Abstract
The bacterial second messenger c-di-GMP is used in many species to control essential processes that allow the organism to adapt to its environment. The c-di-GMP riboswitch (GEMM) is an important downstream target in this signaling pathway and alters gene expression in response to changing concentrations of c-di-GMP. The riboswitch selectively recognizes its second messenger ligand primarily through contacts with two critical nucleotides. However, these two nucleotides are not the most highly conserved residues within the riboswitch sequence. Instead, nucleotides that stack with c-di-GMP and that form tertiary RNA contacts are the most invariant. Biochemical and structural evidence reveals that the most common natural variants are able to make alternative pairing interactions with both guanine bases of the ligand. Additionally, a high resolution (2.3 Å) crystal structure of the native complex reveals that a single metal coordinates the c-di-GMP backbone. Evidence is also provided that after transcription of the first nucleotide on the 3′-side of the P1 helix, which is predicted to be the molecular switch, the aptamer is functional for ligand binding. Although large energetic effects occur when several residues in the RNA are altered, mutations at the most conserved positions, rather than at positions that base pair with c-di-GMP, have the most detrimental effects on binding. Many mutants retain sufficient c-di-GMP affinity for the RNA to remain biologically relevant, which suggests that this motif is quite resilient to mutation.
The symmetric dinucleotide bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) acts as a second messenger signaling molecule in many bacterial species. It regulates processes central to bacterial adaptability including motility, biofilm formation, and virulence (1–5). Diverse environmental or intracellular signals trigger either the production of c-di-GMP by diguanylate cyclase (DGC) enzymes or its degradation to pGpG and/or GMP by specific phosphodiesterases (PDEs) (1, 6). In order to generate a phenotypic response to changing c-di-GMP concentrations, macromolecular targets within the cell interact with this second messenger. Several families of protein receptors have been discovered that use different mechanisms to alter bacterial behavior in response to c-di-GMP (1, 6–17).
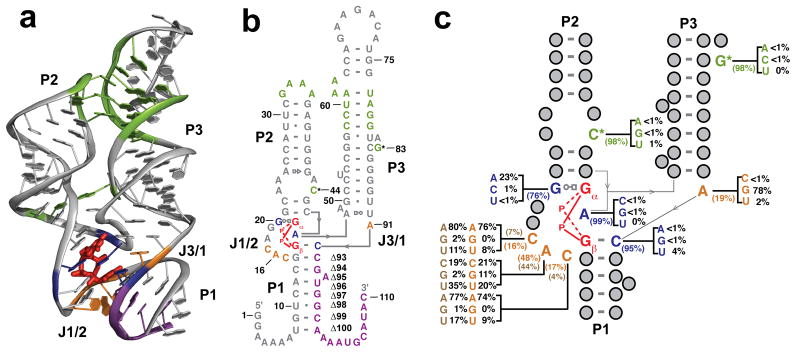
In addition to protein receptors, the RNA c-di-GMP riboswitch (GEMM riboswitch) has been implicated as an important c-di-GMP target. This riboswitch resides in the 5′ untranslated region (UTR) of genes involved in many pathways that are regulated by c-di-GMP signaling (18, 19). Like other riboswitch classes, these non-coding RNA segments contain a domain that binds the ligand and is followed by a second element that modulates gene expression at the transcriptional or translational level (20). We have recently reported the atomic resolution structure of the ligand binding (aptamer) domain of a c-di-GMP riboswitch from Vibrio cholerae bound to c-di-GMP at 2.7 Å resolution (Figure 1a) (21). The RNA is composed of three helices and binds its ligand at the three-helix junction. The tertiary architecture is supported by a tetraloop/tetraloop receptor (TL/TLR) interaction between the P2 and P3 helices, as well as a universally conserved base pair between C44 and G83 that bridges these two stems (Figure 1).
Figure 1.
Structure of the Vc2 c-di-GMP riboswitch aptamer from V. cholerae highlighting regions studied by mutagenesis. a. Crystal structure representation of the aptamer. The cocrystallization protein U1A has been omitted for clarity. The regions highlighted in green represent tertiary contacts that will be disrupted in this study. The purple region shows the nucleotides on the 3′ end of the P1 helix that will be truncated. The orange nucleotides are those mutated in the study of the J1/2 region. Blue nucleotides directly contact c-di-GMP, which is shown in red. b. Secondary structure representation of the 110 Vc2 c-di-GMP aptamer construct used for biochemical analysis. Coloring is the same as in part a. c. Representation of the three-helix junction of the aptamer showing phylogenetic conservation of nucleotides studied by mutagenesis. Coloring is the same as in parts a and b. The orange values for nucleotides in the J1/2 region represent the distribution of nucleotides when nucleotide 91 is an A; the brown values represent the distribution of nucleotides when nucleotide 91 is a G.
At the same time that our structure was reported, a second structure of the same RNA was published by Kulshina et al. at 3.2 Å resolution (22). The observed global architecture, as well as the interactions involved in ligand recognition, is consistent between the two structures. Additionally, these authors performed SAXS and nuclease protection experiments. They concluded that major conformational changes occur in the riboswitch upon c-di-GMP binding, including stabilization of the binding pocket, formation of the P1 helix and interaction between helices P2 and P3 (22). Despite the existence of these two structures, a high resolution native structure has not yet been reported that would elucidate the potential role of Mg2+ in c-di-GMP binding. In our previously reported structure we observed metals coordinated to the phosphodiester backbone of c-di-GMP (21). However, the structure was of an iridium hexammine derivative and therefore we could not definitively establish the presence and location of metals in the native state. No metals were reported in the second structure of the c-di-GMP riboswitch described, most likely due to the moderate resolution of that structure (22). Here we report the 2.3 Å crystal structure of the native complex and describe the nature of the metal ion interactions with the c-di-GMP backbone.
In both of the published structures, the guanine bases of c-di-GMP are specifically and asymmetrically recognized by two nucleotides. G20 makes contacts to the Hoogsteen face of the top guanine base, Gα, while C92 forms a standard Watson-Crick base pair with the lower guanine, Gβ. A universally conserved adenosine at position 47 stacks between the two bases (Figure 1) (21, 22). The interactions formed between the riboswitch and c-di-GMP contribute to an extremely tight binding affinity (Kd approximately 10 pM) for this complex, the highest affinity of any c-di-GMP receptor identified and the tightest RNA-small molecule interaction reported to date. This high affinity and the resulting slow off-rate suggests that this is a kinetically controlled riboswitch, the activity of which is modulated primarily by the on-rate of ligand binding (21).
Phylogenetic analysis reveals an unanticipated pattern of sequence conservation within the riboswitch. Nucleotides that directly interact with the guanine bases of c-di-GMP are not the most highly conserved residues in the structure. The most invariant bases are instead those that form stacking interactions with the ligand and participate in tertiary interactions within the RNA (Figure 1c). This phylogenetic curiosity suggests that it may be possible to form alternative interactions with the bases of c-di-GMP and retain a functional riboswitch. Here we present biochemical and structural characterization of the most commonly observed sequence variants as well as mutational analysis of several regions of the RNA to define the sequence flexibility of the riboswitch. Specifically, we have systematically mutated the nucleotides involved in ligand recognition, those involved in forming critical tertiary contacts and nucleotides in the J1/2 region that form the edge of the binding pocket (Figure 1a,b). We have also measured kon and koff for select mutants to establish which step in the binding process is affected.
Biochemical and structural evidence suggests that the P1 helix forms when c-di-GMP binds (19, 21, 22). This is likely to be the molecular trigger for riboswitch signaling and provides a potential mechanism by which ligand binding can alter gene expression. It is not clear whether P1 must be present in order for c-di-GMP to bind. Here we establish the 3′-portion of the P1 helix that is required for c-di-GMP binding to define the point in the RNA transcript at which the aptamer becomes functional.
Materials and Methods
Materials
RNA molecules were cloned and transcribed as previously reported (21). All mutant RNAs used for biochemical analysis were in the background of the 110 Vc2 RNA (19, 21) (Figure 1b) unless otherwise stated. RNAs used for crystallography included a binding site for the U1A protein and shortened 5′ and 3′ ends as described previously (21). Radiolabeled c-di-GMP was synthesized enzymatically as described (21, 23). Both c-di-GMP and c-di-AMP were chemically synthesized on solid support using p-methoxy protected phosphoramidites purchased from ChemGenes (24). Each compound was purified by semi-preparative high performance liquid chromatography (HPLC) using triethylammonium acetate, pH 6.0 as the stationary phase and acetonitrile as the mobile phase.
Methods
Kd, kon, and koff measurements by gel-shift analysis
Equilibrium dissociation constants were measured by incubating increasing amounts of RNA with trace radiolabeled c-di-GMP and separating bound and free ligand on a native gel as described previously (21). Briefly, riboswitch RNAs were folded in the presence of radiolabeled c-di-GMP and folding buffer (10 mM sodium cacodylate, pH 6.8, 10 mM MgCl2, 10 mM KCl) by heating to 70 ºC for 3 minutes and slow cooling. The binding reactions were then incubated at room temperature until equilibrium was achieved, usually about 1 hour. For high affinity RNAs, the incubation time was increased until no change in the fraction bound was observed, indicating equilibrium had been reached. This was possible for all mutants with an incubation time of ≤ 95 hours. Bound and free c-di-GMP were resolved on a native gel (100 mM Tris/HEPES, pH 7.5, 10 mM MgCl2, 0.1 mM EDTA) at 4 ºC. Because the Kd of the G20A mutant was quite low (210 pM), it was confirmed by taking the ratio of the koff and kon values. kon and koff values were measured as described (21) using a 100-fold excess of unlabeled c-di-GMP as the quench and fit to equations 1 and 2, respectively:
| (1) |
| (2) |
where F=fraction bound, A=amplitude, k=rate, t=time, F0=fraction bound at time=0, F∞=fraction bound at time=∞ and kon was calculated by dividing k from equation 1 by the RNA concentration used in the experiment. ΔGbind and ΔΔGbind values were calculated using the following equations:
| (3) |
| (4) |
where R=gas constant and T=absolute temperature.
Crystallization
The native and G20A mutant complexes were crystallized by hanging drop vapor diffusion as described (21), without soaking with iridium hexammine. The RNA complex contained 100 μM wild-type or G20A RNA, 215 μM c-di-GMP, and 140 μM U1A protein in folding buffer consisting of 10 mM MgCl2, 10 mM KCl, and 10 mM sodium cacodylate, pH 6.8. This solution was mixed in either a 1:1 or 2:3 ratio with well solution, consisting of 22% PEG 550 MME, 5 mM MgSO4, 50 mM MES, pH 6.0, and 300 mM NaCl. Both the C92U mutant complex with c-di-GMP as well as the G20A/C92U mutant complex with c-di-AMP were crystallized under similar conditions with the concentration of PEG 550 MME increased to 25% (v/v). Crystals of the G20A/C92U mutant complex with c-di-GMP were obtained by streak seeding with native crystals after the drops equilibrated for 3–6 hours. The crystallization condition was identical to that used to obtain the C92U mutant crystals but the RNA was at a slightly lower concentration of 70–90 μM. All crystals were stabilized in 30% PEG 550 MME, 5 mM MgSO4, 50 mM MES, pH 6.0, 300 mM NaCl, and 200–500 μM of the appropriate ligand and flash frozen in liquid nitrogen.
Structure determination and refinement
Diffraction data were collected at beamline X25 at the National Syncrotron Light Source (NSLS) at Brookhaven National Laboratory (BNL) and processed using HKL2000 (25). The diffraction pattern was slightly anisotropic, causing the completeness to be less than 100% at the highest resolution. In the case of the native structure, I/σ was approximately 1 at 2.1 Å, but the completeness was only 56% in the highest resolution shell. A completeness of >95% in the highest resolution bin was only observed at resolutions of 2.4 Å or lower. We report the resolution to 2.3 Å, where I/σ is > 2 and completeness is approximately 90% in the highest resolution shell. All structures were solved by molecular replacement using Phaser (26). The search model consisted of the U1A protein used for co-crystallization and the RNA from the structure reported previously (PDB ID 3IRW). Nucleotides in and around the binding pocket were removed from the initial search model in order to remove model bias. Clear density was observed for the ligand in all cases. Rebuilding was done in Coot (27) and refinement was performed in Refmac (28). Figures were made using PyMol (DeLano Scientific) (29).
Results
Structure of the native complex
We determined the native structure of the c-di-GMP bound riboswitch at 2.3 Å resolution (See Table S1 for crystallographic statistics). This higher resolution structure reveals greater detail in P1, including an alternative pairing within the helix, and the presence of one fully hydrated magnesium ion coordinating the backbone of c-di-GMP.
The improved resolution makes it possible to fully establish the pairing interactions in the P1 helix. In the original riboswitch structure, U96 was modeled in a base pair with A12, leaving G97 to pair with C11. G98 was not included in the model (21). The higher resolution electron density maps provide evidence that U96 is instead flipped out of the helix to form a crystal contact with a symmetry related molecule (Figure S1). G97 then forms an AG pair with A12 and G98 a Watson-Crick pair with C11. The conformation of U96 is most likely a result of crystallization and we anticipate that the A12/U96 Watson-Crick pair forms in solution. The 5′ end of P1 also forms crystal contacts with other portions of the RNA. Two GC pairs and a UU pair are formed between G8, G9, and U10 with nucleotides in the U1A binding loop that were inserted for co-crystallization (Figure S1). While the G8 and G9 pairs are likely the result of a having a shorter 3′ than 5′ end of P1 and many, if not all, of these alternative pairings are likely crystallization artifacts, they are indicative of a helix that possesses some measure of instability. The propensity to form alternative pairings with other RNA segments is consistent with its anticipated function as a molecular switch.
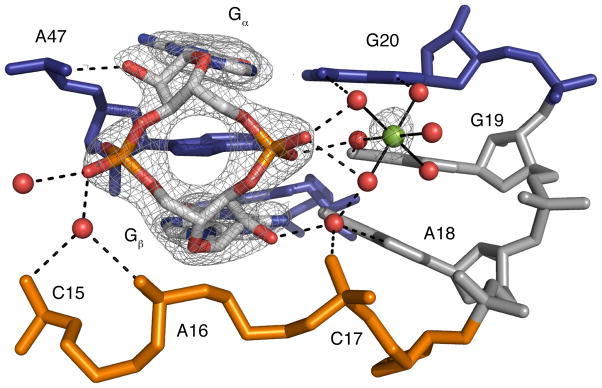
The native structure reveals that the phosphate 5′ of Gα (PGα) is coordinated by a hydrated magnesium ion, while the phosphate 5′ of Gβ (PGβ) is not contacted by any metals (Figure 2). In the iridium hexammine bound structure, a large anomalous signal was observed near PGα, indicating the presence of an iridium hexammine ion. A peak was observed at this position in the native structure in the initial map from molecular replacement (approximately 2σ). A fully hydrated magnesium with the metal placed at the center of the peak fits the density well. This structure reveals that the two non-bridging phosphate oxygens make outer-sphere contacts with three of the water molecules coordinated to the magnesium (Figure 2). Two RNA residues, G19 and G20, also make contact to this metal. Of the nine iridium hexammine ions observed in the previous structure, this is the only case in which a corresponding magnesium ion was detected in the native structure.
Figure 2.
Native complex of the riboswitch with c-di-GMP. Coloring of nucleotides is the same as in Figure 1 and c-di-GMP is colored by atom, with oxygen shown in red, nitrogen in blue, and carbon in white. Magnesium is shown as a green sphere and water molecules are shown as red spheres. 2FO-FC omit density contoured at 1 σ is shown in gray around c-di-GMP and the hydrated magnesium ion. 2FO-FC omit density was calculated by removing c-di-GMP and the hydrated metal prior to map calculation. Hydrogen bonds are shown as black dashed lines.
No density corresponding to metal ions was observed near PGβ. Two waters were observed that make hydrogen bonds to one of the phosphate oxygens, but neither are at a distance consistent with a metal contact. In the iridium hexammine structure, two weak peaks were observed near PGβ (21). The absence of such a peak in the native structure, as well as the lack of any strong density near this phosphate in either of the structures, suggests that metals are not involved in binding PGβ.
As was observed in the derivative structure, asymmetry is present in contacts to the guanine bases, both in type and number (Figure S2). Gα is contacted at N7 and O6 by G20 and at N2 by the 3′-O of A48 and the 2′-OH of C46. No hydrogen bonds are made to N3 and N1 is contacted by a water molecule. Gβ is more extensively contacted. Every hetero-atom of Gβ except N9 is involved in hydrogen bonding with the riboswitch. The Watson-Crick face is contacted by C92 with N2 additionally forming an interaction with N1 of A18. N3 is contacted by N4 of C17 and N7 by the 2′-OH of A47 (Figure S2). This extensive contact made to c-di-GMP by the riboswitch contributes to the extremely high affinity of this RNA for its ligand.
Analysis of nucleotides in direct contact with c-di-GMP
Despite the apparent importance of contacts made to the bases of c-di-GMP, G20 and C92, the nucleotides that interact with Gα and Gβ,, are not universally conserved, but they are conserved as a purine and a pyrimidine, respectively. Instead, the adenosine that intercalates between the bases of the ligand (A47) and the residues that form the base pair between the P2 and P3 helices (C44 and G83) are the most highly conserved nucleotides in the riboswitch (Figure 1c). This observation suggests that other residues may be able to substitute at critical positions and make an alternative set of interactions with the bases of c-di-GMP (Figure 1c). To determine whether this lack of strict conservation leads to an ability to tolerate mutations at these key locations, we mutated G20, C92, and A47 and determined the consequence on c-di-GMP affinity.
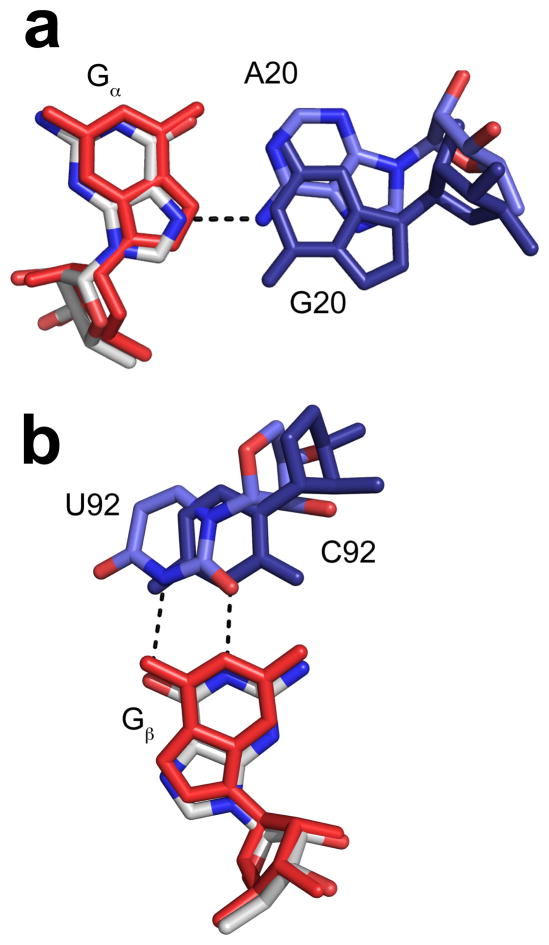
Mutational and structural analysis supports the crystallographic observation that G20 is important for c-di-GMP binding but is consistent with the phylogenetic prediction that an adenosine can be accommodated at this position. Mutation to an adenosine resulted in only a 20-fold loss in binding affinity. To understand this modest effect, we solved the crystal structure (2.9 Å resolution) of the G20A point mutant and determined how A20 interacts with c-di-GMP. The global architecture is unchanged relative to the wild-type RNA. A20 occupies a position similar to G20 but the orientation of the base is shifted slightly, allowing it to make a contact with the N7 of Gα (Figure 3a). This places the N1 of A20 within hydrogen bonding distance of O6 of Gα (2.8 Å), which raises the possibility that at acidic pHs the N1 of A20 may be protonated. In contrast to the modest effect of the G to A transition, mutation of G20 to a C or a U resulted in 400 and >10,000-fold effects, respectively (Table 1).
Figure 3.
Structures of G20A and C92U mutant riboswitches bound to c-di-GMP. a. Overlay of the wild-type structure with the G20A mutant structure. c-di-GMP from the wild-type complex is shown in red. c-di-GMP from the G20A mutant complex is colored by atom with oxygen shown in red, nitrogen in blue, and carbon in white. G20 from the wild-type structure is shown in dark blue and the A20 mutant is colored by atom with oxygen shown in red, nitrogen in blue, and carbon in light blue. The hydrogen bond is shown as a black dashed line. b. Overlay of the wild-type structure with the C92U mutant structure. c-di-GMP from the wild-type complex is shown in red. c-di-GMP from the C92U mutant complex is colored by atom as in a. C92U from the wild-type structure is shown in dark blue and the U92 mutant is colored by atom with oxygen shown in red, nitrogen in blue, and carbon in light blue. Hydrogen bonds are shown as black dashed lines.
Table 1.
Binding affinities for binding pocket mutants.
| RNA | Kd (nM) | ΔΔGbind (kcal/mol) |
|---|---|---|
| Vc2 110 (Wild-type) | 0.011 | 0 |
|
| ||
| G20A | 0.21 ± 0.067 | 1.8 |
| G20C | 3.9 ± 0.18 | 3.5 |
| G20U | 330 ± 150 | 6.1 |
|
| ||
| C92A | 97 ± 15 | 5.4 |
| C92G | 21000 ± 5800 | 8.6 |
| C92Ua | 15 ± 1.1 | 4.3 |
|
| ||
| A47C | 8500 ± 3000 | 8.0 |
| A47G | 4200 ± 1700 | 7.6 |
| A47U | 3500 ± 520 | 7.5 |
All values reported for C92U were reported previously (21).
The base pair between C92 and Gβ can also accommodate mutation although the magnitude of the effects are larger. Mutation of C92 to each of the other three nucleotides resulted in effects ranging from 1500 to 2,000,000-fold (Table 1). The most common natural variant, C92U, is the least destabilizing, resulting in a 1500-fold loss of affinity for c-di-GMP. To define how U92 is able to recognize Gβ, we determined the structure of the C92U mutant riboswitch bound to c-di-GMP (3.0 Å resolution). The only change relative to wild-type is observed in the binding pocket, where U92 forms a GU wobble pair with Gβ (Figure 3b). There is ample room for U92 to shift relative to C92 in order to adopt this conformation. Mutation of C92 to nucleotides not able to form standard base pairs with Gβ resulted in substantial decreases in affinity for c-di-GMP, 10,000-fold and 2,000,000-fold for A and G, respectively (Table 1). In fact, the C92G mutation is the most detrimental of all the mutations made to the binding pocket, resulting in a nearly 9 kcal/mol loss in binding energy. This suggests that in addition to not being able to pair with Gβ, G92 may be detrimental to the fold of the RNA by mispairing with C15 and extending the P1 helix into the binding pocket (see J1/2 data below).
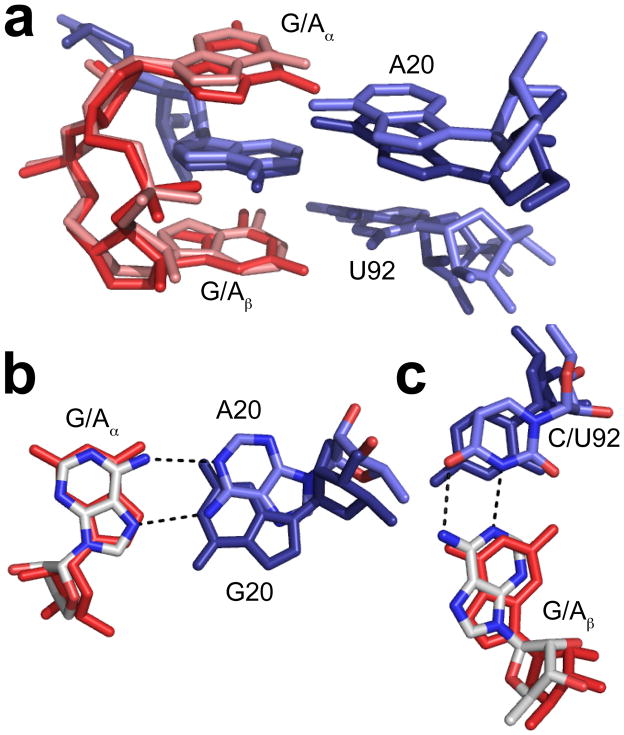
To further investigate the interactions of binding pocket nucleotides with the bases of the ligand, we solved two additional crystal structures. We previously reported that a double mutant riboswitch, G20A/C92U, was able to bind both c-di-GMP and a related molecule c-di-AMP (21). The double mutant has a 4-fold preference for c-di-AMP over c-di-GMP. We determined the structure of this double mutant bound to both c-di-GMP (3.0 Å resolution) and to c-di-AMP (3.2 Å resolution) in order to characterize the nature of these interactions. In both cases, unambiguous density was observed for the cyclic dinucleotides, confirming that they bind to the RNA in the same orientation (Figure 4a). In the c-di-GMP bound structure, Gα interacts with A20 the same as in the single G20A mutant structure described above and Gβ forms a GU wobble pair as observed in the C92U mutant structure. In the case of the c-di-AMP ligand, A20 is able to form two hydrogen bonds with Aα (Figure 4b) and Aβ forms a standard Watson-Crick base pair with U92 (Figure 4c). The presence of two hydrogen bonds between A20 and Aα compared with the single hydrogen bond formed with Gα may account for the 4-fold preference of this mutant for c-di-AMP over c-di-GMP.
Figure 4.
Structures of the G20A/C92U double mutant riboswitch bound to c-di-GMP and c-di-AMP. a. Overlay of the structures of the G20A/C92U mutant riboswitch bound to c-di-GMP and the same RNA bound to c-di-AMP. c-di-GMP is shown in red and c-di-AMP is shown in pink. Binding pocket nucleotides corresponding to the c-di-GMP structure are shown in dark blue and those from the c-di-AMP structure are shown in light blue. b. Overlay of the wild-type structure with the G20A/C92U c-di-AMP bound structure, shown at Aα. c-di-GMP from the wild-type complex is shown in red. c-di-AMP is colored by atom as in Figure 3. G20 from the wild-type structure is shown in dark blue and A20 is colored by atom with oxygen shown in red, nitrogen in blue, and carbon in light blue.. Hydrogen bonds are shown as black dashed lines. c. Overlay of the wild-type structure with the G20A/C92U c-di-AMP bound structure, shown at Aβ. c-di-GMP from the wild-type complex is shown in red. c-di-AMP is colored by atom as in Figure 3. C92 from the wild-type structure is shown in dark blue and U92 is colored by atom with oxygen shown in red, nitrogen in blue, and carbon in light blue. Hydrogen bonds are shown as black dashed lines.
In contrast to the other two residues directly involved in ligand binding, A47 is completely intolerant of mutation, despite the fact that A47 does not directly base pair with either of the Gs of c-di-GMP. All mutations at A47 resulted in extremely large decreases (>100,000-fold) in binding affinity (Table 1). These are among the largest effects observed for any point mutations tested in this study, an observation consistent with the high degree of conservation. We anticipate that the mutational effects are large because of both reduced base stacking efficiency in the case of the pyrimidines and steric clashes with the ligand. The A47G mutation likely creates a clash between the O6 of G47 and one of the non-bridging oxygens of c-di-GMP.
In summary, these data establish that the magnitudes of the mutational effects are closely correlated with the degree of conservation. They also suggest that contacts to Gβ may contribute more to ligand binding than those made to Gα.
Helix packing mediated by the C44/G83 base pair and the TL/TLR
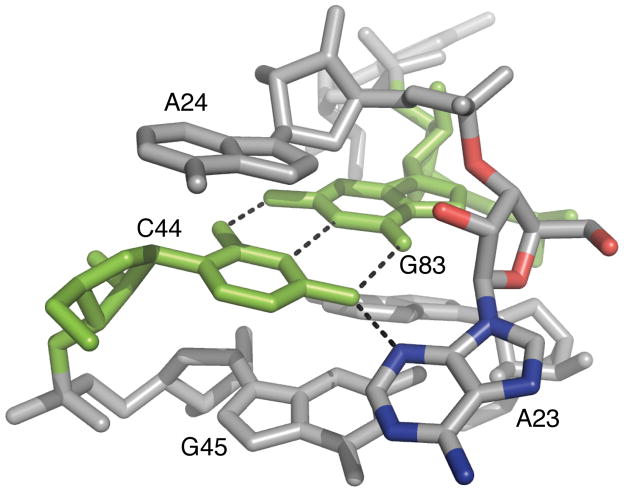
Two nucleotides that show absolute conservation in the c-di-GMP riboswitch consensus secondary structure are C44 and G83. Prior to the structure determination, it was anticipated that C44 might play a direct role in base pairing to one of the c-di-GMP Gs because it is the only absolutely conserved cytidine in the RNA (19). Somewhat surprisingly, the structure revealed that this base was involved in a base pair with G83 and was well-removed from the c-di-GMP binding site (~10 Å). This long-range base pair with G83 stabilizes tertiary packing of the P2 and P3 helices. The high level of conservation suggests that this base pair may play an important role in both orienting the two helices with respect to one another and aiding the proper folding of the binding pocket. To test the importance of this CG base pair, we mutated it to a UG wobble and a UA pair, and also introduced a GG mispair.
All mutations to the C44/G83 base pair had dramatic effects on c-di-GMP affinity. Introduction of a UG wobble pair (C44U) resulted in a >100,000-fold loss in affinity for c-di-GMP, while mutation to a UA pair (C44U/G83A) caused an almost 20,000-fold decrease. These conservative mutations were just as destabilizing as the G-G mispair (C44G) which resulted in a 25,000-fold effect on the Kd (Table 2). The large decrease in affinity upon mutation to a UG or a UA pair was unexpected but is consistent with the absolute conservation of this base pair as a CG pair. Closer inspection of the structure revealed that C44 hydrogen bonds with A23, a highly conserved nucleotide in the bulge in the P2 stem (Figure 5). In the C44U mutant RNA, not only would this interaction be lost but the shifted conformation of the GU wobble pair is likely to create a steric clash with A23. The decrease in affinity from a CG to a UA pair most likely reflects the greater stability of CG pairs as well as the loss of the hydrogen bond between C44 and A23. However, these hypotheses did not fully explain the magnitude of the decrease in affinity from mutation of a CG to a UA pair (5.8 kcal/mol). Because this interaction is involved in forming critical interactions necessary for the fold of the RNA, we decided to further investigate this mutant by measuring the on and off-rates of ligand binding.
Table 2.
Binding affinities for c-di-GMP riboswitch mutants mutants with disrupted tertiary contacts.
| RNA | Kd (nM) | ΔΔGbind (kcal/mol) |
|---|---|---|
| Wild-type | 0.011 | 0 |
|
| ||
| C44U | 1500 ± 460 | 7.0 |
| C44U/G83A | 190 ± 8.6 | 5.8 |
| C44G | 250 ± 5.2 | 5.9 |
|
| ||
| GUAA | 2.2 ± 0.77 | 3.1 |
| Helical | <0.5a | < 2.3 |
| UUCG | 11 ± 1.8 | 4.1 |
This value could not be measured using equilibrium methods and an upper estimate is given.
Figure 5.
C44/G83 base pair that bridges the P2 and P3 helices. C44 and G83 are shown in green, oxygen and nitrogen atoms of A23 are shown in red and blue respectively. Hydrogen bonds are shown as black dashed lines.
The koff of the C44U/G83A mutant is >2000-fold faster while the kon is 10-fold slower than the wild-type value, suggesting that formation of this base pair may be rate-limiting for forming a binding competent RNA (Table 5). One hypothesis is that this base pair is not fully formed in the apo aptamer but must be made during the binding process. This may correlate with SAXS data, which predict this base-pairing interaction is not present in the unbound state (22). The increase in the off-rate is consistent with what was observed previously when the off-rate for the C92U mutant was measured. In that case, the on-rate was equivalent to the wild-type value and the change in affinity was observed as an increase in the off-rate (21).
Table 5.
Rate constants for select binding pocket, tertiary contact, and P1 mutants.
| RNA | Kd (nM) | kon (M−1 min−1) | koff (min−1) |
|---|---|---|---|
| Wild-type | 0.011 | 1.0 ± 0.016 × 106 | 1.1 ± 0.082 × 10−5 |
|
| |||
| C92Ua | 15 ± 1.1 | 8.0 ± 0.99 × 105 | 1.0 ± 0.16 × 10−2 |
| G20A | 0.21 ± 0.067 | 9.1 ± 1.8 × 105 | 2.2 ± 0.077 × 10−4 |
|
| |||
| C44U/G83A | 190 ± 8.6 | 8.7 ± 1.2 × 104 | 2.5 ± 0.25 × 10−2 |
|
| |||
| Vc2 Δ95 | 63 ± 13 | 2 × 105 b | 1.3 ± 0.10 × 10−2 |
| Vc2 Δ96 | 5.4 ± 0.59 | 2 × 106 b | 1.0 ± 0.10 × 10−2 |
| Vc2 Δ98 | 5.9 ± 0.79 | 5.0 ± 1.0 × 105 | 3 × 10−3 b |
| Vc2 Δ99 | < 0.8c | 1.3 ± 0.40 × 106 | - |
| Vc2 Δ100 | < 0.5c | 1.2 ± 0.30 × 106 | - |
All values reported for C92U were reported previously (21).
Calculated from the equation Kd=koff/kon.
These values could not be measured using equilibrium methods and upper estimates are given.
In addition to the C44/G83 base pair, the TL/TLR interaction that occurs between the top of P2 and P3 is likely to play an important role in stabilizing the overall fold of the RNA. This interaction appears to be conserved in c-di-GMP riboswitch sequences, but naturally occurring variations of the TL and TLRs motifs are observed (18, 19). We investigated the effect of disrupting the TL/TLR interaction on c-di-GMP binding by altering the TL as well as the TLR. We first made a conservative mutation in the TL, changing the GAAA sequence to GUAA, which is still in the GNRA TL family and is present in many c-di-GMP riboswitch sequences (18, 19). This mutant lost approximately 200-fold in affinity for c-di-GMP, but retained a Kd in the low nanomolar range. Naturally occurring GAAA loops are rarely found with helical receptors (30) so we also attempted to disrupt the TL/TLR interaction by making the TLR helical by deleting two As from the sequence, A62 and A63. This mutant had an affinity that was too tight to measure by equilibrium methods (<0.5 nM), suggesting that the TL/TLR interaction remained at least partially intact. We then replaced the TL with a stable, but completely different TL, UUCG, that does not interact with 11-nt receptors (30). This mutation resulted in an approximately 1000-fold loss in affinity for c-di-GMP. Although this is a large effect (4.1 kcal/mol), even this most disruptive mutation resulted in an affinity that was still in the low nanomolar range, revealing that the TL/TLR interaction can be mutated without a large loss in affinity for c-di-GMP.
Role of the single stranded J1/2 and J3/1 regions in ligand binding
In addition to residues that make direct and specific contacts to c-di-GMP and those involved in tertiary contacts, other elements may contribute to the riboswitch’s ability to bind this second messenger. Two regions that we investigated are the linker segments, J1/2 and J3/1, that form the edge of the binding pocket (Figure 1). The J1/2 region, specifically nucleotides 15–17, is of particular interest because these nucleotides are located close to c-di-GMP but do not show strong sequence conservation (Figure 1c). Additionally, the orientation of these residues is different in the two reported structures of this riboswitch (21, 22), placing these nucleotides in different proximities to the backbone and bases of c-di-GMP. Such differences leave open the possibility that these nucleotides are flexible in the ligand bound state and/or that a subset of the residues make direct contacts to the ligand. The electron density in this region in the iridium hexammine structure was poorly defined, which made it difficult to assign the exact positions of the bases. The electron density for these nucleotides in the high resolution native structure confirms that the majority of the J1/2 bases are pointing away from the backbone of c-di-GMP, although one nucleotide appears to form a single hydrogen bond with a base of the ligand. C15 and A16 are primarily involved in stacking contacts with the P1 nucleotide A95 (Figure S1) and are far from the ligand, while the exocyclic amine of C17 contacts the N3 of Gβ ((Figure S2).
In order to test if C17 or other residues in J1/2 contribute to ligand binding affinity, we made mutations that reveal a preference against Us and Gs in this region. All mutations were studied in the background of the C44U/G83A double mutant due to the extremely tight affinity of the wild-type RNA for c-di-GMP (10 pM), the slow on-rate, and the consequential slow approach to equilibrium. This RNA binds c-di-GMP with a Kd of 190 nM, allowing us to measure equilibrium binding affinities without an extensive pre-equilibration period (incubation of ≤ 1 hour). We found that mutation of C17 to an A had only a 3-fold effect on binding affinity (Table 3), consistent with its phylogenetic conservation as an A (Figure 1c). However, both the C17U and C17G mutants resulted in at least a 800-fold loss in affinity for the ligand (no binding could be detected up to 150 μM RNA). The unexpectedly large magnitude of these effects led us to determine if mutations elsewhere in J1/2 would produce a similar result. C15U and A16U mutations resulted in a 6-fold and a 100-fold decrease respectively, while the C15G mutation rendered the RNA incapable of binding ligand (Table 3).
Table 3.
Binding affinities for J1/2 mutants.
| RNA | Kd (nM) | Fold decrease | ΔΔGbind (kcal/mol) |
|---|---|---|---|
| Wild-typea | 190 ± 8.6 | 1 | 0 |
|
| |||
| C15A/C17A | 580 ± 57 | 3.1 | 0.66 |
| C17U | > 150,000b | > 800 | > 4.0 |
| C17G | > 150,000b | > 800 | > 4.0 |
| C15U | 1100 ± 71 | 5.8 | 1.0 |
| C15G | > 150,000b | > 800 | > 4.0 |
| A16U | 20,000 ± 460 | 110 | 2.8 |
| A91G | 100 ± 16 | 0.53 | −0.38 |
| A16U/A91G | 1400 ± 58 | 7.4 | 1.2 |
| C17U/A91G | 11,000 ± 500 | 58 | 2.4 |
All J1/2 mutations were made in the background of the C44U/G83A mutation.
No binding was detected at 150 μM RNA.
Although consistent with a direct role for C17 in binding, the > 4.0 kcal/mol loss in binding energy upon mutation of C17 to a U or a G seemed too large to result simply from loss of one hydrogen bond (Table 3). Phylogenetic evidence suggests that the identity of nucleotides in J1/2 is important, not only for contacting c-di-GMP, but also to prevent misfolding of the RNA. A negative covariation between nucleotides in J1/2 and J3/1 is observed in the phylogeny (Figure 1c), specifically a selection against base pairing that could lead to misfolding of the binding pocket. There is a bias against Us in the J1/2 region when there is an A at position 91, as there is in this sequence, whereas there are fewer Cs in J1/2 when position 91 is a G. Guanines are quite rare at all three positions (Figure 1c). This suggests that a G in the J1/2 region could displace Gβ by forming a Watson-Crick base pair with C92 and continuing the P1 helix into the binding pocket. This observation could explain the result with the C17G and C15G mutations which had no detectable affinity for c-di-GMP. A similar hypothesis would also explain the large effect of the C92G mutation (see above).
We tested for possible extension of the P1 helix by mispairing of J1/2 with residue 91 in J3/1. We specifically asked if the effects of the U mutations at position 15–17 could be rescued by making a non-compensatory A91G mutation in J3/1. In isolation the A91G mutant showed a 2-fold greater affinity for c-di-GMP compared to the wild-type (Table 3). Consistent with this hypothesis, both the A16U/A91G and C17U/A91G mutants resulted in affinity for c-di-GMP that was increased by at least 14-fold relative to the A16U and C17U single mutants (Table 3). However, this does not explain why the A91G mutation does not cause a decrease in affinity in the wild-type sequence where there are Cs present at positions 15 and 17. This suggests there is another source of J1/2 misfolding. The most obvious candidate is the highly conserved A47 which could base pair with Us introduced into the J1/2 segment; however, a potential interaction between these residues could not be tested due to the low affinity of A47 mutants.
These data are consistent with the conclusion that the J1/2 nucleotides are important primarily for maintaining the integrity of the binding pocket. Mutagenesis is consistent with the observed contact between C17 and Gβ Both C and A are able to function at this position, providing evidence that the exocyclic amine is the important functional group for binding. However, the anti-covariation observed between J1/2 nucleotides and position 91, the large effects seen in mutants not predicted to contact c-di-GMP, and the rescue observed when 91 is mutated to a non-complementary nucleotide support the hypothesis that the identity of these nucleotides are selected primarily to prevent misfolding of the RNA.
Effects of P1 length on ligand affinity
When the c-di-GMP riboswitch was discovered, it was predicted to be composed of two helices, now termed P2 and P3, and single-stranded flanking sequences to the 5′ and 3′ sides (18, 19). These single-stranded regions became less flexible upon c-di-GMP binding (19). The crystal structure revealed that they form a third helix, P1, when the ligand is bound (21). By analogy to other riboswitch classes (31), formation of this helix is likely to serve as the genetic switch for gene expression control. If this is the case, the riboswitch would only be functional once the RNA polymerase has transcribed a sufficient number of nucleotides on the 3′ end for the formation of this helix. Defining the role of this stem in c-di-GMP binding, as well as the position at which it is stably formed, would establish at what point during transcription a functional aptamer is created. In order to simulate the growing RNA transcript, we tested the binding affinity of a series of RNAs with systematic truncations of the nucleotides on the 3′ end of P1 (These RNAs are named according to the first nucleotide deleted, i.e. Δ93 corresponds to an RNA in which nucleotide 93 and all residues 3′ of 93 have been removed).
The c-di-GMP aptamer is functional after the first nucleotide of the P1 helix is released from the RNA polymerase. When the entire 3′ end of P1 was deleted (Vc2 Δ93) the aptamer lost nearly 500,000-fold (7.5 kcal/mol) in affinity for c-di-GMP (Table 4). However, when C93 was included, the affinity increased substantially, resulting in a nanomolar Kd. When the second base pair of P1 was included (Vc2 Δ95), the binding affinity increased an additional 4-fold. Inclusion of the bulged nucleotide, A95, improved the affinity for c-di-GMP a further 16-fold, but the addition of the next two nucleotides had no effect on the Kd. Further extension of the P1 helix by one or two of the three terminal base pairs (Vc2 Δ99 and Vc2 Δ100) resulted in an affinity that could no longer be measured by equilibrium methods and appears to be in the picomolar range.
Table 4.
Binding affinities for P1 truncation mutants.
| RNA | Kd (nM) | ΔΔGbind (kcal/mol) |
|---|---|---|
| Wild-type | 0.011 | 0 |
|
| ||
| Vc2 Δ93 | 3400 ± 160 | 7.5 |
| Vc2 Δ94 | 280 ± 59 | 6.0 |
| Vc2 Δ95 | 63 ± 13 | 5.1 |
| Vc2 Δ96 | 5.4 ± 0.59 | 3.7 |
| Vc2 Δ97 | 4.6 ± 0.25 | 3.6 |
| Vc2 Δ98 | 5.9 ± 0.79 | 3.7 |
| Vc2 Δ99 | < 0.8a | < 2.5 |
| Vc2 Δ100 | < 0.5a | < 2.3 |
These values could not be measured using equilibrium methods and upper estimates are given.
These data show that the first two nucleotides of the P1 helix are required for low nanomolar affinity for c-di-GMP and additional residues serve to make binding irreversible. From the measurements of koff for P1 truncation mutations (see below), it appears that after the two base pairs directly below c-di-GMP form, the complex will not dissociate before the genetic decision is made. These results support the hypothesis that c-di-GMP binding positions the 3′ end of the RNA as it forms a base pair with C92. This positioning and the favorable stacking network formed then encourages the formation of the P1 helix as additional nucleotides are added to the 3′ end of the RNA.
Effect of mutations on kon and koff
We previously determined that the c-di-GMP riboswitch is a kinetically controlled riboswitch and that the response of this effector is modulated primarily through the on-rate (21). The binding rate constants of the C92U and C44U/G83A mutants suggested that the majority of the effect in Kd is observed as a change in the off-rate and that the on-rate can vary by as much as 10-fold. In order to determine if this was a general phenomenon, we measured kon and koff for several additional mutant riboswitches (Table 5).
Regardless of where in the RNA the mutation occurs, the change in affinity that results from mutation is observed primarily as an increase in the off-rate. Rates were measured for two binding pocket mutants, G20A and the previously measured C92U. In both cases, the binding rate constant was within two-fold of the wild-type kon but the koff increased by the same magnitude as the Kd. In addition to these and the C44U/G83A mutant, kon was measured for three P1 truncation mutants and koff was measured for two different P1 mutants. These mutants have on-rates within 5-fold of the wild-type value, while koff values were 300–1000-fold faster.
In all mutants studied, the loss in affinity appears to primarily translate into a shorter half-life of the c-di-GMP-RNA complex. In only one mutant, the C44U/G83A double mutant discussed above, is the change in kon larger than 5-fold.
Conclusions
The c-di-GMP riboswitch is an important macromolecular target that alters gene expression upon binding of c-di-GMP. We previously determined that this riboswitch is able to selectively recognize its second messenger ligand primarily through contacts with two critical nucleotides (21). However, the phylogeny revealed that the nucleotides that directly interact with c-di-GMP are not invariant. A47, which forms stacking interactions with the ligand, as well as the C44/G83 base pair that appears necessary for forming the tertiary architecture of the aptamer are instead the least variable residues. This study reveals that although large effects are seen when several regions in the RNA are altered, mutations at the most conserved positions, rather than at positions that pair with the bases of c-di-GMP, have the most detrimental effects on binding.
Because the affinity of the wild-type sequence for c-di-GMP is so high, many of the mutations have large effects, but the Kd remains in the nanomolar range. Due to the slow on-rate, we calculate that c-di-GMP concentrations in the high nanomolar to low micromolar may be necessary to elicit a response on the biological time scale (21). Therefore, many mutations with large energetic effects may have no significant effect on riboswitch function, suggesting that this motif may be particularly resilient to mutation. This hypothesis would explain the incomplete conservation of binding pocket nucleotides. In addition, changes in affinity primarily result in faster off-rates, leaving the on-rate within approximately 10-fold of the wild-type value. It is possible that as the Kds of these variant riboswitches approach the cellular concentration of c-di-GMP, their regulation could switch from kinetic to thermodynamic control.
Supplementary Material
Acknowledgments
We thank A. Héroux for collection of native data; S. Meyers and the beamline staff at X25 and X12C at the National Syncrotron Light Source at Brookhaven National Laboratory; M. Strickler and the Yale Center for Structural Biology core staff; J. Wang for data processing advice; E. Smith for phylogenetic analysis; E. Butler, E. Moore, and N. Carrasco; D. Hiller and other members of the Strobel lab for helpful conversations.
Abbreviations
- c-di-GMP
bis-(3′-5′)-cyclic dimeric guanosine monophosphate
- TL
tetraloop
- TLR
tetraloop receptor
- PEG
polyethylene glycol
- MES
2-(N-morpholino)ethanesulfonic acid
Footnotes
Supported by National Institutes of Health Grants GM087906 to A.L.L and GM022778 to S.A.S.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org, under accession codes 3MXH, 3MUM, 3MUR, 3MUT, and 3MUV.
Supporting Information Available
Two figures and one table as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 2.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 3.Römling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 7.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP Binding Protein YcgR Controls Flagellar Motor Direction and Speed to Affect Chemotaxis by a “Backstop Brake” Mechanism. Mol Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. Second Messenger-Mediated Adjustment of Bacterial Swimming Velocity. Cell. 2010;141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 9.Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. Vibrio cholerae VpsT Regulates Matrix Production and Motility by Directly Sensing Cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri J Bacteriol. 2009;191:7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin KH, Lee YC, Tu ZL, Chen CH, Tseng YH, Yang JM, Ryan RP, McCarthy Y, Dow JM, Wang AHJ, Chou SH. The cAMP Receptor-Like Protein CLP is a Novel c-di-GMP Receptor Linking Cell-Cell Signaling to Virulence Gene Expression in Xanthomonas campestris. J Mol Biol. 2010;396:646–662. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 12.Tao F, He YW, Wu DH, Swarup S, Zhang LH. The cNMP Domain of Xanthomonas campestris Global Regulator Clp Defines a New Class of C-di-GMP Effectors. J Bacteriol. 2010;192:1020–1029. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newell PD, Monds RD, O’Toole GA. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci USA. 2009;106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 18.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, Neph S, Tompa M, Ruzzo WL, Breaker RR. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith K, Lipchock S, Ames T, Wang J, Breaker R, Strobel S. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulshina N, Baird N, Ferré-D’Amaré A. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiburu I, Shurer A, Yan L, Sintim HO. A simple solid-phase synthesis of the ubiquitous bacterial signaling molecule, c-di-GMP and analogues. Mol Biosyst. 2008;4:518–520. doi: 10.1039/b719423d. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 29.DeLano WL. DeLano Scientific LLC. Palo Alto, CA, USA: 2008. [Google Scholar]
- 30.Costa M, Michel F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.