Abstract
The phototrophic microbial mat community of Mushroom Spring, an alkaline siliceous hot spring in Yellowstone National Park, was studied by metatranscriptomic methods. RNA was extracted from mat specimens collected at four timepoints during light-to-dark and dark-to-light transitions in one diel cycle, and these RNA samples were analyzed by both pyrosequencing and SOLiD technologies. Pyrosequencing was used to assess the community composition, which showed that ∼84% of the rRNA was derived from members of four kingdoms Cyanobacteria, Chloroflexi, Chlorobi and Acidobacteria. Transcription of photosynthesis-related genes conclusively demonstrated the phototrophic nature of two newly discovered populations; these organisms, which were discovered through metagenomics, are currently uncultured and previously undescribed members of Chloroflexi and Chlorobi. Data sets produced by SOLiD sequencing of complementary DNA provided >100-fold greater sequence coverage. The much greater sequencing depth allowed transcripts to be detected from ∼15 000 genes and could be used to demonstrate statistically significant differential transcription of thousands of genes. Temporal differences for in situ transcription patterns of photosynthesis-related genes suggested that the six types of chlorophototrophs in the mats may use different strategies for maximizing their solar-energy capture, usage and growth. On the basis of both temporal pattern and transcript abundance, intra-guild gene expression differences were also detected for two populations of the oxygenic photosynthesis guild. This study showed that, when community-relevant genomes and metagenomes are available, SOLiD sequencing technology can be used for metatranscriptomic analyses, and the results suggested that this method can potentially reveal new insights into the ecophysiology of this model microbial community.
Keywords: metatranscriptome, photosynthesis, phototrophy, cyanobacteria, chlorophyll, reaction centers
Introduction
The chlorophototrophic microbial mats of alkaline siliceous hot springs in Yellowstone National Park have served for decades as model systems for understanding the composition, structure and function of microbial communities (Brock, 1978; Ward et al., 1998, 2002, 2006, 2011; Ward and Castenholz, 2000; Klatt et al., 2011). Members of the kingdoms Cyanobacteria (mostly Synechococcus spp.) and Chloroflexi (family Chloroflexales; filamentous anoxygenic phototrophs (FAPs), mostly Roseiflexus and Chloroflexus spp.) (Pierson and Castenholz, 1974; van der Meer et al., 2010; Ward et al., 2011) are the dominant organisms in these mat communities (Table 1). Cultivation and DNA sequencing methods have produced complete or nearly complete consensus genome sequences for several chlorophototrophic organisms that occur in these mats, including Synechococcus spp. strains A and B′, which are close relatives of indigenous populations that co-occur in the 60 °C mat (Ferris and Ward, 1997; Allewalt et al., 2006; Bhaya et al., 2007); Roseiflexus sp. strain RS-1 (van der Meer et al., 2010; Bryant et al., 2011); and Chloroflexus aurantiacus strains Y-400-fl and 396-1 (Madigan and Brock, 1977; Bryant et al., 2011). The application of metagenomic methods, in combination with traditional microbial cultivation methods, recently led to the identification of a previously unknown chlorophototroph, Candidatus Chloracidobacterium thermophilum, from the kingdom Acidobacteria as an additional member of these mats (Bryant et al., 2007). The genome of this organism has also recently been completed (Bryant et al., 2011; AM Garcia Costas, Z Liu, LP Tomsho, SC Schuster, DM Ward, and DA Bryant, in preparation). As documented in the accompanying paper (Klatt et al., 2011) and in previous studies (Bryant et al., 2007), metagenomics analyses additionally suggested the existence of other chlorophototrophic members of this community, which were previously undescribed and are currently uncultivated organisms from the kingdoms Chloroflexi and Chlorobi (Table 1). Collectively, the availability of these genomes was instrumental in analyzing metagenomes from these mat communities (Klatt et al., 2011).
Table 1. Relationship between predominant phototrophic phylogenetic groups and functional guilds.
| Phylogenetic group | Chlorophylls | Carbon metabolism | Reaction center gene expressiona | Chl gene expressiona | Functional guild |
|---|---|---|---|---|---|
| Synechococcus spp. A-like | Chl a | Autotrophy | Day | Day | Oxygenic phototrophs (600–700 nm) |
| Synechococcus spp. B′-like | Chl a | Autotrophy | Day | Day | Oxygenic phototrophs (600–750 nm) |
| Roseiflexus spp. | BChl a | Mixotrophy | Morning low light | Evening | 850–950 nm mixotrophs |
| Anaerolineae-like Chloroflexi | BChl ab | Unknown | Evening | Evening | BChl a/near-IR mixotrophs |
| Chloroflexus spp. | Major: BChl c, Minor: BChl a | Mixotrophy | Morning low light | Evening | 700–750 nm mixotrophs |
| C. thermophilum | Major: BChl c, Minor: BChl a, Chl a | Heterotrophy | Evening | Day | 700–750 nm heterotrophs |
| Chlorobiales | Major: BChl c, Minor: BChl a, Chl a | Mixotrophy | Evening | Day | 700–750 nm mixotrophs |
Period of maximal gene expression among times sampled to date.
Insufficient evidence currently exists to determine whether this organism can synthesize other chlorophylls.
Understanding transcription and its regulation in natural microbial communities has been an intriguing, although technically challenging goal, mainly because of the broad biodiversity of most such communities and the inherent difficulties in working with RNA. Microarrays (Parro et al., 2007) and cloning of randomly amplified complementary DNAs (cDNAs) (Poretsky et al., 2005) have been successfully applied to metatranscriptomic studies. Because of recent developments in sequencing technologies, metatranscriptomic analyses by intensive RNA sequencing have recently emerged as a much more powerful approach to the study of gene transcription in microbial communities. Moreover, there is no prerequisite to obtain genome sequences as required for microarray analyses, and such approaches can rapidly generate enormous amounts of information. Several pioneering studies using pyrosequencing technology on marine and soil microbial communities (Frias-Lopez et al., 2008; Urich et al., 2008; Gilbert et al., 2008) have not only demonstrated the feasibility of such an approach, but have also demonstrated its advantages in defining community structure (Urich et al., 2008) and its power in identifying mRNAs of known or novel genes (Gilbert et al., 2008). Several studies (Gilbert et al., 2008; Hewson et al., 2009; Poretsky et al., 2009) compared metatranscriptomes obtained under different environmental conditions, and these studies were able to detect differential gene expression. However, compared with the number of genes occurring in a microbial community (at least tens of thousands, if not millions), the number of mRNA sequences generated in the above-mentioned pyrosequencing studies (from ∼5000 to ∼160 000) were insufficient to provide satisfactory coverage or to infer statistically meaningful transcription patterns under different conditions for most genes occurring in the members of a complex community. For this reason previous studies have mainly focused on the most dominant members of a given microbial community, and comparisons of transcriptional behaviors of different phylogenetic and physiological groups within the same community have not yet been described. Therefore, one of the main objectives of this study was to test the feasibility of using so-called Next-Generation (NextGen) sequencing methodologies in metatranscriptomic studies.
In a complementary study to that of Klatt et al. (2011), two different approaches to transcription profiling of the chlorophototrophic microbial mat community of Mushroom Spring were compared. In the first approach, pyrosequencing (Roche GS FLX, 454 Life Sciences, Branford, CT, USA) of cDNAs produced from RNA extracted from mat samples collected at 60 °C for four different time points in a diel cycle was carried out. Samples were collected from light-to-dark and dark-to-light transition periods, because these have been shown to be important times for changes in mat physiology and metabolism, such as changes in anoxygenic photosynthesis (van der Meer et al., 2005) and nitrogen fixation (Steunou et al., 2006, 2008). The ∼225-bp sequences obtained by this approach primarily provided information on the composition of the community based on stable (that is, rRNA) sequences. In a second approach, and as a pilot study to determine the feasibility of using NextGen sequencing to conduct a full hourly diel-cycle metatranscriptomic analysis, extensive SOLiD-3 (Applied Biosystems, Carlsbad, CA, USA) sequencing was carried out using cDNAs produced from the same four RNA samples used in the pyrosequencing analysis. After in silico filtering to remove stable rRNA sequences, this approach provided 50-bp sequences derived from expressed sequences (that is, mRNAs) that provided detailed information about the global patterns of transcription for predominant members of the mat community. The combination of these two approaches validated and refined conclusions about the mat composition derived from metagenomic analyses (Klatt et al., 2011), and accurately reproduced previous observations concerning the expression of Synechococcus spp. genes for photosynthesis and nitrogen fixation (Steunou et al., 2006, 2008). This study further showed that, if suitable reference genome sequences or extensive metagenomic data are available, NextGen sequencing can be used to obtain unprecedented insights into the in situ physiological properties of community members as inferred from transcription profiles.
Materials and methods
Sample collection
Samples of microbial mats were collected from Mushroom Spring in the Yellowstone National Park on 10–11 July 2008 at a temperature of 61 to 64 °C. All samples were collected from the same, localized region of the mat, but the ambient temperature was slightly lower at sunset (2100 hours) and slightly higher in the morning because of wind and flow conditions. Mat core samples (∼0.5 cm2 area, #4 cores) were collected at 2100 hours (at sunset), 0515 hours (just before sunrise), 0640 hours (indirect sunlight on the mat) and 0840 hours (direct sunlight on the mat) at the same site. The light intensities at these four sampling times were ∼15, 0.05, 46 and 960 μmol photons m−2s−1, respectively. The top ∼2-mm thick layer containing most mat chlorophototrophs (Bauld and Brock, 1973; Ruff-Roberts et al., 1994) was removed with a razor blade, placed in a 2.0 ml screw-cap tube containing 0.5 g of sterile glass beads (∼100–200 μm diameter), and immediately immersed in liquid nitrogen. Samples were kept frozen until used for RNA extraction.
Methods for the extraction of total RNA, synthesis of cDNA, preparation of cDNA libraries, construction of rRNA and gene, and protein databases, analytical methods for the pyrosequencing and SOLiD databases, mapping validation and other statistical analyses are described in detail in the Supplementary Material. The sequences collected in this study have been deposited under accession number SRA018112 in the NCBI Sequence Read Archive.
Results and discussion
Pyrosequencing and composition analysis of the microbial mat samples
Table 2 summarizes the data obtained by pyrosequencing of cDNAs derived from the phototrophic microbial mat samples collected at four different times in a single diel cycle from Mushroom Spring. The total number of sequences ranged from 59 283 for the sample collected at sunset (2100 hours) to 128 208 for the sample collected in the low-light morning period (0640 hours) before the mat was exposed to full sunlight. As expected, most sequences were derived from rRNAs, which accounted for 88.9–94.4% of the total sequences. The percentage of sequences assignable as mRNAs differed for the four samples. The percentage of mRNA sequences was lowest (2.4%) just before sunrise (0515 hours) and was highest (8.1%) when the mat was fully illuminated (0840 hours). The percentage of sequences that were not identified as rRNA or mRNA was essentially constant in the four samples at 2.4–3.0%.
Table 2. Summary of cDNA sequences using pyrosequencing.
| Sunset 2100 hours | Pre-dawn 0515 hours | Morning, low light 0640 hours | Morning, high light 0840 hours | |
|---|---|---|---|---|
| Total cDNA sequences | 59 283 | 108 948 | 128 208 | 113 128 |
| Ribosomal RNAs | 54 660 (92.2%) | 102 829 (94.4%) | 119 654 (93.3%) | 100 566 (88.9%) |
| Messenger RNAs | 3006 (5.1%) | 3154 (2.4%) | 5536 (4.3%) | 9167 (8.1%) |
| Unidentified RNAs | 1617 (2.7%) | 2965 (2.7%) | 3018 (2.4%) | 3395 (3.0%) |
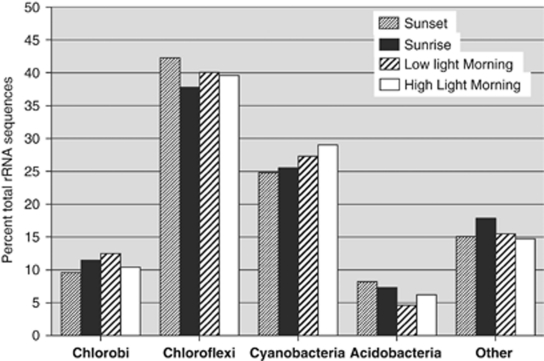
Figure 1 summarizes the kingdom-level composition of the sequences that were identified as rRNAs (both large subunit and small subunit). These data show that ∼84% of the rRNA sequences were derived from members of only four kingdoms: Chloroflexi (∼40%), Cyanobacteria (∼27%), Chlorobi (∼11%) and Acidobacteria (∼6%). The remaining sequences (∼16%) assigned as rRNAs were widely distributed across the domain Bacteria (Supplementary Figure S1). No sequences obtained from the upper mat layer could definitively be assigned to members of the domain Archaea. In agreement with Klatt et al. (2011), the next most abundant sequences were assigned to the kingdoms Firmicutes and Bacteriodetes, which represented less than 2% of the rRNA sequences. Overall, the best matches to these remaining sequences were distributed across hundreds of different bacterial sequence entries. As can be seen in Figure 1, only small differences in the percentage composition values were observed for the samples collected at four different times during the light-to-dark and dark-to-light transition periods. Although mat Synechococcus spp. exhibit phototaxis (Ramsing et al., 1997) and members of Chloroflexi have exhibited the most dramatic evidence of vertical mobility in field studies (Boomer et al., 2000), the movements of such organisms are relatively slow. It took 24 h of shading the mat to observe ‘puffs' of filaments migrating to the surface in Octopus Spring (Boomer et al., 2000). The small differences in sequence composition are unlikely to arise because of the movements of organisms up or down between the upper 2 mm and deeper mat layers (not sampled) during the diel cycle. However, it is possible that some of these differences reflect minor variations in the rRNA content of the cells as a function of the time of day. Assuming that the cells of all samples were lysed with equal efficiency in all samples, it is more likely that these differences merely reflect the slight compositional variation of the mat at the nearby but obviously slightly different sampling locations. Whatever the case, the composition of the rRNA sequences recovered from the mat at the kingdom level was essentially constant across the diel cycle within the narrow temperature range (61–64 °C) and localized region of the mat that was sampled. These observations allow the samples to be compared in order to analyze changes in transcripts as a function of the diel cycle.
Figure 1.
Kingdom-level composition of the mat community based on rRNA sequences. Only RNA sequences generated by pyrosequencing were analyzed. Data shown here are the sum of the aligned sequences for the large subunit and small subunit rRNA sequences.
The conclusion that the upper photic layer of the mat is mostly comprised of organisms from four kingdoms agrees with results from metagenomic analyses (Klatt et al., 2011). On the basis of recruitment of sequences to relevant reference genomes, metagenomic analyses of high- and low-temperature metagenomes of Mushroom Springs suggested that Cyanobacteria, Chloroflexi, Acidobacteria and Chlorobi accounted for about 41.7, 32.1, 7.6 and 3.4% of the sequences, respectively (Klatt et al., 2011). Several factors might be responsible for the observed abundance differences inferred from the metagenomic versus the metatranscriptomic data. First, these methods are not quantitative and very little is known about the relative cellular contents of DNA, RNA and proteins of the organisms comprising the mat. Second, the samples were not collected from the same mat location, and moreover, the samples were collected at different times of the year (mid-summer versus late fall). These differences could have influenced the percentage of metabolically active cells, which would in turn affect the yields of DNA and rRNA. Finally, although the Synechococcus spp. cells are much larger in volume than those of the members of the Chloroflexi in the mat, the exact effects of these differences on DNA and ribosome content per cell are unknown. For all of these reasons, the precise composition of the mat cannot accurately be determined by either method.
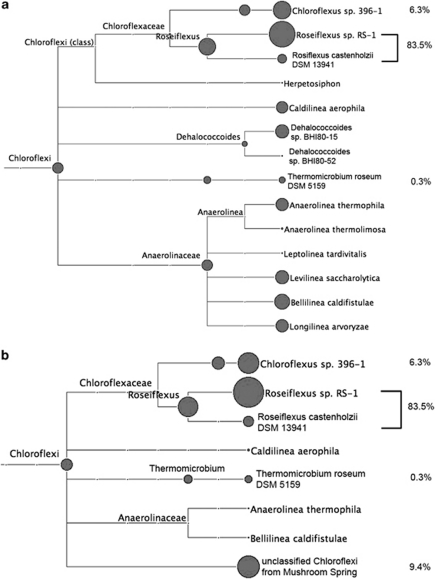
Because of the diversity exhibited by the members of the four major kingdoms, especially Chloroflexi and Acidobacteria, which include phototrophs and non-phototrophs (Ward et al., 2009; Bryant et al., 2011), in-depth analyses of the 16S rRNA sequences of these kingdoms were necessary. These analyses were carried out using reference sequences with clearer taxonomic affiliations and with higher resolving power to discriminate among similar sequence alignments (see Supplementary Information for details). Figure 2 shows the results of one such analysis for the kingdom Chloroflexi for the sequences derived from the sample collected at sunset (2100 hours). The majority (83.5%) of the sequences in this analysis were assigned as Roseiflexus spp. and 6.3% of the sequences were assigned to Chloroflexus spp. Roseiflexus sp. RS-1 and Chloroflexus sp. 396–1 were the closest sequenced representatives from these two genera, respectively. Sequences derived from close relatives of the aerobic chemo-organotroph, Thermomicrobium roseum, which was reported to be present in similar mat systems in Yellowstone (Wu et al., 2009; Klatt et al., 2011), were also detected, although at a very low level (0.3%). As shown in Figure 2a, about 10% of the remaining sequences were assigned to three different families, mainly Anaerolineaceae. To probe the phylogenetic affiliation of these remaining sequences in greater detail, the 16S rRNA sequence of a novel, unclassified member of the Chloroflexi from the mat metagenome was used as additional reference. Figure 2b shows that almost all of the remaining sequences not assigned as Roseiflexus spp. or Chloroflexus spp. were assigned to this added sequence derived from the metagenome (Klatt et al., 2011). Thus, this unclassified member of the Chloroflexi and its close relatives probably accounted for ∼10% of Chloroflexi or ∼4% of the total rRNA sequences. On the basis of analyses of the metagenome of Mushroom and Octopus Springs, Klatt et al. (2011) have found strong evidence that this high-abundance, unclassified and uncultivated Chloroflexi population (most closely, but still distantly related to members of the class Anaerolineae) is highly likely to be a previously unrecognized chlorophototroph. This is consistent with the observation that all other high-abundance populations in this mat community are also chlorophototrophs.
Figure 2.
In-depth composition analyses of Chloroflexi 16S rRNA sequences based on the high-resolution Chloroflexi 16S rRNA database (a) and after addition of partial 16S rRNA sequence of the putatively phototrophic, currently uncultivated Chloroflexi population/cluster/scaffolds from Mushroom Spring (b) in one sample (sunset). Only RNA sequences generated by pyrosequencing were analyzed. The sizes of the circles are proportional to the relative abundances of rRNA sequences assigned to a particular taxon. Figure generated by MEGAN (Huson et al., 2007) (absolute score cutoff=71, relative cutoff=0% of best hit).
As shown in Supplementary Figure S2 and as judged by the sequence similarity distribution of the 16S rRNA sequences derived from the sunset sample, members of the genus Synechococcus spp. accounted for 99.3% of the rRNA sequences assigned to the kingdom Cyanobacteria (Supplementary Figure S2A), and not surprisingly, the previously characterized, mat-derived Synechococcus sp. strain A and Synechococcus sp. strain B′ best represented those sequences (Bhaya et al., 2007). Because no mat representative from the kingdom Chlorobi has yet been cultured or had its genome completely sequenced, the situation for this taxon was more complex. Among well-characterized Chlorobiales strains, the closest relative of the Chlorobiales-like mat population is Chp. thalassium (Gibson et al., 1984). Approximately 45% of the rRNA sequences were assigned to this genus, and the remaining sequences assigned to the Chlorobiales were widely distributed across the other three genera included in the analysis (Supplementary Figure S2B). However, when the 16S rRNA sequences of the Chlorobiales population was recovered from the mat metagenome and used in a similar analysis, all of the Chlorobiales-like rRNA sequences were assigned to this single sequence type (Supplementary Figure S2C). Candidatus C. thermophilum accounted for 99.8% of the sequences assigned to the kingdom Acidobacteria (Supplementary Figure S2D). The same in-depth analysis was carried out on all samples, and the results for all four samples were similar (data not shown for the other three samples). Collectively, the rRNA sequence data suggested that the organisms belonging to the kingdom Chloroflexi are significantly more phylogenetically heterogeneous than those that are affiliated with the Cyanobacteria, Chlorobi and Acidobacteria. More importantly, the organisms affiliated with these four kingdoms, which collectively accounted for ∼84% of the rRNA sequences detected from the mat community, are known to be, or are highly likely to be, chlorophototrophs.
Pyrosequencing and mRNA analysis of the microbial mat samples
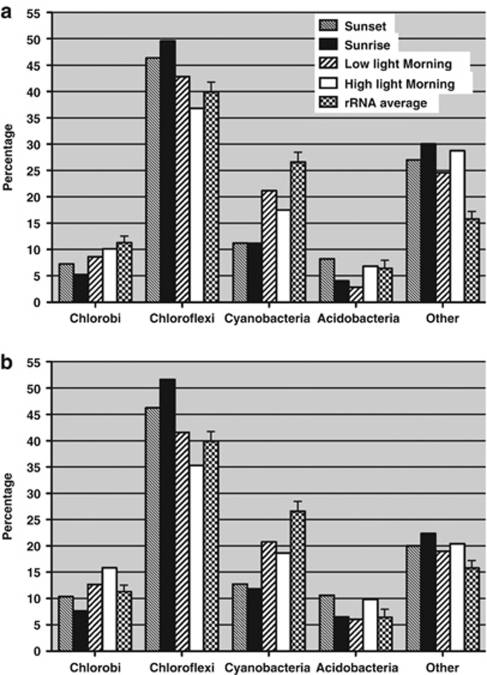
Figure 3a shows a compositional analysis of the mRNA sequences derived from pyrosequencing. These data have been compared with the average percentage composition value for each major taxon based on rRNA, which were derived from the data in Figure 1. For each of the major kingdoms, the highest percentage of mRNA sequences assigned by comparison with the reference metagenome for the four samples was comparable to their average percent composition based on the rRNA sequences. The percentage of sequences identified as mRNAs but assigned to the ‘Others' category was consistently higher than the average rRNA content. The main reason behind this observation was the incomplete identification of sequences of the seven major chlorophototrophic populations in the reference metagenome. Small scaffolds in the metagenome were assigned as ‘Others' (see Supplementary Materials for more Supplementary Information) because it is very difficult to assign them to a phylogentic group with high confidence. Many of these small scaffolds probably belong to the seven major populations. For example, some of these scaffolds contain fragments of highly expressed genes, such as photosynthetic reaction centers and nitrogenase genes. Another observation was that the percentages of Chloroflexi mRNA sequences were higher than the average rRNA percentage for this kingdom, whereas the opposite was true for the Cyanobacteria. This is expected because the Chloroflexi populations have larger genomes (more than 4000 genes each) than the Synechococcus spp. (∼2800 genes each). (Table 3) Although the relationship between mRNA sequence abundances and genome sizes is probably not linear, it is likely that cells with larger genomes will have more mRNA sequences than cells with smaller genomes when both are compared under similar metabolic activity levels.
Figure 3.
Composition of mRNA sequences at the kingdom level (major organisms only) in both data sets, pyrosequencing (a) and SOLiD-3 (b) compared with respective average rRNA sequence percentages. Mean±s.d. is shown for rRNA sequence percentages for all four samples. mRNA sequences of seven predominant phototrophic phylogenetic groups and functional guilds are counted. Percentages were calculated based on total mRNA sequences aligned to reference metagenome.
Table 3. Comparison of cDNA sequences assigned to major reference genomes using two sequencing methods.
| Phylogenetic groupsa | Number of genes in reference metagenome | Number of genes in a comparable genomeb |
Number of mRNA sequences assignedc |
Number of genes coveredd |
Number of genes significantly regulatede |
|||
|---|---|---|---|---|---|---|---|---|
| 454 | SOLiD | 454 | SOLiD | 454 | SOLiD | |||
| Rosei | 4733 | 4517 | 6446 | 784685 | 1879 | 4575 | 28 | 2123 |
| Chlor | 2003 | 4044 | 210 | 36549 | 126 | 1736 | 0 | 265 |
| Anaer | 2614 | Not Available | 778 | 77958 | 358 | 2326 | 2 | 535 |
| SyneA + SyneB′ | 5433 | 5622 | 2908 | 356910 | 1204 | 5226 | 13 | 2695 |
| Cab | 2847 | 3285 | 1025 | 193815 | 465 | 2776 | 5 | 1781 |
| GSB | 2775 | Not Available | 1560 | 274210 | 513 | 2553 | 7 | 1708 |
| Sum of above | 20405 | Not Available | 12927 | 1724127 | 4545 | 19192 | 55 | 9107 |
Rosei, Roseiflexus sp.; Chlor, Chloroflexus sp.; Anaer, novel Anaerolineae-like Chloroflexi population; SyneA, Synechococcus sp. strain A; SyneB′, Synechococcus sp. strain B′ Cab, Candidatus Chloracidobacterium thermophilum; GSB, a Chlorobiales population.
Genomes of strains isolated in the same or similar microbial communities: Roseiflexus sp. RS-1 (complete); Synechococcus sp. strain A (complete); Synechococcus sp. strain B′ (complete); Candidatus Chloracidobacterium thermophilum (complete); Chloroflexus sp. 396–1 (draft).
Total of uniquely assigned cDNA sequences in the four samples.
Genes with at least one assigned cDNA sequence in the four samples.
Genes, expression levels of which are statistically different (P<0.001) between at least one pair of samples.
After careful consideration of the factors described above, large changes in the mRNA percentages among different timepoints were nevertheless observed for two of the four kingdoms. The mRNAs assigned to Cyanobacteria were significantly less abundant than their average rRNA content at sunset and pre-dawn, but the mRNA levels for the Cyanobacteria increased dramatically when the mat was fully illuminated in the morning. The sequences identified as mRNAs from the Acidobacteria were highest at sunset and in high-intensity light in the morning but were significantly lower in the other two samples. The percentages of mRNA sequences were often dictated by expression patterns for the most highly expressed genes. The gene with the most mRNA sequences usually accounted for 5–10% of the mRNA sequences assigned to a particular taxon, and sometimes accounted for up to 25% of the mRNA sequences in a single sample. For all of the chlorophototrophs in the community, genes related to photosynthesis, especially genes encoding reaction center polypeptides, were consistently at or near the top of the lists of genes with most mRNA sequences (Supplementary Table 1). Thus, the change in the percentages of mRNA sequences of Cyanobacteria and Acidobacteria reflected how their photosynthesis-related genes are regulated and expressed (see further discussion below).
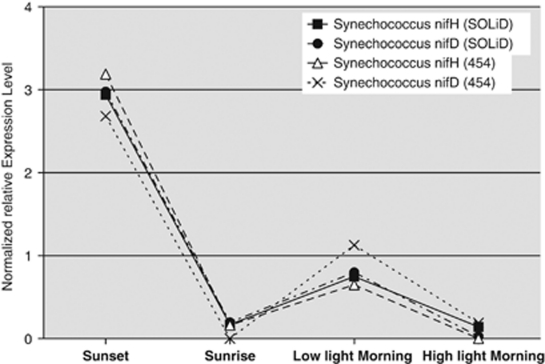
The total numbers of sequences assigned to specific coding sequences (that is, as derived from mRNAs) in the pyrosequencing approach were rather low (∼3000–9000; Table 2), especially in relationship to the number of genes collectively encoded in the reference genomes of the seven major taxa (>20 000; Table 3). Although the coverage and resolution power of the data obtained through this approach were obviously limited, it was still possible to identify statistically meaningful changes in gene expression across the four time points and to draw meaningful conclusions from a few highly expressed genes. For example, it was previously shown that the nifH and nifD genes, encoding nitrogenase in Synechococcus spp. populations in this mat system, exhibited maximal transcription levels at sunset and a secondary rise in transcription levels in the low-light early morning period (Steunou et al., 2008). This was clearly evident in the pattern of expression of the Synechococcus spp. nifH and nifD genes detected in this analysis (Figure 4). The mRNA abundances for the nif genes was highest in the sunset sample, were equivalently low in the pre-dawn and morning high-light samples, and showed a smaller but nevertheless significant increase in the morning low-light sample. Although these data helped to validate the cDNA sequencing approach to transcription profiling, the low number of total mRNA sequences led us to explore NextGen sequencing of cDNAs by using SOLiD-3 methodology to determine whether this method could be used to assess global transcription patterns in the members of the mat community.
Figure 4.
Relative transcript abundances of two Synechococcus spp. nitrogenase genes in two different data sets. Number of mRNA sequences assigned to Synechococcus sp. strain A-like and Synechococcus sp. strain B′-like nifH and nifD genes or partial genes were summed, normalized and compared.
SOLiD-3 sequencing and transcription of photosynthesis genes
Table 4 summarizes the data obtained from the sequencing of cDNA samples prepared from the same four RNA samples described above, but by using the SOLiD-3 methodology. Overall, about 10–12 million sequences were obtained for each of the four cDNA samples. Obviously, because of the large amount of sequence information, the BLAST-based approach used for the data generated from pyrosequencing became computationally untenable, and instead the BWA rapid alignment tool was used (Li and Durbin, 2009). The 50-bp sequences mapped to rRNA databases were undoubtedly much less informative phylogenetically than their ∼225-bp counterparts generated by pyrosequencing. Thus, the rRNA sequences obtained from the SOLiD-3 data were not analyzed further and will not be discussed further here. Simulations performed by the method of Richter et al. (2008) showed that the upper limit for unique mapping errors, when allowing three to five mismatches for 50-bp sequences, was ∼0.007 and 3.12% for non-uniquely mapped sequences (Supplementary Material and Supplementary Table S3). These are estimated upper limits because the simulated data includes rRNA sequences and other repeat sequences (for example, insertion sequences, transposases and other repeated sequences) that were actually removed (or were not counted) by the transcript mapping procedure used in this study.
Table 4. Summary of cDNA sequences using SOLiD technology.
| Sunset 2100 hours | Pre-dawn 0515 hours | Morning, low light 0640 hours | Morning, high light 0840 hours | |
|---|---|---|---|---|
| Total mapped sequences | 12 520 192 | 10 840 071 | 9 974 349 | 10 574 300 |
| Sequences mapped to CDS | 743 902 (5.9%) | 318 205 (2.9%) | 477 362 (4.8%) | 1 057 433 (10.0%) |
| Sequences uniquely mapped to CDS | 623 239 (5.0%) | 261 838 (2.4%) | 393 655 (3.9%) | 883 799 (8.4%) |
Although the sequences in SOLiD data sets were different in length from the ones in pyrosequencing data sets and were aligned using different softwares, the results were generally very similar in multiple ways. In all, 2.9–10.0% of the cDNA sequences (that is, 318 205 to 1 057 433 total cDNA sequences) could be mapped to coding sequences in the reference metagenomes. For a few sequences, it was not possible to map these sequences uniquely to a single gene (for example, for some transcripts encoding members of the psbA multigene family found in Synechococcus spp.). Relatively few instances of this type were encountered, and in general these few cases did not interfere with the global interpretation of the results. Similar to the results obtained by pyrosequencing (Table 2), the sequences that were assigned as mRNAs were lowest in abundance in the pre-dawn sample and were highest in abundance in the morning sample obtained after the mat received full sunlight. Figure 3b shows the compositions of mRNA sequences among four kingdoms. When compared with the pyrosequencing results in Figure 3a, it was very obvious that the pattern of changes of mRNA sequences of each kingdom across the four timepoints was almost identical, although some differences in the actual percentages were observed. For example, the percentage of Chlorobi and Acidobacteria mRNA sequences were consistently higher in SOLiD data sets than that in pyrosequencing data sets. These differences might have arisen from different biases in sequence amplification procedures, because the libraries were prepared using different kits as prescribed by the manufacturers of the two platforms. However, these biases would not affect gene expression patterns among four timepoints inferred from those two data sets because the biases should be consistently present within each platform among the four different timepoint samples. The expression patterns of genes derived from the two data sets were indeed very similar. As shown in Figure 4, the SOLiD-3 and pyrosequencing methods produced essentially identical expression patterns for nifH and nifD, and as mentioned above, this pattern also matches that obtained by reverse transcription-PCR (Steunou et al., 2008). For 55 genes that were detected as significantly regulated genes in the pyrosequencing data sets (Table 3), the normalized expression patterns for the two data sets were compared to determine how well the two data sets were correlated. The correlation coefficients for 49 of the 55 genes were 0.8 or greater for the two expression profiles. These observations established the consistency and the credibility of both data sets and validated the analytical approaches used.
It was also obvious that the SOLiD-3 data sets were superior in providing insights into gene regulation. As a result of the much greater sequencing depth at a similar cost (>100-fold greater sequencing depth), the SOLiD-3 data allowed transcripts to be detected for more than 90% of the >20 000 genes in the reference data set. Furthermore, the resolution power resulting from the large numbers of mRNA sequences for each gene allowed statistically meaningful comparisons to be made for a much larger number of genes. The SOLiD-3 sequencing approach detected transcripts for 19 192 genes, of which nearly half (9107, 47%) were differentially expressed in a statistically meaningful manner (Table 3).
Because the mat community members showing highest abundances were chlorophototrophs, photosynthesis genes were analyzed first. Photosynthetic reaction center genes, which have tentatively been assigned by phylogenetic analyses to the unclassified and uncultured Anaerolineae-like members of the Chloroflexi (Supplementary Information), were among the top genes with most abundant mRNA sequences (Supplementary Table S1). This suggests that these genes have an important role in the biological processes of this population, and thus provides further very strong support for the hypothesis that this population represents a previously unidentified and uncultured chlorophototroph. This is also the case for the novel Chlorobiales spp. Although it was previously expected to be a chlorophototroph because all other known Chlorobiales spp. are photolithoautotrophs (Bryant et al., 2011), the recent discovery of a non-phototrophic member of the Chlorobi, Ignavibacterium album (Iino et al., 2009), made this assumption questionable. These findings about its photosynthesis genes being highly transcribed strongly suggest that Chlorobiales spp. in the mat are chlorophototrophs.
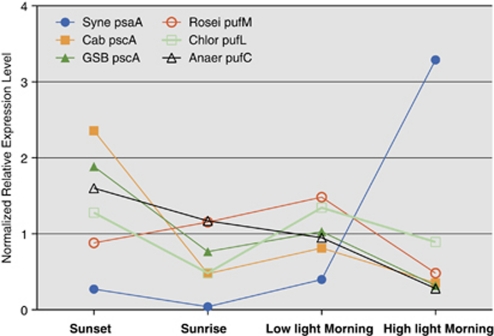
Figure 5 shows the expression patterns for the genes encoding the subunits of the type-1 or type-2 photochemical reaction centers of six major chlorophototrophs (Table 1) in the mat community (for this analysis we did not differentiate here between A-like and B′-like Synechococcus spp. sequences; Table 1). Consistent with the chlorophototrophic lifestyles of the organisms belonging to these four kingdoms, in each case the genes encoding reaction centers were either the most highly expressed, or among the group of most highly expressed, genes (Supplementary Table S1). For the Synechococcus spp., the mRNA levels for psaA, which encodes one of the two large core subunits of the photosystem I reaction center (Grotjohann and Fromme, 2005), were lowest in the pre-dawn sample but rose very steeply as the light intensity increased in the morning. Overall, the transcript abundance for psaA increased more than 50-fold in the morning, and similar increases were observed for other psa genes, as well as the psb genes encoding subunits of photosystem II (data not shown; Steunou et al., 2008). In contrast, the transcript abundance for photosynthetic reaction center genes of all anoxygenic phototrophs decreased when the mat was fully illuminated. The transcription patterns for the pscA genes, which encode subunits of type-1 reaction centers of Candidatus C. thermophilum and the Chlorobiales-like populations in the mat (Bryant et al., 2007), were very similar. For these two organisms, the pscA transcripts were most abundant at sunset and were least abundant when the mat was fully illuminated in the morning. Transcription patterns for the pufM, pufL and pufC genes, associated with the type-2 reaction centers of three different Chloroflexi populations, were inconsistent, but transcript levels for these genes seemed to decrease as the light intensity increased in the morning. Because the changes in the transcript levels for these genes in the Chloroflexi were small (∼two-fold changes) for the time points sampled, we are currently unsure whether these differences reflect real changes in transcript levels or merely statistical variation (Figure 5).
Figure 5.
Relative transcript abundances of photosynthetic reaction center genes of six different phototrophs. The relative transcript abundances of the indicated photosynthetic reaction center genes with most assigned mRNA sequences (SOLiD data set only) are plotted for six different phototrophs. Syne, Synechococcus sp. strain A and B′ Cab, Candidatus Chloracidobacterium thermophilum; GSB, a Chlorobiales population; Rosei, Roseiflexus sp.; Chlor, Chloroflexus sp.; Anaer, novel Anaerolineae-like Chloroflexi population.
With the obvious caveat that patterns of transcription obviously do not necessarily correlate with protein levels and enzyme activity patterns, these differences in transcription patterns for reaction center proteins nevertheless strongly imply that the organisms belonging to the major taxa in the mats likely use different strategies for light capture and usage (Table 1). Because it is unlikely that the specific absorption properties of individual cells in the mat change much during the diel cycle, the observed differences in transcription of the various chlorophototrophs suggest that it is the removal of inhibitory factors (for example, oxygen) or the availability of required limiting nutrients for photoheterotrophs (for example, organic acids or amino acids) that ultimately controls when transcription of a given population will be maximal. Thus, for each type of organism the specific light-harvesting strategy must be to supply an adequate energy supply when the light intensity is not necessarily maximal. This could explain why chlorosomes, with their very large numbers of chromophores and low energy costs for production (Bryant and Frigaard, 2006), are the preferred antenna complexes for most of the anoxygenic phototrophs in the mats.
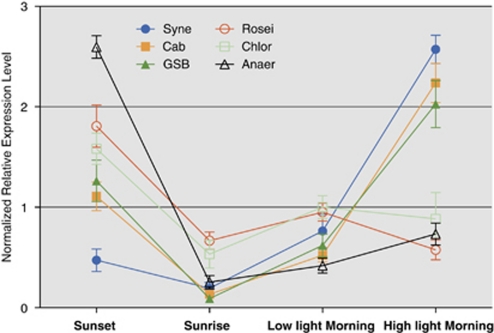
The biogenesis of the photosynthetic apparatus requires cells to synthesize chlorophylls and/or bacteriochlorophylls in addition to the proteins that bind these prosthetic groups. Figure 6 summarizes the sequence results for bacteriochlorophyll biosynthesis genes, expression patterns of which were statistically meaningful in the four major taxa. The pattern of expression of the chlorophyll biosynthesis genes for Synechococcus spp. was very similar to the pattern of expression for psaA (Figure 5) and other psa and psb genes. Transcript levels increased very sharply as the light intensity increased in the morning. However, for Roseiflexus spp. the expression of genes for bacteriochlorophyll a biosynthesis was highest at sunset and lowest in the morning when the mat received full sunlight. This pattern was consistent with transcripts involved in bacteriochlorophyll biosynthesis from Chloroflexus spp. and the novel Anaerolineae-like population as well, and was generally similar to that for expression of the reaction center gene pufM except that pufM transcription was lower at sunset (compare Figures 5 and 6). This pattern is consistent with the known physiology of characterized Chloroflexi strains, which are facultatively phototrophic. The biosynthesis of bacteriochlorophyll and components of the photosynthetic apparatus is strongly suppressed by oxygen in Chloroflexus aurantiacus (Bryant et al., 2011). The transcription patterns for the bacteriochlorophyll biosynthesis genes for the populations of Candidatus C. thermophilum and the mat Chlorobiales spp. were essentially identical (Figure 6). Transcripts were high at sunset, declined overnight and rose sharply again in the morning and were at the highest levels when the mat received full sunlight (Figure 6). For the organisms belonging to these two populations, transcript levels for genes encoding enzymes of bacteriochlorophyll biosynthesis increased when the mat was fully illuminated, whereas transcript levels for reaction centers decreased. Interestingly, both of these types of organisms produce chlorosomes as light-harvesting antenna structures (Bryant and Frigaard, 2006; Bryant et al., 2007), and it is possible that the synthesis of reaction centers (dependent on bacteriochlorophyll a and chlorophyll a, and maximal at sunset) and chlorosomes, which are dependent on bacteriochlorophyll c biosynthesis (Bryant and Frigaard, 2006; Gomez Maqueo Chew and Bryant, 2007), may occur at different times of the day.
Figure 6.
Relative transcript abundances for bacteriochlorophyll synthesis genes of six different phototrophs. Mean±s.e.m. are shown here for all bacteriochlorophyll synthesis genes of six different phototrophs, expression levels of which are statistically different (P<0.001) between at least a pair of samples. Syne, Synechococcus sp. strain A and B′ Cab, Candidatus Chloracidobacterium thermophilum; GSB, a Chlorobiales population; Rosei, Roseiflexus sp.; Chlor, Chloroflexus sp.; Anaer, novel Anaerolineae-like Chloroflexi population.
To study intra-guild gene expression for A-like and B′-like Synechococcus spp. transcripts, we first identified pairs of orthologous genes (blastp, reciprocal best hit), for which transcripts were both detectable and clearly assignable to one or the other genome type. The data were then examined to identify examples in that transcript levels for the two populations (i) differed in abundance in a statistically significant manner (t-test, P<0.05); or (ii) had different expression patterns (correlation coefficient <0). Examples of both types can be seen in Supplementary Table S2. For example, transcripts for genes encoding several chaperones were much lower in abundance in A-like than in B′-like populations. Moreover, transcripts of several genes for quinone biosynthesis and those for a glycolate oxidase subunit and glycerate kinase were more abundant in B′-like than in A-like populations. The temporal pattern of transcription of several of these genes also differed (Supplementary Table S2, for example, sensor histidine kinase and the Fe/S subunit of glycolate oxidase). Differences in both the abundance and temporal expression pattern were also observed for sensory kinases and response regulators, which suggested that entire regulons might be differentially expressed within members of these two populations. These examples illustrate that substantial transcriptional differences, which presumably reflect metabolic and physiological differences, occur with the A-like and B′-like Synechococcus populations within the guild of oxygenic phototrophs.
Conclusions
The data shown here showed that pyrosequencing and SOLiD-3 sequencing of cDNA samples provided complementary information concerning the composition and physiology/metabolism of the phototrophic microbial mats of alkaline siliceous hot springs. The longer sequences derived from pyrosequencing (∼225 bp) provided detailed information concerning the composition of the microbial mat community. These data showed clearly that the phototrophic mat community is principally comprized of chlorophototrophic organisms derived from four kingdoms: Chloroflexi, Cyanobacteria, Chlorobi and Acidobacteria. The analysis of rRNA sequences provided more evidence for the presence of a previously unknown, unclassified and uncultured, and deeply branching chlorophototrophic member of the Chloroflexi in the mat. Within-guild differences in expression patterns were observed for the oxygenic photosynthesis guild, suggesting that with the current approach, the ability to distinguish the origin of transcripts is at a higher level of phylogenetic resolution than cluster analysis of metagenomic assemblies (Klatt et al., 2011). The analysis of mRNA sequences further demonstrated the feasibility and accuracy of metatranscriptomics by SOLiD NextGen sequencing for the first time. Although only four timepoints were analyzed here, this study suggested that the different phototrophic populations in the mat exhibit temporal differences in their transcriptional patterns (Table 1). Because of the ability to achieve dramatically greater sequencing depth at a reasonable cost, SOLiD-3 sequencing can provide unprecedented details concerning the global, diel patterns of transcription that occur for the organisms of this microbial mat community. Samples have been collected and sequencing has been completed for a complete, hour-by-hour analysis of the diel transcription pattern for this chlorophototrophic mat community. The description of these global diel patterns of transcription, which will provide novel insights into the in situ physiology of this community, will be presented elsewhere.
Acknowledgments
This research was supported by the National Science Foundation Frontiers in Integrative Biology Research Program (EF-0328698), the National Aeronautics and Space Administration Exobiology Program (NAG5-8807), and the US. Department of Energy (DOE), Office of Biological and Environmental Research (BER) as part of BER's Genomic Science Program (GSP). This contribution originates from the GSP Foundational Scientific Focus Area (FSFA) at the Pacific Northwest National Laboratory (PNNL). DAB additionally and gratefully acknowledges support from the National Science Foundation (MCB-0523100), Department of Energy (DE-FG02-94ER20137), and the Joint Genome Institute. The work conducted by the US. Department of Energy Joint Genome Institute is supported by the Office of Science of the US. Department of Energy under Contract No. DE-AC02-05CH11231. Finally, We gratefully acknowledge the support and assistance of National Park Service personnel at Yellowstone National Park.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allewalt JP, Bateson MM, Revsbech NP, Slack K, Ward DM. Effect of temperature and light on growth of and photosynthesis by Synechococcus isolates typical of those predominating in the octopus spring microbial mat community of Yellowstone National Park. Appl Environ Microbiol. 2006;72:544–550. doi: 10.1128/AEM.72.1.544-550.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauld J, Brock TD. Ecological studies of Chloroflexis, a gliding photosynthetic bacterium. Arch Microbiol. 1973;92:267–284. [Google Scholar]
- Bhaya D, Grossman AR, Steunou A, Khuri N, Cohan FM, Hamamura N, et al. Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J. 2007;1:703–713. doi: 10.1038/ismej.2007.46. [DOI] [PubMed] [Google Scholar]
- Boomer SM, Pierson BK, Austinhirst R, Castenholz RW. Characterization of novel bacteriochlorophyll-a-containing red filaments from alkaline hot springs in Yellowstone National Park. Arch Microbiol. 2000;174:152–161. doi: 10.1007/s002030000189. [DOI] [PubMed] [Google Scholar]
- Brock TD. Thermophilic Microorganisms and Life at High Temperatures. Springer Verlag: New York; 1978. [Google Scholar]
- Bryant DA, Costas AM, Maresca JA, Chew AG, Klatt CG, Bateson MM, et al. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic Acidobacterium. Science. 2007;317:523–526. doi: 10.1126/science.1143236. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Frigaard N-U. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 2006;14:488–496. doi: 10.1016/j.tim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Bryant DA, Klatt CG, Frigaard N-U, Liu Z, Li T, Zhao F, et al. 2011Comparative and functional genomics of anoxygenic green bacteria from the taxa Chlorobi, Chloroflexi, and AcidobacteriaIn: Burnap RL, Vermaas W (eds),Advances in Photosynthesis and RespirationVol. 33Functional Genomics and Evolution of Photosynthetic Systems, Springer: Dordrecht, The Netherlands; in press [Google Scholar]
- Ferris MJ, Ward DM. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J, Pfennig N, Waterbury JB. Chloroherpeton thalassium gen. nov. et spec. nov., a non-filamentous, flexing and gliding green sulfur bacterium. Arch Microbiol. 1984;138:96–101. doi: 10.1007/BF00413007. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Huang Y, Edwards R, Li W, Gilna P, et al. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One. 2008;3:e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Maqueo Chew A, Bryant DA. Chlorophyll biosynthesis in bacteria: the origins of structural and functional diversity. Annu Rev Microbiol. 2007;61:113–129. doi: 10.1146/annurev.micro.61.080706.093242. [DOI] [PubMed] [Google Scholar]
- Grotjohann I, Fromme P. Structure of cyanobacterial Photosystem I. Photosynth Res. 2005;85:51–72. doi: 10.1007/s11120-005-1440-4. [DOI] [PubMed] [Google Scholar]
- Hewson I, Poretsky RS, Dyhrman ST, Zielinski B, White AE, Tripp HJ, et al. Microbial community gene expression within colonies of the diazotroph, Trichodesmium, from the Southwest Pacific Ocean. ISME J. 2009;3:1286–1300. doi: 10.1038/ismej.2009.75. [DOI] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T, Mori K, Uchino Y, Nakagawa T, Harayama S, Suzuki KI.2009Ignavibacterium album gen. nov., sp. nov., a moderately thermophilic anaerobic bacterium isolated from microbial mats at a terrestrial hot spring, and proposal of Ignavibacteria classis nov. for a novel lineage at the periphery of the green sulfur bacteria Int J Syst Evol Microbiol 60(part 61376–1382.PMID 19671715. [DOI] [PubMed] [Google Scholar]
- Klatt CG, Wood JM, Rusch DB, Bateson MM, Hamamura N, Heidelberg JF, et al. Community ecology of hot spring cyanobacterial mats: predominant populations and their functional potential. ISME J. 2011;5:1262–1278. doi: 10.1038/ismej.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan MT, Brock TD. ‘Chlorobium-type' vesicles of photosynthetically-grown Chloroflexus aurantiacus observed using negative staining techniques. J Gen Microbiol. 1977;102:279–285. [Google Scholar]
- van der Meer MTJ, Klatt CG, Wood J, Bryant DA, Bateson MM, Lammerts L, et al. Cultivation and genomic, nutritional and lipid biomarker characterization of Roseiflexus sp. strains closely related to predominant in situ populations inhabiting Yellowstone hot spring microbial mats. J Bacteriol. 2010;192:3033–3042. doi: 10.1128/JB.01610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer MTJ, Schouten S, Bateson MM, Nübel U, Wieland A, Kühl M, et al. Diel variations in carbon metabolism by green nonsulfur-like bacteria in alkaline silicious hot spring microbial mats from Yellowstone National Park, USA. Appl Environ Microbiol. 2005;71:3978–3986. doi: 10.1128/AEM.71.7.3978-3986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parro V, Moreno-Paz M, González-Toril E. Analysis of environmental transcriptomes by DNA microarrays. Environ Microbiol. 2007;9:453–464. doi: 10.1111/j.1462-2920.2006.01162.x. [DOI] [PubMed] [Google Scholar]
- Pierson BK, Castenholz RW. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen and sp nov. Arch Microbiol. 1974;100:5–24. doi: 10.1007/BF00446302. [DOI] [PubMed] [Google Scholar]
- Poretsky RS, Bano N, Buchan A, LeCleir G, Kleikemper J, Pickering M, et al. Analysis of microbial gene transcripts in environmental samples. Appl Environ Microbiol. 2005;71:4121–4126. doi: 10.1128/AEM.71.7.4121-4126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11:1358–1375. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- Ramsing NB, Ferris MJ, Ward DM. Light-induced motility of thermophilic Synechococcus isolates from Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1997;63:2347–2354. doi: 10.1128/aem.63.6.2347-2354.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter DC, Ott F, Auch AF, Schmid R, Huson DH. MetaSim—A sequencing simulator for genomics and metagenomics. PLoS One. 2008;3:e3373. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff-Roberts AL, Kuenen JG, Ward DM. Distribution of cultivated and uncultivated cyanobacteria and Chloroflexus-like bacteria in hot spring microbial mats. Appl Environ Microbiol. 1994;60:697–704. doi: 10.1128/aem.60.2.697-704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steunou AS, Bhaya D, Bateson MM, Melendrez MC, Ward DM, Brecht E, et al. In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc Natl Acad Sci USA. 2006;103:2398–2403. doi: 10.1073/pnas.0507513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steunou AS, Jensen SI, Brecht E, Becraft ED, Bateson MM, Kilian O, et al. Regulation of nif gene expression and the energetics of N2 fixation over the diel cycle in a hot spring microbial mat. ISME J. 2008;2:364–378. doi: 10.1038/ismej.2007.117. [DOI] [PubMed] [Google Scholar]
- Urich T, Lanzén A, Qi J, Huson DH, Schleper C, Schuster SC. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One. 2008;3:e2527. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Bateson MM, Ferris MJ, Kühl M, Wieland A, Koeppel A, et al. Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Phil Trans R Soc Lond B Biol Sci. 2006;361:1997–2008. doi: 10.1098/rstb.2006.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Castenholz RW.2000Cyanobacteria in geothermal habitatsIn: Whitton BA, Potts M (eds),Ecology of Cyanobacteria Kluwer Academic Publishers: Dordrecht, The Netherlands; 37–59. [Google Scholar]
- Ward DM, Ferris MJ, Nold SC, Bateson MM. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Klatt CG, Wood J, Cohan FM, Bryant DA.2011Functional genomics in an ecological and evolutionary context: maximizing the value of genomes in systems biologyIn: Burnap RL, Vermaas W (eds),Advances in Photosynthesis and RespirationVol. 33Functional Genomics and Evolution of Photosynthetic Systems, Springer: Dordrecht, The Netherlands; in press. [Google Scholar]
- Ward DM, Papke RT, Nübel U, McKitrick MC.2002Natural history of microorganisms inhabiting hot spring microbial mat communities: clues to the origin of microbial diversity and implications for microbiology and macrobiologyIn: JT Staley and A.-L. Reysenbach (eds),Biodiversity of Microbial Life: Foundations of Earth's Biosphere John Wiley and Sons: New York; 27–48. [Google Scholar]
- Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M, et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol. 2009;75:2046–2056. doi: 10.1128/AEM.02294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Raymond J, Wu M, Chatterji S, Ren Q, Graham JE, et al. Complete genome sequence of the aerobic CO-oxidizing thermophile Thermomicrobium roseum. PLoS ONE. 2009;4:e4207. doi: 10.1371/journal.pone.0004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Zhang H, Hanada S, Nagashima KV, Shimada K, Matsuura K. Structural and spectroscopic properties of a reaction center complex from the chlorosome-lacking filamentous anoxygenic phototrophic bacterium Roseiflexus castenholzii. J Bacteriol. 2005;187:1702–1709. doi: 10.1128/JB.187.5.1702-1709.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.