Abstract
The extent of horizontal gene transfer (HGT) among marine pelagic prokaryotes and the role that HGT may have played in their adaptation to this particular environment remain open questions. This is partly due to the paucity of cultured species and genomic information for many widespread groups of marine bacteria and archaea. Molecular studies have revealed a large diversity and relative abundance of marine planktonic archaea, in particular of Thaumarchaeota (also known as group I Crenarchaeota) and Euryarchaeota of groups II and III, but only one species (the thaumarchaeote Candidatus Nitrosopumilus maritimus) has been isolated in pure culture so far. Therefore, metagenomics remains the most powerful approach to study these environmental groups. To investigate the impact of HGT in marine archaea, we carried out detailed phylogenetic analyses of all open reading frames of 21 archaeal 16S rRNA gene-containing fosmids and, to extend our analysis to other genomic regions, also of fosmid-end sequences of 12 774 fosmids from three different deep-sea locations (South Atlantic and Adriatic Sea at 1000 m depth, and Ionian Sea at 3000 m depth). We found high HGT rates in both marine planktonic Thaumarchaeota and Euryarchaeota, with remarkable converging values estimated from complete-fosmid and fosmid-end sequence analysis (25 and 21% of the genes, respectively). Most HGTs came from bacterial donors (mainly from Proteobacteria, Firmicutes and Chloroflexi) but also from other archaea and eukaryotes. Phylogenetic analyses showed that in most cases HGTs are shared by several representatives of the studied groups, implying that they are ancient and have been conserved over relatively long evolutionary periods. This, together with the functions carried out by these acquired genes (mostly related to energy metabolism and transport of metabolites across membranes), suggests that HGT has played an important role in the adaptation of these archaea to the cold and nutrient-depleted deep marine environment.
Keywords: Thaumarchaeota, marine Euryarchaeota, metagenomics, deep ocean, planktonic archaea, horizontal gene transfer
Introduction
Horizontal gene transfer (HGT) is an important process in microbial evolution (Gogarten et al., 2002; Gogarten and Townsend, 2005). Acquisition of exogenous genes by HGT can allow the recipient species to adapt rapidly to novel environments. This is very well known for species of medical interest that have acquired pathogenicity islands and antibiotic resistance genes through HGT (Hacker and Kaper, 2000; Hastings et al., 2004; Wright, 2007; Juhas et al., 2009). However, much less is known about microorganisms living in natural environments, especially those belonging to the domain Archaea. An interesting example is the acquisition by marine planktonic archaea of a proteorhodopsin gene of likely bacterial origin (Frigaard et al., 2006). This single protein confers an immediate advantage to the cells bearing it: the ability to obtain chemical energy from sunlight (phototrophy) (Béjá et al., 2000a). Despite this outstanding example, quantitative studies of the impact of HGT in natural populations of archaea remain very scarce. A few years ago, we studied a genome fragment of a marine planktonic group I archaeon, DeepAnt-EC39, retrieved from a fosmid library of planktonic DNA from the Antarctic polar front (500 m deep). Detailed phylogenetic analysis of the 41 genes present in this genome fragment revealed that 11 of them had been most likely acquired by HGT from different sources, including bacteria and other unrelated archaea (López-García et al., 2004). This represented 26.8% of the genes analysed, which was a surprisingly high proportion, suggesting that HGT might have been an important process in the evolutionary history of this marine archaeon and opened the possibility for HGT levels higher than suspected in other marine planktonic archaea also.
Marine planktonic archaea belong to three distinct taxonomic groups. Group I archaea were initially considered as members of the Crenarchaeota based on their relationship with the hyperthermophilic Crenarchaeota in 16S ribosomal RNA gene (rDNA) phylogenies. They were first discovered in marine planktonic samples (DeLong, 1992; Fuhrman et al., 1992) but subsequent studies revealed that they are also frequent in other environments, including soils (Jurgens et al., 1997), lakes (Schleper et al., 1997) and both cold and hot terrestrial springs (Barns et al., 1996). So far, only two marine archaea of this group have been studied in detail: the sponge symbiont Cenarchaeum symbiosum (Preston et al., 1996), and the chemolithoautotrophic ammonium oxidiser Candidatus Nitrosopumilus maritimus, the first species to be isolated in pure culture (Könneke et al., 2005). Differences in gene content with the classical hyperthermophilic Crenarchaeota and phylogenetic analysis of conserved genes involved in translation led to propose that group I may define a new archaeal phylum, the Thaumarchaeota, independent from the two phyla recognised until now, the Crenarchaeota and the Euryarchaeota (Brochier-Armanet et al., 2008). In the following, we will refer to group I Crenarchaeota as Thaumarchaeota, independently of it being a third distinct phylum or a sister group to the hyperthermophilic Crenarchaeota.
The other two groups of marine planktonic archaea, groups II and III are phylogenetically related and branch as sister-group of the Thermoplasmatales within the Euryarchaeota in 16S rDNA phylogenetic trees (DeLong, 1992; Fuhrman and Davis, 1997). In contrast with Thaumarchaeota, these euryarchaeotal groups lack any cultured representative and their metabolic capabilities remain unknown, except for the indication of a possible phototrophic metabolism in group II archaea from surface waters (Frigaard et al., 2006). Group II Euryarchaeota usually appear to be relatively more abundant in surface waters compared with Thaumarchaeota (Karner et al., 2001; Ghai et al., 2010), but they may be abundant in deep-sea waters as well, reaching in some oceanic regions even higher proportions than Thaumarchaeota (Martín-Cuadrado et al., 2008). The much more enigmatic group III Euryarchaeota have been almost exclusively detected in deep-sea plankton, and are found in general in lower abundance than Thaumarchaeota and group II Euryarchaeota (Fuhrman and Davis, 1997; Martín-Cuadrado et al., 2008).
The study of the gene repertoire and the quantification of HGT events in these archaea are important to better understand their adaptation to marine pelagic environments. Given the difficulties in isolating and culturing them, metagenomic methods remain the most powerful to get access to the gene and genome sequences of these microorganisms. To evaluate the occurrence of HGT in non-cultured marine planktonic archaea, we used a double approach to study metagenomic fosmid libraries of deep-sea plankton collected at three different locations (South Atlantic and Adriatic Sea at 1000 m, and Ionian Sea at 3000 m). First, we reconstructed phylogenetic trees for all the proteins encoded in 21 fosmids containing 16S rDNAs of Thaumarchaeota and group II and III Euryarchaeota. Second, we carried out phylogenetic analyses of the fosmid-end sequences of 12 774 fosmids from the same libraries (4124 from AD1000, 4549 from KM3 and 4101 from SAT1000). These approaches targeted different genomic regions and allowed us to infer relatively high levels of HGT for the three archaeal groups, between 25% and 40%, and from a variety of donors. Therefore, our results agree with previous observations of the prevalence of HGT in the evolutionary history of marine planktonic Thaumarchaeota (López-García et al., 2004). Interestingly, our analyses show that this phenomenon is not restricted to this group but is also significant in marine planktonic groups II and III Euryarchaeota. This suggests that HGT may have played an important role in the adaptation of archaea to the marine pelagic environment.
Materials and methods
16S rDNA-containing complete fosmid sequences
Archaeal fosmids containing 16S rRNA genes in the different deep-sea metagenomic libraries were identified in a previous work by PCR using specific archaeal primers and fully sequenced (Martín-Cuadrado et al., 2008). Twelve fosmids belonged to Thaumarchaeota and nine to Euryarchaeota (seven to group II and two to group III). The complete set of fosmids contained 681 open reading frames (ORFs).
Fosmid-end sequencing
Bi-directional fosmid-end sequences for 5000 randomly chosen fosmids were already available for the deep-sea plankton metagenomic library KM3 (Ionian Sea, 3000 m depth) (Martín-Cuadrado et al., 2007). We sequenced both ends of 5000 fosmid inserts from each of the two additional metagenomic libraries, SAT1000 (South Atlantic, 1000 m depth) and AD1000 (Adriatic Sea, 1000 m depth). Clones were picked randomly and transferred to 96-well plates containing lysogeny broth medium plus chloramphenicol. Fosmid-end Sanger sequencing was carried out at the Genoscope (Evry, France) using pCC1Fos specific primers. The total number of good-quality sequences retained for this analysis after vector sequence trimming was 7701 (AD1000), 8482 (KM3) and 7630 (SAT1000), with average sequence lengths of 743, 813 and 656 bp, respectively.
Phylogenetic analysis of ORF sequences from complete fosmid sequences
For each ORF of the 21 archaeal fosmids containing 16S rRNA genes (Martín-Cuadrado et al., 2008), we retrieved its homologues in two steps. First, we carried out a BLASTp (Altschul et al., 1997) search on the non-redundant database at NCBI (http://www.ncbi.nlm.nih.gov/) with default parameters, except for the max-target-sequences threshold, which was fixed at 1000. Second, we carried out a similar search but restricted only to the archaeal sequences of the non-redundant database to assure the exhaustive recovery of archaeal homologous sequences. The sequences resulting from the two searches were gathered in a single data set and aligned using Muscle 3.6 (Edgar, 2004) and the resulting alignment was manually inspected and refined using the MUST software (Philippe, 1993). For each ORF a preliminary phylogenetic analysis was conducted using the neighbour-joining method (Saitou and Nei, 1987). Based on the resulting phylogenetic tree, we selected a subset of 80–100 representative sequences, which were re-aligned with Muscle 3.6 and the resulting alignment manually refined. Ambiguously aligned regions were removed before phylogenetic analysis. Maximum likelihood phylogenetic tree reconstruction was carried out on the remaining positions using PHYML (Guindon and Gascuel, 2003) and TREEFINDER (Jobb et al., 2004) applying the Le and Gascuel (LG) model (Le and Gascuel, 2008) with a Gamma correction (four discrete categories) to take into account evolutionary rate variation among sites. Tree robustness was estimated by a non-parametric bootstrap approach using PHYML and the same parameters with 100 replicates of the original data set.
Taxonomic ascription and phylogenetic analysis of fosmid-end sequences
Each fosmid-end DNA sequence was queried using BLASTx (Altschul et al., 1997) against a local sequence database containing all protein sequences encoded by 291 bacterial, 50 archaeal and 53 eukaryotic complete annotated genomes as well as 12 813 amino-acid ORFs predicted from environmental oceanic archaeal BAC and fosmid sequences from various previous works (Béjá et al., 2000b; Hallam et al., 2004; López-García et al., 2004; Moreira et al., 2004; Martín-Cuadrado et al., 2008). An automated analysis of the 15 best BLASTx hits (counting only one result per species) allowed us to determine the most probable taxonomic origin of the corresponding fosmid-end sequence. We attributed a putative archaeal origin if more than 50% of the best hits (counting only one BLAST result per species) corresponded to archaeal sequences or if at least one euryarchaeotal sequence and one crenarchaeotal sequence appeared among those 15 best hits. This type of taxonomic affiliation based on simple sequence similarity criteria can be affected by sequence composition biases or by the different number of genomes available for each taxonomic group in the queried database (see below). Therefore, all fosmids with at least one end of potential archaeal origin were further analysed by phylogenetic analysis, which is much more robust than BLAST to infer evolutionary relationships (Koski and Golding, 2001). In fact, if a given sequence branches within a particular taxonomic group with good statistical support, this is unlikely to be affected by the number of sequences available for the different groups. Thus, to test whether the preliminary BLAST-based taxonomic ascriptions to archaea were indeed correct, we aligned the amino-acid translated sequences of both ends of the corresponding fosmids with the respective 50 best BLASTp hit sequences (keeping only one BLAST result per species) using Muscle 3.6 (Edgar, 2004), and these multiple alignments were used to reconstruct maximum likelihood phylogenetic trees using TREEFINDER (Jobb et al., 2004) with the same model and parameters as for the analysis of ORFs from complete fosmid sequences. All trees were visually inspected to determine the taxonomic group to which each fosmid end belonged. As control, the same analysis was also done on the completely sequenced genomes of two thaumarchaeotal (C. symbiosum and Candidatus N. maritimus) and two bacterial (the gammaproteobacterium Alteromonas macleodii and the planctomycete Rhodopirellula baltica) species, which were cut in pieces of ∼40 kb long to simulate environmental fosmids, generating 1000 fragments per genome.
Results and Discussion
Phylogenetic analysis of complete fosmid sequences
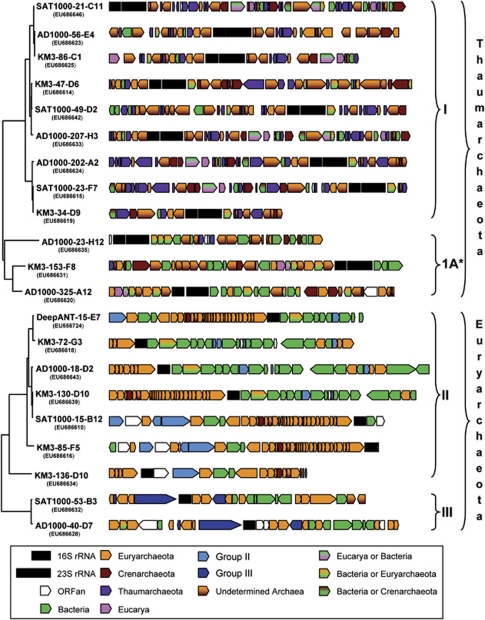
The 21 archaeal fosmids containing 16S rRNA genes used for our analysis are those recently described by Martín-Cuadrado et al. (2008). Twenty of them were retrieved by phylogenetic screening of three fosmid libraries constructed from deep-sea planktonic samples: AD1000 (Adriatic Sea, 1000 m depth, seven fosmids), KM3 (Ionian Sea, 3000 m depth, eight fosmids) and SAT1000 (South Atlantic, 1000 m depth, five fosmids). We also included one fosmid from the DeepANT library (Antarctic polar front, 500 m depth). Phylogenetic analysis of their 16S rDNA sequences revealed that 12 of them belonged to Thaumarchaeota (including three from the divergent group 1A, related to the environmental sequence pSL12 from Yellowstone, see Barns et al., 1996 and DeLong et al., 2006), and the remaining nine belonged to Euryarchaeota species (seven from group II and two from group III) (Martín-Cuadrado et al., 2008). Altogether, these 21 fosmids encoded 681 ORFs (399 for Thaumarchaeota and 282 for groups II/III Euryarchaeota). However, because these three archaeal groups have most likely a single 16S rRNA gene (Martín-Cuadrado et al., 2008), all these fosmids encompassed the same genomic region. Therefore, several homologous genes were present in different fosmids. More precisely, the 681 ORFs represented 289 gene families (200 in Thaumarchaeota and 90 in group II/III Euryarchaeota, one family being found in both archaeal groups), so that the number of different genes was much smaller than the total number of ORFs.
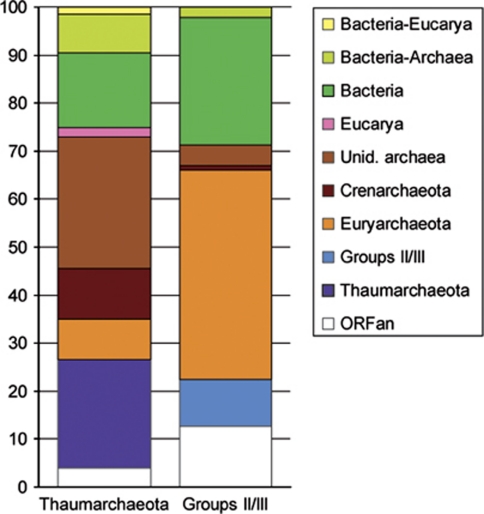
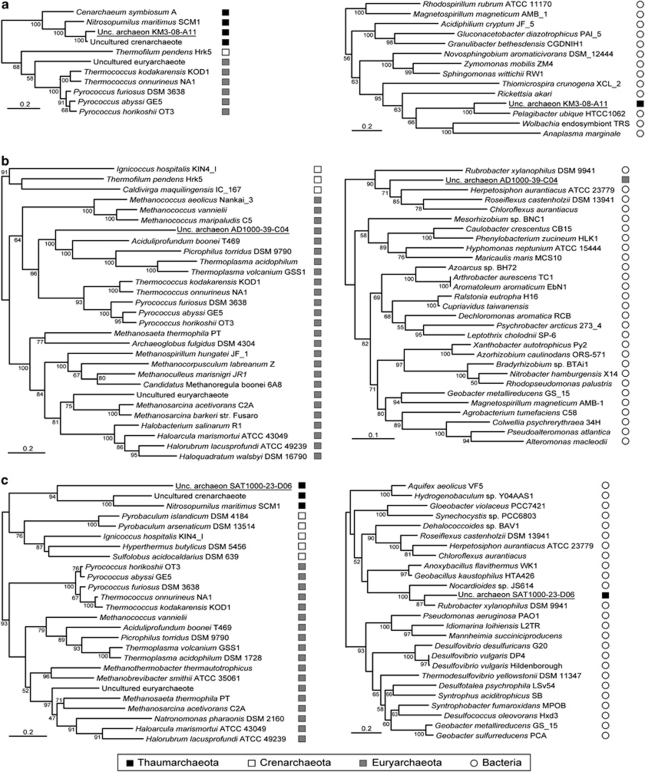
Maximum likelihood phylogenetic analyses of the ORFs found in the 12 thaumarchaeotal fosmids showed that most of them (73–81%, depending on each fosmid) were of archaeal origin (Figures 1 and 2). A large majority of them had closely related homologues in C. symbiosum and Candidatus N. maritimus genomes, confirming the taxonomic affiliation of those fosmids to Thaumarchaeota. Moreover, 45 (22%) of those genes were exclusively found in Thaumarchaeota representatives (Figures 1 and 2 and Supplementary Figure 1a), likely corresponding to ancient gene innovations in this archaeal lineage. For those having homologues in other archaea, a clear euryarchaeotal or crenarchaeotal phylogenetic relationship could be inferred only for a few ORFs (Figures 1 and 2). This would be in agreement with Thaumarchaeota representing an evolutionary lineage distinct from classical hyperthermophilic Crenarchaeota and Euryarchaeota. Among the total 200 phylogenetic trees of the protein families present in the thaumarchaeotal fosmids, Thaumarchaeota sequences branched among the hyperthermophilic Crenarchaeota or within Euryarchaeota in only 21 and 17 phylogenetic trees, respectively (Figures 1 and 2 and Supplementary Figures 1b and c). Thus, HGTs from these groups to Thaumarchaeota appeared to be relatively rare. In contrast, we observed a higher frequency of HGTs involving bacterial donors (31 cases) (Figures 1 and 2). As expected from their high relative abundance in deep-ocean ecosystems (DeLong et al., 2006; Martín-Cuadrado et al., 2007), most of these HGT involved proteobacterial donors but also, and more surprisingly, Firmicutes and Chloroflexi (Figures 3a and b). Although much less frequent than Proteobacteria, both groups are relatively abundant in deep waters of the Mediterranean basin (Martín-Cuadrado et al., 2007; Quaiser et al., 2011). These genes of bacterial origin, which are scattered in the genomic fragments analysed (Figure 1), are often shared by a variety of thaumarchaeotal species, including C. symbiosum and N. maritimus, suggesting that the HGT events are ancient. This was also the case for three ORFs of eukaryotic origin (Figures 1, 2 and 3c).
Figure 1.
Genomic organisation of 21 fosmids obtained from deep-sea plankton containing archaeal 16S rRNA genes. The corresponding phylogenetic trees of their 16S rRNA genes are shown on the left. Small and large black rectangles correspond to 16S and 23S rRNA genes, respectively, whereas ORFs are represented by arrowed boxes. The evolutionary origin of each ORF is indicated by a colour code. The different archaeal groups studied are indicated on the right. The symbol ‘*' denotes the thaumarchaeotal group 1A corresponding to the divergent pSL12-like clade.
Figure 2.
Histogram representing the phylogenetic origin of the ORFs present in the 21 analysed fosmids. The percentages were derived from individual gene phylogenetic analysis of 200 ORFs for Thaumarchaeota and 90 ORFs for group II/III Euryarchaeota.
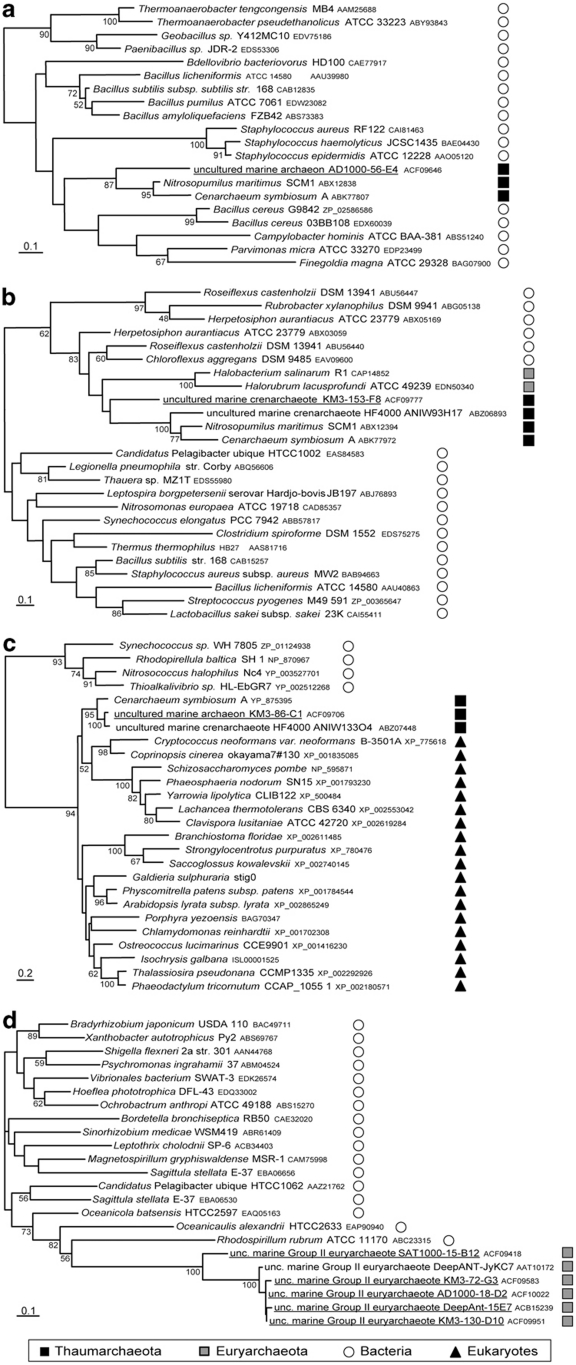
Figure 3.
Maximum likelihood phylogenetic trees illustrating HGT events from diverse donors to Thaumarchaeota and group II Euryarchaeota. (a) HGT from a firmicute donor; (b) HGT from a Chloroflexi donor; (c) HGT from a eukaryotic donor; (d) HGT from a proteobacterial donor. Sequences from our fosmids are underlined. The scale bar represents the average number of substitutions per site. Numbers at nodes correspond to bootstrap values (only those >50% are shown).
Our results support previous observations showing a relatively high HGT frequency in Thaumarchaeota (López-García et al., 2004). The main difference with previous analyses concerns the proportion of ORFan genes, which decreases from 41.5% in DeepAnt-EC39 to 2.4%, and, conversely, the proportion of ORFs specific of Thaumarchaeota, which grows from 4.9% in DeepAnt-EC39 to 34.1% in the fosmids of this study. This is largely due to the fact that more sequence data for Thaumarchaeota are now available, in particular the complete genome sequences of C. symbiosum and N. maritimus. Nevertheless, a significant frequency of gene acquisition from bacterial and, to a less extent, archaeal donors is still observed for Thaumarchaeota, suggesting that HGT has actually played an important role in the adaptation of these organisms to their environment.
In the case of the seven group II euryarchaeotal fosmids, synteny was remarkably conserved. This was partly, but not exclusively, due to the presence of the very well conserved spectinomycin operon encoding several ribosomal proteins upstream of the 16S rDNA (Moreira et al., 2004; Martín-Cuadrado et al., 2008). In contrast with group I fosmids, we detected very few ORFs (9.5%) specific to Group II and shared by at least 2 of its representatives (Figure 1 and Supplementary Figure 1d). The majority of the other ORFs (49.2%) have homologues in other archaea and most (44.4%) showed a clear euryarchaeotal origin (Figures 1 and 2b), in agreement with the phylogenetic position of group II inferred from 16S rDNA analysis. The remaining ORFs (27%) appeared to have been acquired by HGT from different bacterial donors (Figures 1 and 3d). In contrast with the genes of bacterial origin in thaumarchaeotal fosmids, these ORFs concentrate in the region downstream the 16S rDNAs (Figure 1). The opposite region is occupied in all group II fosmids by the large spectinomycin operon of ribosomal proteins, which are know to be reluctant to HGT (Matte-Tailliez et al., 2002) or recombination events that might disrupt the operon organisation. Therefore, the presence of this large operon is likely responsible for the lack of observable HGT events in this genomic region. However, in those fosmids for which we have sequences further upstream the spectinomycin operon (SAT1000-15-B12, KM3-85-F5, KM3-136-D15 and DeepAnt-15-E5), several cases of HGT from bacteria are apparent (Figure 1). Hence, if we exclude this highly conserved operon, the levels of HGT inferred upstream and downstream of it suggest a very large amount of HGT in this archaeal lineage.
The analysis of the two fosmids from group III Euryarchaeota, which showed a well conserved synteny, displayed an intermediate picture: 13 genes (41.9%) supported a euryarchaeotal origin whereas eight ORFs (25.8%) were apparently of bacterial origin (Figure 1). In contrast to group II Euryarchaeota, these genes acquired by HGT did not cluster in a particular genomic region.
Analysis of fosmid-end sequences
A potential bias inherent to the study of 16S rDNA-containing fosmids may derive from the fact that all of them target the same genomic region, especially if we take into account that these fosmids most likely belong to archaeal species with a single copy of rRNA genes (Martín-Cuadrado et al., 2008). In the absence of data for other genomic regions, one cannot exclude that the 16S rDNA genomic neighbourhood had an anomalous frequency of HGT and that the values observed do not represent the whole genome, although this would be at odds with the general idea that this region is enriched in conserved and highly expressed genes (Krawiec and Riley, 1990). In order to overcome this potential limitation, we tried a novel approach based on the phylogenetic study of pairs of fosmid-end sequences belonging to the same fosmid clone. If both insert ends of a given fosmid show the same and well supported phylogenetic origin (that is bacterial or archaeal), it may be hypothesised that it reflects the phylogenetic affiliation of the whole genomic fragment. On the contrary, if a different phylogenetic affiliation (for example, archaeal for one end and bacterial for the other) is strongly supported for each end, it may be hypothesised that an HGT event is affecting one of the two ends. In that case, the affiliation of the whole fosmid itself (as archaeal or bacterial) would be difficult to discern based on the fosmid-end sequences only. However, a number of recent observations provide clues to decide on the phylogenetic affiliation of such genome fragments. First, several complete fosmid sequences from marine prokaryotes, from both plankton and sediment, have been published in the last years, showing no case of direct HGT from archaea to bacteria (see, for example, Nesbo et al., 2005; Moreira et al., 2006; Quaiser et al., 2008). This supports the idea that interdomain HGT frequencies are much smaller in bacterial than in archaeal fosmids. This contrasts with the fact that genes of archaeal origin seem to be frequent in certain non-planktonic bacterial groups, such as the Thermotogales (Zhaxybayeva et al., 2009). Second, a similar absence of archaeal genes is observed in typical marine planktonic bacteria for which complete genome sequences are available, such as Pelagibacter ubique (Giovannoni et al., 2005) or Alteromonas macleodii (Ivars-Martínez et al., 2008), which possess an exceedingly rare number of genes of archaeal origin. In contrast, a relatively high level of HGT from bacterial donors can be estimated in the genomes of Cenarchaeum symbiosum and Candidatus N. maritimus (see below). Therefore, it seems reasonable to hypothesise that fosmids containing a mixture of end sequences of archaeal and bacterial affiliation are most probably archaeal with a certain proportion of bacterial genes acquired by HGT.
Based on this, we analysed the fosmid-end sequences of 12 774 fosmids from the three metagenomic libraries to which our completely sequenced fosmids belong (4124 from AD1000, 4549 from KM3 and 4101 from SAT1000). We attributed a first taxonomic affiliation to their end sequences by studying the taxonomy of the 15 best hits for each sequence identified by BLASTx search (see Materials and methods). For those fosmids having at least one end sequence suggesting an archaeal affiliation, we tested it by maximum likelihood phylogenetic analysis of the corresponding translated amino-acid sequence. We identified in this way 590 fosmids with at least one end sequence of probable archaeal origin (Supplementary Table 1). From the 1180 end sequences of these fosmids, only 27 had homologues in more than one fosmid. Therefore, our end sequence data set had very little redundancy, validating our initial idea of using this approach to analyse a wide and random representation of genes from these uncultured marine archaea. In all 355 of these fosmids had both end sequences supporting an archaeal affiliation (2.77% of all fosmids in our libraries) and 255 had ends supporting different taxonomic origins (that is one archaeal and one bacterial, 1.99% of all fosmids). KM3 was the library with the highest frequency of those mixed cases (116), followed by AD1000 (84) and SAT1000 (48). According to the above reasoning, we interpreted these mixed cases as HGT from bacterial donors into archaeal recipients. They represented 43.22% of the 590 fosmids tentatively attributed to archaeal species, indicating that ∼21% of the ORFs encoded by the corresponding fosmid ends were acquired from bacteria (Figure 4a). This estimate was remarkably congruent with the frequencies of bacteria-to-archaea HGT that we deduced by the phylogenetic analyses of all ORFs from 21 complete fosmid sequences (ranging from 16 to 30%, see above), which suggested that the genomic region containing the rRNA genes had an HGT frequency comparable to that of other parts of these archaeal genomes.
Figure 4.
(a) Histogram showing the different types of archaeal fosmids found in our libraries according to the taxonomic affiliation of their two fosmid-end sequences. Arch: archaea of uncertain taxonomy; Bact: bacteria; Cren: Crenarchaeota; Eury: Euryarchaeota; Thaum: Thaumarchaeota. (b) Taxonomic ascription of the bacterial donors involved in HGT events detected by phylogenetic analysis of fosmid-end sequences.
To test whether this method overestimated the number of HGT events detected, we carried out a simulation using the first complete genome sequences of thaumarchaeotal species, C. symbiosum (Hallam et al., 2006) and Candidatus N. maritimus (Walker et al., 2010) (no complete genome sequence for any group II or III Euryarchaeota is available yet). We generated 1000 artificial fosmid sequences by cutting randomly these genome sequences in fragments of 40 kbp. We then applied the same procedure described above to the two 800 bp end sequences of each of these pseudo-fosmids (after exclusion of C. symbiosum and Candidatus N. maritimus from the BLAST database). We retrieved 51% (C. symbiosum) and 48.7% (N. maritimus) of the pseudo-fosmids with both end sequences of archaeal affiliation, and 49% (C. symbiosum) and 51% (N. maritimus) with at least one of bacterial affiliation. Among them, a relatively small number of cases (∼10% in each genome) showed bacterial affiliation for both end sequences (data not shown). These simulations suggested that these planktonic archaea have indeed acquired a relatively high percentage of genes, ∼25%, from bacteria by HGT. At any rate, because we inferred comparable HGT frequencies for our fosmids and for the complete genomes of C. symbiosum and Candidatus N. maritimus, our approach did not appear to overestimate the amount of HGT in the fosmids from our three libraries. Moreover, this suggests that it provided relatively good estimates of HGT frequencies, at least for the Thaumarchaeota and probably also for the group II and III Euryarchaeota. As an additional control, we applied the same strategy on two complete genome sequences of typical marine bacteria: the gammaproteobacterium A. macleodii and the planctomycete Rhodopirellula baltica. In the case of A. macleodii we did not detect any sequence of archaeal origin in this way, whereas we detected only two in R. baltica (the hypothetical protein RB12247, GenBank GI 32477404, and the X-pro-aminopeptidase PepQ2, GenBank GI 32476507, data not shown). These values were significantly smaller than those found for C. symbiosum, Candidatus N. maritimus and our archaeal fosmids, supporting the idea that most interdomain HGT events in marine plankton occur from bacteria to archaea. Nevertheless, it could be argued that our results might be biased by the small amount of sequence data available for marine planktonic archaea in comparison with marine bacteria. However, although this bias might affect the results obtained for A. macleodii, belonging to a bacterial group (the Gammaproteobacteria) with dozens of complete genome sequences available, it should not affect the results obtained for R. baltica, one of the rare planctomycete genome sequences available, and the only one from marine environments. Moreover, our results agreed with previous analyses of planctomycete genomes showing a very small number of genes of archaeal origin (Fuchsman and Rocap, 2006).
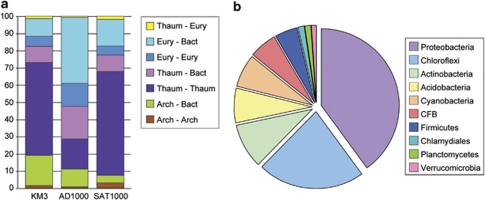
The results of our phylogenetic analyses were not accurate enough to determine the precise phylum at the origin of 142 fosmid-end sequences of bacterial affiliation (∼56% of the total 255 sequences of bacterial origin). For the rest, the identified donors belonged to very diverse bacterial phyla, with a clear dominance of Proteobacteria, which were the donors in 64 cases (Figure 4b). Among them, when a particular subdivision was unambiguously identified as donor, Alphaproteobacteria was the most frequent one (19 cases, Figures 4b and 5a), followed by Delta- and Gammaproteobacteria (6 and 3 cases, respectively). This trend was also observed from our phylogenetic analyses of genes from complete fosmid sequences. Proteobacteria, in particular Alphaproteobacteria, are very common inhabitants of marine plankton (Giovannoni et al., 1990; Rappé et al., 2000; López-García et al., 2001) so that, quantitatively, they can be considered as very likely donors in HGT exchanges. The fact of retrieving Chloroflexi as the second most frequent donor (19 cases, Figures 4b and 5b), in particular in libraries AD1000 and KM3 (10 and 8 cases, respectively), but also in whole fosmid gene phylogenies, might appear more surprising. Nevertheless, as mentioned above, this phylum is relatively abundant in the deep Mediterranean Sea (Martín-Cuadrado et al., 2007; Quaiser et al., 2011), which makes it a likely alternative donor. Other groups (Actinobacteria, Acidobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Planctomycetes and Verrucomicrobia) appeared to have also contributed with several genes to the genomes of planktonic marine archaea (Figures 4b and 5c).
Figure 5.
Maximum likelihood trees illustrating different evolutionary histories of ORFs in fosmid-end sequences. (a) Fosmid with a Thaumarchaeota end (left) and an HGT from Alphaproteobacteria (right); (b) fosmid with a group II/III end (left) and an HGT from Chloroflexi (right); (c) fosmid with a Thaumarchaeota end (left) and an HGT from Actinobacteria (right). Sequences from our fosmids are underlined. The scale bar represents the average number of substitutions per site. Numbers at nodes correspond to bootstrap values (only those >50% are shown).
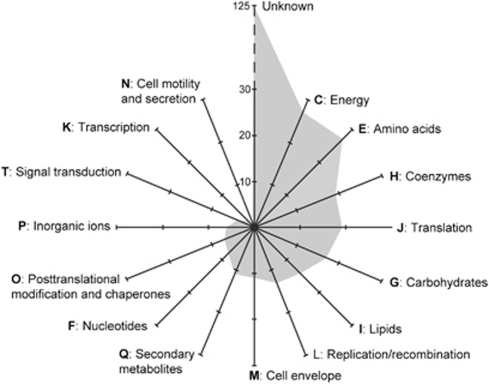
In addition to their phylogenetic origin, we also studied the function of the 255 genes presumably acquired by these archaea from bacterial donors by querying them against the COG database (Tatusov et al., 2003). Among these genes, 71 had no homologues in the COG database and 54 corresponded to hypothetical proteins of unknown function (Figure 6). COG classification of the remaining genes suggested that most cases of HGT concerned genes involved in various metabolisms. Among them, NADH:ubiquinone and Fe-S oxidoreductases in COG category C (energy production and conversion), and amino-acid ABC transporters and carbohydrate transporters (COG categories E and G, amino acid transport and metabolism and carbohydrate transport and conversion) were the most represented. A previous analysis of metagenomic data from the KM3 site showed that transporters, namely amino acid and carbohydrate transporters, are overrepresented in the deep-planktonic community (Martín-Cuadrado et al., 2007), indicating that they are important for living in this nutrient-depleted environment. Because of their functional importance and high frequency, they constitute suitable targets to be involved in HGT events. The dominance of metabolic genes in the HGT pool agreed with studies showing that metabolic genes are among those being most frequently involved in HGT events (Jain et al., 1999; Gogarten and Townsend, 2005). However, we also identified a number of HGT events involving genes belonging to the ‘Information storage and processing' COG categories, which are commonly thought to be much less prone to HGT (Jain et al., 1999). Nevertheless, most of the translation proteins that we identified as HGT acquisitions were aminoacyl-tRNA synthetases, which are known to be frequently transferred even across different domains, in particular under antibiotic selective pressure (Brown et al., 1998; Wolf et al., 1999; Brochier et al., 2002). This may be surprising for planktonic organisms as antibiotics can hardly exert any pressure on very diluted environments like plankton. However, previous work suggests that some of these marine archaea, in particular members of group II Euryarchaeota, probably live attached to particles (Martín-Cuadrado et al., 2008), where antibiotics may exert a strong selective pressure in a high competition environment. Interestingly, we showed in a previous study that the only protein with traces of positive selection among those encoded in our archaeal complete fosmid sequences was an informational one, the ribosomal protein S3 found in the spectinomycin operon of group II Euryarchaeota fosmids, which further suggests that antibiotics may exert a selective pressure on these archaea (Martín-Cuadrado et al., 2008). In the case of the proteins related to DNA processing, the most represented were DNA helicases and glycosylases involved in DNA repair. Within the third major family of COG categories (‘Cellular processes'), we observed several HGT cases for the category M (cell envelope biogenesis, outer membrane, 11 cases) but very few for the others (see Figure 6).
Figure 6.
Functional classification of sequences of bacterial origin in archaeal fosmids according to COG families. These sequences were detected in 255 fosmids carrying one end sequence of archaeal phylogenetic ascription and the other one of bacterial phylogenetic ascription.
Concluding remarks
Studying the evolutionary history of genes found in marine planktonic archaea can help to better characterise these still poorly known lineages and to better understand their adaptations to oceanic environments. In particular, the role that HGT may have played in this process remains an open question. To address it, we studied the phylogeny of 681 ORFs from 21 rDNA-containing archaeal fosmid sequences, as well as the phylogeny of the end sequences of 590 fosmids containing archaeal genome fragments as a proxy for the random exploration of other genome regions of these organisms. Our results not only support previous observations showing a relatively high HGT frequency in Thaumarchaeota (López-García et al., 2004) but also extend them to the group II and III Euryarchaeota, which showed even a higher frequency of genes acquired by HGT from bacteria (27% and 25.8%, respectively). In fact, 57% the archaeal fosmids with one fosmid-end sequence of probable bacterial origin belonged to group II/III Euryarchaeota, in contrast to 43% belonging to Thaumarchaeota (Figure 4a). Moreover, in our three libraries, fosmids with mixed bacterial-euryarchaeotal ends were much more frequent than fosmids with two euryarchaeotal ends, in contrast with the thaumarchaeotal fosmids, dominated by those with two thaumarchaeotal ends (except in the AD1000 library, see Figure 4a). These results reinforce the idea that HGT may be more prevalent in the group II/III Euryarchaeota.
The acquisition of bacterial genes appears to have been a convergent evolutionary process in all major groups of marine planktonic archaea (that is, with parallel independent acquisitions from a variety of donors), likely having a wide impact on the evolution of their genomes. Some of the genes acquired from bacteria, such as the amino-acid and carbohydrate transporters, are most likely important in the adaptation to the mesophilic oligotrophic marine environment. Several of the HGT events that we detected in the different marine archaeal groups appear to be relatively ancient because they are shared by fosmids belonging to distant species, as deduced from their distance in phylogenetic trees (Figure 3). This suggests that those genes acquired by HGT were fixed in the genomes of these archaea for long evolutionary times and that they are essential for these organisms. A similar situation has been described in another archaeal group, the Thermoplasmatales, a group of Euryarchaeota that acquired a large number of genes from hyperthermophilic crenarchaeotal donors belonging to the Sulfolobales (Ruepp et al., 2000). At an even larger evolutionary scale, all species of the bacterial group of the Thermotogales appear to have acquired many genes from archaeal donors (Zhaxybayeva et al., 2009). In both cases, acquired genes are involved in functions important for the adaptation of these organisms to their respective environments. Similarly, Thaumarchaeota and group II/III Euryarchaeota may represent two cases of convergent acquisition of a significant amount of bacterial genes during their adaptation to the same natural environment. Additional genome sequence data from these groups will be necessary to allow a more detailed characterisation of this phenomenon and its evolutionary consequences.
Acknowledgments
This work was funded by the French National Agency for Research (EVOLDEEP project, contract number ANR-08-GENM-024-002). C Brochier-Armanet is supported by an ‘Action Thématique et Incitative sur Programme' (ATIP) of the French Centre National de la Recherche Scientifique (CNRS).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barns SM, Delwiche CF, Palmer JD, Pace NR. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béjá O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000a;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Béjá O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP, et al. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol. 2000b;2:516–529. doi: 10.1046/j.1462-2920.2000.00133.x. [DOI] [PubMed] [Google Scholar]
- Brochier C, Bapteste E, Moreira D, Philippe H. Eubacterial phylogeny based on translational apparatus proteins. Trends Genet. 2002;18:1–5. doi: 10.1016/s0168-9525(01)02522-7. [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- Brown JR, Zhang J, Hodgson JE. A bacterial antibiotic resistance gene with eukaryotic origins. Curr Biol. 1998;8:R365–R367. doi: 10.1016/s0960-9822(98)70238-6. [DOI] [PubMed] [Google Scholar]
- DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard NU, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- Fuchsman CA, Rocap G. Whole-genome reciprocal BLAST analysis reveals that planctomycetes do not share an unusually large number of genes with Eukarya and Archaea. Appl Environ Microbiol. 2006;72:6841–6844. doi: 10.1128/AEM.00429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Davis AA. Widespread Archaea and novel Bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar Ecol Prog Ser. 1997;150:275–285. [Google Scholar]
- Fuhrman JA, McCallum K, Davis AA. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- Ghai R, Martín-Cuadrado AB, Molto AG, Heredia IG, Cabrera R, Martín J, et al. Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J. 2010;4:1154–1166. doi: 10.1038/ismej.2010.44. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, et al. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Rosenberg SM, Slack A. Antibiotic-induced lateral transfer of antibiotic resistance. Trends Microbiol. 2004;12:401–404. doi: 10.1016/j.tim.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Ivars-Martínez E, Martin-Cuadrado AB, D'Auria G, Mira A, Ferriera S, Johnson J, et al. Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J. 2008;2:1194–1212. doi: 10.1038/ismej.2008.74. [DOI] [PubMed] [Google Scholar]
- Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G, Lindstrom K, Saano A. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol. 1997;63:803–805. doi: 10.1128/aem.63.2.803-805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J Mol Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- Krawiec S, Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990;54:502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- López-García P, Brochier C, Moreira D, Rodríguez-Valera F. Comparative analysis of a genome fragment of an uncultivated mesopelagic crenarchaeote reveals multiple horizontal gene transfers. Environ Microbiol. 2004;6:19–34. doi: 10.1046/j.1462-2920.2003.00533.x. [DOI] [PubMed] [Google Scholar]
- López-García P, López-López A, Moreira D, Rodríguez-Valera F. Diversity of free-living prokaryotes from a deep-sea site at the Antarctic Polar Front. FEMS Microbiol Ecol. 2001;36:193–202. doi: 10.1016/s0168-6496(01)00133-7. [DOI] [PubMed] [Google Scholar]
- Martín-Cuadrado AB, López-García P, Alba JC, Moreira D, Monticelli L, Strittmatter A, et al. Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS ONE. 2007;2:e914. doi: 10.1371/journal.pone.0000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Cuadrado AB, Rodríguez-Valera F, Moreira D, Alba JC, Ivars-Martinez E, Henn MR, et al. Hindsight in the relative abundance, metabolic potential and genome dynamics of uncultivated marine archaea from comparative metagenomic analyses of bathypelagic plankton of different oceanic regions. ISME J. 2008;2:865–886. doi: 10.1038/ismej.2008.40. [DOI] [PubMed] [Google Scholar]
- Matte-Tailliez O, Brochier C, Forterre P, Philippe H. Archaeal phylogeny based on ribosomal proteins. Mol Biol Evol. 2002;19:631–639. doi: 10.1093/oxfordjournals.molbev.a004122. [DOI] [PubMed] [Google Scholar]
- Moreira D, Rodriguez-Valera F, Lopez-Garcia P. Analysis of a genome fragment of a deep-sea uncultivated Group II euryarchaeote containing 16S rDNA, a spectinomycin-like operon and several energy metabolism genes. Environ Microbiol. 2004;6:959–969. doi: 10.1111/j.1462-2920.2004.00644.x. [DOI] [PubMed] [Google Scholar]
- Moreira D, Rodriguez-Valera F, Lopez-Garcia P. Metagenomic analysis of mesopelagic Antarctic plankton reveals a novel deltaproteobacterial group. Microbiology. 2006;152:505–517. doi: 10.1099/mic.0.28254-0. [DOI] [PubMed] [Google Scholar]
- Nesbo CL, Boucher Y, Dlutek M, Doolittle WF. Lateral gene transfer and phylogenetic assignment of environmental fosmid clones. Environ Microbiol. 2005;7:2011–2026. doi: 10.1111/j.1462-2920.2005.00918.x. [DOI] [PubMed] [Google Scholar]
- Philippe H. MUST, a computer package of Management Utilities for Sequences and Trees. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CM, Wu KY, Molinski TF, DeLong EF. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaiser A, Lopez-Garcia P, Zivanovic Y, Henn MR, Rodriguez-Valera F, Moreira D. Comparative analysis of genome fragments of Acidobacteria from deep Mediterranean plankton. Environ Microbiol. 2008;10:2704–2717. doi: 10.1111/j.1462-2920.2008.01691.x. [DOI] [PubMed] [Google Scholar]
- Quaiser A, Zivanovic Y, Moreira D, López-García P. Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J. 2011;5:285–304. doi: 10.1038/ismej.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé MS, Vergin K, Giovannoni SJ. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol Ecol. 2000;33:219–232. doi: 10.1111/j.1574-6941.2000.tb00744.x. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW, et al. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000;407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schleper C, Holben W, Klenk HP. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl Environ Microbiol. 1997;63:321–323. doi: 10.1128/aem.63.1.321-323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases—analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, Swithers KS, Lapierre P, Fournier GP, Bickhart DM, DeBoy RT, et al. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc Natl Acad Sci USA. 2009;106:5865–5870. doi: 10.1073/pnas.0901260106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.