Abstract
This is the second paper in a series of three that investigates eukaryotic microbial diversity and taxon distribution in the Cariaco Basin, Venezuela, the ocean's largest anoxic marine basin. Here, we use phylogenetic information, multivariate community analyses and statistical richness predictions to test whether protists exhibit habitat specialization within defined geochemical layers of the water column. We also analyze spatio-temporal distributions of protists across two seasons and two geographic sites within the basin. Non-metric multidimensional scaling indicates that these two basin sites are inhabited by distinct protistan assemblages, an observation that is supported by the minimal overlap in observed and predicted richness of sampled sites. A comparison of parametric richness estimations indicates that protistan communities in closely spaced—but geochemically different—habitats are very dissimilar, and may share as few as 5% of total operational taxonomic units (OTUs). This is supported by a canonical correspondence analysis, indicating that the empirically observed OTUs are organized along opposing gradients in oxidants and reductants. Our phylogenetic analyses identify many new clades at species to class levels, some of which appear restricted to specific layers of the water column and have a significantly nonrandom distribution. These findings suggest many pelagic protists are restricted to specific habitats, and likely diversify, at least in part due to separation by geochemical barriers.
Keywords: protists, diversity, anoxic, 18S rRNA approach
Introduction
Protists are pivotal members of aquatic microbial communities. Through grazing on prokaryotic and other eukaryotic prey, they regenerate nutrients and modify or remineralize organic matter (Taylor, 1982; Jumars et al., 1989; Sherr and Sherr, 2002). In addition, they can affect the quantity, activity and physiological state of their prey (Madsen et al., 1991; Frias-Lopez et al., 2009). Through direct and indirect effects, protists help determine metabolic potentials of microbial communities and influence aquatic carbon cycling. Bacterial grazing is principally performed by small flagellated protists and ciliates (Sherr and Sherr, 2002; Frias-Lopez et al., 2009). Grazing releases inorganic nutrients that often limit primary production, and makes organic carbon available to higher trophic levels (Berman et al., 1987; Caron, 1994, 2000; Sherr and Sherr, 2002; Pernthaler, 2005; Jurgens and Massana, 2008). In recognition of their importance in aquatic communities, protists are now considered in numerical models of carbon cycling and in paradigms of surface and deep-ocean microbial ecology (Aristegui et al., 2009). However, relatively little is still known about protistan diversity and distributions, particularly in the deep sea, and the degree to which specific habitats select for unique communities.
A century ago Beijerinck (1913) suggested that, in the microbial world, ‘everything is everywhere', implying that microorganisms are limitless in their dispersal abilities. This idea became a topic of debate (Finlay and Fenchel, 1999; Finlay, 2002; Richards and Bass, 2005; Martiny et al., 2006; Bass et al., 2007). For example, one view holds (Finlay and Clarke, 1999) that a soil sample under 0.1 cm2, can contain at least 78% of all globally found species of examined protists (the Genus Paraphysomonas), implying that liters of sea water examined here might contain the bulk of protistan diversity described to date. A logical extension of this view is a proposition that protistan species that are abundant locally should also be abundant globally, and vice versa (Finlay et al., 2001). An alternative view is the ‘moderate endemicity' model, proposed by Foissner (2006). The latter posits that some protistan species have cosmopolitan distributions, whereas other species have restricted distributions, a hypothesis gaining experimental support (Bass et al., 2007).
Determining whether microbial eukaryotes have distinctive biogeographies is far from trivial. Given the high abundances and reproductive potentials of microorganisms, high dispersal rates could allow many species to physically permeate almost all environments (Finlay, 2002; Cavalier-Smith, 2004; Fenchel and Finlay, 2004), but a number of empirical studies do not support this idea (Foissner, 1999; Rutherford et al., 1999; Lawley et al., 2004; Richards and Bass, 2005; Boenigk et al., 2006; Telford et al., 2006; Soininen and Heino, 2007; Fuhrman et al., 2008). Two examples are studies of longitudinal richness gradients of some marine foraminiferans (Rutherford et al., 1999) and bacteria (Fuhrman et al., 2008), suggesting that temperature explains a large portion of microbial endemicity. A handful of studies have addressed habitat specialization of marine protists but have either focused on specific groups (Guillou et al., 2008) or are based on a relatively small number of samples and sequences (Edgcomb et al., 2002, 2009; Stoeck et al., 2003, 2006, 2007; Countway et al., 2005; Zuendorf et al., 2006; Alexander et al., 2009).
The Cariaco Basin, Venezuela, harbors the world's largest truly marine body of permanently anoxic water, and provides a convenient model to study the role of geochemical gradients as possible barriers for active and passive protistan migration and colonization. Temporal changes in the biogeochemistry of the basin have been well studied (Scranton et al., 2001; Muller-Karger et al., 2001a; Astor et al., 2003) as well as the spatio-temporal dynamics of bacterial populations (Lin et al., 2008). Seasonal shifts in local upwelling intensity, rates of fluvial discharge, trade wind intensity and lateral intrusions of oxic waters influence primary production, microbial biomass and metabolic rates (Muller-Karger et al., 2001a; Taylor et al., 2001a; Astor et al., 2003; Lin et al., 2008). However, little is known about these effects on protistan populations. The Cariaco's water column transitions from fully oxic to sulfidic across a temporally varying boundary between 250 m and 350 m of depth. Within the redox transition zone lies strong gradients in O2, NO3−, H2S, NH4+, NO2−, PO43− and CH4, and enrichments in S2O32−, SO32−, S0, Mn2+ and Fe2+ that select for specific prokaryotic phylotypes at different depths (Taylor et al., 2001a; Lin et al., 2006, 2007, 2008; Li et al., 2008). Within the redoxcline, peaks in prokaryotic metabolic activity and cell numbers are observed, which often coincide with peaks in protistan cell numbers (Taylor et al., 2001a, 2006). Lin et al. (2008) showed significant vertical variations in bacterial community structure between oxic, transition and anoxic zones, as well as horizontally between different sites. Vertical, seasonal and geographic patterns in prokaryotic community distribution have also been documented in other locations (see Treusch et al., 2009, for recent synopsis). Recently, genetic approaches have begun to document marine protistan diversity (see Edgcomb et al. in this issue for a detailed discussion). However, spatio-temporal analyses of protistan communities have been limited (for example, Countway et al., 2007; Not et al., 2007). Studies have thus far been generally constrained to fewer than 2000 total small subunit ribosomal rRNA gene sequences from any single environment or sample, which in light of observed diversity makes it difficult to assess spatio-temporal differences in protistan communities. Here, we attempt to provide a more comprehensive approach. Although we did not collect multiple samples at each sampling point, we went substantially beyond earlier investigations by collecting samples spanning the major biogeochemical habitats of the Cariaco Basin, revisiting these habitats in different seasons and undertaking the largest Sanger-based 18S rRNA gene sequencing effort within a given oceanographic regime to date. Our data provide a unique opportunity to examine species richness and distribution within an oceanographic regime.
Materials and methods
More extensive information is in the Supplementary Materials and Methods in the companion paper in this issue by Edgcomb et al.
Sample collection
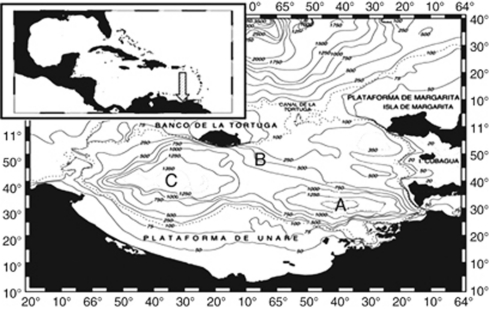
Samples were collected from three sites in the Cariaco Basin, Venezuela: Site A (10.50°N, 64.66°W), Site B (10.40°N, 64.46°W) and Site C (10.40°N, 65.35°W) between January and May of 2005 (Figure 1). Samples for DNA extractions were collected at depths corresponding to 40 m above the oxic/anoxic interface, within the oxic/anoxic interface, 40 m below the interface and at 900 m. The oxic/anoxic interface was defined as the depth at which oxygen became undetectable. Thus, a total of 24 water samples were collected (2 months × four depths × three sites). Samples were withdrawn from Niskin bottles (General Oceanics, Miami, FL, USA) under N2 pressure and captured on 47 mm Durapore membranes (Millipore, Billerica, MA, USA) (0.45 μ pore size) onboard, with little to no sample exposure to the atmosphere. Depending on cell concentration in each sample, 2.5–2.8 l samples were filtered under gentle vacuum (<25 cm Hg) until no passage of water through filters was observed. At least two filters from each sample were used for DNA extractions. Membranes were stored in cryovials containing DNA extraction buffer (Stoeck et al., 2003) at −20 °C until further processing.
Figure 1.
Map of sample sites in the Cariaco Basin, Venezuela. Site A represents the site of the US–Venezuelan CARIACO biogeochemical time series program in the eastern sub-basin. Site B is a shallower site south of the Tortuga channel, the source of incoming Caribbean waters. Site C is centered in the less productive western sub-basin to the east of Canal Centenela.
DNA preparation and PCR amplification of 18S rDNA
DNA was prepared using previously described methods (Stoeck et al., 2003, 2007). Depending on yield, DNA from 2–3 filters per depth at each site was combined before PCR. The 18S rRNA gene was amplified using four different primer combinations; Euk528F (5′-CGGTAATTCCAGCTCC-3′) paired with U1492R (5′-GGTTACCTTGTTACGACTT-3′), U1391R (5′-GGGCGGTGTGTACAARGR-3′) or with U1517R 5′-ACGGCTACCTTGTTACGACTT-3′) and Euk360F (5′-CGGAGARGGMGCMTGAGA-3′) paired with U1492R. All primer combinations were applied to all samples, and PCR products from ⩾three reactions were pooled per primer pair for each sample.
Clone library construction and sequencing
To constrain costs and allow for deeper sequencing of individual libraries, PCR products for sites B and C were pooled from the same depths before clone library construction (reducing the number of analyzed sites to two, and the total number of analyzed samples from 24 to 16). Separate clone libraries were constructed for each of the four depths from the eight-site B/C samples (four from May 2005 and four from January 2005) and from the eight-site A samples. Processing of the data used PHRED and PHRAP (Ewing and Green, 1998; Ewing et al., 1998), and a pipeline script to call bases from chromatograms and perform quality control procedures. The sequences were checked for chimeras using the Bellerophon Chimera Check and the Check_ Chimera utilities (Ribosomal Database Project) (Cole et al., 2003). After removal of short sequences (<800 bp), putative chimeras, bacterial, archaeal and metazoan sequences, the remaining sequences were grouped into operational taxonomic units (OTUs) based on 90, 95, 98 and 99% rRNA gene sequence similarity levels. This was achieved by first making all possible pairwise sequence alignments by using ClustalW (Thompson et al., 1994), calculating percentage sequence identities, followed by clustering the sequences by using the unweighted pair group method with arithmetic mean, as implemented in the OC clustering program (http://www.compbio.dundee.ac.uk/Software/OC/oc.html). All protistan sequences have been deposited in the Genbank, see the accompanying paper in this issue, Edgcomb et al., for details.
Phylogenetic analyses
For our phylogenetic analyses, we used ‘centroid' sequences, each representing OTUs sharing 95% identity (for practical considerations). The centroid sequence was defined as the least dissimilar sequence relative to all sequences within a cluster at the 95% threshold. A centroid representative from each OTU was compared against the GenBank-nt database using BLASTn in search of their closest sequence relatives. The highest scoring cultured and uncultured sequence relatives of each centroid were retrieved, and aligned using ARB (Ludwig et al., 2004).
Statistical analyses of clone library data for richness predictions
For each sample, we obtained ‘frequency count' data, that is, the numbers of OTUs registered in the corresponding clone library only once, twice and so on, at the 90, 95, 98 and 99% levels of sequence identity. This data was used to estimate, at each % identity level, the total number of OTUs, representing the sum of seen and unseen. The clustering of the entire data set was used to test the fit of a wide range of parametric and non-parametric models for predicting total protistan diversity in the Cariaco Basin (see Edgcomb et al., this issue for a complete description), a methodology that has been used in previous studies (Hong et al., 2006; Jeon et al., 2006) for the estimation of microbial richness. This method was implemented here using the software package CatchAll (Bunge, 2011). The entire data set was then separated by sites (A vs combined B and C) and the four depths from site A (40 m above the oxic/anoxic interface, interface, 40 m below and 670–900 m) and clustered separately. These data were used to predict taxon richness of the different environments based on clustering data at 99% sequence similarity (for details see Supplementary Materials and Methods in Edgcomb et al., this issue).
Canonical correspondence analysis, multi-response permutation procedure and non-metric multidimensional scaling
Canonical correspondence analysis (CCA) is an ordination method that allows for the exploration of community responses to environmental gradients by reducing the dimensionality of the analysis such that the axes reflect a linear combination of the environmental variables and the OTU data (McCune and Grace, 2002). CCA was used here to elucidate the relationships between protistan community structure and concentrations of dissolved O2 and sulfide. Multi-response permutation procedure (MRPP) is a tool used for testing the hypothesis of no effect of a variable on two different groups of entities and can help determine whether a variable exerts a statistically significant influence on sampled communities (McCune and Grace, 2002). We used MRPP to assess the effect of sulfide, oxygen, season, site and depth on the distribution of OTUs. Cases in which species composition is influenced by external factors such as oxygenated water or terrestrial sediment intrusions from seasonal riverine inputs instead of simply internal factors or environmental gradients, non-metric multidimensional scaling (NMS) is an ordination method that might be more appropriate (McCune and Grace, 2002). We ran a NMS analysis using PC_ORD v4 (MJM Software Design, Gleneden Beach, OR, USA) and selected the autopilot run with the slow and thorough option (McCune and Grace, 2002). Stress values were used as a measure of goodness of fit and values <15 indicate a low probability of drawing the wrong inferences from the results. Monte Carlo tests were used to identify dimensions with solutions that were significantly different from those arising strictly by chance. CCA, NMS and MRPP analyses were applied to our sequence data set, clustered at the 95% sequence identity threshold.
Urn model hypothesis test
As a further statistical test of equal distribution of any particular OTU across the sampled depths, we used an ‘urn model', which assumes that the occurrence of each sequence is equally likely in all urns (every depth in this case). The probability of 0, 1, 2 or 3 empty urns was determined by a well-established formula (Kolchin and Chistyakov, 1975). This allows computation of the probability that at least one urn will be empty (OTU does not appear at any one depth), thus obtaining the P-value for the test of the null hypothesis of random distribution.
Results
A total of 64 clone libraries were produced after applying the different primer sets to DNA extracted from each of the samples (16 samples × 4 primer sets). From each of the 64 libraries, we sequenced an average of 250 clones, resulting in over 16 000 rRNA gene sequences. Rigorous culling of confirmed or suspected metazoan, chimeric and short (<800 bases) sequences left 6498 high quality (average length 1100 bases) 18S rRNA protistan gene sequences (for details on the number of rRNA gene sequences analyzed in this study classified by season, biogeochemical habitat, site and PCR primer pair from each sample, see Supplementary Table 2 in Edgcomb et al., this issue). Clustering our data set at the 95% and 99% sequence identity thresholds produced a total of 822 and 2106 protistan centroids, respectively. The percentages of these centroids affiliated with different higher-level taxonomic groups of protists based on BLASTn searches are presented in Supplementary Figure S1 for each depth sampled. Rhizaria, followed by Stramenopiles, dominate our clone libraries at all depths, whereas the Ciliophora, Dinoflagellata, Syndiniales and Euglenozoa represent progressively less abundant clones in the libraries.
Predicting OTU richness
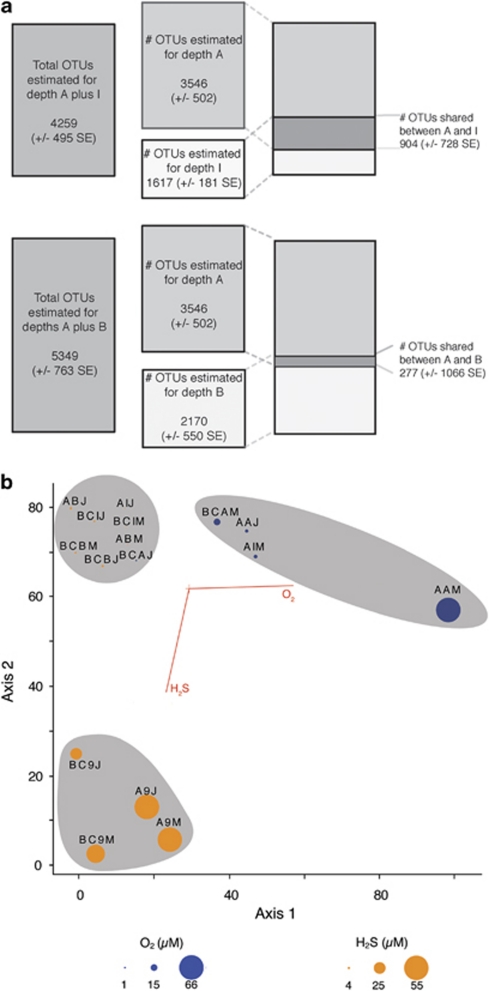
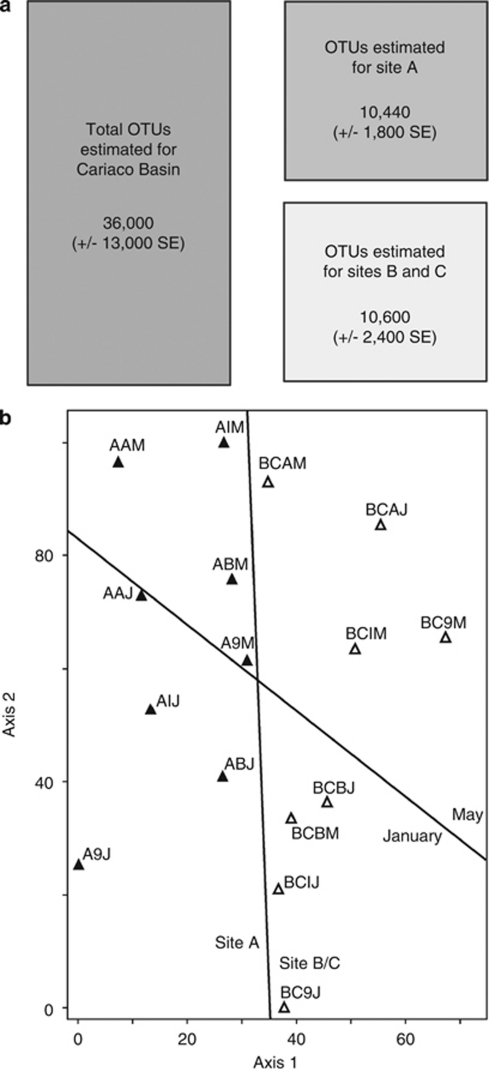
For a complete discussion of the results of OTU clustering see Edgcomb et al., (this issue). The overlap of empirically observed OTUs at the 99% identity level between the sampled depths and sites is presented in Supplementary Figure S3. Between the communities above and within the interface only 9% of OTUs are shared, and less overlap (8%) exists between the communities above and below the interface. In addition, over 90% of OTUs present at site A are not found at sites B and C and vice versa. Figure 2a presents the statistical predictions of total OTUs at 99% sequence identity for sampled depths from 40 m above the oxic/anoxic interface, the interface and 40 m below, as well as pooled data for the first and second, and second and third depths. These three depths were selected for presentation because they are in closest proximity (∼40 m) to one another. The logic behind this analysis is that if one habitat contains X species, the other habitat contains Y species and the two habitats combined contain Z species, then ((X+Y)−Z) is the maximum number of species shared by the two habitats. The results show a predicted overlap of 904 (±728) OTUs between communities above the interface and within the interface (Figure 2a). Less overlap is predicted for the communities above and below the interface (277±1066 OTUs). Following the same logic, we obtained the richness estimated for site A and the combined estimation for sites B and C. These roughly sum to equal the lower bound of the s.e. for the overall basin prediction of diversity, suggesting that many OTUs are unique between these sites (Figure 3a).
Figure 2.
(a) Total protist richness predictions for selected environments (A, 40 m above interface, I, interface and B, 40 m below interface) of the chemically stratified water column in the Cariaco Basin at Site A. (b) A biplot generated from a canonical correspondence analysis (CCA) of our 18S rRNA data set clustered at the 95% sequence identity level. Environmental variables sulfide and oxygen are represented by arrows. Sampled depths are represented by circles, the size and color of which indicate the detected amount of either oxygen or sulfide in that sample. Samples that cluster together on the biplot are highlighted in gray.
Figure 3.
(a) Predicted richness of protistan assemblages in two different geographic locations with in the Cariaco Basin compared with that predicted for the entire basin. (b) Non-metric multidimensional scaling (NMS) ordination of our 18S rRNA data set clustered at the 95% sequence identity level. Triangles represent the 16 samples taken; white and black triangles correspond to combined samples from sites B and C, and site A, respectively. A sample name ending in M or J (that is BCIM, AIJ and so on) corresponds to May and January samples, respectively. Lines are drawn onto the biplot to separate samples that cluster together.
Canonical correspondence analysis, multi-response permutation procedure and non-metric multidimensional scaling
CCA identified environmental parameters that may explain a large proportion of the observed protistan community distribution pattern. The analysis shows a separation of protistan communities within samples from oxygenated, microoxic and sulfidic waters of the Cariaco Basin along O2 and H2S gradients (Figure 2b). The species–environment correlations were 0.971 and 0.990 for axes 1 and 2, respectively, and the two-dimensional plot shows that axes 1 and 2 explained 34.6 and 28.4% of the variance in OTU distribution, respectively. A Monte Carlo test for significance of the Eigenvalues provided a P-value ⩽0.02.
NMS ordination indicates that there are distinct assemblages of protists recovered from site A samples vs sites B/C and between the two seasons (Figure 3b). After 43 iterations, the stress of the final two-dimensional solution was 14.1 (below the cutoff of 15) and therefore the solution was considered stable. Axes 1 and 2 explain 28% and 20% of the variance, respectively. A MRPP was preformed for season, site, sulfide, oxygen and depth, which were all found to have a significant effect (⩽0.05) on the observed distribution of OTUs.
Phylogenetic analyses
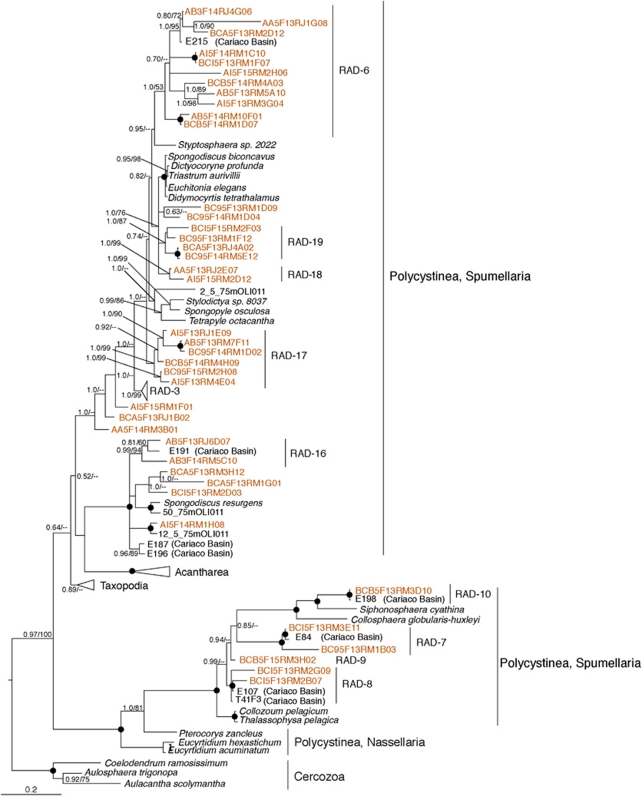
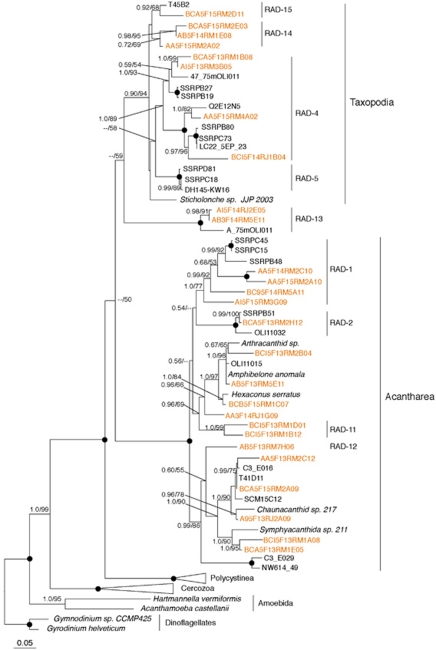
Among rhizarians, Radiolaria were by far the most abundant phylum represented by 2216 sequences and 150 centroids. Over 50% of these fell into one of the five Radiolarian sequence clades (RAD 1–5) described from the Sargasso Sea by Not et al. (2007), principally into the RAD-3 clade, detected from all depths, both seasons and sites (Table 1 ). In addition to these five ‘RAD' clades, several other Radiolarian centroids from our data set form 14 lineages (Figures 4 and 5) labeled RAD 6–19. We identified two new lineages (RAD-11 and 12) within the Acantharea and two new lineages within the Taxopodia (RAD-13, 14). Four RAD lineages (7, 8, 9 and 10) were related to the Polycystinea and do not contain sequences from oxygenated samples. The RAD-9 lineage contained 77 sequences that derive from 7 of the 12 total samples from the anoxic water column.
Table 1. The number of occurrences of various environmental clades detected in this study at each depth sampled.
| Environmental clade | A seq (samp) | I seq (samp) | B seq (samp) | D seq (samp) | Total seq (samp) |
|---|---|---|---|---|---|
| RAD-1 | 2 (1) | 2 (1) | 0 | 2 (1) | 6 (3) |
| RAD-3 | 693 (4) | 333 (4) | 50 (4) | 142 (4) | 1361 (16) |
| RAD-4 | 2 (2) | 3 (1) | 1 (1) | 0 | 6 (4) |
| RAD-6* | 5 (2) | 19 (3) | 157 (4) | 1 (1) | 182 (10) |
| RAD-7* | 0 | 2 (1) | 1 (1) | 1 (1) | 4 (3) |
| RAD-8* | 0 | 2 (2) | 2 (2) | 0 | 4 (4) |
| RAD-9* | 0 | 12 (1) | 57 (3) | 8 (3) | 77 (7) |
| RAD-10* | 0 | 0 | 3 (2) | 0 | 3 (2) |
| RAD-11* | 0 | 2 (2) | 0 | 0 | 2 (2) |
| RAD-12* | 1 (1) | 1 (1) | 2 (1) | 0 | 4 (3) |
| RAD-13* | 0 | 2 (1) | 2 (2) | 1 (1) | 5 (4) |
| RAD-14* | 10 (2) | 1 (1) | 1 (1) | 0 | 12 (4) |
| RAD-15* | 2 (1) | 0 | 1 (1) | 0 | 3 (2) |
| RAD-16* | 3 (2) | 5 (2) | 2 (2) | 18 (1) | 28 (7) |
| RAD-17* | 2 (2) | 0 | 2 (2) | 2 (2) | 6 (6) |
| RAD-18* | 38 (4) | 11 (3) | 1 (1) | 1 (1) | 51 (9) |
| RAD-19* | 125 (2) | 62 (1) | 2 (1) | 82 (1) | 271 (5) |
| MAST-1 | 9 (3) | 10 (2) | 5 (3) | 5 (2) | 29 (10) |
| MAST-3 | 7 (4) | 17 (4) | 19 (4) | 20 (4) | 63 (16) |
| MAST-4 | 0 | 0 | 2 (2) | 6 (1) | 8 (3) |
| MAST-5 | 2 (2) | 3 (1) | 2 (1) | 0 | 7 (5) |
| MAST-7 | 5 (2) | 3 (3) | 3 (1) | 1 (1) | 12 (7) |
| MAST-8 | 12 (3) | 8 (3) | 23 (3) | 28 (4) | 71 (13) |
| MAST-9 | 6 (3) | 14 (2) | 28 (2) | 4 (1) | 52 (8) |
| MAST-12 | 1 (1) | 1 (1) | 0 | 0 | 2 (2) |
| MAST-14* | 0 | 5 (2) | 0 | 0 | 5 (2) |
| MAST-15* | 0 | 0 | 1 (1) | 0 | 1 (1) |
| MAST-16* | 0 | 2 (1) | 2 (1) | 0 | 4 (2) |
| MAST-17* | 6 (2) | 2 (2) | 1 (1) | 0 | 9 (5) |
| MAST-18* | 1 (1) | 0 | 0 | 0 | 4 (2) |
| MAST-19* | 0 | 7 (1) | 2 (2) | 4 (1) | 13 (4) |
| NA1.1 | 0 | 7 (2) | 1 (1) | 2 (2) | 10 (5) |
| NA1.2 | 10 (4) | 30 (3) | 13 (2) | 2 (1) | 45 (10) |
| NA1.3 | 6 (4) | 6 (2) | 10 (3) | 0 | 22 (9) |
| NA1.4 | 21 (4) | 40 (4) | 47 (4) | 25 (2) | 134 (14) |
| NA1.5 | 0 | 0 | 0 | 1 (1) | 1 (1) |
| NA1.6 | 0 | 0 | 4 (1) | 2 (2) | 6 (3) |
| NA1.7 | 5 (2) | 0 | 0 | 0 | 5 (2) |
| NA1.8 | 2 (1) | 3 (3) | 4 (2) | 0 | 9 (6) |
| NA1.9* | 0 | 3 (1) | 1 (1) | 3 (1) | 7 (3) |
| NA1.4A* | 1 (1) | 0 | 2 (2) | 0 | 3 (3) |
| NA1.4B* | 0 | 1 (1) | 4 (2) | 0 | 5 (3) |
| Putative novel euglenozoa class* | 0 | 4 (1) | 25 (4) | 21 (5) | 50 (10) |
| Symbiontida sister clade A* | 0 | 1 (1) | 0 | 9 (1) | 10 (2) |
| Symbtiontida sister clade B* | 0 | 4 (2) | 18 (6) | 10 (2) | 32 (10) |
Abbreviations: MAST, marine stramenopiles; RAD, radiolarian sequence clade; NA, Novel Alveolate sequence clade.
‘seq' indicates the overall number of occurrences and ‘samp' is the number of samples in which the clade was detected. Clades marked with an asterisk are unique to this study.
Figure 4.
Phylogenetic relationships of selected Polycystinea (Radiolaria). The tree was constructed under Bayesian inference using an alignment of 925 unambiguous positions under the General Time Reversible (GTR)+I+Gamma model of sequence evolution. Bootstrap (RAxML) and posterior probability values greater than 50% are shown at the nodes in the order PP/ML. Black circles at nodes represent full posterior probability and bootstrap support. Radiolarian clades are labeled in accordance with the naming scheme established by Not et al. (2007). Centroids from our survey are in orange font. The color reproduction of this figure is available on the html full text version of the Manuscript.
Figure 5.
Phylogenetic relationships of selected Acantharea and Taxopodia (Radiolaria). The tree was constructed under Bayesian inference using an alignment of 887 unambiguous positions under the General Time Reversible (GTR)+I+Gamma model of sequence evolution. Clades are labeled in accordance with the naming scheme established by Not et al. (2007). For more information see Figure 4.
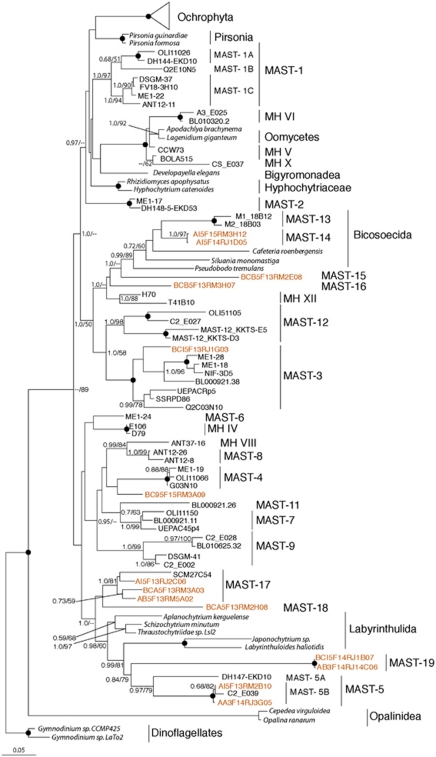
Approximately, 14% of our data set was affiliated with the Stramenopiles, which formed 148 centroids. On our phylogenetic tree of all Stramenopile centroids (data not shown), 339 sequences clustered with the heterotrophic Stramenopiles. Most lineages detected were uncultured marine stramenopiles (MAST) (Massana et al., 2004; Kolodziej and Stoeck, 2007). The number of sequences within each sample affiliated with MAST clades is organized by depth in Table 1. We discovered six lineages, MAST 14–19, that did not group with the 13 established MAST clades (Massana et al., 2004; Kolodziej and Stoeck, 2007) or the ‘mystery heterokont' groups (Richards and Bass, 2005) (Figure 6). Several of these new lineages were not detected in any oxygenated samples (Table 1). The MAST-14, 15 and 16 lineages were affiliated with the Bicosoecida, although the latter two are supported weakly. In contrast, MAST 17 and 19 branch within the Thraustrochytrids and Labyrinthulids, respectively, with strong support from both methods.
Figure 6.
Phylogenetic relationships of selected marine stramenopiles (MAST). The tree was constructed under maximum likelihood using an alignment of 927 unambiguous positions under the General Time Reversible (GTR)+I+Gamma model of sequence evolution. The tree contains full-length sequence representatives from all (except MAST-10) MAST clades and ‘mystery heterokont' clades (Richards and Bass, 2005). A separate RAxML tree based on a smaller number of characters to accommodate the MAST-10 clade (data not shown) confirmed that this was not a member of any of the new MAST clades described in this study. MAST clades are labeled in accordance with the naming scheme established by Massana et al. (2004). For more information see Figure 4.
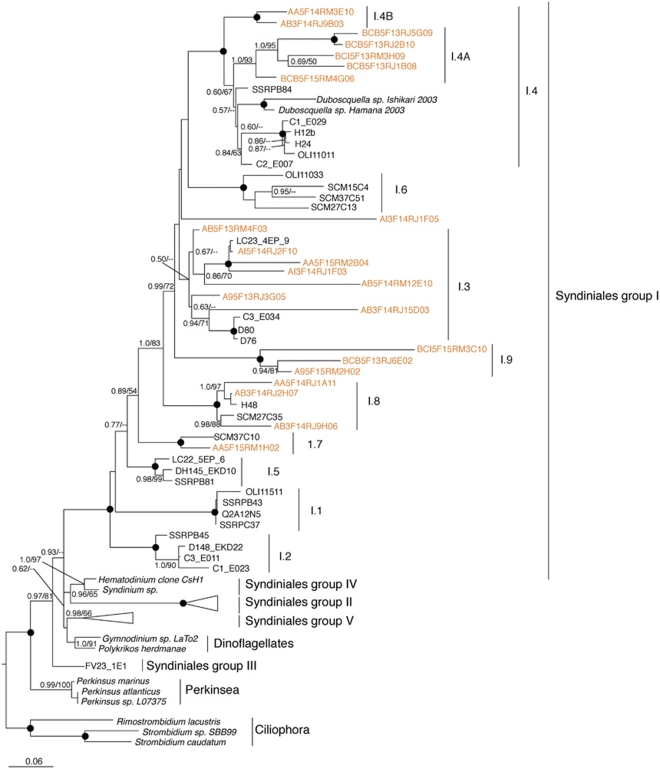
Sequences affiliated with the Syndiniales (Alveolata) represent approximately 16% of our clone library sequences (Supplementary Figure S1). We detected representatives from all five groups of Syndiniales (Guillou et al., 2008) with most of the sequences being related to groups I (247 sequences) and II (140 sequences). Within group I, we identified a new clade (100% support from both methods) labeled I.9 (Figure 7) that does not cluster with the established 8 group-I sub-clades of Syndiniales (Guillou et al., 2008). Within the Novel Alveolate (NA) 1.4 clade, seven centroids from our data set form two supported sub-clades (1.0 posterior probability and 93% bootstrap). Both I.9 and I.4A were detected in three of our anoxic samples and were absent from our oxygenated water column samples (Table 1). Clade 1.4A was detected only from anoxic waters in the pooled samples from sites B/C and was absent from all site A samples.
Figure 7.
Phylogenetic relationships of selected Syndiniales. The tree was constructed under Bayesian inference using an alignment of 869 unambiguous positions under the General Time Reversible (GTR)+I+Gamma model of sequence evolution. The new Novel Alveolate (NA) 1.9 clade is labeled in accordance with the naming scheme established by Guillou et al. (2008). For more information see Figure 4.
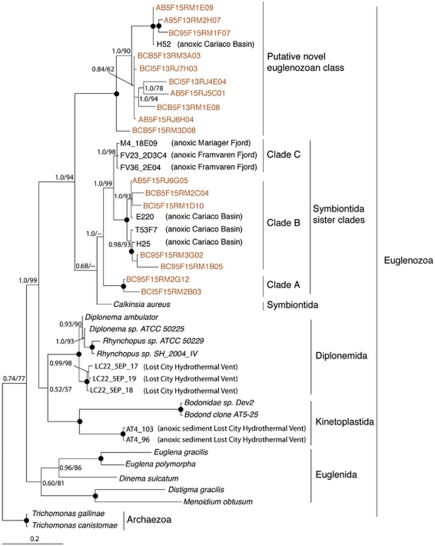
We detected 90 sequences affiliated with the Euglenozoa that fell into 17 centroids at the 95% identity threshold, with the majority detected only in the 900 m anoxic samples (Supplementary Figure S1). A total of 10 centroids, along with a sequence reported from an earlier survey of the Cariaco Basin (Stoeck et al., 2003) form a clade with 100% supported separation from all other major groups of Euglenozoa, and is presented here as a ‘putative novel euglenozoan class' (Figure 8). This clade was detected in 10 out of 13 anoxic samples (9 from our survey plus 1 from Stoeck et al. (2003)) and it was absent from all oxygenated samples (Table 1). The closest named relative to this group of sequences is the recently described Calkinsia aureus (Yubuki et al., 2009). Three sister clades (labeled Symbiontida clades A, B and C) to Calkinsia aureus were also identified; all with strong support on our trees. Clades A and B contain sequences from anoxic samples of the Cariaco Basin only. Clade C consists of sequences from anoxic Framvaren and Mariager Fjords (Behnke et al., 2006; Zuendorf et al., 2006).
Figure 8.
Phylogenetic relationships of selected Euglenozoa. The tree was constructed under Bayesian inference using an alignment of 766 unambiguous positions under the General Time Reversible (GTR)+I+Gamma model of sequence evolution. For more information see Figure 4.
Having observed a number of protistan clades apparently restricted to specific geochemical regimes, we chose representative clades from several Kingdoms to statistically estimate whether their association with the respective regime could be because of chance using the Urn Model Test (Supplementary Figure S2). In all cases, such probability proved infinitesimally low, down to for example, 10−10 in case of the novel rhizarian clade RAD-9.
Discussion
Spatio-temporal distribution patterns and responses to environmental variables
Using phylogenetic and multivariate community analyses coupled with parametric richness estimations, we show that specific environmental variables impact the distribution of protistan taxa, thus, leading to their habitat preference. Three of the sampled habitats are particularly interesting for this type of analysis because they are sufficiently close to each other (40 m), and the water density gradient is sufficiently weak between these layers to allow, in principle, for frequent redistribution of the individual species by advective water mixing or active motility. The null hypothesis is that given no physical barrier to dispersal, mixing would ensure that each species is present throughout these closely spaced layers of the water column. On the basis of results from several parametric statistical analyses, we reject the null hypothesis and show that protistan communities in closely spaced—but geochemically different—habitats are dissimilar and may share as little as 5% of species (Figure 2a). This conclusion is supported by a direct comparison of empirically observed OTUs at these depths showing that over 90% of detected OTUs are unique (Supplementary Figure S3). Furthermore, canonical correspondence analysis of our data set showed that many of the empirically observed OTUs were organized along oxygen and sulfide gradients (Figure 2b), indicating these environmental variables are responsible, in part, for the observed OTU distribution in our data set. We note that distributions of protistan ecotypes most likely are shaped by integrated responses to the complex chemical milieu and the composition/activity of prokaryotic/eukaryotic prey communities and cannot be solely linked to a specific chemical species. In this study, we track distributions of protistan clades through the geochemically stratified water column, recognizing that oxygen and sulfide distributions correlate with numerous chemical variables that define biogeochemical habitats. We also acknowledge that sampling limitations inherent to microbial oceanography studies may mask many spatio-temporal patterns. However, we argue that the relatively large sampling effort made here, across multiple seasons, sites and geochemical habitats, reveals gross patterns in community structure of pelagic protists.
Our NMS ordination indicates that there are distinct assemblages of protists recovered from site A vs sites B/C, and clear divisions in most (but not all) samples between the two seasons (Figure 3b). A MRPP P-value of 0.012 for the test of significance of geographic sites in the observed distribution of OTUs lends support to this conclusion. These results are corroborated by the minimal overlap in OTU numbers estimated and empirically observed for the locations compared (Figure 3a, Supplementary Figure S3), supporting the notion that spatial separation and local biogeochemistry drives, in part, the distribution of these protistan assemblages. The separation of samples from site A vs the combined B and C samples may be due to the fact that sites B/C are closer to channels providing input of oxygenated surface water from the Caribbean Sea. Site A is closest to riverine inputs from coastal Venezuela, which may drive community divergence among sites. Lin et al. (2008) found that among-site variation (site A vs sites B/C) in bacterial communities within the redoxcline was greater than the vertical variation between depths, possibly due to differences in primary production and lateral intrusions of oxic water at the different sites. The western basin has lower surface primary production than the eastern basin (Richards, 1975; Scranton et al., 1987; Muller-Karger et al., 2001a) and such differences likely contribute to variations in microbial communities (Lin et al., 2008).
Water column productivity varies seasonally and spatially across the basin because of nutrient upwelling from late January through June due to intensifying trade winds (Muller-Karger et al., 2001b). Seasonal upwelling events would explain the fact that the site A samples from May in the eastern sub-basin are distinct from the samples taken at the same location in January (Figure 3b). It also explains why a 100% seasonal distinction is not observed for the combined samples from sites B/C because site B is in the eastern sub-basin and site C is in the western sub-basin.
Significant temporal shifts in bacterial community composition along the redoxcline were observed over a 2-year period at the same three sites (Lin et al., 2008). Trophic interactions between aquatic protists and bacteria, better known as the ‘microbial loop,' are thought to have a major role in controlling bacterial dynamics (Taylor, 1982; Azam et al., 1983). Phagotrophic protists can chemically sense and congregate at aggregations of prey (Fenchel and Blackburn, 1999). Their growth rates can keep pace with those of their bacterial prey (Fenchel, 1987; Sherr and Sherr, 1994, 2002; Finlay and Fenchel, 2001), and as bacterial population size and composition changes from site to site and season to season, it is logical to expect local protist populations to exhibit similar variations as the local habitat changes and potential prey species select for community dominance by different protists. Thus, trophic responses could be contributing to the relatively high standard error in some of our estimates of richness (Figure 3a).
Many OTUs were observed to occur exclusively in a particular geochemical regime. Among the 19 Radiolarian clades, 5 were detected in only one habitat (RAD 7, 8, 9, 10 and 11), but most of these are underrepresented in our data set, and are perhaps members of the ‘rare biosphere'. The RAD 9 clade was never observed in the basin's oxygenated waters, but appeared 77 times in over 50% of all samples from oxygen-depleted and sulfidic depths. The probability of such a distribution being due to chance is essentially zero (Supplementary Figure S2). Thus, we conclude that RAD 9, and possibly several other Radiolarian clades, are indeed restricted to anoxic regimes.
The distributions of some clades within the alveolate Order Syndiniales were confined to suboxic and/or anoxic habitats similar to the RAD clades described above, perhaps indicating interesting ecological relationships. The Syndiniales is a diverse group of picoeukaryotes (Moon-van der Staay et al., 2001; Guillou et al., 2008) exhibiting a parasitic lifestyle (Skovgaard et al., 2005; Chambouvet et al., 2008), representing 30% of all dinoflagellate sequences in public databases, and have only been retrieved from marine environments (Guillou et al., 2008). The distribution of Syndiniales may be constrained by the observed endemic distribution of their hosts (Guillou et al., 2008), among them the Radiolaria (Dolven et al., 2007). Indeed, the Syndiniales clade I.9 does seem to be absent from oxygenated waters of the Cariaco Basin, and present exclusively under suboxic and anoxic conditions, mirroring the habitat specialization of several RAD clades (Figures 4, 5; Table 1).
Further examples of apparent habitat specialization can be found among Stramenopiles. Although several of the more frequently encountered MAST taxa seems to be distributed rather homogeneously throughout the water column (for example, MAST 1, 3 and 8), notable exceptions were evident; MAST 4, 14, 15, 16 and 19 were absent from oxygenated waters. In addition, some of these MAST clades appear to prefer anoxic waters immediately below oxic/anoxic interface, which coincidentally exhibits peaks in prokaryotic biomass and production (Taylor et al., 2001b; Stoeck et al., 2003). This distribution is interesting because MAST species have been reported from anoxic environments in the past (Stoeck et al., 2003; Behnke et al., 2006; Zuendorf et al., 2006) and because at least some MAST species are known to be phagotrophic (Massana et al., 2006, 2009). Considering our observations, the possibility that some MAST species are anaerobic bacterivores, and thus involved in regulating bacterial dynamics within and below the redoxcline is suggested.
One of the strongest cases of apparent habitat specialization is represented by the novel Euglenozoa clades we discovered. Some of the newly detected OTUs appear to form a novel class level clade that affiliates strongly with the H52 clone from an earlier survey of the deep anoxic Cariaco Basin (Stoeck et al., 2003) (Figure 8). As importantly, this clade separates from all other Euglenozoa OTUs described to date from other anoxic marine environments (Behnke et al., 2006; Zuendorf et al., 2006; Lopez-Garcia et al., 2007; Lara et al., 2009). Additional euglenozoan signatures detected here form two sister clades to Symbiontida, a recently proposed new class of Euglenozoa represented by only a single species, Calkinsia aureus (Bernhard et al., 2000; Yubuki et al., 2009). Unique α-taxonomic as well as genomic characteristics support the separation of Symbiontida from the three traditional euglenozoan classes Euglenids, Kinetoplastids and Diplonemids (Yubuki et al., 2009). Many euglenozoan 18S rRNA gene sequences revealed here seem unique to suboxic, anoxic or sulfidic habitats (Table 1), and the probability of such skewed distributions being exclusively due to chance is essentially zero (Supplementary Figure S2).
This study expands the taxonomic groups recovered in smaller-scale previous studies of the Cariaco Basin protist community (Stoeck et al., 2003, 2006). Those studies also reported a differential recovery of particular taxonomic groups among depths from one site, but the depth of sequencing in those studies did not allow further testing of these observations.
Relevance to questions regarding protistan distributions
Clades unique to our study are highly suggestive of local speciation. The number of unique clades is high (Figures 4, 5, 6, 7 and 8, Table 1), and these are spread over multiple Kingdoms. A potentially important example is the new putative euglenozoan class (Figure 8), which is distinct from all other euglenozoan taxa described so far from all locations. It is abundant in our sequence collection (a total of 50 individual sequences forming 10 centroids), and is apparently distributed throughout the basin's anoxic waters (detected in over 80% of all the anoxic samples), and does not contain related sequences from the growing number of surveys of anoxic environments reported to date (Edgcomb et al., 2002, 2009; Stoeck et al., 2003, 2009, 2010; Behnke et al., 2006; Zuendorf et al., 2006; Alexander et al., 2009). Numerous other examples of apparent geographic distribution patterns include 14 novel lineages of Rhizaria, a new group-I uncultured Novel Alveolate clade and 6 new MAST Stramenopile clades. Also noteworthy is the absence of representatives from the Sargasso Sea in all of the novel clades reported here, as the Sargasso Sea has been sampled heavily (Venter et al., 2004; Countway et al., 2007; Not et al., 2007; Piganeau and Moreau, 2007; Piganeau et al., 2008) and lies just northeast of the Caribbean Sea. As different sites within even the basin itself harbor distinct species assemblages (Figure 3), it is not unexpected that the Cariaco Basin and Sargasso sea likely contain widely divergent communities. The simplest interpretation of these observations is that at least some microbial eukaryotes speciate, diversify and become restricted to a specific biogeochemical niche, and do not successfully disperse to either similar or dissimilar environments elsewhere on the globe. (We acknowledge that species of protists whose 18S rRNA genes have not yet been sequenced may bias our observations and interpretations regarding the potential for the global distribution of selected protist groups due to their underrepresentation in sequence databases).
At the same time, over 250 rhizarian, Dinoflagellate, Syndiniales, Stramenopile and various fungal OTUs (95% identity) appearing in our Cariaco Basin 18S rRNA libraries have previously been detected elsewhere (data not shown), under both similar and dissimilar biogeochemical conditions (Lopez-Garcia et al., 2001; Moon-van der Staay et al., 2001; Edgcomb et al., 2002; Massana et al., 2002; Stoeck et al., 2003, 2006; Not et al., 2007). This indicates that some protist taxa are dispersed widely, and are perhaps even cosmopolitan in their distribution.
Size-related dispersal of protists
The influence of microbial body size on dispersal has been debated, with arguments made for (Finlay, 2002) and against (Heino and Soininen, 2006; Soininen and Heino, 2007; Heino et al., 2010) the existence of a connection between the two. One hypothesis (Fenchel and Finlay, 2004) asserts that with an increase in organism size there is a decrease in dispersal rate, whereas microbes of smaller sizes are more likely to have a wider dispersal range, leading to cosmopolitan distribution. We analyzed the apparent cosmopolitan and ‘endemic' clades in our data set to see whether a particular body size range dominated either type of clades. We defined clades detected in the Cariaco Basin as likely cosmopolitan if their representatives had been registered in at least three locations worldwide (including our survey), and as potentially endemic if all the clade's representatives were so far unique to the basin. To make realistic size assignments, all clades must fulfill an additional criterion: to be closely related (>90% 18S rRNA gene identity) to a described taxonomic group, the latter with known and relatively narrow size range of its members. Screening our sequence collection lead to identification of 32 likely cosmopolitan and 36 possibly endemic clades that satisfied the above criteria, which we then placed into one of three size groups based on the length and/or diameter of their known relatives: 1–20, 25–50 and 80–200 μm. The results of this classification are presented in Supplementary Table 1. Clearly, our endemic group contains as many clades within the smallest and largest among our size categories. Similarly, the smallest clades are equally distributed between ‘endemic' or ‘cosmopolitan' groups. In spite of uncertainties about definitions of endemicity and size assignments, our analysis does not show a clear connection between the size of a cell and its dispersal potential. This in line with the findings of Heino and Soininen (2006) who, using freshwater diatoms as a model to study microbial dispersal capacity, did not observe strong evidence for a relationship between dispersal and body size.
Conclusions
An in-depth survey of the protistan community in the Cariaco Basin reveals an apparent restriction of some protists to different sites in the basin, as well as oxygenated, suboxic or anoxic/sulfidic environments. Phylogenetic and multivariate community analyses along with parametric estimations of richness imply that geographic location, seasonality and geochemical gradients define the community structure of the marine protists in the Cariaco Basin. We conclude that substantially different communities can exist in close proximity to one another, which permits speciation to proceed differently at different sites and depths in the basin. These analyses support the moderate endemicity model (Foissner, 2006; Bass et al., 2007), which views protists as an assemblage of species, some of which are cosmopolitan and others endemic, and thus biogeographically similar to macroorganisms.
Acknowledgments
We thank the captain and crew of the B/O Hermano Gines and the staff of the Fundación La Salle de Ciencias Naturales, Margarita Island for their assistance during our field work in Venezuela, particularly Yrene Astor. We are grateful to Mary I Scranton and the dedicated researchers of the CARIACO biogeochemical time series program without whom this work would not have been possible. We would like to thank Valentin Ilyin (Northeastern University), David Mark Welch (MBL) and Jed Goldstone (WHOI) for helpful discussions on bioinformatics approaches, Hilary Morrison and Rich Fox (MBL) for the use of their pipeline scripts for sequence data processing, and the Academic Research Computing User Group at Northeastern University for permitting us to use the Opportunity computer cluster. We thank Vladimir Bondarev and Anne-Christin Girnth for assistance with preliminary quality control of our Sanger data set. Comments from two anonymous reviewers significantly improved the manuscript. This research was conducted using the resources of the Cornell University Center for Advanced Computing, which receives funding from Cornell University, New York State, the National Science Foundation and other leading public agencies, foundations and corporations. This research was supported by grants from NSF (MCB-0348341 and DEB-0816840 to SSE, DEB-0816638 to JB, MCB-0348407 to VE and OCE 03-26175 and MCB-03-47811 to GTT) and from Venezuela FONACIT (no. 2000001702 to RV). This is a contribution # 269 from the Marine Science Center, Northeastern University, Nahant, MA, USA.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alexander E, Stock A, Breiner HW, Behnke A, Bunge J, Yakimov MM, et al. Microbial eukaryotes in the hypersaline anoxic L′Atalante deep-sea basin. Environ Microbiol. 2009;11:360–381. doi: 10.1111/j.1462-2920.2008.01777.x. [DOI] [PubMed] [Google Scholar]
- Aristegui J, Gasol JM, Duarte CM, Herndl GJ. Microbial oceanography of the dark ocean's pelagic realm. Limnol Oceanogr. 2009;54:1501–1529. [Google Scholar]
- Astor Y, Muller-Karger F, Scranton MI. Seasonal and interannual variation in the hydrography of the Cariaco Basin: implications for basin ventilation. Cont Shelf Res. 2003;23:125–144. [Google Scholar]
- Azam F, Fenchel T, Field J, Gray J, Meyer-Reil L, Thingstad F. The ecological role of water column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- Bass D, Richards TA, Matthai L, Marsh V, Cavalier-Smith T. DNA evidence for global dispersal and probable endemicity of protozoa. BMC Evol Biol. 2007;7:162. doi: 10.1186/1471-2148-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke A, Bunge J, Barger K, Breiner HW, Alla V, Stoeck T. Microeukaryote community patterns along an O2/H2S gradient in a supersulfidic anoxic fjord (Framvaren, Norway) Appl Environ Microbiol. 2006;72:3626–3636. doi: 10.1128/AEM.72.5.3626-3636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijerinck MW. De Infusies En De Ontdekking Der Backteriën. Jaarboek Van De Koninklijke Akademie Voor Wetenschappen. Müller: Amsterdam, The Netherlands; 1913. [Google Scholar]
- Berman T, Nawrocki M, Taylor GT, Karl DM. Nutrient flux between bacteria, bacterivorous nanoprotozoans and algae. Mar Microb Food Webs (now Aquat Microb Ecol) 1987;2:69–82. [Google Scholar]
- Bernhard JM, Buck KR, Farmer MA, Bowser SS. The Santa Barbara Basin is a symbiosis oasis. Nature. 2000;403:77–80. doi: 10.1038/47476. [DOI] [PubMed] [Google Scholar]
- Boenigk J, Pfandl K, Garstecki T, Harms H, Novarino G, Chatzinotas A. Evidence for geographic isolation and signs of endemism within a protistan morphospecies. Appl Environ Microbiol. 2006;72:5159–5164. doi: 10.1128/AEM.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge J. Estimating the number of species with CatchAll. Proceedings of the 2011 Pacific Symposium on Biocomputing. 2011. [DOI] [PubMed]
- Caron DA. Inorganic nutrients, bacteria, and the microbial loop. Microb Ecol. 1994;28:295–298. doi: 10.1007/BF00166820. [DOI] [PubMed] [Google Scholar]
- Caron DA.(ed.) (2000Protistan Herbivory and Bacterivory Academic Press: London; 289–315. [Google Scholar]
- Cavalier-Smith T. Only six kingdoms of life. Proc Biol Sci. 2004;271:1251–1262. doi: 10.1098/rspb.2004.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambouvet A, Morin P, Marie D, Guillou L. Control of toxic marine dinoflagellate blooms by serial parasitic killers. Science. 2008;322:1254–1257. doi: 10.1126/science.1164387. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Countway PD, Gast RJ, Dennett MR, Savai P, Rose JM, Caron DA. Distinct protistan assemblages characterize the euphotic zone and deep sea (2500 m) of the western North Atlantic (Sargasso Sea and Gulf Stream) Environ Microbiol. 2007;9:1219–1232. doi: 10.1111/j.1462-2920.2007.01243.x. [DOI] [PubMed] [Google Scholar]
- Countway PD, Gast RJ, Savai P, Caron DA. Protistan diversity estimates based on 18S rDNA from seawater incubations in the Western North Atlantic. J Eukaryot Microbiol. 2005;52:95–106. doi: 10.1111/j.1550-7408.2005.05202006.x. [DOI] [PubMed] [Google Scholar]
- Dolven JK, Lindqvist C, Albert VA, Bjorklund KR, Yuasa T, Takahashi O, et al. Molecular diversity of alveolates associated with neritic north atlantic radiolarians. Protist. 2007;158:65–76. doi: 10.1016/j.protis.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Edgcomb V, Orsi W, Leslin C, Epstein SS, Bunge J, Jeon S, et al. Protistan community patterns within the brine and halocline of deep hypersaline anoxic basins in the eastern Mediterranean Sea. Extremophiles. 2009;13:151–167. doi: 10.1007/s00792-008-0206-2. [DOI] [PubMed] [Google Scholar]
- Edgcomb VP, Kysela DT, Teske A, de Vera Gomez A, Sogin ML. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc Natl Acad Sci USA. 2002;99:7658–7662. doi: 10.1073/pnas.062186399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred.I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fenchel T. Ecology of Protozoa. Springer: Berlin; 1987. [Google Scholar]
- Fenchel T, Blackburn N. Motile chemosensory behaviour of phagotrophic protists: mechanisms for and efficiency in congregating at food patches. Protist. 1999;150:325–336. doi: 10.1016/S1434-4610(99)70033-7. [DOI] [PubMed] [Google Scholar]
- Fenchel T, Finlay BJ. The ubiquity of small species: patterns of local and global diversity. Bioscience. 2004;54:777–784. [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Clarke KJ. Apparent global ubiquity of species in the protist genus Paraphysomonas. Protist. 1999;150:419–430. doi: 10.1016/S1434-4610(99)70042-8. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Esteban GF, Clarke KJ, Olmo JL. Biodiversity of terrestrial protozoa appears homogeneous across local and global spatial scales. Protist. 2001;152:355–366. doi: 10.1078/1434-4610-00073. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Fenchel T. Divergent perspectives on protist species richness. Protist. 1999;150:229–233. doi: 10.1016/S1434-4610(99)70025-8. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Fenchel T. Protozoan community structure in a fractal soil environment. Protist. 2001;152:203–218. doi: 10.1078/1434-4610-00060. [DOI] [PubMed] [Google Scholar]
- Foissner W. Protist diversity: estimates of the near-imponderable. Protist. 1999;150:363–368. doi: 10.1016/S1434-4610(99)70037-4. [DOI] [PubMed] [Google Scholar]
- Foissner W. Biogeography and dispersal of micro-organisms: a review emphasizing protists. Acta Protozool. 2006;45:111–136. [Google Scholar]
- Frias-Lopez J, Thompson A, Waldbauer J, Chisholm SW. Use of stable isotope-labelled cells to identify active grazers of picocyanobacteria in ocean surface waters. Environ Microbiol. 2009;11:512–525. doi: 10.1111/j.1462-2920.2008.01793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA. 2008;105:7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, Viprey M, Chambouvet A, Welsh RM, Kirkham AR, Massana R, et al. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata) Environ Microbiol. 2008;10:3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- Heino J, Mauricio Bini L, Karjalainen SM, Mykra H, Soininen J, Vieira LCG, et al. Geographical patterns of micro-organismal community structure: are diatoms ubiquitously distributed across boreal streams. Oikos. 2010;119:129–137. [Google Scholar]
- Heino J, Soininen J. Regional occupancy in unicellular eukaryotes: a reflection of niche breadth, habitat availability or size-related dispersal capacity. Freshwater Biol. 2006;51:672–685. [Google Scholar]
- Hong SH, Bunge J, Jeon SO, Epstein SS. Predicting microbial species richness. Proc Natl Acad Sci USA. 2006;103:117–122. doi: 10.1073/pnas.0507245102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SO, Bunge J, Stoeck T, Barger KJ, Hong SH, Epstein SS. Synthetic statistical approach reveals a high degree of richness of microbial eukaryotes in an anoxic water column. Appl Environ Microbiol. 2006;72:6578–6583. doi: 10.1128/AEM.00787-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumars PA, Penry DL, Baross JA, Perry MJ, Frost BW. Closing the the microbial loop: dissolved carbon pathway to heterotrophic bacteria from incomplete ingestion, digestion and absorption in animals. Deep-Sea Res. 1989;36:483–495. [Google Scholar]
- Jurgens K, Massana R.(eds). (2008Protistan Grazing on Marine Bacterioplankton Wiley: New York; 383–441. [Google Scholar]
- Kolchin VF, Chistyakov VP. Combinatorial problems of probability theory. J Math Sci. 1975;4:217–243. [Google Scholar]
- Kolodziej K, Stoeck T. Cellular identification of a novel uncultured marine stramenopile (MAST-12 Clade) small-subunit rRNA gene sequence from a Norwegian estuary by use of fluorescence in situ hybridization scanning electron microscopy. Appl Environ Microbiol. 2007;73:2718–2726. doi: 10.1128/AEM.02158-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara E, Moreira D, Vereshchaka A, Lopez-Garcia P. Pan-oceanic distribution of new highly diverse clades of deep-sea diplonemids. Environ Microbiol. 2009;11:47–55. doi: 10.1111/j.1462-2920.2008.01737.x. [DOI] [PubMed] [Google Scholar]
- Lawley B, Ripley S, Bridge P, Convey P. Molecular analysis of geographic patterns of eukaryotic diversity in Antarctic soils. Appl Environ Microbiol. 2004;70:5963–5972. doi: 10.1128/AEM.70.10.5963-5972.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XN, Taylor GT, Astor Y, Scranton M. Relationship of sulfur speciation to hydrographic conditions and chemoautotrophic production in the Cariaco Basin. Marine Chem. 2008;112:53–64. [Google Scholar]
- Lin X, Wakeham SG, Putnam IF, Astor YM, Scranton MI, Chistoserdov AY, et al. Comparison of vertical distributions of prokaryotic assemblages in the anoxic Cariaco Basin and Black Sea by use of fluorescence in situ hybridization. Appl Environ Microbiol. 2006;72:2679–2690. doi: 10.1128/AEM.72.4.2679-2690.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XJ, Scranton MI, Chistoserdov AY, Varela R, Taylor GT. Spatiotemporal dynamics of bacterial populations in the anoxic Cariaco Basin. Limnol Oceanogr. 2008;53:37–51. [Google Scholar]
- Lin XJ, Scranton MI, Varela R, Chistoserdov A, Taylor GT. Compositional responses of bacterial communities to redox gradients and grazing in the anoxic Cariaco Basin. Aquat Microb Ecol. 2007;47:57–72. [Google Scholar]
- Lopez-Garcia P, Rodriguez-Valera F, Pedros-Alio C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Vereshchaka A, Moreira D. Eukaryotic diversity associated with carbonates and fluid-seawater interface in Lost city hydrothermal field. Environ Microbiol. 2007;9:546–554. doi: 10.1111/j.1462-2920.2006.01158.x. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EL, Sinclair JL, Ghiorse WC. In situ biodegradation: microbiological patterns in a contaminated aquifer. Science. 1991;252:830–833. doi: 10.1126/science.2028258. [DOI] [PubMed] [Google Scholar]
- Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Massana R, Castresana J, Balague V, Guillou L, Romari K, Groisillier A, et al. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl Environ Microbiol. 2004;70:3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Guillou L, Diez B, Pedros-Alio C. Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the ocean. Appl Environ Microbiol. 2002;68:4554–4558. doi: 10.1128/AEM.68.9.4554-4558.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Terrado R, Forn I, Lovejoy C, Pedros-Alio C. Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ Microbiol. 2006;8:1515–1522. doi: 10.1111/j.1462-2920.2006.01042.x. [DOI] [PubMed] [Google Scholar]
- Massana R, Unrein F, Rodriguez-Martinez R, Forn I, Lefort T, Pinhassi J, et al. Grazing rates and functional diversity of uncultured heterotrophic flagellates. ISME J. 2009;3:588–596. doi: 10.1038/ismej.2008.130. [DOI] [PubMed] [Google Scholar]
- McCune B, Grace JB. Analysis of Ecological Communities. MJM Software Design. Gleneden: Oregon; 2002. [Google Scholar]
- Moon-van der Staay SY, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- Muller-Karger F, Varela R, Thunell R, Scranton M, Bohrer R, Taylor GT, et al. Annual cycle of primary production in the Cariaco Basin: response to upwelling and implications for vertical transport. J Geophys Res. 2001a;106:4527–4542. [Google Scholar]
- Muller-Karger F, Varela R, Thunell R, Scranton M, Bohrer R, Taylor GT, et al. Annual cycle of primary production in the Cariaco Basin: response to upwelling and implications for vertical export. J Geophys. Res. 2001b;106:4527–4542. [Google Scholar]
- Not F, Gausling R, Azam F, Heidelberg JF, Worden AZ. Vertical distribution of picoeukaryotic diversity in the Sargasso Sea. Environ Microbiol. 2007;9:1233–1252. doi: 10.1111/j.1462-2920.2007.01247.x. [DOI] [PubMed] [Google Scholar]
- Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol. 2005;3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- Piganeau G, Desdevises Y, Derelle E, Moreau H. Picoeukaryotic sequences in the Sargasso sea metagenome. Genome Biol. 2008;9:R5. doi: 10.1186/gb-2008-9-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau G, Moreau H. Screening the Sargasso Sea metagenome for data to investigate genome evolution in Ostreococcus (Prasinophyceae, Chlorophyta) Gene. 2007;406:184–190. doi: 10.1016/j.gene.2007.09.015. [DOI] [PubMed] [Google Scholar]
- Richards F. The Cariaco Basin (Trench) Oceanogr Mar Bio Ann Rev. 1975;13:11–67. [Google Scholar]
- Richards TA, Bass D. Molecular screening of free-living microbial eukaryotes: diversity and distribution using a meta-analysis. Curr Opin Microbiol. 2005;8:240–252. doi: 10.1016/j.mib.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Rutherford S, D′Hondt S, Prell W. Environmental controls on the geographic distribution of zooplankton diversity. Nature. 1999;400:749–753. [Google Scholar]
- Scranton M, Astor Y, Bohrer R, Ho T, Muller-Karger F. Controls on temporal variability of the geochemistry of the deep Cariaco Basin. Deep-Sea Res I. 2001;48:1605–1625. [Google Scholar]
- Scranton M, Sayles F, Bacon M, Brewer P. Temporal changes in the hydrography and chemistry of the Cariaco Trench. Deep-Sea Res I. 1987;34:945–963. [Google Scholar]
- Sherr EB, Sherr BF. Bacterivory and herbivory: key roles of phagotrophic protists in pelagic food webs. Microb Ecol. 1994;28:223–235. doi: 10.1007/BF00166812. [DOI] [PubMed] [Google Scholar]
- Sherr EB, Sherr BF. Significance of predation by protists in aquatic microbial food webs. Antonie Leeuwenhoek. 2002;81:293–308. doi: 10.1023/a:1020591307260. [DOI] [PubMed] [Google Scholar]
- Skovgaard A, Massana R, Balague V, Saiz E. Phylogenetic position of the copepod-infesting parasite Syndiniumturbo (Dinoflagellata, Syndinea) Protist. 2005;156:413–423. doi: 10.1016/j.protis.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Soininen J, Heino J. Variation in niche parameters along the diversity gradient of unicellular eukaryote assemblages. Protist. 2007;158:181–191. doi: 10.1016/j.protis.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Bass D, Nebel M, Christe R, Jones MDH, Breiner H-W, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Behnke A, Christen R, Amaral-Zettler L, Rodriguez-Mora MJ, Chistoserdov A, et al. Massively parallel tag sequencing reveals the complexity of anaerobic marine protistan communities. BMC Biol. 2009;7:72. doi: 10.1186/1741-7007-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Hayward B, Taylor GT, Varela R, Epstein SS. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist. 2006;157:31–43. doi: 10.1016/j.protis.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Stoeck T, Kasper J, Bunge J, Leslin C, Ilyin V, Epstein S. Protistan diversity in the arctic: a case of paleoclimate shaping modern biodiversity. PLoS ONE. 2007;2:e728. doi: 10.1371/journal.pone.0000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Taylor GT, Epstein SS. Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean Sea) Appl Environ Microbiol. 2003;69:5656–5663. doi: 10.1128/AEM.69.9.5656-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GT. The role of pelagic heterotrophic protozoa in nutrient cycling: a review. Ann Inst Oceanogr (Suppl.), Paris. 1982;58:227–241. [Google Scholar]
- Taylor GT, Iabichella-Armas M, Varela R, Müller-Karger F, Lin X, Scranton MI.2006Microbial ecology of the Cariaco basin's redoxclineIn: Neretin NL (ed.).Past and Present Water Column Anoxia Springer: Dordrecht, The Netherlands; 473–499. [Google Scholar]
- Taylor GT, Scranton MI, Iabichella I, Ho T-Y, Thunell RC, Muller-Karger F, et al. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol Oceanogr. 2001a;46:148–163. [Google Scholar]
- Taylor GT, Scranton MI, Iabichella M, Y HT, Thunell RC, Muller-Karger F, et al. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol Oceanogr. 2001b;46:148–163. [Google Scholar]
- Telford RJ, Vandvik V, Birks HJ. Dispersal limitations matter for microbial morphospecies. Science. 2006;312:1015. doi: 10.1126/science.1125669. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgens DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;33:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3:1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Yubuki N, Edgcomb VP, Bernhard JM, Leander BS. Ultrastructure and molecular phylogeny of Calkinsiaaureus: cellular identity of a novel clade of deep-sea euglenozoans with epibiotic bacteria. BMC Microbiol. 2009;9:16. doi: 10.1186/1471-2180-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuendorf A, Bunge J, Behnke A, Barger KJ, Stoeck T. Diversity estimates of microeukaryotes below the chemocline of the anoxic Mariager Fjord, Denmark. FEMS Microbiol Ecol. 2006;58:476–491. doi: 10.1111/j.1574-6941.2006.00171.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.