Abstract
Quantum dots (QDs) are semiconductor nanocrystals with extensive imaging and diagnostic capabilities, including the potential for single molecule tracking. Commercially available QDs offer distinct advantages over organic fluorophores, such as increased photostability and tunable emission spectra, but their cadmium selenide (CdSe) core raises toxicity concerns. For this reason, replacements for CdSe-based QDs have been sought that can offer equivalent optical properties. The spectral range, brightness and stability of InP QDs may comprise such a solution. To this end, LANL/CINT personnel fabricated moderately thick-shell novel InP QDs that retain brightness and emission over time in an aqueous environment. We are interested in evaluating how the composition and surface properties of these novel QDs affect their entry and sequestration within the cell. Here we use epifluorescence and transmission electron microscopy (TEM) to evaluate the structural properties of cultured Xenopus kidney cells (A6; ATCC) that were exposed either to commercially available CdSe QDs (Qtracker® 565, Invitrogen) or to heterostructured InP QDs (LANL). Epifluorescence imaging permitted assessment of the general morphology of cells labeled with fluorescent molecular probes (Alexa Fluor® ® phalloidin; Hoechst 33342), and the prevalence of QD association with cells. In contrast, TEM offered unique advantages for viewing electron dense QDs at higher resolution with regard to subcellular sequestration and compartmentalization. Preliminary results show that in the absence of targeting moieties, InP QDs (200 nM) can passively enter cells and sequester nonspecifically in cytosolic regions whereas commercially available targeted QDs principally associate with membranous structures within the cell. Supported by: NIH 5R01GM084702.
Keywords: Xenopus, CdSe, InP, cell culture, epifluorescence, semiconductor nanocrystals, fluorescence, quantum dots, transmission electron microscopy (TEM)
1. INTRODUCTION
On the forefront of research in nanobiotechnology is the exploration of potential applications for quantum dots in basic and biomedical research. Quantum dots (QDs) are fluorescent semiconductor nanocrystals that originated in the field of materials science, but their unique properties make them desirable tools for many disciplines [1]. In the life sciences, it is hoped that their applications will permit new ways of observing, quantifying, and manipulating cellular processes that occur at the nanoscale. Much of current research is focused on using QDs to understand intracellular communication and signaling, and to create high resolution cellular images that can be used for long term in vivo studies as well as diagnostics [2]. QDs are particularly attractive for cancer research since they are inclined to accumulate preferentially in cancer tissue [3, 4]. Tumors are highly vascularized and tend to have high retention due to their lack of lymphatic drainage, allowing them to be an ideal destination for nanoparticles [3]. In addition to a tool for diagnosis and detection, fluorescent QDs have been suggested as a possible treatment for cancer when conjugated to an antibody (cetuximab- C225) for human epidermal growth factor receptor (EGFR-1) and combined with radiofrequency field exposure. This treatment has been shown to significantly reduce cancer cell viability [5]. For these reasons it is critical to evaluate the suitability of QDs as molecular probes and their potential effects on cells in culture, including how and where nanomaterials are internalized and ultimately, how they can be safely excreted.
QDs, compared to traditional organic fluorophores, do not photobleach as quickly over extended observation times, exhibit higher quantum yields, and offer tunable emission spectra that prevent overlap between other markers and cellular autofluorescence [2, 6–9]. QDs also tend to be brighter than traditional dyes by an order of magnitude and can be excited by any wavelength ranging from UV to red [7, 10]. In biological systems, QDs offer the advantage of probe stability because they are not readily degraded or metabolized in physiological environments [7]. Although this can be beneficial for signal preservation, the heavy metal cores of QDs raise toxicity concerns. For example, cadmium exposure has been associated with kidney damage and cancer [11, 12]. It is currently believed that these disorders are caused by cadmium’s ability to replace zinc in biochemical reactions [11, 12].
Since the most common type of commercially available QDs are made with cadmium cores, replacements for CdSe-based QDs that can provide equivalent or superior optical properties are highly desirable and sought after [13]. In addition to variation in core composition, QDs can be chemically engineered with a wide variety of surface modifications, matched to the particular application. When QDs designed for biological systems are fabricated with a water-soluble, protective, biocompatible coat (usually PEG or gelatin) but without reactive side chains, they are considered non-targeted [6]. In contrast, targeted QDs are those used as probes for specific biomolecules in cells when ligands are attached to their outer coat as reactive side chains [7, 9, 13–17].
We are interested in evaluating the potential of heterostructured InP QDs to serve as an alternative to CdSe-based QDs for biological applications. Although InP QDs have not yet been tested extensively for biocompatibility, the related InGaP QDs have been shown to be ~10-fold less toxic than CdSe QDs in porcine kidney cells [18]. We designed this pilot study to determine the consequences of exposure to moderately thick-shell InP/ZnS core/shell QDs for cultured Xenopus kidney cells (A6; ATCC). Amphibian kidney cells were selected for analysis because of the aforementioned effects of cadmium on the kidney, and also because amphibians are frequently used to assess environmental quality, including toxic effects of pollutants. The non-targeted InP QDs used in our experiments were engineered to retain brightness and emission in an aqueous environment. We compared the effects of these non-targeted InP QDs with those of commercial non-targeted QDs that previous work has shown do not enter A6 cells at exposure concentrations up to 100 nM [6]. We reasoned that if non-targeted InP QDs have similar properties, they are good candidates for the design of targeted biomolecular probes through surface modification.
One of the most appealing aspects of using QDs as molecular probes is their capacity for visualization in both light and electron microscopes [15, 16]. Accordingly, we used epifluorescence and transmission electron microscopy (TEM) to evaluate morphological and ultrastructural properties of A6 cells exposed either to novel heterostructured InP QDs (LANL/CINT) or to commercially available CdSe QDs (Qtracker® 565 targeted; Qtracker® 565 non-targeted QDs; Invitrogen). Epifluorescence imaging permitted assessment of the general morphology of cells labeled with fluorescent molecular probes (Alexa Fluor® phalloidin; Hoechst 33342), and identification of any cells that appear associated with QDs (either internally or on the cell plasma membrane). TEM offered the unique advantage of visualizing the electron dense QDs at a higher resolution and provided the optics necessary to analyze the subcellular sequestration and compartmentalization of QDs [13]. Our preliminary results show that cells exposed to InP QDs at concentrations up to 200 nM show comparable morphology to that of cells grown in the absence of QDs. Moreover non-targeted InP QDs can be detected in the cytosol of very few cells when delivered passively in the bath at a 200 nM concentration Taken together, our findings support the notion that these InP QDs show promise as probes for cell surface and intracellular bimolecular targets.
2. METHODOLOGY
2.1 QD fabrication
2.1.1 Chemicals
Indium (III) acetate (99.99%) was purchased from Strem Chemicals; myristic acid (99.5+%) and 1-octadecene (ODE; 90%, tech grade) were procured from Acros Organics. Zinc stearate (ZnO 12.5–14%) came from Alfa Aesar, while oleylamine (70%, tech grade), sulfur (99.999%), tris(trimethylsilyl)phosphine ((TMS)3P; 95%) were from Aldrich and toluene and ethanol were from Fisher Scientific. All of the chemicals were used without further purification. Shell precursor stock solutions (0.2 M) were prepared by heating and degassing zinc stearate and sulfur in ODE, respectively.
2.1.2 InP/ZnS core-shell synthesis
InP/ZnS core-shell QDs were synthesized using a modification of a previously published one-pot synthesis protocol [19]. In a glovebox, 0.4 mmol indium acetate, 1.54 mmol myristic acid, and 4 g ODE were mixed in a 100 mL round-bottomed flask. This mixture was heated to 100 °C under vacuum before the temperature was raised to 188 °C under argon flow. A syringe containing 1 mL of a 0.2 M stock of (TMS)3P in ODE and 0.4 mL oleylamine was prepared in the glovebox and rapidly injected into the hot indium solution. The temperature was decreased to 178 °C and maintained for one hour before the core solutions were cooled to room temperature. To deposit ZnS shells on the InP cores, the core reactions were heated to 150 °C under argon and 0.6 mL of the 0.2 M zinc stearate precursor solution was added dropwise. Ten minutes later, the same volume of sulfur in ODE was added and the temperature was raised to 220 °C. Following an initial 75 minute anneal, the zinc and sulfur precursors were added alternatingly at 15 minute intervals. The six successive cation/anion injection steps added 0.8, 1.0, 1.2, 1.4, 1.6, and 1.8 mL of each precursor. The resulting reaction solution was mixed with toluene and precipitated with ethanol. The QDs were pelleted with centrifugation and resuspended in toluene. The water transfer of the QD was achieved by ligand exchange with mercaptoundecanoic acid using tetramethylammonium hydroxide as the transfer reagent (from toluene to water). Even after several months stored at 4 °C, both the organic and aqueous solutions of the InP/ZnS QDs exhibited quantum yields between 20 and 25%. (See Fig. 1 for photoluminescence spectrum).
Figure 1.
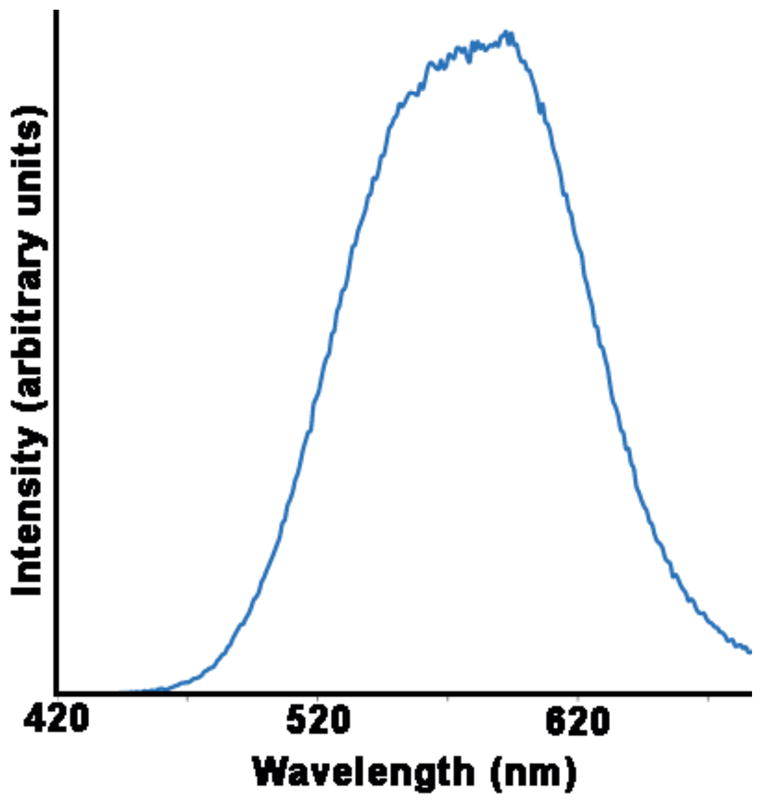
Photoluminescence spectrum of InP/ZnS core/shell NQDs in water.
2.2 Cell lines and reagents
Xenopus laevis kidney cells (A6) and fetal bovine serum were acquired from American Type Culture Collection (Manassas, VA). NCTC-109 media, 200 mM L-glutamine, 0.25% Trypsin-EDTA, and phosphate buffered saline (PBS) were obtained from Sigma-Aldrich (St. Louis, MO). Commercial QDs, Qtracker® 565 non-targeted quantum dots and Qtracker® 565 Cell Labeling Kit (targeted), Alexa Fluor® 680 phalloidin, Hoechst 33342, and SlowFade® Antifade Kit were obtained from Invitrogen (Carlsbad, CA). Heterostructured QDs were provided by LANL/CINT (Los Alamos, NM) as described in 2.1. Glutaraldehyde, osmium tetroxide (OSO4), uranyl acetate, lead citrate, the Embed 812 Kit, and formvar/carbon film grids (100 mesh) were obtained from Electron Microscopy Sciences (Hatfield, PA). Cells were grown on dishes or culture slide chambers depending on whether they were designated for epifluorescence or TEM imaging. Cells for TEM were cultured and processed in poly-d-Lysine coated glass bottom dishes (35 mm) with 14 mm microwells purchased from MatTek Corp. (Ashland, MA). Cells intended for epifluorescence imaging were grown and processed in four chambered slides, Lab Tek II, purchased from VWR (West Chester, PA).
2.3 Cell culture
Cells were grown in 25 cm2 flasks and passaged at least once before plating for exposure to QDs. A6 cells were grown in NCTC-109 media supplemented with 10% fetal bovine serum and 2 mM L-glutamine, and 18 MΩ water. All solutions were purchased sterile or sterilized by 0.2 μm filtration. Media was changed every 48 hours and cell health was observed using an inverted Nikon Diaphot TMD phase-contrast microscope. Cells were subcultured by partial digestion using 0.25% trypsin-EDTA. After trypsinization, cell concentrations were determined using a hemocytometer, cells were then centrifuged, resuspended, and plated at a final density of 8 × 104 cells per cm2. According to the manufacturer’s specifications, 100 μL of cells were allowed to incubate for 1hr at 26° C on each of the MatTek glass bottom wells before adding additional media. This allowed cells time to settle and adhere to the desired region. All cells were allowed to grow for 24 hrs at 26°C and 5% CO2 prior to QD exposure.
2.4 Incubation with QDs
Experiments were designed to permit epifluorescence and TEM imaging of all 6 treatment paradigms. A chamber and a dish of cells was assigned to each of the following treatments: (1) autofluorescence, to provide a background signal control; (2) InP (LANL/CINT) QD solution control (without QDs), which consisted of ultrapure water, 1% isopropanol, and highly dilute mercaptoundecanoic acid (MUA; 1:1000) in media. This treatment controlled for the presence of any reagents that were used for precipitation and ligand capping and that might be present in the InP QD solution; (3) 50 nM InP (LANL/CINT) QDs solution in media; (4) 200 nM InP (LANL/CINT) QDs solution in media; (5) 200 nM Invitrogen 565 non-targeted QDs solution (we chose the non-targeted invitrogen 565 QDs as a suitable comparison to the InP QDs because both lack targeting moieties); (6) 20 nM Invitrogen targeted 565 QDs (positive control). The 20 nM, 50 nM and 200 nM concentrations were selected to represent a 10 fold range beginning with 20 nM, a maximal concentration typically used for targeted QD applications.
2.5 Fluorescent probes and signal detection
All treatment groups, with the exception of the autofluorescence control, were stained with Alexa Fluor® 680 phalloidin (Figs. 2A2; 2B2; 2C2; 3A2; 3B2) to show general cytoskeletal morphology and with Hoechst 33342 to identify nuclei (Figs. 2A3; 2B3; 2C3; 3A3; 3B3). These fluorophores allowed the quantification of the percentage of cells that were associated with QDs under epifluorescence. Alexa Fluor® 680 phallodin was chosen to achieve spectral separation from the heterostructed or commercial QDs. QD association was determined using an inverted Nikon Eclipse TE-2000-S microscope. Cell structure and QD association were analyzed by differential interface contrast (DIC) and epifluorescence microscopy (Figs. 2–3). Fluorescent probe detection and images were acquired using a 20x objective (N.A., 0.50) and appropriate filter cubes. QDs were detected with a G-2E/C-TRITC filter set. Hoechst 33342 and Alexa Fluor® 680 phalloidin were detected with the filter sets UV-2A and CY5HQ, respectively. The percentage of cells that showed QD association was determined using MetaMorphR Offline Imaging System Version 6.0r5 (Universal Imaging Corporation, Downingtown, PA). Cells that appeared to have QDs associated with them were counted and the number was divided by the total number of cells (Hoechst 33342 counts) in the visual field of a 20x air objective.
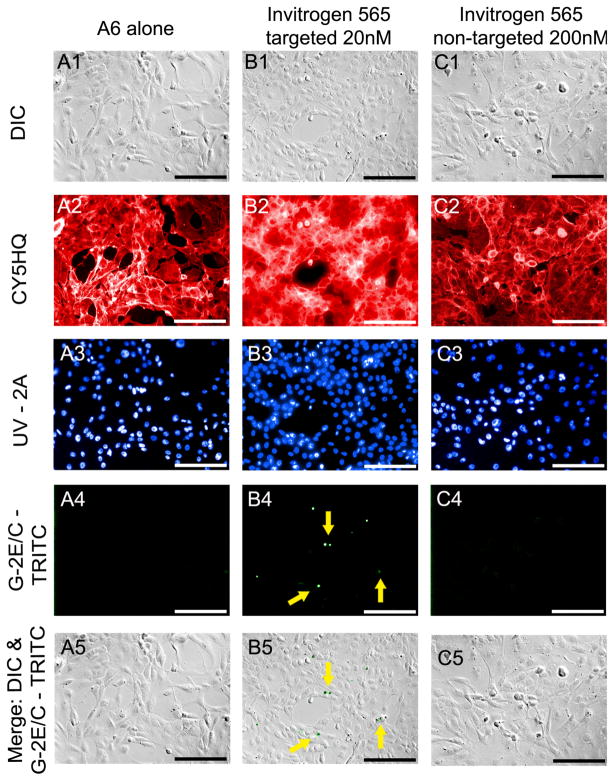
Figure 2. Epifluorescence and DIC images of commercial QDs.
Epifluorescence imaging permitted the qualitative and quantitative comparison of cells not incubated with QDs (A) and cells incubated for 24 hours with either 20nM Qtracker 565 (B) or 200nM Qtracker 565nm nontargeted QDs (C). A qualitative assessment of cellular morphology was completed by evaluating DIC (A1-C1) and epifluorescence (A2-C2) images of Alexa Fluor® 680 phalloidin. A quantitative assessment of cellular association with QDs was carried out by dividing the total number of cells associated with QDs detected in merged images (A5-C5) by the total number of cell nuclei labeled with Hoechst 33342 (A3-C3). All images were captured using a 20x air objective, NA= 0.50. Scale bar = 50μm.
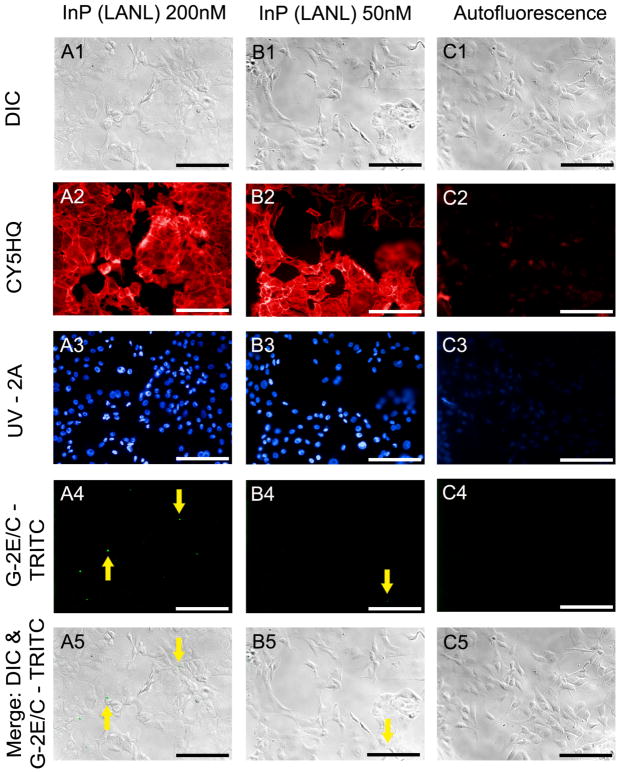
Figure 3. Epifluorescence and DIC images of heterostructured InP QDs.
Epifluorescence imaging permitted the qualitative and quantitative comparison of cells incubated for 24 hours with either 200nM of InP (LANL) 578 QDs (A), 50nM of InP (LANL) 578 QDs (B), and control cells not incubated with QDs (C). A qualitative assessment of cellular morphology was completed by evaluating DIC (A1-C1) and epifluorescence (A2, B2) images of Alexa Fluor® 680 phalloidin. A quantitative assessment of cellular association with QDs was carried out by dividing the total number of cells associated with QDs detected in merged images (A5-C5) by the total number of cell nuclei labeled with Hoechst 33342 (A3, B3). All images were captured using a 20x air objective, NA= 0.50. Scale bar = 50 μm.
Although the InP QDs could be seen with the UV-2A filter set along with the stained nuclei, they were easily and solely identifiable with the G-2E/C-TRITC filter set. Images were captured using a CoolSNAP™ HQ CCD camera (Roper Scientific, Tucson, AZ) and acquired using MetaVue™ Imaging System Version 6.0r5 (Universal Imaging Corpoation, Downingtown, PA). Image color and merges were assigned and created using Adobe Photoshop CS4.
2.6 Fixation and staining for epifluorescence microscopy
Upon completion of the 24 hr exposure, cells were rescued from QD contact and rinsed with serum free media. All treatment groups were initially fixed with 3.7% formaldehyde for 10 minutes at room temperature. The cells in the dishes intended for TEM were covered in PBS and placed in a 4°C refrigerator until further processing. Cell culture slides designated for epifluorescence imaging were rinsed 2x with PBS, then permeabilized with 0.2% Triton X100 for 20 minutes at room temperature. Cells were rinsed 2x with PBS for 2 minutes. Alexa Fluor® 680 phalloidin with 1% bovine serum albumin in PBS was added to the slide chambers, and the cells were incubated in the dark for 20 minutes at room temperature according to manufacturer’s specifications. Alexa Fluor® 680 Phalloidin (165 nM) was used for the commercial (565) and InP QDs (578) as well as those cells exposed to LANL’s QD control solution. Cells were rinsed 2x with PBS for 2 minutes. Hoechst 33342 (2 μg/ml) was then added to each of these chambers and incubated in the dark for 10 minutes at room temperature according to manufacturer’s specifications. Cells were again rinsed 2x in PBS for 2 minutes.
Culture slides were prepared for imaging by adding AntiFade® Solution C (SlowFade® Antifade Kit) for 15 minutes at room temperature. AntiFade Solution C was removed and a drop of AntiFade® Solution A (SlowFade® Antifade Kit) was added prior to applying a glass cover slip over the slide. Those culture slide chambers designated to serve as autofluorescence controls received all of the above treatments with the exception of the fluorescent dyes (Alexa Fluor® phalloidin; Hoechst 33342), in which case, they received the same amount of buffer (1% bovine serum albumin in PBS and PBS, respectively) as the treatment samples. Slide and cover slip were sealed together using nail polish. Digital images used as controls for background autofluorescence were taken at the same or average exposure times as those that yielded fluorescence in labeled samples.
2.7 TEM preparation
Following the formaldehyde fixation utilized for epifluorescence imaging, cells were fixed overnight in 2.5% glutaraldehyde in PBS at 4° C. Cells were rinsed thoroughly in PBS and post-fixed for 90 minutes in 2% osmium tetroxide (1:1 PBS and 4% OsO4) [20]. Cells were rinsed once with dH2O for 15 minutes and then sequentially exposed to 50% then 80% ethanol/dH2O twice for 10 minutes each, followed by 100% ethanol twice for 15 minutes. Cells then were incubated twice (30 minutes followed by overnight) with resin (EMbed 812 Kit). A final change of Embed 812 was allowed to cure in a 60° C oven for 24 hours. Samples then were removed from the MatTek dishes using pliers and liquid nitrogen to break away the dish (polystyrene and glass) from the resin block. The sample-containing resin block was then double embedded into the resin remaining from the previous day and allowed to cure for 48 hrs. The double embedment allowed for the monolayer of cells to be protected and evenly cut from both sides.
Ultra thin sections ~70nm were cut using a Leica UC6 ultramicrotome. Sections were placed on copper 100 mesh carbon-formvar grids and post stained with uranyl acetate and lead citrate. TEM images were taken using a model H-7650 TEM (Hitachi High-Technologies, Pleasanton, CA) at an accelerating voltage of 80 kV. Digital images were acquired with an integrated CCD camera system (AMT Corp., Danvers, MA). Ultrathin sections were cut and placed to fill 3 grids per treatment group.
3. RESULTS
3.1 Epifluorescence of commercial dots
A6 cells grown in the absence of QDs and labeled with fluorescent molecular probes (Alexa Fluor® 680 phalloidin; Hoechst 33342) demonstrate the morphology characteristic of this cell line when grown in our laboratory using the conditions recommended by the vendor (Fig. 2A). QD association did not appear to induce a change in the general morphology or viability of the cells as indicated by the similarity in cytoskeletal structure (Figs. 2A2-2C2) and in nuclear staining (Figs. 2A3-2C3) between cells grown in the presence or absence of QDs.
As expected, Invitrogen’s Qtracker® 565 CdSe targeted QDs appeared associated with cells at 20 nM after a 24 hr exposure (Figs. 2B4–5). Epifluoresence microscopy showed these samples emitting a fluorescence signal that was observable above that of autofluorescence in the filter set used for to detect QD emission (Fig. 2B4). Under differential interference contrast, cells exposed to targeted commercial dots (Fig. 2B) maintained a similar morphology to those serving as a control (Fig. 2A). Results also suggest the maintenance of a viable cytoskeletal structure as seen under epifluorescence microscopy with the Alexa Fluor® phalliodin staining (Fig. 2B2).
A6 cells exposed to 200 nM of non-targeted commercial dots for a period of 24 hrs showed no visible signs of QD association (Figs. 2C4–5). This result is consistent with previous work [6]. Under differential interference contrast, cells exposed to non-targeted commercial dots at 200 nM appear to maintain a viable morphology (Figs. 2C1 & 2C5).
3.2 Epifluorescence of heterostructured InP QDs
Differential interference contrast images of cells in the autofluorescence control group demonstrate a lack of identifiable morphological differences between cells exposed to QDs (Figs. 3A & B) and those grown in their absence (Fig. 3C). The low contribution of background autofluorescence to epifluorescence images is shown by the general lack of signal in images taken from cells not exposed to the fluorescent dyes (Fig. 3C) under the same exposure time and conditions as those labeled with fluorophores (Figs. 3A & B).
A6 cells incubated for 24 hrs with 50 nM of InP (LANL/CINT) 578 QDs typically did not show detectable QD association. Occasionally, regions in the cell monolayer exhibited a bright fluorescent signal typical of QD emission (Fig. 3B4). However, these occurrences were rare.
Cell morphology and viability appear similar to those cells grown in the absence of QDs (Fig. 2A). Cells exposed to 200 nM of InP (LANL/CINT) 578 QDs for a period of 24 hrs show low, but detectable, incidence of QD association. Quantitative assessment showed that approximately 5.9% of cells were associated with QDs. Cell morphology and viability did not appear to be affected by the presence of the QD solution with or without QDs as indicated by the similarity in structure of cells grown in 200 nM (Fig. 3A) to those grown the absence of QDs (Fig. 2A) or QD control solution (data not shown).
3.3 TEM of commercial QDs
The physical properties of quantum dots facilitate imaging with both light and electron microscopy [16]. Epifluorescence images can be used to determine whether QDs are associated with cells but cannot distinguish whether QDs are internalized or adherent to the cell surface. To this end, TEM was used to assess whether quantum dots were associated with the cell surface membrane or if internalized, the intracellular location.
Invitrogen’s Qtracker® 565 CdSe targeted QDs at 20 nM associated with membranous structures in cells after a 24 hr exposure. QDs appear along the outer membrane of the cell as well as within a lysosomal or endosomal structure within the cell (Fig. 4B). This result is consistent with previous work [7, 21]. According to the manufacturer, QDs were conjugated with an arginine peptide to aid in cellular entry [21, 22]. QDs appear to maintain their integrity as demonstrated by the apparent lack of aggregates. As a result, a higher magnification was necessary to view individual QDs (accelerated voltage = 100kV).
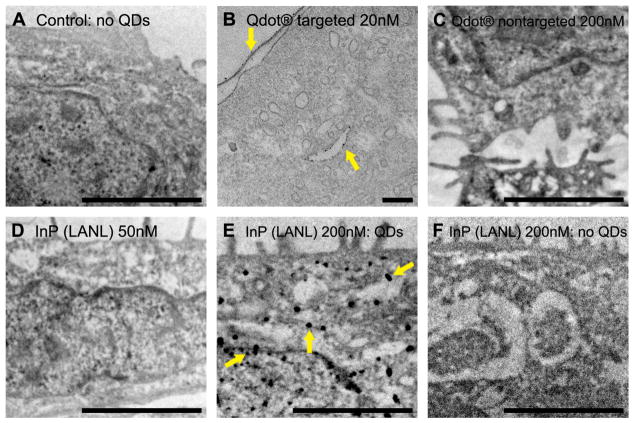
Figure 4. Transmission electron micrographs of all treatment groups.
(A) Cells grown in the absence of QDs provide a control and are free of electron dense clusters. A6 cells were incubated for 24 hrs with one of the following: (B) 20nM Qtracker 565 targeted QDs; (C) 200nM Qtracker 565nm nontargeted QDs; (D) 50nM InP (LANL) QDs; (E & F) 200nM InP (LANL) QDs. Cells exposed to 20 nM Qtracker 565 targeted QDs show electron dense nanoparticles associating with membranous structures within the cell (B). Cells exposed to 200 nM Qtracker 565nm nontargeted (C) and 50 nM of InP (LANL) QDs (D) appear to be clear of electron dense nanoparticles. Cells exposed to 200 nM of InP (LANL) QDs show large aggregations of electron dense material in some cells (E), while other cells in the same treatment group appear free of aggregates (F). A, C, D, E, F: Scale bar = 2 μm; images acquired at an accelerated voltage of 10 kV. B: Scale bar = 200 nm; image acquired at an accelerated voltage of 100 kV.
Consistent with previous work, Invitrogen’s non-targeted CdSe QDs do not appear to enter the cells at 200 nM (Fig. 4C) [6]. Cells under this condition appear to have morphology similar to those not exposed to QDs (Fig. 4A).
3.4 TEM of heterostructured InP QDs
In cells exposed to 50 nM of InP (LANL/CINT) QDs, electron dense clusters were not detected using electron microscopy (Fig. 4D). This result is consistent with the epifluorescent images, which suggest that although some QD association may occur, the rate of entry is <1%. In contrast, TEM images of cells exposed to 200 nM of InP (LANL/CINT) QDs show electron dense material in the cytosol either dispersed (Fig. 4F) or in aggregates (Fig. 4E).
4. DISCUSSION
The intent of this study was to establish baseline conditions for the passive entry into cells of heterostructured InP QDs (LANL/CINT) such that future work could focus on conjugating targeting moieties that direct them to specific cellular locations. Since QDs are brighter than traditional organic fluorophores and have a tunable emission spectra, they could potentially be used for multiplexing and detecting molecules that occur at relatively low densities within cells or cellular membranes [2, 6–8].
In this preliminary study we compared the cellular sequestration of commercially available CdSe QDs (Invitrogen) and heterostructured InP QDs (LANL/CINT), paying particular attention to their association with, and sequestration by, A6 cells. Fluorophores and QDs chosen for this work were selected based on their compatibility with available optics and our ability to attain spectral separation during imaging. The heterostuctured InP QDs fabricated at LANL/CINT have a peak emission wavelength of 578 nM (Fig. 1). Thus, Invitrogen’s commercially available QDs with a peak emission wavelength of 565 nM (targeted and non-targeted) were selected as a suitable comparison as both QDs could be visualized using the same G-2E/C - TRITC filter set. Additionally, Invitrogen’s 565 QDs had previously been selected for use in our laboratory due to their intermediate size and brightness as compared with other commercially available QDs [22]. Alexa Fluor® 680 phalloidin was chosen as a probe for cytoskeletal actin because its absorption and emission wavelengths were separated from those of the QDs by approximately 100 nM and it was easily visualized using our CY5HQ filter set. Although both types of QDs are detected with the UV – 2A filter set along with the nuclear (Hoechst 33342) stain, QD emissions could be separated from the Hoechst signal in the G-2E/C filter set.
As expected and consistent with previous studies, Invitrogen’s commercially available non-targeted QDs were not detected either associated with, or sequestered by, A6 cells at concentrations up to 200 nM (Figs. 2C4–5 & 4C) [6]. While non-targeted QDs are coated with PEG to reduce non-specific binding, they are not outfitted with reactive side chains that would increase electrostatic interactions and absorption [22, 23].
Results indicated that following a 24 hr incubation period, A6 cells can internalize Invitrogen’s CdSe targeted QDs at 20 nM (Fig. 4B) and LANL/CINT’s InP QDs at 200 nM (Fig. 4E). TEM findings are consistent with observations using epifluorescence microscopy (Figs. 2B4, 3A4). As previously demonstrated, Invitrogen’s CdSe targeted QDs are sequestered in cellular structures that appear to be endosomes or lysosomes. (Fig. 4B) [7, 21]. LANL/CINT’s InP QDs were detected in cytosolic regions either dispersed (Fig. 4F) or in aggregates (Fig. 4E). Although an occasional fluorescent signal was observed in the cells exposed to 50 nM of InP QDs using epiflourescence microscopy (Fig. 3B4), we could not detect QDs within cells at this concentration using TEM (Fig. 4D). This is likely due to the infrequent entry of QDs into cells at this concentration.
It was encouraging that the InP (LANL/CINT) QDs only marginally associated with cells at concentrations much higher than the typical passive delivery concentration of 20 nM. Impressively, the concentration of the InP QDs had to be increased by an order of magnitude from 20 nM to 200 nM before it was possible to detect low levels of association with, and sequestration by, A6 cells (5.9%). In order for single particle tracking to be possible using the InP QDs, it must be said with some degree of certainty that the QDs are largely entering the cell in response to a targeting moiety and not due to nonspecific endocytosis [24]. It is advantageous that these InP QDs, which lacked a targeting moiety, showed very low cellular entry at a high (200 nM) concentration since this characteristic will facilitate their use in future studies where specific targeting moieties will be attached.
5. CONCLUSION
With the addition of specialized targeting moieties, the potential uses of QDs in bioscience research are extensive. However, this study and others demonstrate that a number of challenges remain until this prospect is realized. Some of the major obstacles include the fabrication of QDs that are less toxic than traditional commercially available CdSe core QDs, as well as production of QDs that are photostable and have biocompatible surface coats that do not aggregate or degrade over time. Furthermore, QDs ideally should enter cells in a directed and specific manner, for example via a targeting moiety, and be capable of maintaining spectral emission and stability once inside the cell. In addition to reduced toxicity, researchers exploring in vivo methods may also desire a means by which cells could safely excrete and clear the QDs while sustaining biocompatibility during experimentation.
As novel QDs are synthesized that attempt to address the issues presented above, it will be of critical importance to carefully characterize how their physical and chemical properties (e.g. core composition, shell size, attached targeting moieties) affect cellular entry, sequestration and long-term fate. This information is necessary to be able to fabricate biocompatible QDs that will accept the attachment of a diverse set of targeting moieties in an aqueous biological environment while maintaining even dispersal, efficient fluorescence, and photostability [10].
The results shown here are encouraging towards meeting this goal because the heterostructured InP QDs (LANL/CINT) did not easily enter cells or associate with the cellular membrane. Significantly, their optical properties (emission wavelength and efficiency) were shown to be stable for at least several months when stored as aqueous solutions. Once the InP QDs are outfitted with a custom and specific targeting motif, an additional challenge will be to ensure their entry into cells. As a result, we intend to investigate alternative methods for intracellular delivery of QDs such as particle bombardment (biolistics). This technique may eliminate some of the challenges associated with membrane permeability, and allow for the addition of highly specific targeting moieties while eliminating the need for a membrane-penetrating moiety.
Acknowledgments
We would like to thank Dr. Peter Cooke and the NMSU Electron Microscopy Laboratory for assistance with TEM, as well as V. Bleu Knight for editorial assistance. This research was made possible by a National Institutes of Health award to JAH (PI) (NIH 5R01GM084702) and EES (consortium PI) and a Holloman Air Force Base Officers Spouses Club Scholarship to EMR. This work was performed, in part, at the Center for Integrated Nanotechnologies, a U.S. Department of Energy, Office of Basic Energy Sciences user facility at Los Alamos National Laboratory (Contract DE-AC52-06NA25396) and Sandia National Laboratories (Contract DE-AC04-94AL85000).
References
- 1.Alivisatos AP. Semiconductor clusters, nanocrystals, and quantum dots. Science. 1996;271(5251):933–937. [Google Scholar]
- 2.Michalet X, Pinaud FF, Bentolila LA, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307(5709):538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park K, Lee S, Kang E, et al. New generation of multifunctional nanoparticles for cancer imaging and therapy. advanced functional materials. 2009;19(10):1553–1566. [Google Scholar]
- 4.Gao XH, Cui YY, Levenson RM, et al. In vivo cancer targeting and imaging with semiconductor quantum dots. Nature Biotechnology. 2004;22(8):969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 5.Glazer ES, Curley SA. Radiofrequency field-induced thermal cytotoxicity in cancer cells treated with fluorescent nanoparticles. Cancer. 2010;116(13):3285–3293. doi: 10.1002/cncr.25135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight VB, Serrano EE. The accumulation of nontargeted quantum dots in cultured human embryonic kidney cells. Proceedings of SPIE. 2006;6096(60960Y):1–11. [Google Scholar]
- 7.Jaiswal JK, Mattoussi H, Mauro JM, et al. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nature Biotechnology. 2003;21(1):47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 8.Yong KT, Roy I, Ding H, et al. Biocompatible near-infrared quantum dots as ultrasensitive probes for long-term in vivo imaging applications. Small. 2009;5(17):1997–2004. doi: 10.1002/smll.200900547. [DOI] [PubMed] [Google Scholar]
- 9.Pinaud F, Clarke S, Sittner A, et al. Probing cellular events, one quantum dot at a time. Nature Methods. 2010;7(4):275–285. doi: 10.1038/nmeth.1444. [DOI] [PubMed] [Google Scholar]
- 10.Dubertret B, Skourides P, Norris DJ, et al. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298(5599):1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 11.Il’yasova D, Schwartz GG. Cadmium and renal cancer. Toxicology and Applied Pharmacology. 2005;207(2):179–186. doi: 10.1016/j.taap.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Kolonel LN. Association of cadmium with renal cancer. Cancer. 1976;37(4):1782–1787. doi: 10.1002/1097-0142(197604)37:4<1782::aid-cncr2820370424>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 13.Deerinck TJ. The application of fluorescent quantum dots to confocal, multiphoton, and electron microscopic imaging. Toxicologic Pathology. 2008;36(1):112–116. doi: 10.1177/0192623307310950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serrano EE, Knight VB. Nanobiophotonics and Biomedical Applications II. SPIE- The International Society for Optical Engineering; San Jose: 2005. Multiphoton imaging of quantum dot bioconjugates in cultured cells following Nd:YLF laser excitation. [Google Scholar]
- 15.Deerinck TJ, Giepmans BNG, Smarr BL, et al. Light and electron microscopic localization of multiple protiens using quantum dots. Humana Press; 2007. pp. 43–53. [DOI] [PubMed] [Google Scholar]
- 16.Giepmans BNG, Deerinck TJ, Smarr BL, et al. Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nature Methods. 2005;2(10):743–749. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- 17.Nisman R, Dellaire G, Ren Y, et al. Application of quantum dots as probes for correlative fluorescence, conventional, and energy-filtered transmission electron microscopy. Journal of Histochemistry & Cytochemistry. 2004;52(1):13–18. doi: 10.1177/002215540405200102. [DOI] [PubMed] [Google Scholar]
- 18.Stern ST, Zolnik BS, McLeland CB, et al. Induction of autophagy in porcine kidney cells by quantum dots: A common cellular response to nanomaterials? Toxicological Sciences. 2008;106(1):140–152. doi: 10.1093/toxsci/kfn137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pradhan N, Reifsnyder D, Xie R, et al. Surface ligand dynamics in growth of nanocrystals. Journal of the American Chemical Society. 2007;129(30):9500–9509. doi: 10.1021/ja0725089. [DOI] [PubMed] [Google Scholar]
- 20.Sabatini DD, Bensch K, Barrnett RJ. Cytochemistry and electron microscopy – preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. Journal of Cell Biology. 1963;17(1):19. doi: 10.1083/jcb.17.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagerholm BC, Wang MM, Ernst LA, et al. Multicolor coding of cells with cationic peptide coated quantum dots. Nano Letters. 2004;4(10):2019–2022. [Google Scholar]
- 22.Knight VB, Serrano EE. Tissue and species differences in the application of quantum dots as probes for biomolecular targets in the inner ear and kidney. Ieee Transactions on Nanobioscience. 2006;5(4):251–262. doi: 10.1109/tnb.2006.886551. [DOI] [PubMed] [Google Scholar]
- 23.Chung YC, Chen IH, Chen CJ. The surface modification of silver nanoparticles by phosphoryl disulfides for improved biocompatibility and intracellular uptake. Biomaterials. 2008;29(12):1807–1816. doi: 10.1016/j.biomaterials.2007.12.032. [DOI] [PubMed] [Google Scholar]
- 24.Delehanty JB, Mattoussi H, Medintz IL. Delivering quantum dots into cells: strategies, progress and remaining issues. Analytical and Bioanalytical Chemistry. 2009;393(4):1091–1105. doi: 10.1007/s00216-008-2410-4. [DOI] [PubMed] [Google Scholar]