Abstract
Microbial diversity and distribution are topics of intensive research. In two companion papers in this issue, we describe the results of the Cariaco Microbial Observatory (Caribbean Sea, Venezuela). The Basin contains the largest body of marine anoxic water, and presents an opportunity to study protistan communities across biogeochemical gradients. In the first paper, we survey 18S ribosomal RNA (rRNA) gene sequence diversity using both Sanger- and pyrosequencing-based approaches, employing multiple PCR primers, and state-of-the-art statistical analyses to estimate microbial richness missed by the survey. Sampling the Basin at three stations, in two seasons, and at four depths with distinct biogeochemical regimes, we obtained the largest, and arguably the least biased collection of over 6000 nearly full-length protistan rRNA gene sequences from a given oceanographic regime to date, and over 80 000 pyrosequencing tags. These represent all major and many minor protistan taxa, at frequencies globally similar between the two sequence collections. This large data set provided, via the recently developed parametric modeling, the first statistically sound prediction of the total size of protistan richness in a large and varied environment, such as the Cariaco Basin: over 36 000 species, defined as almost full-length 18S rRNA gene sequence clusters sharing over 99% sequence homology. This richness is a small fraction of the grand total of known protists (over 100 000–500 000 species), suggesting a degree of protistan endemism.
Keywords: protists, diversity, species richness, anoxic, pyrosequencing, 18S rRNA approach
Introduction
The history of research on protistan diversity and taxonomy spans almost two centuries. Protists attained their status as a separate kingdom even before the role of the nucleus was first proposed (and so protists included bacteria as a result; Owen, 1860; Haeckel, 1866). Centuries later, it is still essentially unknown whether all the major protistan groups have or have not been discovered, whether or not protistan species are globally distributed (Finlay and Fenchel, 1999; Foissner, 1999, 2006; Finlay, 2002; Baldauf, 2003; Cavalier-Smith, 2004), or how many species are present in a given environment (Jeon et al., 2006).
One of the main reasons for such a lamentable state of affairs is the extent of protistan diversity. The number of protistan life forms appears to overwhelm our current abilities to completely inventory all species in a single sample from most environments, let alone get a complete representation of the community present in the ecosystem from which this sample was drawn. Over the past 10 years, numerous studies have applied ribosomal RNA (rRNA) gene sequencing in attempts to describe the surprising diversity of microbial eukaryotes (for example, Diez et al., 2001; Lopez-Garcia et al., 2001; Moon-van der Staay et al., 2001; Moreira and Lopez-Garcia, 2002). Most of these studies were limited to <1000 clones sequenced (Christen, 2008), forcing researchers to focus on single samples. As shown by the preponderance of sequences registered only once in the library, and by statistical modeling (Hong et al., 2006; Jeon et al., 2006, 2008), current practices have universally resulted in only partial recovery of any single sample's diversity and richness, have provided little to no sample replication within a particular environment, and have proved insufficient to settle biogeographic arguments regarding the global scale of microbial eukaryotic diversity, their degree of endemicity, and even the species richness of a single sample. High-throughput pyrosequencing technology promised to address methodological shortcomings by recovering uncommon, perhaps even exceedingly rare species (Sogin et al., 2006; Huber et al., 2007), but the short read lengths of 454 sequences made it necessary to rely on the existing long rRNA gene sequences to establish taxonomic identities. Also, concerns remain about the role that sequencing errors may play in producing an artifactual picture of the sample's richness (Kunin et al., 2010). To complicate matters further, the design of PCR primers used for both Sanger and pyrosequencing approaches may be significantly biased in their recovery of protists, possibly creating a distorted view of the extant richness and diversity. Under these circumstances, comparing two species lists is difficult and these problems are compounded by uncertainties about nonparametric statistical methods commonly used to estimate the size of the species pool.
In these companion papers, we attempt to address at least some of the limitations of the studies published to date. Our experimental plan was to (i) focus on an environment rather than on a sample, (ii) scale up sequencing efforts proportionally to the large number of samples required to survey this environment, (iii) employ a dual sequencing (Sanger/pyrosequencing) strategy, (iv) minimize the biases of both approaches (v) and analyze the molecular diversity data using statistical tools we developed for the purpose of estimating microbial richness. With these goals, we established in 2004 a multi-year Microbial Observatory in the Cariaco Basin off the coast of Venezuela. The Cariaco Basin is the world's largest, truly marine anoxic system and it offers a large number of contrasting biogeochemical habitats that can be sampled in a single hydrographic cast. We sampled the Basin extensively at three stations, in two contrasting seasons, each time at four depths and across strong biogeochemical gradients spanning fully oxygenated layers to deep, highly sulfidic habitats. We believe that these samples represent fairly broad coverage of the Basin's geochemistry, its protists, their spatial heterogeneity and seasonal dynamics. In this first paper, we present the largest, to date, set of protistan gene sequences from an environment >800-nt-long each (16 000 total, 6489 protistan) for the purpose of providing reliable statistical estimates of total protistan richness within our samples, with meaningful standard error. To minimize bias, we employed a multiple PCR primer strategy, and also complemented our Sanger clone libraries with ∼82 000 quality protistan sequences using massively parallel pyrosequencing of amplicons from the V9 region. This dual sequencing approach enabled the first direct comparison of environmental 18S rRNA gene sequence diversity obtained by these two methods. The two sequence technologies showed substantial similarity in the types of protists recovered, and in their relative proportions. This tentatively indicated that both approaches represented the target diversity reasonably well.
Materials and methods
For more details see Supplementary Materials and methods.
Sample collection
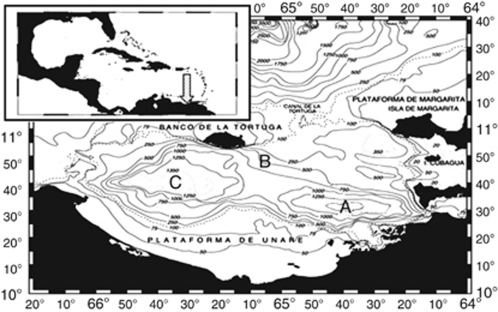
Samples were collected from three stations in the Cariaco Basin, Venezuela: Station A (10.50°N, 64.66°W), Station B (10.40°N, 64.46°W) and Station C (10.40°N, 65.35°W) (Figures 1 and 2, Supplementary Table S1). Samples for DNA extractions were collected at depths corresponding to 40 m above the oxic/anoxic interface, the oxic/anoxic interface, 40 m below the interface and at 900 m. The oxic/anoxic interface was defined as the depth at which oxygen became undetectable. This depth usually corresponded to a particle density maximum detected by transmissometry. Samples were withdrawn from Niskin bottles under N2 pressure into sterile, intravenous bags immediately after retrieval and stored at in situ temperatures. Samples were then filtered onto 47 mm Durapore membranes (Millipore, Billerica, MA, USA) (0.45 μ pore size) onboard, with little to no exposure of the sample to the atmosphere. Depending on the cell concentration in each sample, 2.5–2.8 l of seawater were filtered per sample under gentle vacuum (<25 cm Hg) until no passage of water through filters was observed. The variable filtration volume (filtering until passage of water through the filter ceased) does not completely control for differences in population abundances between our samples, but it likely reduces biases that may be introduced by potentially unequal sampling efforts. Filtration was conducted under a gentle vacuum (<25 cm Hg). Membranes were stored in cryovials containing DNA extraction buffer (Stoeck et al., 2003) at −20 °C until further processing.
Figure 1.
Map of study site. Station A represents the site of the US–Venezuelan Cariaco biogeochemical time series program Station A in the eastern sub-basin. Station B is a shallower station south of the Tortuga channel, the source of incoming Caribbean waters. Station C is centered in the less productive western sub-basin.
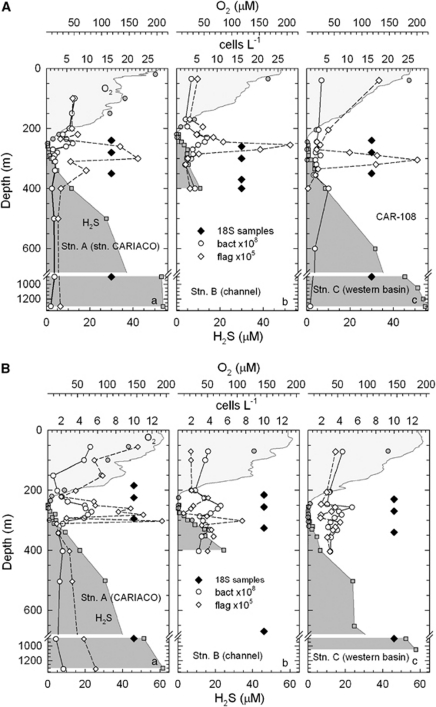
Figure 2.
Vertical profiles of oxygen, hydrogen sulfide and bacterial and flagellate abundances at the time of sampling for protistan DNA in (A) January and (B) May 2005; modified from Taylor et al. (2001) and Scranton et al. (2006).
DNA preparation and PCR amplification of 18S recombinant DNA
High-molecular-weight DNA was prepared using previously described methods (Stoeck et al., 2003, 2007). DNA quality was checked by agarose gel electrophoresis (1%), and the DNA yield was quantified using a Nanodrop ND-1000 UV–Vis spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). The DNA yield from a minimum of two filters per depth at each station was combined before PCR. The 18S rRNA gene was amplified using four different primer combinations; Euk528F (5′-CGGTAATTCCAGCTCC-3′) paired with U1492R (5′-GGTTACCTTGTTACGACTT-3′), U1391R (5′-GGGCGGTGTGTACAARGR-3′) or U1517R (5′-ACGGCTACCTTGTTACGACTT-3′) and Euk360F (5′-CGGAGARGGMGCMTGAGA-3′) paired with U1492R. All primer combinations were applied to all samples, and PCR products from a minimum of three reactions per primer pair were pooled.
Clone library construction and sequencing
To constrain cost, PCR products for stations B and C were pooled from the same depths before clone library construction. This admittedly caused loss of some information on distribution of protists, but allowed a more in-depth sequencing of samples, better covering the Basin. Separate clone libraries were constructed for each of four depths from the eight pooled B/C samples (four from January and four from May 2005) and from the eight station A samples, producing 64 different clone libraries. Inserts were sequenced bi-directionally using an Applied Biosystems (Carlsbad, CA, USA) 3730XL capillary sequencer. Processing of the data used PHRED, PHRAP (Ewing and Green, 1998; Ewing et al., 1998) and a pipeline script to call bases from chromatograms and perform quality control procedures. The sequences were checked for chimeras using the Bellerophon Chimera Check and the Check Chimera utilities (Ribosomal Database Project) (Cole et al., 2003). After removal of short sequences (<800 bp), putative chimeras, bacterial, archaeal and metazoan sequences, remaining sequences were grouped into operational taxonomic units (OTUs) based on 90%, 95%, 98% and 99% rRNA gene sequence similarity levels. This was achieved by first making all possible pair-wise sequence alignments by using ClustalW (Thompson et al., 1994), calculating % sequence identities, followed by clustering the sequences by using the unweighted pair group method with arithmetic mean as implemented in the OC clustering program (http://www.compbio.dundee.ac.uk/ Software/OC/oc.html). The number of OTUs and their frequencies at each cutoff value became the subject of statistical analyses. All protistan sequences have been deposited in GenBank under the accession numbers GU819239–GU825728.
Statistical analyses of clone library data
For each sample, we obtained ‘frequency count' data at the 90%, 95%, 98% and 99% sequence identity levels, that is, the number of OTUs registered in the corresponding clone library only once (the ‘singletons'), or twice (the ‘doubletons'), and so on. On the basis of these frequency count data, we estimated the total number of OTUs at each % identity level, representing the sum of seen (empirically registered) and unseen OTUs (present but undetected due to limited sequencing effort). To do this, we used the program CatchAll (Bunge, 2011) to compute each of five nonparametric (Good-Turing, Chao1, ACE, ACE1 and Chao-Bunge) and eight parametric (Poisson; negative binomial; inverse Gaussian, Pareto and lognormal-mixed Poisson; and mixtures of one, two, or three geometrics) estimators, at every right-truncation point of the data (that is, eliminating outliers at every possible cutoff point in the data), as described previously (Hong et al., 2006, 2009; Jeon et al., 2006, 2008). For the nonparametric analyses, we used a fixed right-truncation point (maximum frequency) equal to 10, and selected optimal analyses as recommended in the statistical literature (we also compared nonparametric results at higher truncation points, but these are typically nonsensical; Bunge and Barger, 2008). For the parametric analyses, we first eliminated all results with an asymptotically corrected goodness-of-fit hypothesis test P-value less than 0.01. We then selected the analysis at each right-truncation point with a minimum value of AICc (Akaike's information criterion, corrected for sample size). Finally, we eliminated analyses with a standard error greater than 1/2 of the total richness estimate. From the remaining results, we selected the one at the highest right-truncation point (so as to use the maximum amount of data) such that an uncorrected χ2 goodness-of-fit hypothesis test P-value exceeded 0.01 (or if this was not possible, the maximum corresponding P-value). This yielded one nonparametric and one parametric estimate of total richness for each sample at each % identity level.
Massively parallel 454 pyrosequencing
The PCR primers we used for 454 sequencing the V9 hypervariable region were 1,391F (5′-GTACACACCGCCCGTC-3′) and EukB (reverse) (5′-TGATCCTTCTGCAGGTTCACCTAC-3′). For each individual environmental DNA extract (each station, depth and season combination), we ran three independent PCR reactions, which were pooled and cleaned using the MinElute PCR purification kit (Qiagen, Valencia, CA, USA). Pyrosequencing (Roche GS FLX Sequencing) was performed by MWG Biotech, Huntsville, AL, USA. In total, we recovered 251 648 sequence reads that were subjected to quality control, leaving us with 82 484 protistan sequences for further consideration. Tag sequences have been deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive under the accession number SRP003469. Annotated tag sequences are available from the authors.
Bioinformatic analysis of tag sequences
We proceeded with our analyses by following a pipeline developed earlier for inspecting and quality checking pyrosequencing reads (Stoeck et al., 2009). Sequences shorter than 100 nucleotides, as well as those with at least one ambiguous position (N), were removed from consideration. To make taxonomic assignments, we first built a reference database containing all small subunit rRNA gene sequences in public databases longer than 500 nucleotides. Each sequence tag was compared with these reference sequences using similarity searches with the program BLASTN and requesting up to 30 best hits, using the following BLAST parameters: -m 7 -r 3 -q -2 -G 6 -E 6 (Stoeck et al., 2010). We then parsed the BLAST output to extract taxonomic assignments at a series of thresholds for sequence similarity (70%, 75%, 80%, 85%, 90%, 92%, 95%, 96%, 97%, 98%, 99% and 100%). Sequence similarity was calculated as the sum of identities for non-overlapping (if any) high scoring pairs (see the BLAST documentation) divided by the length of the query sequence. For each query, we used only the most similar target sequence for which a good taxonomic assignment was provided (that is, longer description in the OC field in the EMBL entries). To enable direct comparison between taxonomic assignments of 454- and Sanger-produced sequences, the above BLAST analyses were repeated for nearly full-length 18S rRNA gene sequences, and separately for their V9 domains.
Results
Overall protistan diversity in the Cariaco Basin
We examined the extent of protistan diversity in the Basin in two ways. First, we amplified the 18S rRNA gene from community DNA, followed by cloning and sequencing the amplicons. From each of the 64 libraries, we sequenced an average of 250 clones, resulting in over 16 000 rRNA gene sequences, or at a minimum, 672 sequences for each of the 16 samples. Culling of confirmed or suspected metazoan, chimeric and short (<800 bp) sequences left 6498 good quality target 18S protistan sequences for further analyses. Supplementary Table S2 presents the final number of sequences, grouped into OTUs sharing over 99% of gene sequence identity, and categorized by PCR primer pair, season, depth and site. Second, we used the same DNA extracts as templates for 16 separate PCR reactions followed by pyrosequencing of the V9 region of the 18S rRNA gene. This produced a collection of over quarter of a million reads, with 173 723 reads meeting our quality control criteria, 52% of which were metazoan and other non-target sequences.
It is important to demonstrate the agreement between the taxonomic assignments established from the nearly full-length rRNA gene sequences vs the V9 tags contained therein. We achieved this by extracting the V9 region from 3107 of our clone sequences that contained this region, and comparing the taxonomic assignments made by BLAST. Supplementary Table S3 shows a general global agreement, as well as several minor differences. The proportion of sequences falling into the unassigned/unclassified categories is larger for the V9 tags and this effect appears stronger in Alveolata. We note that for taxonomic assignment of the pyrosequencing tags, we chose the ⩾70% sequence similarity threshold because at this level fewer tags remained unassigned than at higher thresholds.
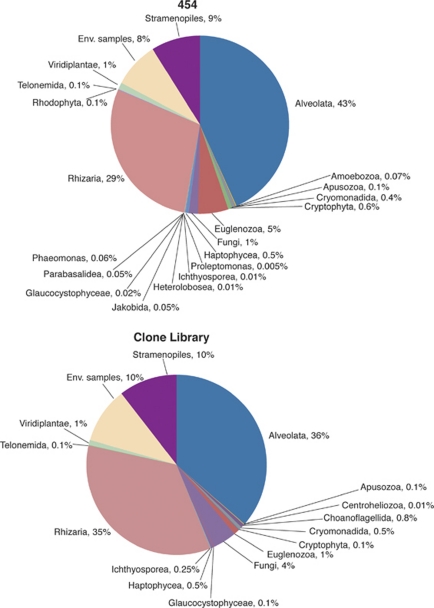
At the highest level of taxonomic assignment, approximately corresponding to kingdoms and phyla, the frequencies of different rRNA gene sequences appear similar between the conventional clone library/Sanger- and pyrosequencing-based methods (Figure 3). Both collections were dominated by Alveolata, Rhizaria, Stramenopiles and unclassified 18S rRNA gene sequences, and in the same order of dominance, followed in both cases by Fungi and Euglenozoa. The most visible difference between Sanger- and pyrosequencing-based methods at this level of taxonomic assignment is the number of rarer taxa constituting <1% of respective sequence collections: 9 vs 14, respectively.
Figure 3.
Phylum- and kingdom-level assignments of the 18S rRNA gene sequence collections obtained by conventional cloning/Sanger sequencing and 454 pyrosequencing approaches.
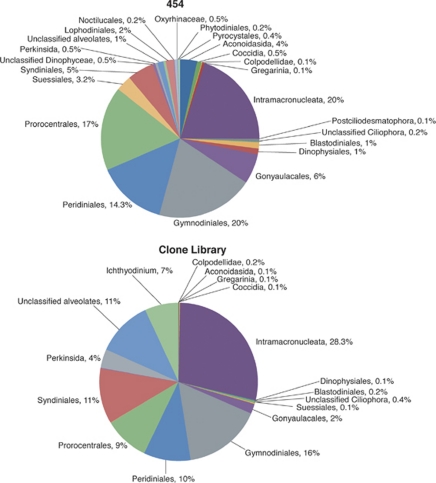
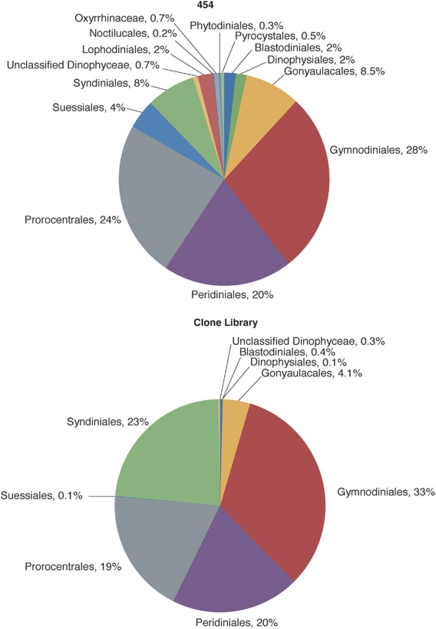
These gross similarities in the overall recovery of protists by the two methods were observed at lower levels of taxonomic assignment as well. For example, in both sequence collections, the Alveolata were dominated by the ciliate subphylum Intramacronucleata and four dinoflagellate orders (Gymnodiniales, Prorocetrales, Syndiniales and Gonyaulacales), as well as unclassified alveolates (Figure 4). Similarly, Stramenopiles were dominated by the same taxa regardless of the method used to detect them (data not shown). Expectedly, pyrosequencing detected more of the rare taxa (again defined as those comprising <1% of the entire collection): 11 vs 8 rare stramenopile taxa registered using clone libraries/Sanger sequencing. Classification to order was the lowest taxonomic level possible for the majority of our tag sequences. At this level, Sanger sequencing and pyrosequencing again recovered similar groups at similar frequencies (Robertson and Burke, 1989). For example, within the dinoflagellate class Dinophyceae, almost three quarters of all sequences were represented by three orders present at essentially the same proportions in our Sanger- and pyrosequencing-based data sets (Gymnodiniales, Peridiniales and Prorocentrales). The other abundantly represented orders were also in the same proportions (Syndiniales and Gonyaulacales) (Figure 5). We note that these gross similarities between the two libraries appear independent of the cutoff level of query/hit similarity used to assign V9 sequences to taxa by BLAST, as raising this cutoff to 80% did not affect the overall pattern (data not shown).
Figure 4.
Subphylum and class level assignments of the alveolate 18S rRNA gene sequence collections obtained by conventional cloning/Sanger sequencing and 454 pyrosequencing approaches.
Figure 5.
Order assignments of the Dinophyceae 18S rRNA gene sequence collections obtained by conventional cloning/Sanger sequencing and 454 pyrosequencing approaches.
Biases of the rRNA gene sequence collection
To generate conventional 18S rRNA gene clone libraries, we used four different PCR primer sets applied to each DNA extract obtained. Table 1 provides a representative subset of these data summarizing the number of occurrences of ciliates by primer set. We chose to present data for this group because we recovered roughly the same number of rRNA gene sequences (≈100) for each of the four PCR primer sets, which simplified the comparison of their respective performances. Also, the relatively large size of the ciliate 18S rRNA gene sequence collection minimized the influence of stochastic variation on the pattern of recovery. Differential recovery of several ciliate clades illustrates the substantial biases of the PCR primer pairs used, such as an apparent discrimination against Euplotida by the 360F/1492R pair, or discrimination for Grossglockneriidae and Choreotrichida by the 528F/1391R pair.
Table 1. Differential recovery of ciliate taxa by four different PCR primer sets.
| 3F14R | 5F13R | 5F14R | 5F15R | |
|---|---|---|---|---|
| Choreotrichida | 0 | 1 | 0 | 0 |
| Colpodidae | 0 | 11 | 0 | 0 |
| Cryptocaryon | 1 | 0 | 0 | 0 |
| Epalxellidae | 1 | 0 | 0 | 0 |
| Euplotida | 0 | 4 | 7 | 10 |
| Furgasoniidae | 0 | 1 | 0 | 0 |
| Grossglockneriidae | 9 | 74 | 3 | 46 |
| Halteriidae | 1 | 0 | 0 | 0 |
| Hausmanniellidae | 0 | 12 | 0 | 0 |
| Nassulidae | 1 | 0 | 1 | 0 |
| Parameciidae | 0 | 1 | 0 | 0 |
| Philasterida | 0 | 3 | 0 | 4 |
| Pleuronematida | 8 | 0 | 31 | 11 |
| Prorodontidae | 0 | 0 | 0 | 1 |
| Stichotrichida | 79 | 0 | 58 | 32 |
| Strombidiidae | 4 | 0 | 5 | 13 |
| Tintinnida | 0 | 0 | 1 | 0 |
| Total | 104 | 107 | 106 | 117 |
Total protistan richness in the Cariaco Basin
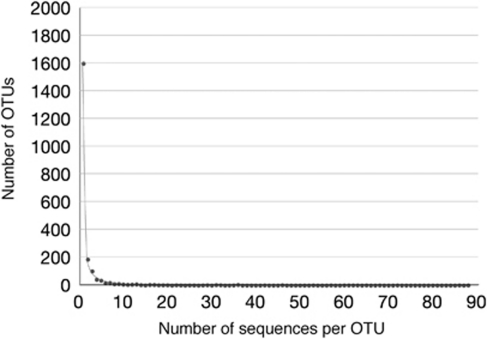
We clustered protistan 18S rRNA gene sequences recovered from our clone libraries into OTUs at different levels of sequence identity. The 6498 individual sequences fell into 2099, 1496, 767 and 392 OTUs at 99%, 98%, 95% and 90% identity levels, respectively. We then reconstructed the OTU frequency distributions (Figure 6), and modeled these distributions to estimate the size of microbial richness that must exist in our samples, and therefore in the Basin to account for the empirical data collected, using the program CatchAll (Bunge, 2011). In accordance with previous experience (Bunge and Barger, 2008; Jeon et al., 2008; Hong et al., 2009), the best-fitting models are finite mixtures of one, two or three geometric distributions, while the Poisson, negative binomial, inverse Gaussian-, Pareto- and lognormal-mixed Poisson provided inferior fits or presented other statistical anomalies (data not shown). Typically, higher order mixtures are required to produce acceptable richness estimates and goodness-of-fit assessments that satisfy our criteria (see Materials and methods). For purely comparative purposes, Table 2 also presents the richness predictions based on nonparametric richness lower bound Chao1 and the nonparametric estimator ACE1; these are shown only for comparison with the parametric estimates.
Figure 6.
The fit of the three-mixed exponential distribution (solid line) to the empirical data points (filled circles, OTUs clustered at 99% 18S rRNA gene sequence identity).
Table 2. Predicted richness of protistan assemblages in the Cariaco Basin, with associated statistics.
| % Identity of sequences within OTU | Statistic | Best parametric model | Nonparametric estimators | |
|---|---|---|---|---|
| ⩾99 | Three-mixed exponential | Chao1 | ACE1 | |
| Estimated total number of OTUs | 35968 | 8960 | 17335 | |
| s.e. | 13123 | 613 | 1490 | |
| CI | 18373–72596 | 7860–10269 | 14684–20545 | |
| GOF | 0.1396 | NA | NA | |
| ⩾98 | Three-mixed exponential | |||
| Estimated total number of OTUs | 12480 | 4351 | 7198 | |
| s.e. | 3489 | 282 | 601 | |
| CI | 7480–21666 | 3850–4959 | 6137–8503 | |
| GOF | 0.8204 | NA | NA | |
| ⩾95 | Two-mixed exponential | |||
| Estimated total number of OTUs | 4039 | 1951 | 2799 | |
| s.e. | 798 | 156 | 280 | |
| CI | 2815–6015 | 1685–2300 | 2323–3427 | |
| GOF | 0.0598 | NA | NA | |
| ⩾90 | Three-mixed exponential | |||
| Estimated total number of OTUs | 1143 | 748 | 1029 | |
| s.e. | 162 | 74 | 130 | |
| CI | 887–1532 | 630–924 | 821–1338 | |
| GOF | 0.6788 | NA | NA | |
Abbreviations: CI, confidence interval; GOF, goodness-of-fit; NA, not applicable; OTU, operational taxonomic unit.
GOF (P-value for the corrected Pearson χ2 goodness-of-fit test).
The estimates of the total protistan richness in our samples based on two- and three-mixed exponential distributions show that they may harbor thousands of protistan genera (defined as OTUs formed at 90–95% gene sequence similarity), and up to tens of thousands of protistan species (defined as OTUs formed at 98–99% gene sequence similarity), indicating that our empirical collection sampled about 20–30% of all the genera, and about 5–10% of all the species in our samples. By extension, these are minimal estimates of protistan richness in the entire Basin.
Discussion
The first of our companion papers offers a general description of the Cariaco Basin, the overall view of protistan diversity we detected, and a comparison of the 18S rRNA gene sequence collections produced by the tandem approach of two alternative sequencing technologies (Sanger and pyrosequencing). The Cariaco Basin is an excellent model environment to study protistan species richness, phylogenetic diversity, habitat specialization, community structure, its dynamics and global uniqueness. This is because the Basin is a mosaic of dramatically different biogeochemical niches, and has existed as such for millions of years (Robertson and Burke, 1989), though it likely went through periods of oxidation (Peterson et al., 2000). The Cariaco water column progresses from fully oxic to sulfidic across a temporally varying boundary between 250 and 350 m. Within the redox transition zone lie strong gradients in O2, NO3−, H2S, NH4+, NO2−, PO43− and CH4, and enrichments in S2O32−, SO32−, S0, Mn2+ and Fe2+ that select for specific bacterial and archaeal phylotypes (Taylor et al., 2001; Scranton et al., 2006; Li et al., 2008; Lin et al., 2008). This transition zone exhibits a peak in prokaryotic metabolic activity and cell numbers, which often coincides with peaks in protist cell numbers.
Protists are important members of aquatic microbial communities because of their autotrophic and heterotrophic activities. Their grazing of prokaryotic and other eukaryotic microbes regenerates nutrients, modifies or re-mineralizes organic matter (for example, Taylor, 1982; Taylor et al., 1985; Sherr and Sherr, 2002), and can affect the quantity, activity and physiological state of their prey (Madsen et al., 1991; Frias-Lopez et al., 2009). Grazing releases inorganic nutrients that often limit primary production, and makes organic carbon available to higher trophic levels (Berman et al., 1987). In our companion paper (Orsi et al., 2011), we identify many new clades at species to class levels, some of which appear restricted to specific layers of the water column and have a significantly nonrandom distribution. We also use multivariate community analyses coupled with parametric richness estimates for selected regions of the Basin to demonstrate that distinct communities of protists exist within close proximity (40 m) to each other. These findings suggest many pelagic protists are specialized to specific habitats, and likely diversify, at least in part due to separation by geochemical barriers. For a discussion of the influence of biogeochemical factors on protistan community structure, see Orsi et al. (2011).
Overall protistan diversity in the Cariaco Basin
Owing to the scale of sequencing efforts, we anticipated and demonstrated recovery of the expectedly large diversity that contains representatives of all major clades (Figure 3). The remarkable similarity between the results produced by the two sequencing approaches employed came as a surprise. Indeed, at all three hierarchical levels examined, both sequence collections were dominated by the same taxa, often present at essentially the same frequencies (Figures 3,4,5). Such comparisons are rare, and, to the best of our knowledge, ours is the first published report of such a comparison for protists. Although similarity at the highest level of taxonomic hierarchy (Figures 3, 4) is not counterintuitive, we find it remarkable that the overall similarity between compositions of the dominant clades holds in many cases down to the level of protistan orders (Figure 5). For example, three dinoflagellate orders, Gymnodiniales, Peridiniales and Prorocentrales comprised 72% of either sequence collection, at essentially the same frequencies. We did not extend these analyses to the levels of family or genus because this level of assignment is not currently reliable for pyrosequencing data. The differences between the two sequence collections do not appear until we consider relatively rare taxa with frequencies in single percentage points of the total respective databases. Phyla represented by less than 1% of the total number of sequences are more numerous in the collection produced by pyrosequencing than Sanger sequencing (Figure 3), which we attribute to the sequencing depth afforded by the former method. We note, however, that the impact of pyrosequencing errors on recovered diversity has not yet been fully established (Kunin et al., 2010), and we interpret the rare taxa with caution.
Biases of the rRNA gene approach and their minimization
One of our goals was to produce a minimally biased molecular survey of protists, and we attempted to minimize the PCR primer biases (Suzuki and Giovannoni, 1996; Polz and Cavanaugh, 1998; Jeon et al., 2008; Hong et al., 2009) by using a multiple-primer approach (Stoeck et al., 2006). Recently, our group showed statistically that no tested pair of conserved primers could provide amplification of all templates present in the DNA extract, and that a combination of PCR reactions each using different primers led to qualitatively richer clone libraries for both protists (Jeon et al., 2008) and bacteria (Hong et al., 2009). The difference in performance of the four primer pairs used here is illustrated in Table 1, which gives examples of organisms completely missed from the clone libraries created by all but one PCR primer set, or organisms universally detected by all but one primer pair. Note that for Table 1 we chose to present representatives of the phylum Ciliophora. Such a choice is conservative because PCR primers have been designed using a significant number of ciliate sequences, potentially leading to less discrimination against these organisms compared with less studied groups.
We tentatively conclude that the multiple PCR primer approach provided a more balanced view of microbial composition in the Cariaco Basin, and that this study is less biased than the majority of previous protistan surveys (including, for example, our own earlier study of the Cariaco Basin (Stoeck et al., 2003). Coupled with the size of the data set, we are well-positioned to model distributions in order to estimate the total number of protists in the Basin, including those missed by our libraries.
Total protistan richness in the Cariaco Basin
We used a new statistical approach (Hong et al., 2006) to model OTU frequency distributions and to provide estimates of the total protistan richness in our samples, and thus conservatively in the entire Basin (The estimate of total taxonomic richness for our samples is an estimate of the total richness that would be observed if sampling were carried out using the same procedures, but with infinite effort, that is, exhaustively. It is also an approximation of the total richness of the Basin, under the assumption that the conditions occurring in the individual samples are ‘representative', that is, that the conditions of the individual samples represent an approximately random sample within a given biogeochemical regime extant in the Basin. The locations for sample collection were carefully selected based on a decade of previous Cariaco time series data showing the three sites and four depths we sampled to be representative of the range of biogeochemical conditions in the Basin. It is, however, possible that distributions of OTUs in the Basin are even patchier than we detected because of the additional variations in conditions that we did not sample. This would cause an unknown number of OTUs to be missed in our samples. If this is the case, our estimates of the total diversity are likely to be conservative). Our group developed this method as a general tool for richness estimation (Hong et al., 2006) and later applied it to microbial eukaryotes (Jeon et al., 2006; Stoeck et al., 2007; Edgcomb et al., 2009). Here we produced a sequence collection large enough to be modeled by our parametric statistical method, with an appropriate goodness-of-fit, and most importantly to obtain a standard error sufficiently low as to not render the estimate uninformative. We compared essentially all known parametric and nonparametric estimates for the total richness, along with associated statistics, including standard errors and goodness-of-fit diagnostics. Our data also illustrated a potential underestimation of total richness by nonparametric estimators. Table 2 shows that, at the level of OTUs based on 99% gene sequence identity, the OTUs empirically observed in our samples constitute about 10% of all predicted to be present, and the difference between richness predictions based on parametric vs nonparametric approaches is substantial. As OTUs become more inclusive, and the portion of the sampled richness increases, such differences become smaller; for OTUs based on 90% gene sequence identity, the parametric modeling- and nonparametric estimators-based richness predictions converge on essentially same values.
Our approach enabled us to predict how many OTUs must be present in our samples, and thus conservatively in the Basin: 35 968±13 123 (s.e.) and 4039±798 (s.e.) for OTUs comprised of 18S rRNA gene sequences sharing >99% and >95% identity, respectively. These OTU cutoffs are roughly comparable to species and genera. We argue that this is the first estimate of total protistan richness that is relatively unbiased, and with realistic standard errors. We note that, although individual clone libraries sampled the respective environments to varying degrees (that is, similarly-sized libraries cover diversity differently if the target habitats have different richness), this has no bearing on our estimates. Indeed, the sample size does not influence the value of the sample's predicted richness, only the standard error of its estimate. Some bias in the estimators of total richness may be present for small samples, but this is not a major factor here. Furthermore, differences in sampling intensity may have been minimized by the variable filtration volumes we applied (filtering until filters clogged—see above).
It is instructive to compare our results to the previous richness estimates obtained using a single 3-l sample from the Cariaco Basin (395±153 (s.e.) and 164±40 (s.e.) at 99% and 95% OTU clustering, respectively; Jeon et al., 2006). Samples in this study covered temporal, spatial and biogeochemical variations in the Basin, and resulted in estimates of diversity that are 2–3 orders of magnitude higher. This indicates a very patchy distribution of species over the volume of the Basin, and tentatively suggests the possibility of a high degree of habitat specialization.
The debate over the degree to which protist communities exhibit global distribution has not been fully resolved (Foissner, 1999, 2006; Lawley et al., 2004; Boenigk et al., 2006; Bass et al., 2007; Caron, 2009). We would like to emphasize that all existing knowledge of microbial diversity comes from samples, and, strictly speaking, is limited to the collection of samples analyzed so far—rather than environments from which they were obtained. Under these circumstances, a reasonable analysis would be to compare a proxy for global richness, such as the total number of protistan species known from samples collected to date, to a proxy for richness in a large and varied environment, such as a collection of samples covering its multitude of habitats. This collection should be reasonably large because, as Caron (2009) and Ramette and Tiedje (2006) argue, a species could be globally distributed even if it is not found in each and every sample. The express purpose of our study was to overcome a common limitation of microbial diversity studies, typically based on one to a few samples per environment. We did so by focusing on a large system, and revisiting multiple locations in different seasons, and sampling across its principal biogeochemical regimes, without sacrificing the per sample depth of sequence coverage. It should be noted that not all protist populations were sampled in this study because we intentionally excluded the photic zone. There is an additional important caveat: the total richness, whether in a single sample or collection of samples, can only be estimated statistically, as even these pyrosequencing efforts do not allow for a complete coverage of sample(s) richness. The statistical tools typically used in microbial diversity studies do not necessarily work well for the purpose (Chao and Bunge, 2002). The state-of-the-art statistical tool kit we developed, coupled with a large-scale survey of an environment as large and chemically diverse as the Cariaco Basin, meet the above requirements, and provide us with what is needed for a proxy for total protistan richness in the aphotic zone of the Cariaco Basin. Our analyses show that this proxy, or the number of species of microbial eukaryotes predicted to exist in the collection of our samples, appears to be a substantial figure, but it falls far short of the total number of protistan species described to date (100 000–500 000 species (Corliss, 1982; Adl et al., 2007)) by being 3 to 15 times lower, suggesting a degree of endemism in this environment.
Conclusions
As part of the Cariaco Microbial Observatory, we collected the largest, and possibly the least biased, collection of protistan molecular signatures to date from a single environment. Two sequencing approaches produced globally similar pictures of protistan diversity, which contain representatives of all major protistan clades known today. Parametric statistical modeling predicted the total number of species in samples spanning different locations, seasons and a range of diverse habitats within the Cariaco Basin. This number is far lower than the global known richness, indicating a degree of endemicity in the distribution of the majority of microbial eukaryotes.
Acknowledgments
We thank the captain and crew of the B/O Hermano Gines and the staff of the Fundación La Salle de Ciencias Naturales, Margarita Island for their assistance during our field work in Venezuela, particularly Yrene Astor. We are grateful to the dedicated researchers of the CARIACO biogeochemical time series program, without whom this work would not have been possible. We would like to thank Valentin Ilyin (Northeastern University), David Mark Welch (MBL) and Jed Goldstone (WHOI) for helpful discussions on bioinformatics approaches, Hilary Morrison and Rich Fox (MBL) for the use of their pipeline scripts for sequence data processing, and the Academic Research Computing User Group at Northeastern University for permitting us to use the Opportunity computer cluster. We thank Linda Woodard for supervising the statistical computations. Comments from two anonymous reviewers significantly improved this manuscript. This research was conducted using the resources of the Cornell University Center for Advanced Computing, which receives funding from Cornell University, New York State, the National Science Foundation, and other leading public agencies, foundations and corporations. This research was supported by grants from NSF (MCB-0348341 and DEB-0816840 to SSE, DEB-0816638 to JB, MCB-0348407 to VE and OCE 03-26175 and MCB-03-47811 to GTT), from ANR-Biodiversité project Aquaparadox and the ERA-net project BiodivERsA BioMarKs program to RC, and from Venezuela FONACIT (nos. 96280221 and 2000001702 to RV). This is a contribution # 268 from the Marine Science Center, Northeastern University, Nahant, MA, USA.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adl SM, Leander BS, Simpson AG, Archibald JM, Anderson OR, Bass D, et al. Diversity, nomenclature, and taxonomy of protists. Syst Biol. 2007;56:684–689. doi: 10.1080/10635150701494127. [DOI] [PubMed] [Google Scholar]
- Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- Bass D, Richards TA, Matthai L, Marsh V, Cavalier-Smith T. DNA evidence for global dispersal and probable endemicity of protozoa. BMC Evol Biol. 2007;7:162. doi: 10.1186/1471-2148-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman T, Nawrocki M, Taylor GT, Karl DM. Nutrient flux between bacteria, bacterivorous nanoprotozoans and algae. Mar Microb Food Webs (now Aquat Microbial Ecol) 1987;2:69–82. [Google Scholar]
- Boenigk J, Pfandl K, Garstecki T, Harms H, Novarino G, Chatzinotas A. Evidence for geographic isolation and signs of endemism within a protistan morphospecies. Appl Environ Microbiol. 2006;72:5159–5164. doi: 10.1128/AEM.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge J, Barger K. Parametric models for estimating the number of classes. Biometrical J. 2008;50:971–982. doi: 10.1002/bimj.200810452. [DOI] [PubMed] [Google Scholar]
- Bunge J. Proceedings of the 2011 Pacific Symposium on Biocomputing. Kohala Coast, Hawaii; 2011. Estimating the Number of Species with CatchAll; pp. 121–130. [DOI] [PubMed] [Google Scholar]
- Caron DA. Past president's address: protistan biogeography: why all the fuss. J Eukaryot Microbiol. 2009;56:105–112. doi: 10.1111/j.1550-7408.2008.00381.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Only six kingdoms of life. Proc Biol Sci. 2004;271:1251–1262. doi: 10.1098/rspb.2004.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Bunge J. Estimating the number of species in a stochastic abundance model. Biometrics. 2002;58:531–539. doi: 10.1111/j.0006-341x.2002.00531.x. [DOI] [PubMed] [Google Scholar]
- Christen R. Global sequencing: a review of current molecular data and new methods available to assess microbial diversity. Microbes Environ. 2008;23:253–268. doi: 10.1264/jsme2.me08525. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, et al. The ribosomal database project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corliss JO. Numbers of species comprising the phyletic groups assignable to the kingdom Protista. J Protozool. 1982;29:499. [Google Scholar]
- Diez B, Pedros-Alio C, Massana R. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol. 2001;67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgcomb V, Orsi W, Leslin C, Epstein SS, Bunge J, Jeon S, et al. Protistan community patterns within the brine and halocline of deep hypersaline anoxic basins in the eastern Mediterranean Sea. Extremophiles. 2009;13:151–167. doi: 10.1007/s00792-008-0206-2. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Fenchel T. Divergent perspectives on protist species richness. Protist. 1999;150:229–233. doi: 10.1016/S1434-4610(99)70025-8. [DOI] [PubMed] [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Foissner W. Protist diversity: estimates of the near-imponderable. Protist. 1999;150:363–368. doi: 10.1016/S1434-4610(99)70037-4. [DOI] [PubMed] [Google Scholar]
- Foissner W. Biogeography and dispersal of microorganisms: a review emphasizing protists. Acta Protozool. 2006;45:111–136. [Google Scholar]
- Frias-Lopez J, Thompson A, Waldbauer J, Chisholm SW. Use of stable isotope-labelled cells to identify active grazers of picocyanobacteria in ocean surface waters. Environ Microbiol. 2009;11:512–525. doi: 10.1111/j.1462-2920.2008.01793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel E.1866Generelle Morphologie der OrganismenVol. II.Georg Reimer: Berlin [Google Scholar]
- Hong S, Bunge J, Leslin C, Jeon S, Epstein SS. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 2009;3:1365–1373. doi: 10.1038/ismej.2009.89. [DOI] [PubMed] [Google Scholar]
- Hong SH, Bunge J, Jeon SO, Epstein SS. Predicting microbial species richness. Proc Natl Acad Sci USA. 2006;103:117–122. doi: 10.1073/pnas.0507245102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Jeon S, Bunge J, Leslin C, Stoeck T, Hong S, Epstein SS. Environmental rRNA inventories miss over half of protistan diversity. BMC Microbiol. 2008;8:222. doi: 10.1186/1471-2180-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SO, Bunge J, Stoeck T, Barger KJ, Hong SH, Epstein SS. Synthetic statistical approach reveals a high degree of richness of microbial eukaryotes in an anoxic water column. Appl Environ Microbiol. 2006;72:6578–6583. doi: 10.1128/AEM.00787-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Lawley B, Ripley S, Bridge P, Convey P. Molecular analysis of geographic patterns of eukaryotic diversity in Antarctic soils. Appl Environ Microbiol. 2004;70:5963–5972. doi: 10.1128/AEM.70.10.5963-5972.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XN, Taylor GT, Astor Y, Scranton MI. Sulfur speciation in the Cariaco Basin with reference to chemoautotrophic production. Mar Chem. 2008;112:53–64. [Google Scholar]
- Lin X, Scranton MI, Chistoserdov AY, Varela R, Taylor GT. Spatiotemporal dynamics of bacterial populations in the anoxic Cariaco Basin. Limnol Oceanogr. 2008;53:37–51. [Google Scholar]
- Lopez-Garcia P, Rodriguez-Valera F, Pedros-Alio C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- Madsen EL, Sinclair JL, Ghiorse WC. In situ biodegradation: microbiological patterns in a contaminated aquifer. Science. 1991;252:830–833. doi: 10.1126/science.2028258. [DOI] [PubMed] [Google Scholar]
- Moon-van der Staay SY, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- Moreira D, Lopez-Garcia P. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 2002;10:31–38. doi: 10.1016/s0966-842x(01)02257-0. [DOI] [PubMed] [Google Scholar]
- Orsi W, Edgcomb V, Jeon SO, Leslin C, Bunge J, Taylor GT, et al. 2011Protistan microbial observatory in the Cariaco Basin, Caribbean. IIHabitat specialization. ISME J(in press). [DOI] [PMC free article] [PubMed]
- Owen R. Palaeontology. Adam and Charles Black: Edinburgh; 1860. [Google Scholar]
- Peterson LC, Haug GH, Hughen KA, Rohl U. Rapid changes in the hydrologic cycle of the tropical Atlantic during the last glacial. Science. 2000;290:1947–1951. doi: 10.1126/science.290.5498.1947. [DOI] [PubMed] [Google Scholar]
- Polz MF, Cavanaugh CM. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A, Tiedje JM. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb Ecol. 2006;53:197–207. doi: 10.1007/s00248-005-5010-2. [DOI] [PubMed] [Google Scholar]
- Robertson P, Burke K. Evolution of southern Caribbean Plate boundary, vicinity of Trinidad and Tobago. AAPG Bulletin. 1989;73:490–509. [Google Scholar]
- Scranton MI, Taylor GT, Astor Y, Muller-Karger F.2006Temporal variability in the nutrient chemistry of the Cariaco BasinIn: Neretin LN (ed).Past and Present Water Column Anoxia, NATO Sci Ser Springer: The Netherlands; 139–160. [Google Scholar]
- Sherr EB, Sherr BF. Significance of predation by protists in aquatic microbial food webs. Antonie van Leeuwenhoek. 2002;81:293–308. doi: 10.1023/a:1020591307260. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored ‘rare biosphere'. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Bass D, Nebel M, Christe R, Jones MDH, Breiner H-W, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Behnke A, Christen R, Amaral-Zettler L, Rodriguez-Mora MJ, Chistoserdov A, et al. Massively parallel tag sequencing reveals the complexity of anaerobic marine protistan communities. BMC Biol. 2009;7:72. doi: 10.1186/1741-7007-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Hayward B, Taylor GT, Varela R, Epstein SS. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist. 2006;157:31–43. doi: 10.1016/j.protis.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Kasper J, Bunge J, Leslin C, Ilyin V, Epstein S. Protistan diversity in the arctic: a case of paleoclimate shaping modern biodiversity. PLoS ONE. 2007;2:e728. doi: 10.1371/journal.pone.0000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Taylor GT, Epstein SS. Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean Sea) Appl Environ Microbiol. 2003;69:5656–5663. doi: 10.1128/AEM.69.9.5656-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GT. The role of pelagic protozoa in nutrient cycling: a review. Ann Inst Oceanogr (Suppl), Paris. 1982;58:227–241. [Google Scholar]
- Taylor GT, Iturriaga R, Sullivan CW. Interactions of bactivorous grazers and heterotrophic bacteria with dissolved organic matter. Mar Ecol Prog Ser. 1985;23:129–141. [Google Scholar]
- Taylor GT, Scranton MI, Iabichella M, Tung-Yuan H, Thunell RC, Muller-Karger F, et al. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol Oceanogr. 2001;46:148–163. [Google Scholar]
- Thompson JD, Higgens DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;33:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.