Abstract
Elucidating how and to what extent CpG islands (CGIs) are methylated in germ cells is essential to understand genomic imprinting and epigenetic reprogramming1-3. Here, we present the first integrated epigenomic analysis of mammalian oocytes, identifying over a thousand CGIs methylated in mature oocytes. We show that these CGIs depend on DNMT3A and DNMT3L4-5, but are not distinct at the sequence level, including in CpG periodicity6. They are preferentially located within active transcription units and are relatively depleted in H3K4me3, supporting a general transcription-dependent mechanism of methylation. Very few methylated CGIs are fully protected from post-fertilisation reprogramming but, surprisingly, the majority exhibits incomplete demethylation in E3.5 blastocysts. Our study shows that CGI methylation in gametes is not entirely related to genomic imprinting, but is a strong factor in determining methylation status in preimplantation embryos, suggesting a need to reassess mechanisms of post-fertilization demethylation.
Because DNA methylation in oocytes occurs in meiotically arrested cells3,7, it represents a uniquely informative system for investigating requirements and mechanisms of de novo methylation. These mechanisms, especially at CGIs, are poorly understood, mainly because of the very limited number of methylated CGIs identified so far in germ cells. To obtain genome-wide information on DNA methylation in oocytes, as well as in sperm, we performed Reduced Representation Bisulphite Sequencing (RRBS), using a protocol optimised for low amounts of DNA (Supplementary Fig. 1). RRBS combines the base-pair resolution and quantitative assessment of bisulphite sequencing with high enrichment for CGIs8-9. The fidelity of the method was demonstrated by detection of the expected methylation of known maternal germline differentially methylated regions (DMRs) at imprinted loci (Supplementary Fig. 2).
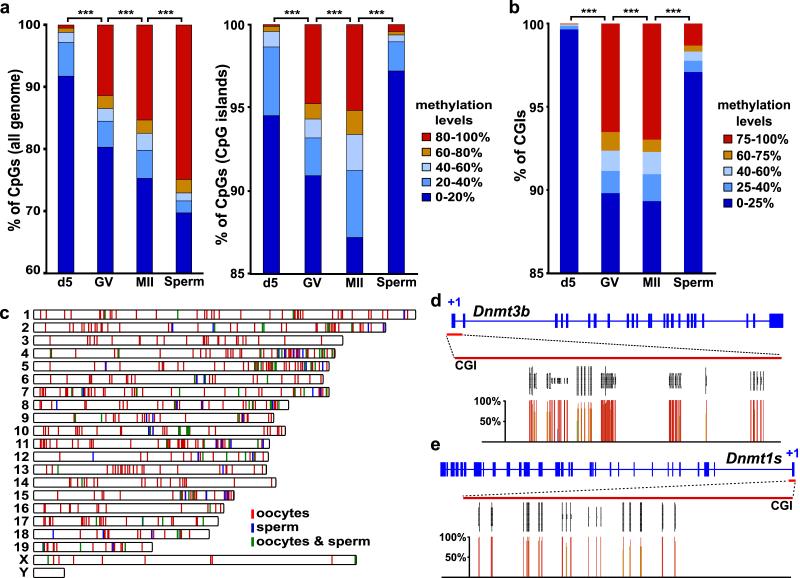
CpG methylation overall, and in CGI and repetitive element contexts, showed a dynamic profile during oocyte growth: 0.5% of all CpGs assessed by RRBS were highly methylated in d5 oocytes (≥80% methylation), 11.3% in d20 germinal vesicle (GV) and 15.3% in ovulated metaphase II (MII) oocytes. CpG methylation was lower overall in mature oocytes than sperm (24.9% of CpGs highly methylated in sperm), consistent with previous observations on repetitive elements10; methylation in a CGI context, irrespective of location with respect to genes, was markedly lower in sperm (Fig. 1a, Supplementary Fig. 3a-b & 4). Using a threshold for scoring CGIs that reads should cover ≥10% of the CpGs per CGI (see Methods for a full account), we obtained information on ~15,000 (~65%) of the extended set of CGIs recently identified by CAP-Seq11, and identified 1062 methylated CGIs (≥75% methylation) in mature oocytes (Fig 1b-c; Supplementary Table 1). By extrapolation, there may be ~1600 fully methylated CGIs in mature oocytes. Of interest, we found that the CGIs associated with the major promoters of Dnmt3b and Dnmt1 (Dnmt1s) were methylated (Fig. 1d,e, Supplementary Fig. 9). Eighty-nine CGIs identified as methylated in MII oocytes were not fully methylated in GV oocytes, demonstrating that CGIs acquire methylation at different rates during oocyte growth, as reported for germline DMRs12-13 (Supplementary Table 2). In sperm, we identified 185 fully methylated CGIs, 58 of which were methylated exclusively in sperm and 100 were also methylated in mature oocytes (27 of the CGIs methylated in sperm were not informative in mature oocyte datasets) (Fig 1b-c; Supplementary Table 1). For subsequent analyses, we considered CGIs scored with ≥75% methylation as fully methylated and those with ≤25% methylation as unmethylated; this does not exclude that CGIs scored between these cut-offs possess methylation.
Figure 1. DNA methylation landscape in oocytes and sperm determined by RRBS.
a-b, Distribution of CpG methylation levels across the genome (a, left), within CGIs (a, right), and CGI methylation (b), in immature (d5), mature (GV and MII) oocytes and sperm (***: p<0.001, χ2 test). The number of CpGs and CGIs analysed is indicated in Suppl. Fig 1b. c, Chromosome distribution of the 1062 CGIs methylated in oocytes and 185 CGIs methylated in sperm (100 CGIs in both). d, CpG methylation levels (percentage of all cytosines called methylated) at the Dnmt3b and Dnmt1s promoter CGIs in GV oocytes. The grey vertical lines represent the sequencing read depth of individual cytosines; below, percentage methylation of the corresponding CpGs is represented by coloured vertical lines.
Having identified the extent of CGI methylation in gametes, we tested whether they possessed distinctive sequence properties. In comparison with unmethylated CGIs, higher proportions of CGIs methylated in oocytes and sperm were intragenic (Supplementary Fig 3a, 3c), similar to findings of CGI methylation in somatic tissues11,14-15. CGIs methylated in oocytes and sperm were shorter than unmethylated CGIs, with lower GC content and CpG density (Supplementary Fig. 5a-d). These properties may reflect the enrichment in intragenic CGIs, which are shorter and less CpG dense (data not shown), and this trend was also observed in comparing methylated intragenic and promoter CGIs (Supplementary Fig. 5e). Contrary to reports implicating tandem repeats in DMR methylation16, the content of this repetitive element class was similarly low in germline methylated and unmethylated CGIs (Supplementary Fig. 5f). A systematic analysis of sequence properties performed using EpiGRAPH17 identified sequence attributes enriched in methylated CGIs (Supplementary Table 3), but was not able to predict the methylation status of CGIs based on these. A key observation implicating intrinsic sequence properties in de novo methylation of DMRs is the tetrameric association of DNMT3A and DNMT3L, the factors responsible for DMR methylation4-5,18, which results in the two DNMT3A catalytic sites being separated by a spacing corresponding to 8-10bp of DNA. This spacing was described as the dominant CpG periodicity unit of maternal germline DMRs6. Importantly, we did not observe any differences in the CpG periodicity of CGIs methylated in oocytes or sperm, or unmethylated CGIs (Supplementary Fig. 5g). Overall, these results indicate that CGIs methylated in gametes do not appear to have strong discriminating sequence features.
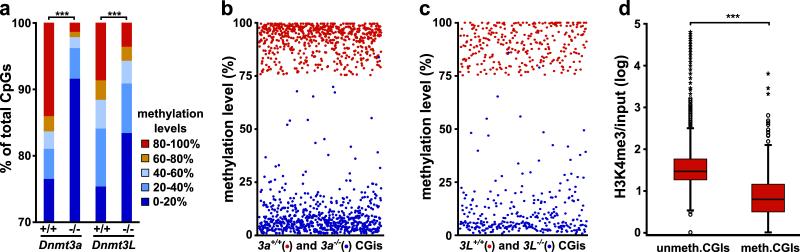
We next examined whether CGIs methylated in oocytes depend on DNMT3A and DNMT3L, by performing RRBS on GV oocytes genetically depleted in these factors. Both Dnmt3a−/− and Dnmt3L−/− oocytes exhibited a gross, genome-wide reduction in CpG methylation, including at repetitive elements and CGIs independent of their genic location (Fig. 2a, Supplementary Fig. 6). Of 654 CGIs methylated in Dnmt3a+/+ oocytes with coverage in Dnmt3a−/− oocytes, the vast majority (96%) were unmethylated in Dnmt3a−/− oocytes (Fig. 2b). We also had information on 301 methylated CGIs in Dnmt3L+/+ oocytes and their Dnmt3L−/− counterparts; similarly, 92% were unmethylated in Dnmt3L−/− oocytes (Fig. 2c). Interestingly, the CGIs remaining methylated (≥75%) in Dnmt3L−/− oocytes were also highly methylated in Dnmt3a−/− oocytes, sperm and d5 oocytes, suggesting that they are incompletely demethylated during primordial germ cell reprogramming19 (Supplementary Table 1). Overall, this demonstrates the genome-wide role of DNMT3A and DNMT3L in CGI methylation beyond genomic imprinting.
Figure 2. Mechanism of DNA methylation establishment in oocytes.
a, Distribution of CpG methylation levels across the genome in Dnmt3a−/− and Dnmt3L−/− oocytes and their wild-type counterparts (+/+); the number of CpGs analysed is indicated in Suppl. Fig 1b (***: p<0.001, χ2 test). b-c, Methylation levels of CGIs in Dnmt3a−/− and Dnmt3L−/− oocytes; only those CGIs for which methylation was ≥75% in the corresponding wild-type oocytes are displayed. d, Overall correlation between H3K4me3 enrichment determined in d15 oocytes by ChIP-seq and methylation status of CGIs (all CGIs irrespective of genomic location; ***: p<0.001, Mann-Whitney U test).
Recent reports highlight the link between active transcription and DNA methylation. We previously showed that transcription across the DMRs of the imprinted Gnas locus is required for their methylation in oocytes12. The H3K4 demethylase KDM1B, which is associated with active gene bodies, is required for methylation of a subset of DMRs in oocytes20-21. In addition, interaction of DNMT3A and DNMT3L with chromatin is inhibited by H3K4 methylation22-23, whereas DNMT3A binds H3K36me324, a transcriptional elongation mark. To explore the general relationship between transcription and CGI methylation in oocytes, we undertook mRNA-Seq in d10 oocytes (onset of de novo methylation) (Supplementary Fig. 7). This showed that methylated CGIs annotated as intragenic were more likely to be within transcription units active in oocytes compared to unmethylated intragenic CGIs (75% versus 28%, p<0.001, χ2 test). Furthermore, methylated CGIs overlapping annotated promoters were more frequently within overlapping transcripts compared to unmethylated promoter CGIs (39% versus 8%, p<0.001, χ2 test). In addition, mRNA-Seq identified alternative, upstream promoters for 35% of the methylated CGIs, versus 10% of unmethylated CGIs (p<0.001, χ2 test) (Supplementary Fig. 7). These observations strengthen an association between location within active transcription units and probability of methylation. We also performed H3K4me3 ChIP-Seq in d15 oocytes (early phase of de novo methylation) (Supplementary Fig. 7). This revealed that CGIs methylated in oocytes were relatively depleted in H3K4me3: 6.4% of methylated CGIs had significant H3K4me3 enrichment compared with 60.9% of unmethylated CGIs (13% versus 65% for CGIs associated with annotated promoters, 2% versus 34% for intragenic CGIs and 4.5% versus 36% for intergenic CGIs) (Fig. 2d; p<0.001, Mann-Whitney U test). H3K4me3 enrichment at a large proportion of intra- and intergenic unmethylated CGIs has been described in other cell types, including ES cells11.
Having identified a large set of methylated CGIs in mature oocytes, we asked what the biological role of such methylation is. The function of genes associated with methylated CGIs as assessed by gene ontology analysis is diverse (data not shown). To assess the impact of methylation on gene expression in oocytes, we compared expression in fully-grown GV oocytes with d10 growing oocytes by mRNA-Seq, but did not observe marked differences in expression levels of transcripts associated with methylated CGIs, including those with methylated promoter CGIs (Fig. 3a). This suggests that most such transcripts accumulate during oocyte growth before the onset of methylation, or that these genes are transcribed from alternative, unmethylated promoters. Such alternative promoters may ensure stringent control of these genes by oocyte-specific factors or environment, and this control might also necessitate that their ‘somatic’ promoters are silenced. If CGI methylation does not contribute significantly to regulation of the maternal mRNA store in oocytes, it may nevertheless have important repercussions for expression of the associated genes post-fertilisation.
Figure 3. Biological significance and fate of CGI methylation in oocytes.
a, mRNA expression levels in d10 and GV oocytes of the genes associated with methylated CGIs, either promoter (red, n=410) or intragenic (blue, n=555). b, Methylation levels in blastocysts of the CGIs identified as methylated in mature oocytes; twelve known germline DMRs with informative coverage are displayed in red (range 45.2%-58.7%). c, Range of methylation in blastocysts of the CGIs methylated specifically in oocytes (n=803) or sperm (n=51), methylated in both oocytes and sperm (n=86) and unmethylated in gametes (n=11512). d, Bisulphite sequencing in GV oocytes, sperm and C57BL/6JxCAST/Ei hybrid E3.5 blastocysts of the Syt2 CGI. Bisulphite sequence profiles from the maternal (mat) and paternal (pat) alleles in blastocysts were discriminated by polymorphisms between C57BL/6J and CAST/Ei. Open circles represent unmethylated CpGs and filled circles methylated CpGs.
Following fertilisation, methylation is comprehensively reprogrammed (except for imprinted genes): the maternal genome is passively demethylated between the zygote and morula and the paternal genome actively in the zygote; this is followed by the establishment of new methylation landscapes2. To evaluate the significance of gamete-derived CGI methylation, we performed RRBS on blastocysts (E3.5). This was validated by the expected degree of methylation at twelve known maternal germline DMRs (range 45.2%-58.7%). Consistent with genome-wide erasure, there was a substantial reduction in the proportion of methylated CpGs (≥60%) across the genome or within CGIs compared with gametes (Fig. 3b, Supplementary Fig. 8a). Crucially, a minority of CGIs methylated in germ cells showed complete protection from demethylation: only ~15% of CGIs methylated in oocytes retained ≥40% methylation in blastocysts (Fig. 3b, c). This substantial post-fertilisation reprogramming suggests that most CGI methylation in oocytes and sperm is unrelated to imprinting, and argues that maintenance of methylation in preimplantation embryos is a decisive factor in imprinting.
However, we observed that most CGIs methylated in oocytes displayed greater levels of methylation in blastocysts than expected if they were fully subject to passive demethylation, by which methylation should be <2% by the 32-cell stage. This was striking, as very few CGIs are methylated in blastocysts (Fig. 3b, c, Supplementary Fig 8b, c). To examine the degree to which gametic methylation is a factor in CGI methylation in preimplantation embryos, we looked at the dependence of methylation in blastocysts on prior methylation in gametes. Of 280 CGIs displaying intermediate methylation levels (25-40%) in blastocysts, the vast majority (234; 83%; p<0.001, χ2 test) were fully methylated in MII oocytes (including 27 CGIs methylated in both oocyte and sperm) (Supplementary Fig. 8d). In contrast, less than 0.5% of CGIs unmethylated in both gametes are methylated ≥25% in blastocysts (Fig. 3c, Supplementary Table 1). To investigate whether CGI sequence influences the likelihood of maintaining methylation, we checked how the properties of CGIs highly methylated in MII oocytes (≥75%) differed according to methylation level in blastocysts. For most parameters the differences were minor, but there was a tendency for CGIs retaining higher levels of methylation to be shorter and to be intragenically located (Supplementary Fig. 8e, f). To validate CGI methylation allele-specifically, we examined a selection of CGIs in C57BL/6JxCast/Ei hybrid embryos by conventional bisulphite sequencing. As exemplified by the Syt2 locus, the CGI is fully methylated in oocytes and the maternal allele partially retains methylation in blastocysts (Fig. 3d, Supplementary Fig. 9). For CGIs specifically methylated in sperm, there was less evidence for substantial maintenance of methylation in blastocysts (Fig. 3c, Supplementary Fig. 9). These findings extend observations of Borgel et al. who, from MeDIP-chip analysis of promoter methylation in preimplantation embryos, identified some non-imprinted sequences that resist demethylation in preimplantation development25. Thus, CGI methylation status in gametes strongly predisposes towards methylation in blastocysts, either by incomplete post-fertilisation demethylation of methylated CGIs, or because some legacy of gametic methylation instructs their re-methylation in a subpopulation of cells. By either mechanism, mosaicism of CGI methylation patterns between blastomeres is predicted to arise. This does not exclude a contribution of de novo methylation, as some CGIs unmethylated in gametes have become methylated in blastocysts (Fig. 3c, Supplementary Fig. 8d, Supplementary Table 1), including genes involved in trophectoderm development26.
In conclusion, we reveal the extent and dynamics of CGI methylation in oocytes; this provides an important reference by which to judge future studies on mechanisms of de novo methylation in germ cells. A comprehensive account of the differential CGI methylation in male and female gametes is also a prerequisite for defining the full repertoire of imprinted genes and the mechanistic basis of parent-of-origin expression effects in somatic tissues. We also describe an unexpectedly complex fate of gamete-derived methylation after fertilisation. Rather than a binary choice, with DMRs characterised by absolute maintenance and other gametic methylation comprehensively lost through active demethylation or lack of maintenance during the first cleavage divisions, our analysis suggests a greater diversity of methylation choices. This diversity might lead to the establishment of epigenetic mosaicism within the early embryo, which might have the potential to influence first lineage specification27.
Methods
Samples
For methylation analysis, blastocysts (54) were collected from E3.5 and oocytes from 5 day (d5, 1883 oocytes), 20 day (GV, 3586 oocytes) and superovulated 4-5 week (MII, 443 oocytes) old C57BL/6J mice. Dnmt3a−/− GV oocytes (635) were obtained from crosses with ZP3-Cre transgenic males and Dnmt3L−/− GV oocytes (843) from DNMT3L-null homozygous females18; the corresponding control oocytes were collected from wild-type littermates (Dnmt3a+/+ 385 oocytes, Dnmt3L+/+ 809 oocytes). For allelic methylation studies, E3.5 blastocysts were collected from C57BL/6JxCAST/Ei hybrids. For mRNA-Seq, oocytes were collected from 10 day (644 d10 growing oocytes) and 35 day (913 fully grown GV oocytes) old C57BL/6J mice. For the H3K4me3 ChIP-Seq, 6400 d15 growing oocytes (CD1) were collected.
RRBS
Genomic DNA was purified using the Qiaamp DNA Micro kit (Qiagen), followed by MspI digestion (Fermentas), end-repair/A-Tailing (Klenow exo-, Fermentas, dCTPs 1nM, dGTPs, 1nM, dATPs 10nM) and 5mC-adapter ligation (T4 Ligase, Fermentas) performed within the same tube and buffer (Tango 1X, Fermentas), by heat-inactivating each enzymatic step followed by adjustment with the reagents required for the next step. Then bisulphite conversion was performed (Imprint DNA Modification, Sigma) and converted DNA was amplified (6 cycles) using a uracil stalling free polymerase (Pfu Turbo Cx, Stratagene), followed by size selection (150-450bp, Qiagen) and second round amplification (10-13 cycles; Platinium Pfx polymerase, Invitrogen). Libraries were purified (SPRI beads, Agencourt) and sequenced on Illumina GA-IIx. Because of low complexity at the start of each sequence (MspI fragments) Bareback was performed (http://www.bioinformatics.bbsrc.ac.uk/projects). Sequence alignment and methylation calls were performed using Bismark28. CpGs with read depth <5 were discarded. For every analysis, all informative CpGs were used. To score CGI methylation, the following cut-offs were applied. Methylation level was determined for CGIs with information on ≥10% of their total CpGs (with a minimum of 5CpGs), and by averaging individual cytosine methylation levels. CGIs with an average methylation level ≥75% and ≤25% were called methylated and unmethylated, respectively. The unmethylated CGIs group corresponds to CGIs called unmethylated in both oocytes and sperm. A false discovery rate (FDR) for CpG methylation was determined empirically from the calls observed for CpGs within 11 known DMRs and was maximally 0.0439% at the minimum level of observation of 5 reads. p values for all methylated CGIs, based on departure from genome-wide methylation levels, were <0.05% (χ2 test with Bonferroni correction for multiple testing). CGIs with contradictory methylation assignments in replicate libraries were excluded from analysis.
mRNA-Seq
Total RNA was extracted (TriZOL, Invitrogen), mRNA purified using oligo-dT Dynabeads (Invitrogen) and fragmented (Ambion), followed by first strand (Superscript III, Invitrogen) and second strand (DNA Pol.I, NEB) synthesis and reaction purification (Minelute, Qiagen). One-step A-tailing and adapter ligation were performed as for RRBS, followed by size selection (250bp +/−25bp, Qiaquick, Qiagen) and library amplification (15cycles, Platinium Pfx polymerase, Invitrogen). Libraries were purified (SPRI beads, Agencourt) and sequenced on Illumina GA-IIx, either single-read for expression analysis or paired-end for gene structure prediction (d10 oocytes). Sequences were aligned using TopHat.
ChIP-Seq
H3K4me3 immunoprecipitation (39159, Active Motif) was performed as described elsewhere29 with minor modifications. ChIP was performed in duplicate from 3200 oocytes. Illumina Libraries were generated (input and IP) using an NEBNext kit (Set 1, NEB), except that adapter ligation was performed as for RRBS. Sequences were aligned using an ungapped Eland alignment with default stringency parameters. Owing to high background results from the limited starting material, reads from duplicates were combined. Technical assessment was made by comparison with ES cell H3K4me3 ChIP-Seq datasets (GSM594581 and GSM535982)11,30.
Direct Bisulphite Sequencing
DNA was purified by proteinase K digestion and phenol-chloroform extraction, spiked with Lambda DNA and bisulphite treated (Zymo). Each PCR comprised a minimum of 50 oocytes or 2-3 blastocyst equivalents. Cloning and analysis were performed as described elsewhere31, with removal of clones with identical patterns of conversion. Primers used for the amplification of specific CGIs from bisulphite modified DNA are given in Supplementary Table 4.2.
Statistical analysis
For categorical data, such as distribution of CpGs or CGIs methylation, χ2 tests were applied. For quantitative data, Mann-Whitney U tests (between 2 groups) and Kruskall-Wallis tests (between more than 2 groups), were applied.
Additional information
Dataset analysis was based on build NCBIM37/mm9 of the mouse genome and performed using Seqmonk (http://www.bioinformatics.bbsrc.ac.uk/projects). Promoter CGIs were defined as overlapping an annotated TSS (EnsEMBL, Refseq or UCSC); intragenic CGIs as overlapping an annotated gene without its TSS; intergenic CGIs were not overlapping annotated genes or promoters. Promoters were defined as the region 2kb upstream of annotated TSS. For repetitive element analysis, positions of individual instances of LINE, SINE, tandem repeats, long terminal repeats (LTR) and low complexity regions (LCR) were extracted from EnsEMBL. The overlap between full length CGIs and individual repeat types was determined as percentage of the CGI length using a custom Perl script. CpG periodicity was determined as the distribution of inter-CpG distance (from C to C) between all pairs of CpGs in each region, averaged over all of the regions in a particular grouping.
Supplementary Material
Acknowledgements
We thank K. Tabbada for technical assistance with Illumina sequencing, H. Mertani, P. Mollard, W. Dean and W. Reik for input and discussions, and M. Branco and W. Reik for making available the Dnmt3a conditional knock-out line. This work was supported by grants G0800013 and G0801156 from the Medical Research Council to G.K. S.A.S. was supported by the Biotechnology and Biological Sciences Research Council, the Babraham Institute and the Centre for Trophoblast Research.
Footnotes
Accession codes. All sequencing files have been deposited in the Sequence Read Archive under the study accession number ERP000689 (European Nucleotide Archive).
Author Information. The authors declare no competing financial interests.
References
- 1.Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Genes & Development. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Human Molecular Genetics. 2005;14:R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 4.Bourc’his D, Xu G-L, Lin C-S, Bollman B, Bestor TH. Dnmt3L and the Establishment of Maternal Genomic Imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 5.Kaneda M, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 6.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaefer CB, Ooi SKT, Bestor TH, Bourc’his D. Epigenetic Decisions in Mammalian Germ Cells. Science. 2007;316:398–399. doi: 10.1126/science.1137544. [DOI] [PubMed] [Google Scholar]
- 8.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009;48:226–232. doi: 10.1016/j.ymeth.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howlett SK, Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991;113:119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- 11.Illingworth RS, et al. Orphan CpG Islands Identify Numerous Conserved Promoters in the Mammalian Genome. PLoS Genet. 2010;6:e1001134. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chotalia M, et al. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes & Development. 2009;23:105–117. doi: 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucifero D, Mann MRW, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Human Molecular Genetics. 2004;13:839–849. doi: 10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- 14.Illingworth R, et al. A Novel CpG Island Set Identifies Tissue-Specific Methylation at Developmental Gene Loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maunakea AK, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinhart B, Paoloni-Giacobino A, Chaillet JR. Specific Differentially Methylated Domain Sequences Direct the Maintenance of Methylation at Imprinted Genes. Mol. Cell. Biol. 2006;26:8347–8356. doi: 10.1128/MCB.00981-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bock C, Halachev K, Buch J, Lengauer T. EpiGRAPH: user-friendly software for statistical analysis and prediction of (epi)genomic data. Genome Biology. 2009;10:R14. doi: 10.1186/gb-2009-10-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 19.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ciccone DN, et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 21.Fang R, et al. Human LSD2/KDM1b/AOF1 Regulates Gene Transcription by Modulating Intragenic H3K4me2 Methylation. Molecular Cell. 2010;39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ooi SKT, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Research. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhayalan A, et al. The Dnmt3a PWWP Domain Reads Histone 3 Lysine 36 Trimethylation and Guides DNA Methylation. Journal of Biological Chemistry. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borgel J, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 26.Tartakover-Matalon S, et al. Impaired migration of trophoblast cells caused by simvastatin is associated with decreased membrane IGF-I receptor, MMP2 activity and HSP27 expression. Human Reproduction. 2007;22:1161–1167. doi: 10.1093/humrep/del464. [DOI] [PubMed] [Google Scholar]
Methods References
- 27.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat Rev Mol Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 28.Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr167. doi:10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (ChIP) Nat. Protocols. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 30.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomizawa S, et al. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–820. doi: 10.1242/dev.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.