Abstract
Microsomal triglyceride transfer protein (MTP) facilitates the transport of dietary and endogenous fat by the intestine and liver by assisting in the assembly and secretion of triglyceride-rich apolipoprotein B-containing lipoproteins. Higher concentrations of apolipoprotein B lipoproteins predispose individuals to various cardiovascular and metabolic diseases such as atherosclerosis, diabetes, obesity and the metabolic syndrome. These can potentially be avoided by reducing MTP activity. In this article, we discuss regulation of MTP during development, cellular differentiation and diurnal variation. Furthermore, we focus on the regulation of MTP that occurs at transcriptional, post-transcriptional and post-translational levels. Transcriptional regulation of MTP depends on a few highly conserved cis-elements in the promoter. Several transcription factors that bind to these elements and either increase or decrease MTP expression have been identified. Additionally, MTP is regulated by macronutrients, hormones and other factors. This article will address the many ways in which MTP is regulated and advance the idea that reducing MTP levels, rather than its inhibition, might be an option to lower plasma lipids.
Keywords: apolipoprotein B, lipid, lipoprotein, microsomal triglyceride transfer protein, regulation, transcription
Cardiovascular diseases are a major cause of mortality and morbidity in Western countries. A major underlying basis for the high incidence of these diseases is excess plasma lipids that have been associated with a shift in the nutritional balance towards an over-consumption of carbohydrates and lipids. Plasma lipids are carried in apolipoprotein (apo)B-containing lipoproteins. Accumulation of remnant or modified apo B-lipoproteins is considered atherogenic as their deposition in the arterial wall is a major event in the initiation of the disease. The apoB-containing lipoproteins chylomicron and VLDL are assembled and secreted by the intestine and liver, respectively. Besides lipids, biosynthesis of these lipoproteins requires apoB and microsomal triglyceride transfer protein (MTP). ApoB is a structural protein always found associated with these lipoproteins. MTP binds and chaperones lipids to the nascent apoB to prevent aberrant folding and degradation by proteasomes and assists in the intracellular assembly of apoB-lipoproteins [1,2].
Microsomal triglyceride transfer protein is an endoplasmic reticulum resident heterodimeric complex of a unique functional MTP subunit and ubiquitously expressed protein disulfide isomerase subunit that transfers various lipids, interacts with apoB and associates with lipid vesicles [2]. Abetalipoproteinemia subjects that carry mutations in their MTP gene have no plasma apoB-lipoproteins [3]. Because of its role in lipoprotein assembly, MTP inhibition is considered an important modality to treat hyperlipidemia and reduce risk for atherosclerosis [4]. In this regard, several MTP inhibitors have been identified that lower plasma lipids [4,5]; however, they have not reached clinical use due to associated adverse events mainly related to accumulation of lipids in the liver. Therefore, attempts are underway to explore intestine-specific inhibition of MTP to lower plasma lipids. The main purpose of summarizing regulation of MTP is to draw attention to the possibility that MTP inhibition, may be achieved indirectly or in a tissue-specific fashion. It is possible that new knowledge about the regulation of MTP will pave the way for novel approaches to inhibit its activity while avoiding associated side effects.
The liver and intestine are the major organs that express MTP and secrete apoB-containing lipoproteins. Other tissues also express MTP but at much lower levels. For example, placenta, heart, ovary, testis, kidney, pancreas, adipose tissue and retina express measurable MTP mRNA [6–10]. Mechanisms controlling different levels of MTP expression in various tissues are unknown. In general, there is a good agreement between MTP mRNA, protein levels and activity indicating that transcriptional control is the main mechanism of MTP regulation. In this article, we will summarize current knowledge about the MTP promoter, transcriptional control mechanisms and post-transcriptional regulatory events that alter MTP activity and apoB-lipoprotein production.
cis-Elements & transcription factors important for MTP expression
The human MTP gene, MTTP, is approximately 55 kb in length comprised of 18 exons and 17 introns [11]. It has been mapped to q22–24 on chromosome 4 by PCR analysis of rodent/human somatic cell hybrids followed by FISH [11,12], while the mouse MTP gene, Mttp, was localized to a distal region of chromosome 3 by Southern blotting [13]. Studies indicate that mice express two forms of MTP; MTP A and MTP B [14,15]. They arise due to alternate splicing of exon 1. The minor MTP B utilizes an alternate promoter and has two extra N-terminal amino acids. Low levels of MTP expression in antigen-presenting cells might arise from the low activity of the alternate promoter [14,16,17]. A high fat diet in mice with cardiac deficiency of the MTP A isoform results in increased expression of the MTP B form suggesting that both these promoters respond to high fat diet [18].
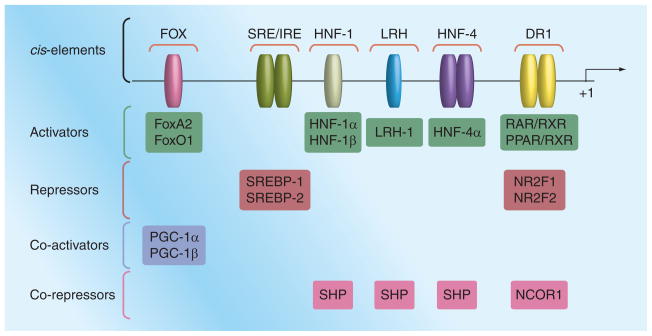
In contrast to mice, humans express one MTP isoform. In vitro promoter-reporter expression studies indicate that the expression of this isoform in the liver and intestine-derived cells depends on approximately 150 bp upstream of the transcription start site [19]. Comparative alignment of these upstream promoter sequences from different species identified several highly conserved motifs [20]. Deletion analysis followed by expression in hepatic cells revealed that the promoter sequence contains critical positive (hepatic nuclear factor [HNF]-1, HNF-4, direct repeat [DR]1 and FOX) and negative regulatory sterol and insulin response element (SRE/IRE) (Figure 1).
Figure 1. A basic microsomal triglyceride transfer protein promoter depicting various cis-elements and transcription factors that bind to these elements.
Various regulatory cis-elements are located in the 600 bp upstream of the transcription start site. These elements bind to various transcription factors (activators) resulting in increased microsomal triglyceride transfer protein promoter activity. They also bind to repressors reducing expression. Additionally, various co-activators and co-repressors modulate microsomal triglyceride transfer protein expression.
DR: Direct repeat; HNF: Hepatic nuclear factor; PGC: Peroxisome proliferator-activated receptor γ coactivator; SHP: Small heterodimeric partner; SRE/IRE: Sterol and insulin response element.
Hepatic nuclear factor-1 element binds to HNF-1α and HNF-1β [Dai K & Hussain M, Unpublished Data], whereas HNF-4 element interacts with the HNF-4α transcription factor [19,21,22]. HNF-4α knockout mice do not express MTP, indicating that it is absolutely required for expression [23]. HNF-1α/β synergistically activate MTP expression along with HNF-4α [22,24]. Hence, HNF-1 and HNF-4 elements and their interactions with HNF-1α/β and HNF-4α transcription factors are critical for basal and synergistic MTP expression.
Several transcription factors bind to the DR1 element. In cells that do not express MTP, this site is occupied by either NR2F1 (also known as COUP-TF1) or NR2F2 (or COUP-TFII). NR2F2 reduces MTP in hepatoma cells [25], whereas NR2F1 suppresses MTP expression in intestinal cells [22]. However, in MTP-expressing cells, RXR-α binds to this site [25]. DR1 sequences are also known to constitute peroxisome proliferator-activated receptor (PPAR) response elements. In fact, MTP expression is increased after treatment with PPARα agonist but not by PPARγ agonist in the liver [26]. Thus, binding of different transcription factors to DR1 might modulate tissue- and cell-specific expression ofMTP.
Overexpression studies [27,28] have identified binding sites for FoxO1 and FoxA2 in the upstream region of the MTP promoter (Figure 1). These transcription factors enhance MTP activity. They respond to insulin signaling and are excluded from the nucleus. Therefore, they might be involved in insulin regulation of MTP (see later).
Microsomal triglyceride transfer protein promoter also harbors a SRE/IRE that negatively regulates MTP expression. Deletion of this element increases MTP promoter activity. Insulin-mediated MTP suppression in HepG2 cells requires this element [19,29]. Sterol regulatory element-binding proteins (SREBPs) bind to this element and reduce MTP expression [29]. However, overexpression of SREBPs in the mouse liver is associated with increased MTP expression [30] most likely due to a secondary effect, as MTP was not identified to be a direct target of SREBPs by microarray analysis [31]. Therefore, it is unclear whether sterol regulation of MTP involves SRE/IRE elements in vivo.
Besides transcription factors that bind to cis-elements in the MTP promoter, several co-activators and co-repressors that bind to these transcription factors and modulate their activity have been identified. Forced expression of PGC-1α and PGC-1β co-activators increases MTP expression [32]. PGC-1β acts as a co-activator with FoxA2 to increase MTP activity [27]. On the other hand, small heterodimeric partner (SHP) represses HNF-4α, LRH-1 and HNF-1α activities by interacting with these factors [21,33,34]. Overexpression and knockdown of SHP reduces and increases MTP activity in mouse liver [33,34]. NCOR1 has been shown to interact with NR2F1 and suppress MTP activity in intestinal cells [22]. Thus, MTP expression is modulated by various co-activators and co-repressors.
In summary, a short MTP promoter with few cis-elements appears to be sufficient for its regulation in cultured cells. HNF-1/HNF-4 and Fox elements are involved in positive regulation of MTP, whereas the SRE/IRE element might be involved in negative regulation. The DR1 element can be engaged to either increase or decrease MTP expression. It remains to be determined whether this small promoter sequence is sufficient for in vivo MTP expression.
Regulation of MTP by macronutrients
MTP gene expression is altered by changes in dietary components. High sucrose diet increases MTP mRNA in the liver but not in the intestine [35] whereas a fructose enriched diet increases MTP mRNA and activity in both the liver and intestine [36]. High saturated fat (hydrogenated coconut oil) [35,37] and cholesterol [38] increase MTP expression in the intestine. In hamsters, long-term high fat diet increases MTP mRNA in the liver and intestine [35]. Increases in MTP activity by saturated fat and cholesterol could potentially involve decreased binding of SREBPs to the SRE/IRE element in the MTP promoter, but this has not been demonstrated in vivo. Recently, Iqbal et al. have proposed that intestinal inositol requiring enzyme 1β (IRE1β) might play a role in this process [38]. In the absence of IRE1β, mice express more MTP in the intestine and develop more hyperlipidemia than control mice when fed a Western diet.
Cell culture experiments suggest regulation of MTP by sterols. Sterol depletion and pravastatin decrease MTP mRNA and protein levels in HepG2 cells [29]. The regulation of MTP by sterols in these cells involves direct interaction of SREBP-2 with the SRE/IRE in the promoter [29]. Oleic acid has been shown to up-regulate MTP in HepG2 cells. This regulation was suggested to not involve SRE/IRE [36]. More studies are needed to determine how MTP is regulated in vitro and in vivo by macronutrients.
Regulation by insulin
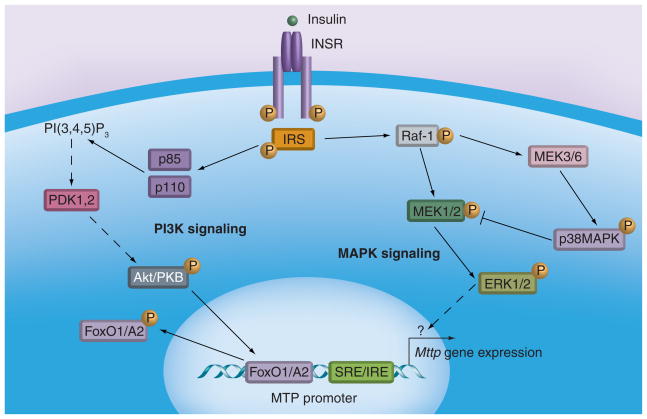
Insulin acutely reduces VLDL secretion. Since MTP is essential for VLDL secretion, it has been speculated that insulin-induced MTP reduction could contribute to decreased VLDL production. Insulin reduces MTP gene transcription in HepG2 cells [19] in a dose- and time-dependent manner [39]. Using different MEK1/2 inhibitors, the dominant negative Raf/MEK1 as well as constitutively active Raf/MEK1 enzymes, Au et al. showed that MTP regulation by insulin involves MAPKerk cascade (Figure 2) [40]. This was confirmed by Allister et al. who also showed that naringenin, a citrus flavonoid, uses the same pathway to suppress MTP expression [41]. Both these studies excluded the PI3-kinase signaling pathway that involves phosphorylation of Akt. It is known that insulin also stimulates MAPKp38, which inhibits the MAPKerk cascade. In fact, inhibition of MAPKp38 increases insulin-mediated MTP suppression involving the MAPKerk cascade [40,41]. The MAPKerk cascade involves phosphorylation and translocation of ERK1/2 (p44/p42) to the nucleus. In the nucleus, these enzymes phosphorylate several transcription factors that ultimately bind to various cis-elements in different genes [42,43]. Subsequent targets of ERK1/2 that ultimately interact with the MTP promoter have not been identified.
Figure 2. Microsomal triglyceride transfer protein regulation by insulin.
Two different pathways regulating microsomal triglyceride transfer protein (MTP) expression have been proposed. In the MAPK signaling pathway, insulin increases phosphorylation of Raf-1, MEK1/2 and ERK1/2, which culminates in reduced expression of MTP in cells. This signaling arm is negatively regulated by MAPKp38. In the PI3K pathway, insulin activates PI3-kinase and Akt/PKB. This leads to increased phosphorylation of forkhead transcription factors, their export from the nucleus and reduced MTP gene transcription.
INSR: Insulin receptor; SRE/IRE: Sterol and insulin response element.
A different type of insulin signaling has been proposed (Figure 2) to involve forkhead transcription factors FoxA2 and FoxO1 [27,28], which probably bind the same binding site within the MTP promoter (Figure 1). These factors are phosphorylated in response to insulin signaling and are excluded from the nucleus. Overexpression of FoxA2 increases MTP expression in ob/ob mice [27]. Its activity is synergistically increased in the presence of PGC1β. Kamagate et al. showed that overexpression of FoxO1 also increases MTP expression and this increased expression is inhibited by insulin or constitutively active Akt [27,28]. Moreover, FoxO1 RNAi decreased hepatic MTP and VLDL production in normal and db/db mice. These data suggest that FoxO1 and FoxA2 play a role in basal hepatic MTP expression.
The above studies indicate that insulin negatively regulates MTP expression. Therefore, it is anticipated that hepatic MTP should be significantly reduced in hyperinsulinemic animals. However, this is not the case. High sucrose fed hamsters show increased MTP expression in the liver [35], whereas fructose-fed hamsters have high hepatic [44] as well as high intestinal [45] MTP. Treatment of fructose-fed hamsters with rosiglitazone improves insulin sensitivity and normalizes MTP expression [46]. Young (6 weeks old) Otsuka Long-Evans Tokushima Fatty (OLETF) rats do not show hyperglycemia and hyperinsulinemia but still have more hepatic MTP [47]. High hepatic MTP levels persist in these rats after the development of hyperglycemia or hyperinsulinemia. Similarly, hyperglycemic and hyperinsulinemic ob/ob mice have high hepatic MTP levels [48]. Also, Zucker obese (fa/fa) rats that show hyperinsulinemia have higher levels of MTP in the liver and intestine [49]. In contrast to these animals, streptozotocin-treated rats [50] and mice [48] or alloxan-treated rabbits [51] show increased intestinal MTP with no change in hepatic MTP expression. A general consensus from these studies is that hyperglycemia and hyperinsulinemia are generally associated with increased, not decreased, MTP expression. This may be the consequence of insulin resistance rather than insulin action. Further studies are necessary to explain the role of insulin and insulin resistance in MTP regulation.
To test the direct effect of insulin on MTP expression, Sparks et al. injected insulin into fasted Apobec1−/− mice that synthesize only apoB100 [52]. Analysis after 2 h revealed reductions in plasma glucose and hepatic FoxO1 and PGC-1α mRNA. There was no significant change in MTP mRNA and activity in these mice suggesting that acute administration of insulin does not decrease MTP expression in these mice, as it does in HepG2 cells. Therefore, it remains to be determined whether insulin is a negative regulator of MTP expression in vivo.
Briefly, MTP expression is regulated by insulin in cultured hepatoma cell lines. Insulin-mediated suppression of MTP in these cells requires the SRE/IRE element. Additionally, loss of the binding of Fox transcription factors to their binding sites also contributes to reduced MTP expression. However, regulation of hepatic MTP by insulin has not been demonstrated in vivo.
Regulation by bile acids
Hirokane et al. have demonstrated that chenodeoxycholate decreases MTP expression in HepG2 cells [21]. Moreover, they showed that the suppressive effect of chenodeoxycholate was mediated by increased SHP expression. Further studies demonstrated that SHP suppresses HNF-4α activity and reduces MTP expression [21]. Thus, bile acids act as negative regulators of MTP expression.
Regulation by leptin
Using various mouse models, Iqbal et al. demonstrated that global leptin receptor deficiency is associated with decreased intestinal MTP expression [53]. Furthermore, they showed that hepatic expression was resilient to leptin receptor deficiency. Increased intestinal MTP expression did not require central leptin signaling in the hypothalamus. Instead, they showed that intestinal cells respond to leptin and regulate MTP levels because they express leptin receptors and its downstream target genes. It remains to be determined how leptin differentially regulates MTP expression in intestinal and hepatic cells and what signaling mechanisms are involved.
Developmental regulation
Expression of MTP is first detected at day 7.5 after gestation [54]. In early development expression mainly occurs in the liver. As the embryo matures the relative expression in the intestine increases compared with the liver and reaches levels higher than liver as seen in the postnatal stage [55]. During embryonic development MTP plays a pivotal role as homozygous knockout mice are not viable and at least suffer from a non-closing anterior neuropore [56]. This phenotype was also observed in apoB knockout mice indicating that MTP is perhaps important for apoB protein stabilization and lipoprotein assembly in embryonic development [57]. Although it cannot be ruled out that homozygous knockout of MTP has a direct intracellular effect on embryonic development it is more likely that visceral endodermal cells that line the inner layer of the yolk sac are involved, since they are embryo derived and produce apoB-containing lipoproteins [58]. These lipoproteins may be used to nurture the embryo and therefore are important for embryonic development. On the other hand, MTP has also been shown to be required for yolk lipid utilization and absorption of dietary lipids in zebrafish larvae [59,60]. However, MTP is not essential for human embryo development as evidenced by the presence of abetalipoproteinemia subjects. Thus, requirement of MTP during embryo development differs in vertebrates.
Although liver is the first organ to express MTP during development, liver-specific MTP knockout mice obtained after crossing with Alb-Cre mice are viable [Khatun I et al., Unpublished Data]. This might be related to late expression of albumin during development. In contrast, deletion of intestine-specific MTP using Villin-Cre transgenic mice does not result in viable progeny. Davidson and associates have obtained intestine-specific knockout mice by postnatal activation of the villin promoter [61]. A clear explanation for a defect in embryonic development in intestine-specific MTP knockout mice is missing, since intestinal function in dietary lipid transport is considered to be important after birth.
Differentiation-dependent induction of MTP in enterocytes
The small intestine is divided anatomically into basal crypts and apical villi. Stem/progenitor cells are present as a layer between crypt and villus cells. Stem/progenitor cells divide, migrate and differentiate into four types of cells; enterocytes, Goblet cells and enteroendocrine cells, all present in the villi, and Paneth cells, at the base of the crypt. Several molecular mechanisms involved in the differentiation of stem cells into villus cells have been identified [62]. An important adaptation during enterocyte maturation is acquisition of the absorptive phenotype necessary for uptake and transport of dietary fat.
To understand differentiation-dependent induction of lipid transport mechanisms, Dai et al. used Caco-2, human colon adenocarcinoma cells [22] that are used extensively to study differentiation and various intestinal functions [63–65] because they possess an inherent genetic ability to become enterocyte-like cells. Quantitative PCR and microarray analyses show strong correlations between gene expression profiles during Caco-2 cell differentiation and that of intestinal epithelia across the crypt–villus axis [66,67]. Differentiation of these cells results in reorganization of cytoskeletal structures, formation of tight junctions, development of cellular polarity and enterocyte-associated functions [65,66,68–70]. Undifferentiated Caco-2 cells do not synthesize or secrete apoB-lipoproteins [71–73]. However, differentiated cells produce chylomicron-size apoB-lipoproteins when media are supplemented with oleic acid [64,71–77]. Dai et al. demonstrated that differentiation-dependent induction of apoB-lipoprotein secretion in Caco-2 cells correlates with the expression of MTP, and not apoB [22]. Mechanistic studies revealed that the MTP promoter engages HNF-4α/HNF-1α in undifferentiated cells and yet is inactive because the DR1 element is occupied by the NR2F1 repressor. During differentiation, NR2F1 expression declines, possibly leading to its decreased binding to the DR1 element and increased expression of MTP. In addition to the transcriptional suppression of MTP by NR2F1, Dai et al. reported that post-transcriptional mechanisms involving IRE1β are also operative in undifferentiated cells [22]. IRE1β expression is high in undifferentiated cells and its levels drop during differentiation. Hence, transcriptional and post-transcriptional mechanisms ensure low expression of MTP in undifferentiated intestinal cells.
To determine whether NR2F1 and IRE1β play a role in MTP expression in mouse intestine, Dai et al. studied the expression of these proteins by immunohistochemistry. There was an inverse relationship in the expression of MTP with NR2F1 and IRE1β along the jejunum to colon axis. Similarly, a reciprocal expression pattern was also seen along the villus to crypt axis in the jejunum. Furthermore, isolated enterocytes expressed higher amounts of MTP and lower amounts of NR2F1 compared with villus cells. They further demonstrated that NR2F1 associates with the MTP promoter in cells that do not express MTP. Indeed, binding of NR2F1 to the MTP promoter was high in crypt but low in villus cells. Therefore, low association of NR2F1 with MTP promoter in villi might contribute to enhanced MTP expression.
In short, these studies have shown that both transcriptional and post-transcriptional mechanisms are involved in differentiation-dependent induction of MTP in enterocytes. In undifferentiated cells, MTP expression is suppressed by the binding of NR2F1 to the DR1 element. Furthermore, high expression of IRE1β might ensure low MTP mRNA levels. During differentiation, expression of these proteins is reduced. Decreased expression of these regulatory proteins increases MTP expression and induces apoB-lipoprotein assembly in differentiated Caco-2 cells.
Circadian regulation
Several biological, physiological and behavioral activities such as feeding, thermogenesis and sleep–wake cycle exhibit circadian rhythms that recur with 24 h intervals and are attuned to sunrise and sunset. Plasma triglyceride and cholesterol concentrations are maintained within a narrow range by balancing lipoprotein production and catabolism and exhibit circadian rhythmicity in humans and rodents [78–82]. Pan and Hussain showed that plasma lipids and MTP expression exhibit synchronized circadian changes in rats and mice and have suggested that changes in intestinal and hepatic MTP might contribute to daily variations in plasma lipids [83]. Changes in MTP and plasma lipids were abrogated when mice were placed in total light or dark for 5 days indicating involvement of light-entrained regulation [84]. Furthermore, surges in both MTP and plasma lipids were altered when mice were subjected to food entrainment [34,83,84]. These diurnal and food-entrained variations were absent in Clock mutant mice that have difficulty in maintaining circadian rhythms. Thus, both light and food entrainment mechanisms regulate plasma lipids and these regulatory mechanisms require normal Clock activity. Pan et al. further showed that Clock regulates diurnal MTP expression at the transcriptional level [34]. MTP gene transcription was high at night and low in the day. Cell culture studies showed that Clock reduces MTP expression at the transcription level [34]. Further studies revealed that Clock increases the expression of SHP, a repressor of MTP [34]. It was further demonstrated that the binding of Clock to the SHP promoter enhances its expression at the onset of light. This leads to increased binding of SHP to the MTP promoter and suppression of its expression during the day. Therefore, SHP is a Clock-controlled gene upregulated by Clock. Higher amounts of SHP suppress MTP expression.
In brief, daily variations in plasma triglyceride have been correlated with changes in intestinal and hepatic MTP. Mechanistic studies revealed that Clock plays an important role in the daily regulation of MTP that is entrained by light as well as food. Clock binds to the SHP promoter and enhances its expression. High levels of SHP negatively regulate MTP expression by interacting and suppressing the activity of different activators.
Post-transcriptional regulation by IRE1β
IRE1β, a homolog of ubiquitously expressed IRE1α, which plays a critical role in unfolded protein response, is primarily expressed in the intestine [85]. Within the intestine, IRE1β protein is detectable in the epithelial cells of stomach, small intestine and colon [22,85]. Its expression increases along the jejunum–colon axis as well as from villus to crypt [22]. Iqbal et al. observed that high cholesterol and Western diets reduce jejunal expression of IRE1β and enhance MTP expression [38]. Ire1b−/− mice express more MTP, absorb more lipids and develop more pronounced hyperlipidemia than controls when challenged with high cholesterol or high fat diets. Mechanistic studies revealed that IRE1β post-transcriptionally cleaves Mttp mRNA initiating its degradation. Hence, IRE1β might act to dampen fat-induced upregulation of intestinal MTP and lipid absorption and act as an anti-lipidemic gene.
Qiu et al. demonstrated that liver-specific ablation of phosphatase and tensin homolog (PTEN) in mice and overexpression of a dominant negative form of PTEN in HepG2 cells lower MTP activity and protein levels by 36–37% [86]. However, under these conditions, reductions in MTP mRNA were modest (−8%). These studies indicate that PTEN might affect MTP protein involving post-transcriptional mechanisms. Further studies are needed to explain how PTEN and AKT lower MTP protein.
Translational & post-translational control
Very little is known about the translational control of MTP. It is known that MTP is a hetero-dimer of 97 and 55 kDa subunits. It is also known that dissociation of the complex leads to irreversible loss of activity. This led to the suggestion that association of these two subunits occurs during or soon after the translation of these peptides. Since, the 55-kDa subunit PDI exists as an independent protein, the translation of this subunit does not require the 97-kDa M subunit. In contrast, the biosynthesis, proper folding and maturation of the 97-kDa subunit requires the 55-kDa subunit.
Pan et al. have demonstrated that CCl4 decreases plasma apoB-lipoproteins and increases hepatic and intestinal lipids in mice in a time-dependent manner [87]. Mechanistic studies revealed that CCl4 decreased apoB-lipoprotein secretion by reducing MTP activity and protein levels without affecting its lipid transfer activity and mRNA levels indicating that CCl4 is not an antagonist and that reductions in MTP activity do not involve transcriptional mechanisms. Furthermore, CCl4 had no effect on MTP biosynthesis but it induced post-translational degradation involving ubiquitinylation and endoplasmic reticulum-associated proteasomal degradation. They also showed that prevention of MTP degradation by proteasomal inhibitors protects against CCl4 toxicity. Further studies showed that free radicals arising from CCl4 by the action of cytochrome P450 oxygenases covalently attach with MTP, inducing its degradation by proteasomes. Thus, MTP degradation after short-term exposure to CCl4 is the major mechanism leading to cellular accumulation of lipids and that covalent modification of MTP leading to its destruction is a key early event in the onset of CCl4-induced steatosis.
The preferential and rapid degradation of MTP after CCl4 exposure suggest that MTP might be involved in the transfer of reactive species and needs further evaluation. It is possible that MTP attempts to transfer damaged lipids and in turn gets damaged. More studies are needed to understand susceptibility of MTP to CCl4 attack. It is unknown whether MTP has residues that are highly susceptible to CCl4 modification.
In short, MTP undergoes post-translational degradation in hepatocytes exposed to CCl4. An unknown metabolite of CCl4 attaches covalently to MTP. This leads to ubiquitinylation and proteasomal degradation of MTP.
Conclusion
In this article, we have discussed regulation of MTP at the transcriptional, post-transcriptional and post-translational levels. The data reviewed indicate that MTP regulation is complex. The major mode of regulation occurs at the transcriptional level. Although several transcription factors, co-activators and co-repressors of MTP have been identified, there is a need for additional information about the mechanisms that control MTP levels in different tissues. This information could be useful in strategically targeting MTP expression and function.
Increased MTP expression during enterocyte differentiation has been shown to involve two proteins, NR2F1 and IRE1β, which negatively regulate MTP expression. The expression of these proteins is high when MTP expression is low. Increases in MTP expression correlate with reduced levels of these proteins.
Bile acid and circadian regulation studies also underscore the importance of repressors in the regulation of MTP. Bile acids increase SHP and reduce MTP expression. Circadian mechanisms also utilize SHP to regulate MTP expression. SHP levels are high in the daytime, which is associated with low MTP expression in rodents. Hence, a major mechanism of MTP regulation involves changes in proteins that suppress MTP expression. It remains to be determined whether these repressors can be augmented to reduce MTP expression.
We have also summarized evidence for the regulation of MTP by lipids, macronutrients and hormones. The exact mechanisms by which these and various other factors exert their effects on MTP levels are unknown. Specifically, molecular mechanisms in the regulation of MTP by insulin, leptin and diurnal variations need to be elucidated.
Future perspective
Direct inhibition of MTP has been associated with undesirable outcomes. There is a need to re-evaluate and come up with new approaches for MTP therapy. The purpose of summarizing MTP regulation was to draw attention to the possibility that MTP reduction, perhaps, can be achieved indirectly or in a tissue-specific fashion involving strategies that do not involve inhibition of lipid transfer activity. If such a mechanism is identified then it might be possible to reduce MTP levels partially in all or a specific tissue to lower plasma lipids and avoid risk of cardiovascular and metabolic diseases. An option to achieve such a goal is to target upstream regulators of MTP. In this regard, transcription factors that regulate MTP expression can be targeted. Perhaps interfering with the synergistic activation of MTP by HNF1/HNF-4a can be used to lower MTP expression. Another possibility is to upregulate proteins that negatively regulate MTP. If MTP levels can be reduced with acceptable side effects, MTP suppressors, besides lowering lipids, might also be useful in weight reduction. Additionally, because of its role in CD1 biosynthesis, MTP suppressors/reducers may be useful in the treatment of natural killer T-cell mediated disorders.
Executive summary.
Microsomal triglyceride transfer protein (MTP) regulation involves a simple promoter that harbors few conserved cis-elements.
Activators, suppressors, co-activators and co-suppressors that regulate MTP have been identified.
MTP is regulated by lipids and hormones; however, the molecular mechanisms involved have not been explained.
MTP is critical for the development of the embryo in mice but not in humans.
MTP gene expression during enterocyte differentiation depends on decreased expression of NR2F1 suppressor and IRE1β endoribonuclease.
CLOCK and small heterodimeric partner play an important role in the diurnal regulation of MTP expression.
CCl4-induced steatosis involves post-translational degradation of MTP.
Acknowledgments
The authors are grateful for critical comments by Janet Sparks.
Footnotes
Financial & competing interests disclosure
This work was supported in part by NIH grants DK46900 and HL95924 to M Mahmood Hussain. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Gordon DA, Jamil H. Progress towards understanding the role of microsomal triglyceride transfer protein in apolipoprotein-B lipoprotein assembly. Biochim Biophys Acta. 2000;1486:72–83. doi: 10.1016/s1388-1981(00)00049-4. [DOI] [PubMed] [Google Scholar]
- 2.Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apolipoprotein B-lipoprotein assembly. J Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- 3.Berriot-Varoqueaux N, Aggerbeck LP, Samson-Bouma M, Wetterau JR. The role of the microsomal triglygeride transfer protein in abetalipoproteinemia. Annu Rev Nutr. 2000;20:663–697. doi: 10.1146/annurev.nutr.20.1.663. [DOI] [PubMed] [Google Scholar]
- 4.Hussain MM, Bakillah A. New approaches to target microsomal triglyceride transfer protein. Curr Opin Lipidol. 2008;19:572–578. doi: 10.1097/MOL.0b013e328312707c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paras C, Hussain MM, Rosenson RS. Emerging drugs for hyperlipidemia. Expert Opin Emerg Drugs. 2010;15:433–451. doi: 10.1517/14728214.2010.481282. [DOI] [PubMed] [Google Scholar]
- 6.Madsen EM, Lindegaard ML, Andersen CB, Damm P, Nielsen LB. Human placenta secretes apolipoprotein B-100-containing lipoproteins. J Biol Chem. 2004;279:55271–55276. doi: 10.1074/jbc.M411404200. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen LB, Veniant M, Boren J, et al. Genes for apolipoprotein B and microsomal triglyceride transfer protein are expressed in the heart: evidence that the heart has the capacity to synthesize and secrete lipoproteins. Circulation. 1998;98:13–16. doi: 10.1161/01.cir.98.1.13. [DOI] [PubMed] [Google Scholar]
- 8.Shoulders CC, Brett DJ, Bayliss JD, et al. Abetalipoproteinemia is caused by defects of the gene encoding the 97 kDa subunit of a microsomal triglyceride transfer protein. Hum Mol Genet. 1993;2:2109–2116. doi: 10.1093/hmg/2.12.2109. [DOI] [PubMed] [Google Scholar]
- 9.Swift LL, Kakkad B, Boone C, et al. Microsomal triglyceride transfer protein expression in adipocytes: a new component in fat metabolism. FEBS Lett. 2005;579:3183–3189. doi: 10.1016/j.febslet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Li CM, Presley JB, Zhang X, et al. Retina expresses microsomal triglyceride transfer protein: implications for age-related maculopathy. J Lipid Res. 2005;46:628–640. doi: 10.1194/jlr.M400428-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Sharp D, Ricci B, Kienzle B, Lin MCM, Wetterau JR. Human microsomal triglyceride transfer protein large subunit gene structure. Biochemistry. 1994;33:9057–9061. doi: 10.1021/bi00197a005. [DOI] [PubMed] [Google Scholar]
- 12.Narcisi TM, Shoulders CC, Chester SA, et al. Mutations of the microsomal triglyceride-transfer-protein gene in Aβ lipoproteinemia. Am J Hum Genet. 1995;57:1298–1310. [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamuta M, Chang BH, Hoogeveen R, Li WH, Chan L. Mouse microsomal triglyceride transfer protein large subunit: cDNA cloning, tissue-specific expression and chromosomal localization. Genomics. 1996;33:313–316. doi: 10.1006/geno.1996.0200. [DOI] [PubMed] [Google Scholar]
- 14.Dougan SK, Rava P, Hussain MM, Blumberg RS. MTP regulated by an alternate promoter is essential for NKT cell development. J Exp Med. 2007;204:533–545. doi: 10.1084/jem.20062006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohler PJ, Zhu MY, Blade AM, Ham AJ, Shelness GS, Swift LL. Identification of a novel isoform of microsomal triglyceride transfer protein. J Biol Chem. 2007;282:26981–26988. doi: 10.1074/jbc.M700500200. [DOI] [PubMed] [Google Scholar]
- 16.Brozovic S, Nagaishi T, Yoshida M, et al. CD1d function is regulated by microsomal triglyceride transfer protein. Nat Med. 2004;10:535–539. doi: 10.1038/nm1043. [DOI] [PubMed] [Google Scholar]
- 17.Dougan SK, Salas A, Rava P, et al. Microsomal triglyceride transfer protein: lipidation and control of CD1d on antigen presenting cells. J Exp Med. 2005;202:529–539. doi: 10.1084/jem.20050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartels ED, Nielsen JM, Hellgren LI, Ploug T, Nielsen LB. Cardiac expression of microsomal triglyceride transfer protein is increased in obesity and serves to attenuate cardiac triglyceride accumulation. PLoS One. 2009;4:e5300. doi: 10.1371/journal.pone.0005300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagan DL, Kienzle B, Jamil H, Hariharan N. Transcriptional regulation of human and hamster microsomal triglyceride transfer protein genes – cell type-specific expression and response to metabolic regulators. J Biol Chem. 1994;269:28737–28744. [PubMed] [Google Scholar]
- 20.Hussain MM, Rava P, Pan X, et al. Microsomal triglyceride transfer protein in plasma and cellular lipid metabolism. Curr Opin Lipidol. 2008;19:277–284. doi: 10.1097/MOL.0b013e3282feea85. [DOI] [PubMed] [Google Scholar]
- 21.Hirokane H, Nakahara M, Tachibana S, Shimizu M, Sato R. Bile acid reduces the secretion of very low density lipoprotein by repressing microsomal triglyceride transfer protein gene expression mediated by hepatocyte nuclear factor-4. J Biol Chem. 2004;279:45685–45692. doi: 10.1074/jbc.M404255200. [DOI] [PubMed] [Google Scholar]
- 22▪▪.Dai K, Khatun I, Hussain MM. NR2F1 and IRE1β suppress MTP expression and lipoprotein assembly in undifferentiated intestinal epithelial cells. Arterioscler Thromb Vasc Biol. 2010;30:568–574. doi: 10.1161/ATVBAHA.109.198135. Describes molecular mechanisms involved in the expression of apolipoproteinB-lipoprotein assembly and secretion during differentiation of Caco-2 cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheena V, Hertz R, Nousbeck J, Berman I, Magenheim J, Bar-Tana J. Transcriptional regulation of human microsomal triglyceride transfer protein by hepatocyte nuclear factor-4α. J Lipid Res. 2005;46:328–341. doi: 10.1194/jlr.M400371-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Kang S, Spann NJ, Hui TY, Davis RA. ARP-1/COUP-TF II determines hepatoma phenotype by acting as both a transcriptional repressor of microsomal triglyceride transfer protein and an inducer of CYP7A1. J Biol Chem. 2003;278:30478–30486. doi: 10.1074/jbc.M304201200. [DOI] [PubMed] [Google Scholar]
- 26.Ameen C, Edvardsson U, Ljungberg A, et al. Activation of peroxisome proliferator-activated receptor α increases the expression and activity of microsomal triglyceride transfer protein in the liver. J Biol Chem. 2005;280:1224–1229. doi: 10.1074/jbc.M412107200. [DOI] [PubMed] [Google Scholar]
- 27▪▪.Wolfrum C, Stoffel M. Coactivation of Foxa2 through Pgc-1β promotes liver fatty acid oxidation and triglyceride/VLDL secretion. Cell Metab. 2006;3:99–110. doi: 10.1016/j.cmet.2006.01.001. Demonstrates that microsomal triglyceride transfer protein (MTP) expression is increased after the forced expression of FoxA2 and that PGC-1β acts as a co-activator with FoxA2 to increase MTP expression. [DOI] [PubMed] [Google Scholar]
- 28▪▪.Kamagate A, Qu S, Perdomo G, et al. FoxO1 mediates insulin-dependent regulation of hepatic VLDL production in mice. J Clin Invest. 2008;118:2347–2364. doi: 10.1172/JCI32914. Shows that overexpression of FoxO1 increases MTP expression. Furthermore, siFoxO1 decreases MTP expression indicating that FoxO1 plays a role in MTP regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato R, Miyamoto W, Inoue J, Terada T, Imanaka T, Maeda M. Sterol regulatory element-binding protein negatively regulates microsomal triglyceride transfer protein gene transcription. J Biol Chem. 1999;274:24714–24720. doi: 10.1074/jbc.274.35.24714. [DOI] [PubMed] [Google Scholar]
- 30.Horton JD, Shimomura I, Brown MS, Hammer RE, Goldstein JL, Shimano H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest. 1998;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horton JD, Shah NA, Warrington JA, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin J, Yang R, Tarr PT, et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1β coactivation of SREBP. Cell. 2005;120:261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Iqbal J, Saha PK, et al. Molecular characterization of the role of orphan receptor small heterodimer partner in development of fatty liver. Hepatology. 2007;46:147–157. doi: 10.1002/hep.21632. [DOI] [PubMed] [Google Scholar]
- 34▪▪.Pan X, Zhang Y, Wang L, Hussain MM. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010;12:174–186. doi: 10.1016/j.cmet.2010.05.014. Shows that CLOCK regulates MTP expression and plasma lipid levels by directly controlling small heterodimeric partner that acts as a repressor of MTP expression. Therefore, high tissue levels of the small heterodimeric partner are correlated with low MTP levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin MCM, Arbeeny C, Bergquist K, Kienzle B, Gordon DA, Wetterau JR. Cloning and regulation of hamster microsomal triglyceride transfer protein – the regulation is independent from that of other hepatic and intestinal proteins which participate in the transport of fatty acids and triglycerides. J Biol Chem. 1994;269:29138–29145. [PubMed] [Google Scholar]
- 36.Qiu W, Taghibiglou C, Avramoglu RK, et al. Oleate-mediated stimulation of microsomal triglyceride transfer protein (MTP) gene promoter: implications for hepatic MTP overexpression in insulin resistance. Biochemistry. 2005;44:3041–3049. doi: 10.1021/bi047803+. [DOI] [PubMed] [Google Scholar]
- 37.Hernandez Vallejo SJ, Alqub M, Luquet S, et al. Short-term adaptation of postprandial lipoprotein secretion and intestinal gene expression to a high-fat diet. Am J Physiol Gastrointest Liver Physiol. 2009;296:G782–G792. doi: 10.1152/ajpgi.90324.2008. [DOI] [PubMed] [Google Scholar]
- 38▪▪.Iqbal J, Dai K, Seimon T, et al. IRE1β inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008;7:445–455. doi: 10.1016/j.cmet.2008.03.005. Identified IRE1β as a tissue-specific endoribonuclease that controls MTP levels and lipid absorption by post-trancriptionally cleaving MTP mRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin MCM, Gordon D, Wetterau JR. Microsomal triglyceride transfer protein (MTP) regulation in HepG2 cells: insulin negatively regulates MTP gene expression. J Lipid Res. 1995;36:1073–1081. [PubMed] [Google Scholar]
- 40.Au WS, Kung HF, Lin MC. Regulation of microsomal triglyceride transfer protein gene by insulin in HepG2 cells: roles of MAPKerk and MAPKp38. Diabetes. 2003;52:1073–1080. doi: 10.2337/diabetes.52.5.1073. [DOI] [PubMed] [Google Scholar]
- 41.Allister EM, Borradaile NM, Edwards JY, Huff MW. Inhibition of microsomal triglyceride transfer protein expression and apolipoprotein B100 secretion by the citrus flavonoid naringenin and by insulin involves activation of the mitogen-activated protein kinase pathway in hepatocytes. Diabetes. 2005;54:1676–1683. doi: 10.2337/diabetes.54.6.1676. [DOI] [PubMed] [Google Scholar]
- 42.Rose BA, Force T, Wang Y. Mitogen-activated protein kinase signaling in the heart: angels versus demons in a heart-breaking tale. Physiol Rev. 2010;90:1507–1546. doi: 10.1152/physrev.00054.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death: is subcellular localization the answer? Cell Cycle. 2009;8:1168–1175. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taghibiglou C, Carpentier A, Van Iderstine SC, et al. Mechanisms of hepatic very low density lipoprotein overproduction in insulin resistance. Evidence for enhanced lipoprotein assembly, reduced intracellular ApoB degradation, and increased microsomal triglyceride transfer protein in a fructose-fed hamster model. J Biol Chem. 2000;275:8416–8425. doi: 10.1074/jbc.275.12.8416. [DOI] [PubMed] [Google Scholar]
- 45.Haidari M, Leung N, Mahbub F, et al. Fasting and postprandial overproduction of intestinally derived lipoproteins in an animal model of insulin resistance. Evidence that chronic fructose feeding in the hamster is accompanied by enhanced intestinal de novo lipogenesis and ApoB48-containing lipoprotein overproduction. J Biol Chem. 2002;277:31646–31655. doi: 10.1074/jbc.M200544200. [DOI] [PubMed] [Google Scholar]
- 46.Carpentier A, Taghibiglou C, Leung N, et al. Ameliorated hepatic insulin resistance is associated with normalization of microsomal triglyceride transfer protein expression and reduction in very low density lipoprotein assembly and secretion in the fructose-fed hamster. J Biol Chem. 2002;277:28795–28802. doi: 10.1074/jbc.M204568200. [DOI] [PubMed] [Google Scholar]
- 47.Kuriyama H, Yamashita S, Shimomura I, et al. Enhanced expression of hepatic acyl-coenzyme A synthetase and microsomal triglyceride transfer protein messenger RNAs in the obese and hypertriglyceridemic rat with visceral fat accumulation. Hepatology. 1998;27:557–562. doi: 10.1002/hep.510270233. [DOI] [PubMed] [Google Scholar]
- 48.Bartels ED, Lauritsen M, Nielsen LB. Hepatic expression of microsomal triglyceride transfer protein and in vivo secretion of triglyceride-rich lipoproteins are increased in obese diabetic mice. Diabetes. 2002;51:1233–1239. doi: 10.2337/diabetes.51.4.1233. [DOI] [PubMed] [Google Scholar]
- 49.Phillips C, Owens D, Collins P, Tomkin GH. Microsomal triglyceride transfer protein: does insulin resistance play a role in the regulation of chylomicron assembly? Atherosclerosis. 2002;160:355–360. doi: 10.1016/s0021-9150(01)00721-3. [DOI] [PubMed] [Google Scholar]
- 50.Gleeson A, Anderton K, Owens D, et al. The role of microsomal triglyceride transfer protein and dietary cholesterol in chylomicron production in diabetes. Diabetologia. 1999;42:944–948. doi: 10.1007/s001250051252. [DOI] [PubMed] [Google Scholar]
- 51.Phillips C, Bennett A, Anderton K, et al. Intestinal rather than hepatic microsomal triglyceride transfer protein as a cause of postprandial dyslipidemia in diabetes. Metabolism. 2002;51:847–852. doi: 10.1053/meta.2002.33350. [DOI] [PubMed] [Google Scholar]
- 52▪.Sparks JD, Chamberlain JM, O’Dell C, Khatun I, Hussain MM, Sparks CE. Acute suppression of apoB secretion by insulin occurs independently of MTP. Biochem Biophys Res Commun. 2011;406:252–256. doi: 10.1016/j.bbrc.2011.02.028. Shows that insulin does not decrease MTP expression in the liver. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iqbal J, Li X, Chang BH, et al. An intrinsic gut leptin-melanocortin pathway modulates intestinal microsomal triglyceride transfer protein and lipid absorption. J Lipid Res. 2010;51:1929–1942. doi: 10.1194/jlr.M005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terasawa Y, Cases SJ, Wong JS, et al. Apolipoprotein B-related gene expression and ultrastructural characteristics of lipoprotein secretion in mouse yolk sac during embryonic development. J Lipid Res. 1999;40:1967–1977. [PubMed] [Google Scholar]
- 55.Shelton JM, Lee MH, Richardson JA, Patel SB. Microsomal triglyceride transfer protein expression during mouse development. J Lipid Res. 2000;41:532–537. [PubMed] [Google Scholar]
- 56.Raabe M, Flynn LM, Zlot CH, et al. Knockout of the abetalipoproteinemia gene in mice: reduced lipoprotein secretion in heterozygotes and embryonic lethality in homozygotes. Proc Natl Acad Sci USA. 1998;95:8686–8691. doi: 10.1073/pnas.95.15.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farese RV, Jr, Ruland SL, Flynn LM, Stokowski RP, Young SG. Knockout of the mouse apolipoprotein B gene results in embryonic lethality in homozygotes and protection against diet-induced hypercholesterolemia in heterozygotes. Proc Natl Acad Sci USA. 1995;92:1774–1778. doi: 10.1073/pnas.92.5.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farese RV, Jr, Cases S, Ruland SL, et al. A novel function for apolipoprotein B. lipoprotein synthesis in the yolk sac is critical for maternal-fetal lipid transport in mice. J Lipid Res. 1996;37:347–360. [PubMed] [Google Scholar]
- 59.Marza E, Barthe C, Andre M, Villeneuve L, Helou C, Babin PJ. Developmental expression and nutritional regulation of a zebrafish gene homologous to mammalian microsomal triglyceride transfer protein large subunit. Dev Dyn. 2005;232:506–518. doi: 10.1002/dvdy.20251. [DOI] [PubMed] [Google Scholar]
- 60.Schlegel A, Stainier DY. Microsomal triglyceride transfer protein is required for yolk lipid utilization and absorption of dietary lipids in zebrafish larvae. Biochemistry. 2006;45:15179–15187. doi: 10.1021/bi0619268. [DOI] [PubMed] [Google Scholar]
- 61.Xie Y, Newberry EP, Young SG, et al. Compensatory increase in hepatic lipogenesis in mice with conditional intestine-specific Mttp deficiency. J Biol Chem. 2006;281:4075–4086. doi: 10.1074/jbc.M510622200. [DOI] [PubMed] [Google Scholar]
- 62.Scoville DH, Sato T, He XC, Li L. Current view: intestinal stem cells and signaling. Gastroenterology. 2008;134:849–864. doi: 10.1053/j.gastro.2008.01.079. [DOI] [PubMed] [Google Scholar]
- 63.Hussain MM, Glick JM, Rothblat GH. In vitro model systems: cell cultures used in lipid and lipoprotein research. Curr Opin Lipidol. 1992;3:173–178. [Google Scholar]
- 64.Hussain MM, Kancha RK, Zhou Z, Luchoomun J, Zu H, Bakillah A. Chylomicron assembly and catabolism: role of apolipoproteins and receptors. Biochim Biophys Acta. 1996;1300:151–170. doi: 10.1016/0005-2760(96)00041-0. [DOI] [PubMed] [Google Scholar]
- 65.Sun H, Chow EC, Liu S, Du Y, Pang KS. The Caco-2 cell monolayer: usefulness and limitations. Expert Opin Drug Metab Toxicol. 2008;4:395–411. doi: 10.1517/17425255.4.4.395. [DOI] [PubMed] [Google Scholar]
- 66.Saaf AM, Halbleib JM, Chen X, et al. Parallels between global transcriptional programs of polarizing Caco-2 intestinal epithelial cells in vitro and gene expression programs in normal colon and colon cancer. Mol Biol Cell. 2007;18:4245–4260. doi: 10.1091/mbc.E07-04-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mariadason JM, Nicholas C, L’Italien KE, et al. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology. 2005;128:1081–1088. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- 68.Engle MJ, Goetz GS, Alpers DH. Caco-2 cells express a combination of colonocyte and enterocyte phenotypes. J Cell Physiol. 1998;174:362–369. doi: 10.1002/(SICI)1097-4652(199803)174:3<362::AID-JCP10>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 69.Halbleib JM, Saaf AM, Brown PO, Nelson WJ. Transcriptional modulation of genes encoding structural characteristics of differentiating enterocytes during development of a polarized epithelium in vitro. Mol Biol Cell. 2007;18:4261–4278. doi: 10.1091/mbc.E07-04-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simon-Assmann P, Turck N, Sidhoum-Jenny M, Gradwohl G, Kedinger M. Invitro models of intestinal epithelial cell differentiation. Cell Biol Toxicol. 2007;23:241–256. doi: 10.1007/s10565-006-0175-0. [DOI] [PubMed] [Google Scholar]
- 71.Hughes TE, Sasak WV, Ordovas JM, Forte TM, Lamon-Fava S, Schaefer EJ. A novel cell line (Caco-2) for the study of intestinal lipoprotein synthesis. J Biol Chem. 1987;262:3762–3767. [PubMed] [Google Scholar]
- 72.Moberly JB, Cole TG, Schonfeld G. Oleic acid stimulation of apolipoprotein B secretion from HepG2 and Caco-2 cells occurs post-transcriptionally. Biochim Biophys Acta. 1990;1042:70–80. doi: 10.1016/0005-2760(90)90058-6. [DOI] [PubMed] [Google Scholar]
- 73.Liao W, Chan L. Apolipoprotein B, a paradigm for proteins regulated by intracellular degradation, does not undergo intracellular degradation in CaCo2 cells. J Biol Chem. 2000;275:3950–3956. doi: 10.1074/jbc.275.6.3950. [DOI] [PubMed] [Google Scholar]
- 74.Luchoomun J, Hussain MM. Assembly and secretion of chylomicrons by differentiated Caco-2 cells: nascent triglycerides and preformed phospholipids are preferentially used for lipoprotein assembly. J Biol Chem. 1999;274:19565–19572. doi: 10.1074/jbc.274.28.19565. [DOI] [PubMed] [Google Scholar]
- 75.Hussain MM. A proposed model for the assembly of chylomicrons. Atherosclerosis. 2000;148:1–15. doi: 10.1016/s0021-9150(99)00397-4. [DOI] [PubMed] [Google Scholar]
- 76.Hussain MM, Kedees MH, Singh K, Athar H, Jamali NZ. Signposts in the assembly of chylomicrons. Front Biosci. 2001;6:D320–D331. doi: 10.2741/hussain. [DOI] [PubMed] [Google Scholar]
- 77.Dashti N, Smith EA, Alaupovic P. Increased production of apolipoprotein B and its lipoproteins by oleic acid in Caco-2 cells. J Lipid Res. 1990;31:113–123. [PubMed] [Google Scholar]
- 78.Hussain MM, Pan X. Clock genes, intestinal transport and plasma lipid homeostasis. Trends Endocrinol Metab. 2009;20:177–185. doi: 10.1016/j.tem.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maillot F, Garrigue MA, Pinault M, et al. Changes in plasma triacylglycerol concentrations after sequential lunch and dinner in healthy subjects. Diabetes Metab. 2005;31:69–77. doi: 10.1016/s1262-3636(07)70169-6. [DOI] [PubMed] [Google Scholar]
- 80.Mondola P, Gambardella P, Santangelo F, Santillo M, Greco AM. Circadian rhythms of lipid and apolipoprotein pattern in adult fasted rats. Physiol Behav. 1995;58:175–180. doi: 10.1016/0031-9384(95)00016-c. [DOI] [PubMed] [Google Scholar]
- 81.Balasubramaniam S, Szanto A, Roach PD. Circadian rhythm in hepatic low-density-lipoprotein (LDL)-receptor expression and plasma LDL levels. Biochem J. 1994;298(Pt 1):39–43. doi: 10.1042/bj2980039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bruckdorfer KR, Kang SS, Khan IH, Bourne AR, Yudkin J. Diurnal changes in the concentrations of plasma lipids, sugars, insulin and corticosterone in rats fed diets containing various carbohydrates. Horm Metab Res. 1974;6:99–106. doi: 10.1055/s-0028-1093890. [DOI] [PubMed] [Google Scholar]
- 83.Pan X, Hussain MM. Diurnal regulation of microsomal triglyceride transfer protein and plasma lipid levels. J Biol Chem. 2007;282:24707–24719. doi: 10.1074/jbc.M701305200. [DOI] [PubMed] [Google Scholar]
- 84.Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J Lipid Res. 2009;50:1800–1813. doi: 10.1194/jlr.M900085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bertolotti A, Wang X, Novoa I, et al. Increased sensitivity to dextran sodium sulfate colitis in IRE1β-deficient mice. J Clin Invest. 2001;107:585–593. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qiu W, Federico L, Naples M, et al. Phosphatase and tensin homolog (PTEN) regulates hepatic lipogenesis, microsomal triglyceride transfer protein, and the secretion of apolipoprotein B-containing lipoproteins. Hepatology. 2008;48:1799–1809. doi: 10.1002/hep.22565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87▪▪.Pan X, Hussain FN, Iqbal J, Feuerman MH, Hussain MM. Inhibiting proteasomal degradation of microsomal triglyceride transfer protein prevents CCl4 induced steatosis. J Biol Chem. 2007;282:17078–17089. doi: 10.1074/jbc.M701742200. First paper to describe that CCl4-induced steatosis involves post-translational degradation of MTP involving ubiquitinylation and proteasomes. [DOI] [PubMed] [Google Scholar]