial in the ongoing transition of clinical cancer care towards personalized molecular medicine. The information contained in the MRS-detected metabolome can be utilized to aid in the diagnosis of cancer, to help select appropriate anticancer therapies, to monitor the tumor response to anticancer treatments, and to develop and evaluate novel targeted molecular therapies. Particularly in the development of novel anticancer therapies that target enzymes in choline and glucose metabolism, MRS techniques are currently proving extremely valuable, with a high potential for clinical translation. Certain MRS-detected metabolites can also serve as biomarkers or surrogate markers for novel molecular therapies that target oncogenic signaling pathways that have a downstream effect on a given MRS-detected metabolite. MRS can easily be integrated with MRI applications that provide complementary information on tumor anatomy.
The limitations of MRS techniques lie in their relatively low detection sensitivity, which can result in insufficient spatial and spectral resolution for clinical cancer detection and treatment monitoring. However, the availability of ever increasing magnetic field strengths and inventions of novel techniques such as hyperpolarization are continuing to push the limits of currently available MRS techniques. With its unmatched ability to provide a multitude of molecular information, MRS applications will undoubtedly play an important role in the future of molecular and personalized cancer care.
5. Acknowledgements
We gratefully acknowledge funding provided by NIH R01 CA134695 and P50 CA103175, and the Norwegian Research Council (183379 and 186479).
Supported by NIH R01 CA134695 and P50 CA103175, and the Norwegian Research Council (183379 and 186479).
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Griffin JL, Kauppinen RA. Tumour metabolomics in animal models of human cancer. J Proteome Res. 2007;6:498–505. doi: 10.1021/pr060464h. [DOI] [PubMed] [Google Scholar]
- 4.Glunde K, Serkova NJ. Therapeutic targets and biomarkers identified in cancer choline phospholipid metabolism. Pharmacogenomics. 2006;7:1109–23. doi: 10.2217/14622416.7.7.1109. [DOI] [PubMed] [Google Scholar]
- 5.Costello LC, Franklin RB. ‘Why do tumour cells glycolyse?’: from glycolysis through citrate to lipogenesis. Mol Cell Biochem. 2005;280:1–8. doi: 10.1007/s11010-005-8841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffin JL, Kauppinen RA. A metabolomics perspective of human brain tumours. FEBS J. 2007;274:1132–9. doi: 10.1111/j.1742-4658.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- 7.Swanson MG, Zektzer AS, Tabatabai ZL, Simko J, Jarso S, Keshari KR, Schmitt L, Carroll PR, Shinohara K, Vigneron DB, Kurhanewicz J. Quantitative analysis of prostate metabolites using 1H HR-MAS spectroscopy. Magn Reson Med. 2006;55:1257–64. doi: 10.1002/mrm.20909. [DOI] [PubMed] [Google Scholar]
- 8.Ross B, Michaelis T. Clinical applications of magnetic resonance spectroscopy. Magn Reson Q. 1994;10:191–247. [PubMed] [Google Scholar]
- 9.Nelson SJ. Analysis of volume MRI and MR spectroscopic imaging data for the evaluation of patients with brain tumors. Magn Reson Med. 2001;46:228–39. doi: 10.1002/mrm.1183. [DOI] [PubMed] [Google Scholar]
- 10.Howe FA, Barton SJ, Cudlip SA, Stubbs M, Saunders DE, Murphy M, Wilkins P, Opstad KS, Doyle VL, McLean MA, Bell BA, Griffiths JR. Metabolic profiles of human brain tumors using quantitative in vivo 1H magnetic resonance spectroscopy. Magn Reson Med. 2003;49:223–32. doi: 10.1002/mrm.10367. [DOI] [PubMed] [Google Scholar]
- 11.Jenkinson MD, Smith TS, Joyce K, Fildes D, du Plessis DG, Warnke PC, Walker C. MRS of oligodendroglial tumors: correlation with histopathology and genetic subtypes. Neurology. 2005;64:2085–9. doi: 10.1212/01.WNL.0000165998.73779.D9. [DOI] [PubMed] [Google Scholar]
- 12.Kurhanewicz J, Vigneron DB, Hricak H, Narayan P, Carroll P, Nelson SJ. Three-dimensional H-1 MR spectroscopic imaging of the in situ human prostate with high (0.24-0.7-cm3) spatial resolution. Radiology. 1996;198:795–805. doi: 10.1148/radiology.198.3.8628874. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs MA, Barker PB, Bottomley PA, Bhujwalla Z, Bluemke DA. Proton magnetic resonance spectroscopic imaging of human breast cancer: a preliminary study. J Magn Reson Imaging. 2004;19:68–75. doi: 10.1002/jmri.10427. [DOI] [PubMed] [Google Scholar]
- 14.Meisamy S, Bolan PJ, Baker EH, Pollema MG, Le CT, Kelcz F, Lechner MC, Luikens BA, Carlson RA, Brandt KR, Amrami KK, Nelson MT, Everson LI, Emory TH, Tuttle TM, Yee D, Garwood M. Adding in vivo quantitative 1H MR spectroscopy to improve diagnostic accuracy of breast MR imaging: preliminary results of observer performance study at 4.0 T. Radiology. 2005;236:465–75. doi: 10.1148/radiol.2362040836. [DOI] [PubMed] [Google Scholar]
- 15.Stanwell P, Gluch L, Clark D, Tomanek B, Baker L, Giuffre B, Lean C, Malycha P, Mountford C. Specificity of choline metabolites for in vivo diagnosis of breast cancer using 1H MRS at 1.5 T. Eur Radiol. 2005;15:1037–43. doi: 10.1007/s00330-004-2475-1. [DOI] [PubMed] [Google Scholar]
- 16.Glunde K, Raman V, Mori N, Bhujwalla ZM. RNA interference-mediated choline kinase suppression in breast cancer cells induces differentiation and reduces proliferation. Cancer research. 2005;65:11034–43. doi: 10.1158/0008-5472.CAN-05-1807. [DOI] [PubMed] [Google Scholar]
- 17.Ramirez de Molina A, Gallego-Ortega D, Sarmentero J, Banez-Coronel M, Martin-Cantalejo Y, Lacal JC. Choline kinase is a novel oncogene that potentiates RhoA-induced carcinogenesis. Cancer research. 2005;65:5647–53. doi: 10.1158/0008-5472.CAN-04-4416. [DOI] [PubMed] [Google Scholar]
- 18.Ramirez de Molina A, Gutierrez R, Ramos MA, Silva JM, Silva J, Bonilla F, Sanchez JJ, Lacal JC. Increased choline kinase activity in human breast carcinomas: clinical evidence for a potential novel antitumor strategy. Oncogene. 2002;21:4317–22. doi: 10.1038/sj.onc.1205556. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Gonzalez A, Ramirez de Molina A, Fernandez F, Lacal JC. Choline kinase inhibition induces the increase in ceramides resulting in a highly specific and selective cytotoxic antitumoral strategy as a potential mechanism of action. Oncogene. 2004;23:8247–59. doi: 10.1038/sj.onc.1208045. [DOI] [PubMed] [Google Scholar]
- 20.Kurhanewicz J, Vigneron DB, Males RG, Swanson MG, Yu KK, Hricak H. The prostate: MR imaging and spectroscopy. Present and future. Radiol Clin North Am. 2000;38:115–38. viii–ix. doi: 10.1016/s0033-8389(05)70152-4. [DOI] [PubMed] [Google Scholar]
- 21.Meisamy S, Bolan PJ, Baker EH, Bliss RL, Gulbahce E, Everson LI, Nelson MT, Emory TH, Tuttle TM, Yee D, Garwood M. Neoadjuvant chemotherapy of locally advanced breast cancer: predicting response with in vivo (1)H MR spectroscopy--a pilot study at 4 T. Radiology. 2004;233:424–31. doi: 10.1148/radiol.2332031285. [DOI] [PubMed] [Google Scholar]
- 22.Baek HM, Chen JH, Nie K, Yu HJ, Bahri S, Mehta RS, Nalcioglu O, Su MY. Predicting pathologic response to neoadjuvant chemotherapy in breast cancer by using MR imaging and quantitative 1H MR spectroscopy. Radiology. 2009;251:653–62. doi: 10.1148/radiol.2512080553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer research. 2004;64:4270–6. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 24.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer research. 1999;59:80–4. [PubMed] [Google Scholar]
- 25.Iorio E, Mezzanzanica D, Alberti P, Spadaro F, Ramoni C, D’Ascenzo S, Millimaggi D, Pavan A, Dolo V, Canevari S, Podo F. Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer research. 2005;65:9369–76. doi: 10.1158/0008-5472.CAN-05-1146. [DOI] [PubMed] [Google Scholar]
- 26.Ackerstaff E, Pflug BR, Nelson JB, Bhujwalla ZM. Detection of increased choline compounds with proton nuclear magnetic resonance spectroscopy subsequent to malignant transformation of human prostatic epithelial cells. Cancer research. 2001;61:3599–603. [PubMed] [Google Scholar]
- 27.Kurhanewicz J, Swanson MG, Nelson SJ, Vigneron DB. Combined magnetic resonance imaging and spectroscopic imaging approach to molecular imaging of prostate cancer. J Magn Reson Imaging. 2002;16:451–63. doi: 10.1002/jmri.10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villeirs GM, Oosterlinck W, Vanherreweghe E, De Meerleer GO. A qualitative approach to combined magnetic resonance imaging and spectroscopy in the diagnosis of prostate cancer. Eur J Radiol. 2010;73:352–6. doi: 10.1016/j.ejrad.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 29.Villeirs GM, De Meerleer GO, De Visschere PJ, Fonteyne VH, Verbaeys AC, Oosterlinck W. Combined magnetic resonance imaging and spectroscopy in the assessment of high grade prostate carcinoma in patients with elevated PSA: A single-institution experience of 356 patients. Eur J Radiol. 2009 Sep 7; doi: 10.1016/j.ejrad.2009.08.007. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs MA, Barker PB, Argani P, Ouwerkerk R, Bhujwalla ZM, Bluemke DB. Combined dynamic contrast and spectroscopic imaging of human breast cancer. J Magn Reson Imaging. 2005;21:23–8. doi: 10.1002/jmri.20239. [DOI] [PubMed] [Google Scholar]
- 31.Katz-Brull R, Lavin PT, Lenkinski RE. Clinical utility of proton magnetic resonance spectroscopy in characterizing breast lesions. J Natl Cancer Inst. 2002;94:1197–203. doi: 10.1093/jnci/94.16.1197. [DOI] [PubMed] [Google Scholar]
- 32.Sinha S, Sinha U. Recent advances in breast MRI and MRS. NMR Biomed. 2009;22:3–16. doi: 10.1002/nbm.1270. [DOI] [PubMed] [Google Scholar]
- 33.Vilanova JC, Barcelo J. Prostate cancer detection: magnetic resonance (MR) spectroscopic imaging. Abdom Imaging. 2007;32:253–61. doi: 10.1007/s00261-007-9191-7. [DOI] [PubMed] [Google Scholar]
- 34.Oshiro S, Tsugu H, Komatsu F, Abe H, Onishi H, Ohmura T, Iwaasa M, Sakamoto S, Fukushima T. Quantitative assessment of gliomas by proton magnetic resonance spectroscopy. Anticancer Res. 2007;27:3757–63. [PubMed] [Google Scholar]
- 35.Server A, Josefsen R, Kulle B, Maehlen J, Schellhorn T, Gadmar O, Kumar T, Haakonsen M, Langberg CW, Nakstad PH. Proton magnetic resonance spectroscopy in the distinction of high-grade cerebral gliomas from single metastatic brain tumors. Acta Radiol. 2010;51:316–25. doi: 10.3109/02841850903482901. [DOI] [PubMed] [Google Scholar]
- 36.Zierhut ML, Ozturk-Isik E, Chen AP, Park I, Vigneron DB, Nelson SJ. (1)H spectroscopic imaging of human brain at 3 Tesla: comparison of fast three-dimensional magnetic resonance spectroscopic imaging techniques. J Magn Reson Imaging. 2009;30:473–80. doi: 10.1002/jmri.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gribbestad IS, Petersen SB, Fjosne HE, Kvinnsland S, Krane J. 1H NMR spectroscopic characterization of perchloric acid extracts from breast carcinomas and non-involved breast tissue. NMR Biomed. 1994;7:181–94. doi: 10.1002/nbm.1940070405. [DOI] [PubMed] [Google Scholar]
- 38.Cheng LL, Chang IW, Louis DN, Gonzalez RG. Correlation of high-resolution magic angle spinning proton magnetic resonance spectroscopy with histopathology of intact human brain tumor specimens. Cancer research. 1998;58:1825–32. [PubMed] [Google Scholar]
- 39.Cheng LL, Chang IW, Smith BL, Gonzalez RG. Evaluating human breast ductal carcinomas with high-resolution magic-angle spinning proton magnetic resonance spectroscopy. J Magn Reson. 1998;135:194–202. doi: 10.1006/jmre.1998.1578. [DOI] [PubMed] [Google Scholar]
- 40.Bolan PJ, Meisamy S, Baker EH, Lin J, Emory T, Nelson M, Everson LI, Yee D, Garwood M. In vivo quantification of choline compounds in the breast with 1H MR spectroscopy. Magn Reson Med. 2003;50:1134–43. doi: 10.1002/mrm.10654. [DOI] [PubMed] [Google Scholar]
- 41.Yeung DK, Cheung HS, Tse GM. Human breast lesions: characterization with contrast-enhanced in vivo proton MR spectroscopy--initial results. Radiology. 2001;220:40–6. doi: 10.1148/radiology.220.1.r01jl0240. [DOI] [PubMed] [Google Scholar]
- 42.Roebuck JR, Cecil KM, Schnall MD, Lenkinski RE. Human breast lesions: Characterization with proton MR spectroscopy. Radiology. 1998;209:269–75. doi: 10.1148/radiology.209.1.9769842. [DOI] [PubMed] [Google Scholar]
- 43.Bakken IJ, Gribbestad IS, Singstad TE, Kvistad KA. External standard method for the in vivo quantification of choline-containing compounds in breast tumors by proton MR spectroscopy at 1.5 Tesla. Magn Reson Med. 2001;46:189–92. doi: 10.1002/mrm.1175. [DOI] [PubMed] [Google Scholar]
- 44.Baik HM, Su MY, Yu H, Nalcioglu O, Mehta R. Quantification of Choline-containing Compounds in Malignant Breast Tumors by (1)H MR Spectroscopy Using Water as an Internal Reference at 1.5 T. Magma. 2006;19:96–104. doi: 10.1007/s10334-006-0032-4. [DOI] [PubMed] [Google Scholar]
- 45.Haddadin IS, McIntosh A, Meisamy S, Corum C, Styczynski Snyder AL, Powell NJ, Nelson MT, Yee D, Garwood M, Bolan PJ. Metabolite quantification and high-field MRS in breast cancer. NMR Biomed. 2009;22:65–76. doi: 10.1002/nbm.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jagannathan NR, Kumar M, Seenu V, Coshic O, Dwivedi SN, Julka PK, Srivastava A, Rath GK. Evaluation of total choline from in-vivo volume localized proton MR spectroscopy and its response to neoadjuvant chemotherapy in locally advanced breast cancer. Br J Cancer. 2001;84:1016–22. doi: 10.1054/bjoc.2000.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danishad KK, Sharma U, Sah RG, Seenu V, Parshad R, Jagannathan NR. Assessment of therapeutic response of locally advanced breast cancer (LABC) patients undergoing neoadjuvant chemotherapy (NACT) monitored using sequential magnetic resonance spectroscopic imaging (MRSI) NMR Biomed. 2010;23:233–41. doi: 10.1002/nbm.1436. [DOI] [PubMed] [Google Scholar]
- 48.Kurhanewicz J, Vigneron DB, Nelson SJ. Three-dimensional magnetic resonance spectroscopic imaging of brain and prostate cancer. Neoplasia. 2000;2:166–89. doi: 10.1038/sj.neo.7900081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mueller-Lisse UG, Swanson MG, Vigneron DB, Hricak H, Bessette A, Males RG, Wood PJ, Noworolski S, Nelson SJ, Barken I, Carroll PR, Kurhanewicz J. Time-dependent effects of hormone-deprivation therapy on prostate metabolism as detected by combined magnetic resonance imaging and 3D magnetic resonance spectroscopic imaging. Magn Reson Med. 2001;46:49–57. doi: 10.1002/mrm.1159. [DOI] [PubMed] [Google Scholar]
- 50.Kurhanewicz J, Vigneron DB, Hricak H, Parivar F, Nelson SJ, Shinohara K, Carroll PR. Prostate cancer: metabolic response to cryosurgery as detected with 3D H-1 MR spectroscopic imaging. Radiology. 1996;200:489–96. doi: 10.1148/radiology.200.2.8685346. [DOI] [PubMed] [Google Scholar]
- 51.Evelhoch JL, Gillies RJ, Karczmar GS, Koutcher JA, Maxwell RJ, Nalcioglu O, Raghunand N, Ronen SM, Ross BD, Swartz HM. Applications of magnetic resonance in model systems: cancer therapeutics. Neoplasia. 2000;2:152–65. doi: 10.1038/sj.neo.7900078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwarz AJ, Maisey NR, Collins DJ, Cunningham D, Huddart R, Leach MO. Early in vivo detection of metabolic response: a pilot study of 1H MR spectroscopy in extracranial lymphoma and germ cell tumours. Br J Radiol. 2002;75:959–66. doi: 10.1259/bjr.75.900.750959. [DOI] [PubMed] [Google Scholar]
- 53.deSouza NM, Soutter WP, Rustin G, Mahon MM, Jones B, Dina R, McIndoe GA. Use of neoadjuvant chemotherapy prior to radical hysterectomy in cervical cancer: monitoring tumour shrinkage and molecular profile on magnetic resonance and assessment of 3-year outcome. Br J Cancer. 2004;90:2326–31. doi: 10.1038/sj.bjc.6601870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng LL, Burns MA, Taylor JL, He W, Halpern EF, McDougal WS, Wu CL. Metabolic characterization of human prostate cancer with tissue magnetic resonance spectroscopy. Cancer research. 2005;65:3030–4. doi: 10.1158/0008-5472.CAN-04-4106. [DOI] [PubMed] [Google Scholar]
- 55.Sitter B, Lundgren S, Bathen TF, Halgunset J, Fjosne HE, Gribbestad IS. Comparison of HR MAS MR spectroscopic profiles of breast cancer tissue with clinical parameters. NMR Biomed. 2006;19:30–40. doi: 10.1002/nbm.992. [DOI] [PubMed] [Google Scholar]
- 56.Martinez-Bisbal MC, Marti-Bonmati L, Piquer J, Revert A, Ferrer P, Llacer JL, Piotto M, Assemat O, Celda B. 1H and 13C HR-MAS spectroscopy of intact biopsy samples ex vivo and in vivo 1H MRS study of human high grade gliomas. NMR Biomed. 2004;17:191–205. doi: 10.1002/nbm.888. [DOI] [PubMed] [Google Scholar]
- 57.Bathen TF, Jensen LR, Sitter B, Fjosne HE, Halgunset J, Axelson DE, Gribbestad IS, Lundgren S. MR-determined metabolic phenotype of breast cancer in prediction of lymphatic spread, grade, and hormone status. Breast Cancer Res Treat. 2007;104:181–9. doi: 10.1007/s10549-006-9400-z. [DOI] [PubMed] [Google Scholar]
- 58.Sitter B, Bathen TF, Singstad TE, Fjosne HE, Lundgren S, Halgunset J, Gribbestad IS. Quantification of metabolites in breast cancer patients with different clinical prognosis using HR MAS MR spectroscopy. NMR Biomed. 2010 Jan 25; doi: 10.1002/nbm.1478. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 59.Arias-Mendoza F, Zakian K, Schwartz A, Howe FA, Koutcher JA, Leach MO, Griffiths JR, Heerschap A, Glickson JD, Nelson SJ, Evelhoch JL, Charles HC, Brown TR. Methodological standardization for a multi-institutional in vivo trial of localized 31P MR spectroscopy in human cancer research. In vitro and normal volunteer studies. NMR Biomed. 2004;17:382–91. doi: 10.1002/nbm.915. [DOI] [PubMed] [Google Scholar]
- 60.Arias-Mendoza F, Payne GS, Zakian KL, Schwarz AJ, Stubbs M, Stoyanova R, Ballon D, Howe FA, Koutcher JA, Leach MO, Griffiths JR, Heerschap A, Glickson JD, Nelson SJ, Evelhoch JL, Charles HC, Brown TR. In vivo 31P MR spectral patterns and reproducibility in cancer patients studied in a multi-institutional trial. NMR Biomed. 2006;19:504–12. doi: 10.1002/nbm.1057. [DOI] [PubMed] [Google Scholar]
- 61.Griffiths JR, Tate AR, Howe FA, Stubbs M. Magnetic Resonance Spectroscopy of cancer-practicalities of multi-centre trials and early results in non-Hodgkin’s lymphoma. Eur J Cancer. 2002;38:2085–93. doi: 10.1016/s0959-8049(02)00389-1. [DOI] [PubMed] [Google Scholar]
- 62.Klomp DW, Wijnen JP, Scheenen TW, Heerschap A. Efficient 1H to 31P polarization transfer on a clinical 3T MR system. Magn Reson Med. 2008;60:1298–305. doi: 10.1002/mrm.21733. [DOI] [PubMed] [Google Scholar]
- 63.Wijnen JP, Scheenen TW, Klomp DW, Heerschap A. (31)P Magnetic resonance spectroscopic imaging with polarisation transfer of phosphomono- and diesters at 3 T in the human brain: relation with age and spatial differences. NMR Biomed. 2010 Jul 28; doi: 10.1002/nbm.1523. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 64.Payne GS, Troy H, Vaidya SJ, Griffiths JR, Leach MO, Chung YL. Evaluation of (31)P high-resolution magic angle spinning of intact tissue samples. NMR Biomed. 2006 doi: 10.1002/nbm.1040. [DOI] [PubMed] [Google Scholar]
- 65.De Silva SS, Payne GS, Thomas V, Carter PG, Ind TE, deSouza NM. Investigation of metabolite changes in the transition from pre-invasive to invasive cervical cancer measured using (1)H and (31)P magic angle spinning MRS of intact tissue. NMR Biomed. 2009;22:191–8. doi: 10.1002/nbm.1302. [DOI] [PubMed] [Google Scholar]
- 66.Gillies RJ, Barry JA, Ross BD. In vitro and in vivo 13C and 31P NMR analyses of phosphocholine metabolism in rat glioma cells. Magn Reson Med. 1994;32:310–8. doi: 10.1002/mrm.1910320306. [DOI] [PubMed] [Google Scholar]
- 67.Ronen SM, Degani H. The application of 13C NMR to the characterization of phospholipid metabolism in cells. Magn Reson Med. 1992;25:384–9. doi: 10.1002/mrm.1910250219. [DOI] [PubMed] [Google Scholar]
- 68.Ronen SM, Rushkin E, Degani H. Lipid metabolism in T47D human breast cancer cells: 31P and 13C-NMR studies of choline and ethanolamine uptake. Biochim Biophys Acta. 1991;1095:5–16. doi: 10.1016/0167-4889(91)90038-y. [DOI] [PubMed] [Google Scholar]
- 69.Ronen SM, Rushkin E, Degani H. Lipid metabolism in large T47D human breast cancer spheroids: 31P- and 13C-NMR studies of choline and ethanolamine uptake. Biochim Biophys Acta. 1992;1138:203–12. doi: 10.1016/0925-4439(92)90039-p. [DOI] [PubMed] [Google Scholar]
- 70.Katz-Brull R, Margalit R, Degani H. Differential routing of choline in implanted breast cancer and normal organs. Magn Reson Med. 2001;46:31–8. doi: 10.1002/mrm.1157. [DOI] [PubMed] [Google Scholar]
- 71.Artemov D, Bhujwalla ZM, Glickson JD. In vivo selective measurement of (1-13C)-glucose metabolism in tumors by heteronuclear cross polarization. Magn Reson Med. 1995;33:151–5. doi: 10.1002/mrm.1910330202. [DOI] [PubMed] [Google Scholar]
- 72.van Zijl PC, Chesnick AS, DesPres D, Moonen CT, Ruiz-Cabello J, van Gelderen P. In vivo proton spectroscopy and spectroscopic imaging of [1-13C]-glucose and its metabolic products. Magn Reson Med. 1993;30:544–51. doi: 10.1002/mrm.1910300504. [DOI] [PubMed] [Google Scholar]
- 73.Ramirez de Molina A, Rodriguez-Gonzalez A, Gutierrez R, Martinez-Pineiro L, Sanchez J, Bonilla F, Rosell R, Lacal J. Overexpression of choline kinase is a frequent feature in human tumor-derived cell lines and in lung, prostate, and colorectal human cancers. Biochem Biophys Res Commun. 2002;296:580–3. doi: 10.1016/s0006-291x(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 74.Ramirez de Molina A, Gutierrez R, Ramos MA, Silva JM, Silva J, Bonilla F, Sanchez JJ, Lacal JC. Increased choline kinase activity in human breast carcinomas: clinical evidence for a potential novel antitumor strategy. Oncogene. 2002;21:4317–22. doi: 10.1038/sj.onc.1205556. [DOI] [PubMed] [Google Scholar]
- 75.Nakagami K, Uchida T, Ohwada S, Koibuchi Y, Morishita Y. Increased choline kinase activity in 1,2-dimethylhydrazine-induced rat colon cancer. Jpn J Cancer Res. 1999;90:1212–7. doi: 10.1111/j.1349-7006.1999.tb00698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakagami K, Uchida T, Ohwada S, Koibuchi Y, Suda Y, Sekine T, Morishita Y. Increased choline kinase activity and elevated phosphocholine levels in human colon cancer. Jpn J Cancer Res. 1999;90:419–24. doi: 10.1111/j.1349-7006.1999.tb00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katz-Brull R, Degani H. Kinetics of choline transport and phosphorylation in human breast cancer cells; NMR application of the zero trans method. Anticancer Res. 1996;16:1375–80. [PubMed] [Google Scholar]
- 78.Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: molecular and biochemical studies. Int J Cancer. 2007;120:1721–30. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 79.Iorio E, Ricci A, Bagnoli M, Pisanu ME, Castellano G, Di Vito M, Venturini E, Glunde K, Bhujwalla ZM, Mezzanzanica D, Canevari S, Podo F. Activation of phosphatidylcholine cycle enzymes in human epithelial ovarian cancer cells. Cancer research. 2010;70:2126–35. doi: 10.1158/0008-5472.CAN-09-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Noh DY, Ahn SJ, Lee RA, Park IA, Kim JH, Suh PG, Ryu SH, Lee KH, Han JS. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett. 2000;161:207–14. doi: 10.1016/s0304-3835(00)00612-1. [DOI] [PubMed] [Google Scholar]
- 81.Foster DA, Xu L. Phospholipase D in cell proliferation and cancer. Mol Cancer Res. 2003;1:789–800. [PubMed] [Google Scholar]
- 82.del Peso L, Lucas L, Esteve P, Lacal JC. Activation of phospholipase D by growth factors and oncogenes in murine fibroblasts follow alternative but cross-talking pathways. Biochem J. 1997;322(Pt 2):519–28. doi: 10.1042/bj3220519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Balsinde J, Winstead MV, Dennis EA. Phospholipase A(2) regulation of arachidonic acid mobilization. FEBS Lett. 2002;531:2–6. doi: 10.1016/s0014-5793(02)03413-0. [DOI] [PubMed] [Google Scholar]
- 84.Laye JP, Gill JH. Phospholipase A2 expression in tumours: a target for therapeutic intervention? Drug Discov Today. 2003;8:710–6. doi: 10.1016/s1359-6446(03)02754-5. [DOI] [PubMed] [Google Scholar]
- 85.Gallazzini M, Ferraris JD, Burg MB. GDPD5 is a glycerophosphocholine phosphodiesterase that osmotically regulates the osmoprotective organic osmolyte GPC. Proc Natl Acad Sci U S A. 2008;105:11026–31. doi: 10.1073/pnas.0805496105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramirez de Molina A, Rodriguez-Gonzalez A, Gutierrez R, Martinez-Pineiro L, Sanchez J, Bonilla F, Rosell R, Lacal J. Overexpression of choline kinase is a frequent feature in human tumor-derived cell lines and in lung, prostate, and colorectal human cancers. Biochem Biophys Res Commun. 2002;296:580–3. doi: 10.1016/s0006-291x(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 87.Momchilova A, Markovska T, Pankov R. Ha-ras-transformation alters the metabolism of phosphatidylethanolamine and phosphatidylcholine in NIH 3T3 fibroblasts. Cell Biol Int. 1999;23:603–10. doi: 10.1006/cbir.1999.0430. [DOI] [PubMed] [Google Scholar]
- 88.Teegarden D, Taparowsky EJ, Kent C. Altered phosphatidylcholine metabolism in C3H10T1/2 cells transfected with the Harvey-ras oncogene. J Biol Chem. 1990;265:6042–7. [PubMed] [Google Scholar]
- 89.Ferretti A, Podo F, Carpinelli G, Chen L, Borghi P, Masella R. Detection of neutral active phosphatidylcholine-specific phospholipase C in Friend leukemia cells before and after erythroid differentiation. Anticancer Res. 1993;13:2309–17. [PubMed] [Google Scholar]
- 90.Podo F, Ferretti A, Knijn A, Zhang P, Ramoni C, Barletta B, Pini C, Baccarini S, Pulciani S. Detection of phosphatidylcholine-specific phospholipase C in NIH-3T3 fibroblasts and their H-ras transformants: NMR and immunochemical studies. Anticancer Res. 1996;16:1399–412. [PubMed] [Google Scholar]
- 91.Wu X, Lu H, Zhou L, Huang Y, Chen H. Changes of phosphatidylcholine-specific phospholipase C in hepatocarcinogenesis and in the proliferation and differentiation of rat liver cancer cells. Cell Biol Int. 1997;21:375–81. doi: 10.1006/cbir.1997.0148. [DOI] [PubMed] [Google Scholar]
- 92.Al-Saffar NM, Troy H, Ramirez de Molina A, Jackson LE, Madhu B, Griffiths JR, Leach MO, Workman P, Lacal JC, Judson IR, Chung YL. Noninvasive magnetic resonance spectroscopic pharmacodynamic markers of the choline kinase inhibitor MN58b in human carcinoma models. Cancer research. 2006;66:427–34. doi: 10.1158/0008-5472.CAN-05-1338. [DOI] [PubMed] [Google Scholar]
- 93.Krishnamachary B, Glunde K, Wildes F, Mori N, Takagi T, Raman V, Bhujwalla ZM. Noninvasive detection of lentiviral-mediated choline kinase targeting in a human breast cancer xenograft. Cancer research. 2009;69:3464–71. doi: 10.1158/0008-5472.CAN-08-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lacal JC. Choline kinase: a novel target for antitumor drugs. IDrugs. 2001;4:419–26. [PubMed] [Google Scholar]
- 95.Ronen SM, Leach MO. Imaging biochemistry: applications to breast cancer. Breast Cancer Res. 2001;3:36–40. doi: 10.1186/bcr268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beloueche-Babari M, Chung YL, Al-Saffar NM, Falck-Miniotis M, Leach MO. Metabolic assessment of the action of targeted cancer therapeutics using magnetic resonance spectroscopy. Br J Cancer. 2010;102:1–7. doi: 10.1038/sj.bjc.6605457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Negendank W. Studies of human tumors by MRS: a review. NMR Biomed. 1992;5:303–24. doi: 10.1002/nbm.1940050518. [DOI] [PubMed] [Google Scholar]
- 98.Podo F. Tumour phospholipid metabolism. NMR Biomed. 1999;12:413–39. doi: 10.1002/(sici)1099-1492(199911)12:7<413::aid-nbm587>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 99.Lucas L, Penalva V, Ramirez de Molina A, Del Peso L, Lacal JC. Modulation of phospholipase D by Ras proteins mediated by its effectors Ral-GDS, PI3K and Raf-1. Int J Oncol. 2002;21:477–85. [PubMed] [Google Scholar]
- 100.Ramirez de Molina A, Rodriguez-Gonzalez A, Penalva V, Lucas L, Lacal JC. Inhibition of ChoK is an efficient antitumor strategy for Harvey-, Kirsten-, and N-ras-transformed cells. Biochem Biophys Res Commun. 2001;285:873–9. doi: 10.1006/bbrc.2001.5250. [DOI] [PubMed] [Google Scholar]
- 101.Ramirez de Molina A, Penalva V, Lucas L, Lacal JC. Regulation of choline kinase activity by Ras proteins involves Ral-GDS and PI3K. Oncogene. 2002;21:937–46. doi: 10.1038/sj.onc.1205144. [DOI] [PubMed] [Google Scholar]
- 102.Ronen SM, Jackson LE, Beloueche M, Leach MO. Magnetic resonance detects changes in phosphocholine associated with Ras activation and inhibition in NIH 3T3 cells. Br J Cancer. 2001;84:691–6. doi: 10.1054/bjoc.2000.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ratnam S, Kent C. Early increase in choline kinase activity upon induction of the H-ras oncogene in mouse fibroblast cell lines. Arch Biochem Biophys. 1995;323:313–22. doi: 10.1006/abbi.1995.9959. [DOI] [PubMed] [Google Scholar]
- 104.Beloueche-Babari M, Jackson LE, Al-Saffar NM, Workman P, Leach MO, Ronen SM. Magnetic resonance spectroscopy monitoring of mitogen-activated protein kinase signaling inhibition. Cancer research. 2005;65:3356–63. doi: 10.1158/10.1158/0008-5472.CAN-03-2981. [DOI] [PubMed] [Google Scholar]
- 105.Yalcin A, Clem B, Makoni S, Clem A, Nelson K, Thornburg J, Siow D, Lane AN, Brock SE, Goswami U, Eaton JW, Telang S, Chesney J. Selective inhibition of choline kinase simultaneously attenuates MAPK and PI3K/AKT signaling. Oncogene. 2010;29:139–49. doi: 10.1038/onc.2009.317. [DOI] [PubMed] [Google Scholar]
- 106.Al-Saffar NM, Jackson LE, Raynaud FI, Clarke PA, Ramirez de Molina A, Lacal JC, Workman P, Leach MO. The phosphoinositide 3-kinase inhibitor PI-103 downregulates choline kinase alpha leading to phosphocholine and total choline decrease detected by magnetic resonance spectroscopy. Cancer research. 2010;70:5507–17. doi: 10.1158/0008-5472.CAN-09-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koul D, Shen R, Kim YW, Kondo Y, Lu Y, Bankson J, Ronen SM, Kirkpatrick DL, Powis G, Yung WK. Cellular and in vivo activity of a novel PI3K inhibitor, PX-866, against human glioblastoma. Neuro Oncol. 2010;12:559–69. doi: 10.1093/neuonc/nop058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Glunde K, Shah T, Winnard PTJ, Raman V, Takagi T, Vesuna F, Artemov D, Bhujwalla ZM. Hypoxia regulates choline kinase expression through hypoxia-inducible factor-1alpha signaling in a human prostate cancer model. Cancer research. 2008;68:172–80. doi: 10.1158/0008-5472.CAN-07-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paris L, Cecchetti S, Spadaro F, Abalsamo L, Lugini L, Pisanu ME, Iorio E, Natali PG, Ramoni C, Podo F. Inhibition of phosphatidylcholine-specific phospholipase C downregulates HER2 overexpression on plasma membrane of breast cancer cells. Breast Cancer Res. 2010;12:R27. doi: 10.1186/bcr2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ross J, Najjar AM, Sankaranarayanapillai M, Tong WP, Kaluarachchi K, Ronen SM. Fatty acid synthase inhibition results in a magnetic resonance-detectable drop in phosphocholine. Mol Cancer Ther. 2008;7:2556–65. doi: 10.1158/1535-7163.MCT-08-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Klawitter J, Anderson N, Klawitter J, Christians U, Leibfritz D, Eckhardt SG, Serkova NJ. Time-dependent effects of imatinib in human leukaemia cells: a kinetic NMR-profiling study. Br J Cancer. 2009;100:923–31. doi: 10.1038/sj.bjc.6604946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sankaranarayanapillai M, Tong WP, Yuan Q, Bankson JA, Dafni H, Bornmann WG, Soghomonyan S, Pal A, Ramirez MS, Webb D, Kaluarachchi K, Gelovani JG, Ronen SM. Monitoring histone deacetylase inhibition in vivo: noninvasive magnetic resonance spectroscopy method. Mol Imaging. 2008;7:92–100. [PubMed] [Google Scholar]
- 113.Warburg O. Über den Stoffwechsel der Carcinomzelle. Naturwissenschaften. 1924;12:1132–7. [Google Scholar]
- 114.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–70. [PubMed] [Google Scholar]
- 115.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB. Akt stimulates aerobic glycolysis in cancer cells. Cancer research. 2004;64:3892–9. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 116.Yeung SJ, Pan J, Lee MH. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell Mol Life Sci. 2008;65:3981–99. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Van Hemelrijck M, Garmo H, Holmberg L, Walldius G, Jungner I, Hammar N, Lambe M. Prostate cancer risk in the Swedish AMORIS study: the interplay among triglycerides, total cholesterol, and glucose. Cancer. doi: 10.1002/cncr.25758. [DOI] [PubMed] [Google Scholar]
- 118.Dong X, Tang H, Hess KR, Abbruzzese JL, Li D. Glucose metabolism gene polymorphisms and clinical outcome in pancreatic cancer. Cancer. 2011 doi: 10.1002/cncr.25612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ikehira H, Hashimoto T, Fukuda H, Ueshima Y, Yamai S, Maki T, Iinuma TA, Tateno Y. Carbon-13 NMR imaging study of in-vivo glucose metabolism. Am J Physiol Imaging. 1990;5:50–4. [PubMed] [Google Scholar]
- 120.Ross BD, Higgins RJ, Boggan JE, Willis JA, Knittel B, Unger SW. Carbohydrate metabolism of the rat C6 glioma. An in vivo 13C and in vitro 1H magnetic resonance spectroscopy study. NMR Biomed. 1988;1:20–6. doi: 10.1002/nbm.1940010105. [DOI] [PubMed] [Google Scholar]
- 121.Kurhanewicz J, Bok R, Nelson SJ, Vigneron DB. Current and potential applications of clinical 13C MR spectroscopy. J Nucl Med. 2008;49:341–4. doi: 10.2967/jnumed.107.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Williams SR, Gadian DG. Tissue metabolism studied in vivo by nuclear magnetic resonance. Q J Exp Physiol. 1986;71:335–60. doi: 10.1113/expphysiol.1986.sp002994. [DOI] [PubMed] [Google Scholar]
- 123.Gillies RJ, Morse DL. In vivo magnetic resonance spectroscopy in cancer. Annu Rev Biomed Eng. 2005;7:287–326. doi: 10.1146/annurev.bioeng.7.060804.100411. [DOI] [PubMed] [Google Scholar]
- 124.Soares DP, Law M. Magnetic resonance spectroscopy of the brain: review of metabolites and clinical applications. Clin Radiol. 2009;64:12–21. doi: 10.1016/j.crad.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 125.Ganslandt O, Stadlbauer A, Fahlbusch R, Kamada K, Buslei R, Blumcke I, Moser E, Nimsky C. Proton magnetic resonance spectroscopic imaging integrated into image-guided surgery: correlation to standard magnetic resonance imaging and tumor cell density. Neurosurgery. 2005;56:291–8. doi: 10.1227/01.neu.0000156782.14538.78. [DOI] [PubMed] [Google Scholar]
- 126.Stadlbauer A, Moser E, Gruber S, Buslei R, Nimsky C, Fahlbusch R, Ganslandt O. Improved delineation of brain tumors: an automated method for segmentation based on pathologic changes of 1H-MRSI metabolites in gliomas. Neuroimage. 2004;23:454–61. doi: 10.1016/j.neuroimage.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 127.Opstad KS, Wright AJ, Bell BA, Griffiths JR, Howe FA. Correlations between in vivo (1)H MRS and ex vivo (1)H HRMAS metabolite measurements in adult human gliomas. J Magn Reson Imaging. 2010;31:289–97. doi: 10.1002/jmri.22039. [DOI] [PubMed] [Google Scholar]
- 128.Tessem MB, Swanson MG, Keshari KR, Albers MJ, Joun D, Tabatabai ZL, Simko JP, Shinohara K, Nelson SJ, Vigneron DB, Gribbestad IS, Kurhanewicz J. Evaluation of lactate and alanine as metabolic biomarkers of prostate cancer using 1H HR-MAS spectroscopy of biopsy tissues. Magn Reson Med. 2008;60:510–6. doi: 10.1002/mrm.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Artemov D, Bhujwalla ZM, Pilatus U, Glickson JD. Two-compartment model for determination of glycolytic rates of solid tumors by in vivo 13C NMR spectroscopy. NMR Biomed. 1998;11:395–404. doi: 10.1002/(sici)1099-1492(199812)11:8<395::aid-nbm536>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 130.Lyon RC, Cohen JS, Faustino PJ, Megnin F, Myers CE. Glucose metabolism in drug-sensitive and drug-resistant human breast cancer cells monitored by magnetic resonance spectroscopy. Cancer research. 1988;48:870–7. [PubMed] [Google Scholar]
- 131.Constantinidis I, Chatham JC, Wehrle JP, Glickson JD. In vivo 13CNMR spectroscopy of glucose metabolism of RIF-1 tumors. Magn Reson Med. 1991;20:17–26. doi: 10.1002/mrm.1910200103. [DOI] [PubMed] [Google Scholar]
- 132.Portais JC, Schuster R, Merle M, Canioni P. Metabolic flux determination in C6 glioma cells using carbon-13 distribution upon [1-13C]glucose incubation. Eur J Biochem. 1993;217:457–68. doi: 10.1111/j.1432-1033.1993.tb18265.x. [DOI] [PubMed] [Google Scholar]
- 133.Singer S, Okunieff P, Gostin C, Thilly WG, Chen LB, Neuringer LJ. 13C- and 31P-NMR studies of human colon cancer in-vitro and in-vivo. Surg Oncol. 1993;2:7–18. doi: 10.1016/0960-7404(93)90039-2. [DOI] [PubMed] [Google Scholar]
- 134.Terpstra M, High WB, Luo Y, de Graaf RA, Merkle H, Garwood M. Relationships among lactate concentration, blood flow and histopathologic profiles in rat C6 glioma. NMR Biomed. 1996;9:185–94. doi: 10.1002/(SICI)1099-1492(199608)9:5<185::AID-NBM414>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 135.Neeman M, Degani H. Metabolic studies of estrogen- and tamoxifen-treated human breast cancer cells by nuclear magnetic resonance spectroscopy. Cancer research. 1989;49:589–94. [PubMed] [Google Scholar]
- 136.Neeman M, Eldar H, Rushkin E, Degani H. Chemotherapy-induced changes in the energetics of human breast cancer cells; 31P- and 13C-NMR studies. Biochim Biophys Acta. 1990;1052:255–63. doi: 10.1016/0167-4889(90)90219-4. [DOI] [PubMed] [Google Scholar]
- 137.Poptani H, Bansal N, Jenkins WT, Blessington D, Mancuso A, Nelson DS, Feldman M, Delikatny EJ, Chance B, Glickson JD. Cyclophosphamide treatment modifies tumor oxygenation and glycolytic rates of RIF-1 tumors: 13C magnetic resonance spectroscopy, Eppendorf electrode, and redox scanning. Cancer research. 2003;63:8813–20. [PubMed] [Google Scholar]
- 138.Ben-Horin H, Tassini M, Vivi A, Navon G, Kaplan O. Mechanism of action of the antineoplastic drug lonidamine: 31P and 13C nuclear magnetic resonance studies. Cancer research. 1995;55:2814–21. [PubMed] [Google Scholar]
- 139.Grinde MT, Moestue S, Borgan E, Risa O, Gribbestad IS. 13C HR MAS MRS reveals differences in the glucose metabolism between two breast cancer xenograft models with different gene expression pattern. Proc Int Soc Magn Res Med. 2010;18 doi: 10.1002/nbm.1683. [DOI] [PubMed] [Google Scholar]
- 140.Wijnen JP, Van der GM, Scheenen TW, Klomp DW, de Galan BE, Idema AJ, Heerschap A. In vivo (13)C magnetic resonance spectroscopy of a human brain tumor after application of (13)C-1-enriched glucose. Magn Reson Imaging. 2010;28:690–7. doi: 10.1016/j.mri.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 141.Halliday KR, Fenoglio-Preiser C, Sillerud LO. Differentiation of human tumors from nonmalignant tissue by natural-abundance 13C NMR spectroscopy. Magn Reson Med. 1988;7:384–411. doi: 10.1002/mrm.1910070403. [DOI] [PubMed] [Google Scholar]
- 142.Golman K, Olsson LE, Axelsson O, Mansson S, Karlsson M, Petersson JS. Molecular imaging using hyperpolarized 13C. Br J Radiol. 2003;76:S118–S27. doi: 10.1259/bjr/26631666. Spec No 2. [DOI] [PubMed] [Google Scholar]
- 143.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A. 2003;100:10158–63. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mansson S, Johansson E, Magnusson P, Chai CM, Hansson G, Petersson JS, Stahlberg F, Golman K. 13C imaging-a new diagnostic platform. Eur Radiol. 2006;16:57–67. doi: 10.1007/s00330-005-2806-x. [DOI] [PubMed] [Google Scholar]
- 145.Yen YF, Kohler SJ, Chen AP, Tropp J, Bok R, Wolber J, Albers MJ, Gram KA, Zierhut ML, Park I, Zhang V, Hu S, Nelson SJ, Vigneron DB, Kurhanewicz J, Dirven HA, Hurd RE. Imaging considerations for in vivo 13C metabolic mapping using hyperpolarized 13C-pyruvate. Magn Reson Med. 2009;62:1–10. doi: 10.1002/mrm.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hu S, Lustig M, Balakrishnan A, Larson PE, Bok R, Kurhanewicz J, Nelson SJ, Goga A, Pauly JM, Vigneron DB. 3D compressed sensing for highly accelerated hyperpolarized (13)C MRSI with in vivo applications to transgenic mouse models of cancer. Magn Reson Med. 2010;63:312–21. doi: 10.1002/mrm.22233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Larson PE, Kerr AB, Chen AP, Lustig MS, Zierhut ML, Hu S, Cunningham CH, Pauly JM, Kurhanewicz J, Vigneron DB. Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging. J Magn Reson. 2008;194:121–7. doi: 10.1016/j.jmr.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Golman K, Zandt RI, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer research. 2006;66:10855–60. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- 149.Nelson SJ, Vigneron D, Kurhanewicz J, Chen A, Bok R, Hurd R. DNP-Hyperpolarized C Magnetic Resonance Metabolic Imaging for Cancer Applications. ApplMagn Reson. 2008;34:533–44. doi: 10.1007/s00723-008-0136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Golman K, Petersson JS, Magnusson P, Johansson E, Akeson P, Chai CM, Hansson G, Mansson S. Cardiac metabolism measured noninvasively by hyperpolarized 13C MRI. Magn Reson Med. 2008;59:1005–13. doi: 10.1002/mrm.21460. [DOI] [PubMed] [Google Scholar]
- 151.Kohler SJ, Yen Y, Wolber J, Chen AP, Albers MJ, Bok R, Zhang V, Tropp J, Nelson S, Vigneron DB, Kurhanewicz J, Hurd RE. In vivo 13 carbon metabolic imaging at 3T with hyperpolarized 13C-1-pyruvate. Magn Reson Med. 2007;58:65–9. doi: 10.1002/mrm.21253. [DOI] [PubMed] [Google Scholar]
- 152.Albers MJ, Bok R, Chen AP, Cunningham CH, Zierhut ML, Zhang VY, Kohler SJ, Tropp J, Hurd RE, Yen YF, Nelson SJ, Vigneron DB, Kurhanewicz J. Hyperpolarized 13C lactate, pyruvate, and alanine: noninvasive biomarkers for prostate cancer detection and grading. Cancer research. 2008;68:8607–15. doi: 10.1158/0008-5472.CAN-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zierhut ML, Yen YF, Chen AP, Bok R, Albers MJ, Zhang V, Tropp J, Park I, Vigneron DB, Kurhanewicz J, Hurd RE, Nelson SJ. Kinetic modeling of hyperpolarized 13C1-pyruvate metabolism in normal rats and TRAMP mice. J Magn Reson. 2010;202:85–92. doi: 10.1016/j.jmr.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Park I, Larson PE, Zierhut ML, Hu S, Bok R, Ozawa T, Kurhanewicz J, Vigneron DB, Vandenberg SR, James CD, Nelson SJ. Hyperpolarized 13C magnetic resonance metabolic imaging: application to brain tumors. Neuro Oncol. 2010;12:133–44. doi: 10.1093/neuonc/nop043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Day SE, Kettunen MI, Gallagher FA, Hu DE, Lerche M, Wolber J, Golman K, Ardenkjaer-Larsen JH, Brindle KM. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007;13:1382–7. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- 156.Ward CS, Venkatesh HS, Chaumeil MM, Brandes AH, Vancriekinge M, Dafni H, Sukumar S, Nelson SJ, Vigneron DB, Kurhanewicz J, James CD, Haas-Kogan DA, Ronen SM. Noninvasive detection of target modulation following phosphatidylinositol 3-kinase inhibition using hyperpolarized 13C magnetic resonance spectroscopy. Cancer research. 2010;70:1296–305. doi: 10.1158/0008-5472.CAN-09-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Keshari KR, Wilson DM, Chen AP, Bok R, Larson PE, Hu S, Van CM, Macdonald JM, Vigneron DB, Kurhanewicz J. Hyperpolarized [2-13C]-fructose: a hemiketal DNP substrate for in vivo metabolic imaging. J Am Chem Soc. 2009;131:17591–6. doi: 10.1021/ja9049355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gallagher FA, Kettunen MI, Hu DE, Jensen PR, Zandt RI, Karlsson M, Gisselsson A, Nelson SK, Witney TH, Bohndiek SE, Hansson G, Peitersen T, Lerche MH, Brindle KM. Production of hyperpolarized [1,4-13C2]malate from [1,4-13C2]fumarate is a marker of cell necrosis and treatment response in tumors. Proc Natl Acad Sci U S A. 2009;106:19801–6. doi: 10.1073/pnas.0911447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wilson DM, Keshari KR, Larson PE, Chen AP, Hu S, Criekinge MV, Bok R, Nelson SJ, Macdonald JM, Vigneron DB, Kurhanewicz J. Multi-compound polarization by DNP allows simultaneous assessment of multiple enzymatic activities in vivo. J Magn Reson. 2010;205:141–7. doi: 10.1016/j.jmr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen AP, Kurhanewicz J, Bok R, Xu D, Joun D, Zhang V, Nelson SJ, Hurd RE, Vigneron DB. Feasibility of using hyperpolarized [1-13C]lactate as a substrate for in vivo metabolic 13C MRSI studies. Magn Reson Imaging. 2008;26:721–6. doi: 10.1016/j.mri.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Smith TA. Facilitative glucose transporter expression in human cancer tissue. Br J Biomed Sci. 1999;56:285–92. [PubMed] [Google Scholar]
- 162.Meadows AL, Kong B, Berdichevsky M, Roy S, Rosiva R, Blanch HW, Clark DS. Metabolic and morphological differences between rapidly proliferating cancerous and normal breast epithelial cells. Biotechnol Prog. 2008;24:334–41. doi: 10.1021/bp070301d. [DOI] [PubMed] [Google Scholar]
- 163.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–20. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 164.Ganapathy V, Thangaraju M, Prasad PD. Nutrient transporters in cancer: relevance to Warburg hypothesis and beyond. Pharmacol Ther. 2009;121:29–40. doi: 10.1016/j.pharmthera.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 165.Pinheiro C, Longatto-Filho A, Scapulatempo C, Ferreira L, Martins S, Pellerin L, Rodrigues M, Alves VA, Schmitt F, Baltazar F. Increased expression of monocarboxylate transporters 1, 2, and 4 in colorectal carcinomas. Virchows Arch. 2008;452:139–46. doi: 10.1007/s00428-007-0558-5. [DOI] [PubMed] [Google Scholar]
- 166.Deberardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–50. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–82. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- 168.Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, Skoldberg F, Husebye ES, Eng C, Maher ER. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der MA, Taschner PE, Rubinstein WS, Myers EN, Richard CW, III, Cornelisse CJ, Devilee P, Devlin B. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–51. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 170.Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomaki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–10. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 171.Barthel A, Okino ST, Liao J, Nakatani K, Li J, Whitlock JP, Jr., Roth RA. Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J Biol Chem. 1999;274:20281–6. doi: 10.1074/jbc.274.29.20281. [DOI] [PubMed] [Google Scholar]
- 172.Majewski N, Nogueira V, Robey RB, Hay N. Akt inhibits apoptosis downstream of BID cleavage via a glucose-dependent mechanism involving mitochondrial hexokinases. Mol Cell Biol. 2004;24:730–40. doi: 10.1128/MCB.24.2.730-740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–83. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 175.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–3. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 176.Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269:23757–63. [PubMed] [Google Scholar]
- 177.Bauer DE, Harris MH, Plas DR, Lum JJ, Hammerman PS, Rathmell JC, Riley JL, Thompson CB. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. Faseb J. 2004;18:1303–5. doi: 10.1096/fj.03-1001fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Walenta S, Wetterling M, Lehrke M, Schwickert G, Sundfor K, Rofstad EK, Mueller-Klieser W. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer research. 2000;60:916–21. [PubMed] [Google Scholar]
- 179.Nelson JA, Falk RE. The efficacy of phloridzin and phloretin on tumor cell growth. Anticancer Res. 1993;13:2287–92. [PubMed] [Google Scholar]
- 180.Ganapathy-Kanniappan S, Vali M, Kunjithapatham R, Buijs M, Syed LH, Rao PP, Ota S, Kwak BK, Loffroy R, Geschwind JF. 3-Bromopyruvate: A New Targeted Antiglycolytic Agent and a Promise for Cancer Therapy. Curr Pharm Biotechnol. 2010;11:510–7. doi: 10.2174/138920110791591427. [DOI] [PubMed] [Google Scholar]
- 181.Hernlund E, Ihrlund LS, Khan O, Ates YO, Linder S, Panaretakis T, Shoshan MC. Potentiation of chemotherapeutic drugs by energy metabolism inhibitors 2-deoxyglucose and etomoxir. Int J Cancer. 2008;123:476–83. doi: 10.1002/ijc.23525. [DOI] [PubMed] [Google Scholar]
- 182.Iaffaioli RV, Tortoriello A, Facchini G, Santangelo M, Bucci L, Fei L, Di MN, Mantovani G, Caponigro F. Phase II study of high-dose epirubicin, lonidamine, alpha 2b interferon in advanced breast cancer. Breast Cancer Res Treat. 1995;35:243–8. doi: 10.1007/BF00665975. [DOI] [PubMed] [Google Scholar]
- 183.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–46. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 184.Loar P, Wahl H, Kshirsagar M, Gossner G, Griffith K, Liu JR. Inhibition of glycolysis enhances cisplatin-induced apoptosis in ovarian cancer cells. Am J Obstet Gynecol. 2010;202:371–8. doi: 10.1016/j.ajog.2009.10.883. [DOI] [PubMed] [Google Scholar]
- 185.Pradelli LA, Beneteau M, Chauvin C, Jacquin MA, Marchetti S, Munoz-Pinedo C, Auberger P, Pende M, Ricci JE. Glycolysis inhibition sensitizes tumor cells to death receptors-induced apoptosis by AMP kinase activation leading to Mcl-1 block in translation. Oncogene. 2010;29:1641–52. doi: 10.1038/onc.2009.448. [DOI] [PubMed] [Google Scholar]
- 186.Hulleman E, Kazemier KM, Holleman A, VanderWeele DJ, Rudin CM, Broekhuis MJ, Evans WE, Pieters R, Den Boer ML. Inhibition of glycolysis modulates prednisolone resistance in acute lymphoblastic leukemia cells. Blood. 2009;113:2014–21. doi: 10.1182/blood-2008-05-157842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gottschalk S, Anderson N, Hainz C, Eckhardt SG, Serkova NJ. Imatinib (STI571)-mediated changes in glucose metabolism in human leukemia BCR-ABL-positive cells. Clin Cancer Res. 2004;10:6661–8. doi: 10.1158/1078-0432.CCR-04-0039. [DOI] [PubMed] [Google Scholar]
- 188.Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–77. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 189.Jordan BF, Black K, Robey IF, Runquist M, Powis G, Gillies RJ. Metabolite changes in HT-29 xenograft tumors following HIF-1alpha inhibition with PX-478 as studied by MR spectroscopy in vivo and ex vivo. NMR Biomed. 2005;18:430–9. doi: 10.1002/nbm.977. [DOI] [PubMed] [Google Scholar]
- 190.Serkova N, Boros LG. Detection of resistance to imatinib by metabolic profiling: clinical and drug development implications. Am J Pharmacogenomics. 2005;5:293–302. doi: 10.2165/00129785-200505050-00002. [DOI] [PubMed] [Google Scholar]
- 191.McKnight TR. Proton magnetic resonance spectroscopic evaluation of brain tumor metabolism. Semin Oncol. 2004;31:605–17. doi: 10.1053/j.seminoncol.2004.07.003. [DOI] [PubMed] [Google Scholar]