Abstract
Extensive genomic characterization of multi-species acid mine drainage microbial consortia combined with laboratory cultivation has enabled the application of quantitative proteomic analyses at the community level. In this study, quantitative proteomic comparisons were used to functionally characterize laboratory-cultivated acidophilic communities sustained in pH 1.45 or 0.85 conditions. The distributions of all proteins identified for individual organisms indicated biases for either high or low pH, and suggests pH-specific niche partitioning for low abundance bacteria and archaea. Although the proteome of the dominant bacterium, Leptospirillum group II, was largely unaffected by pH treatments, analysis of functional categories indicated proteins involved in amino acid and nucleotide metabolism, as well as cell membrane/envelope biogenesis were overrepresented at high pH. Comparison of specific protein abundances indicates higher pH conditions favor Leptospirillum group III, whereas low pH conditions promote the growth of certain archaea. Thus, quantitative proteomic comparisons revealed distinct differences in community composition and metabolic function of individual organisms during different pH treatments. Proteomic analysis revealed other aspects of community function. Different numbers of phage proteins were identified across biological replicates, indicating stochastic spatial heterogeneity of phage outbreaks. Additionally, proteomic data were used to identify a previously unknown genotypic variant of Leptospirillum group II, an indication of selection for a specific Leptospirillum group II population in laboratory communities. Our results confirm the importance of pH and related geochemical factors in fine-tuning acidophilic microbial community structure and function at the species and strain level, and demonstrate the broad utility of proteomics in laboratory community studies.
Keywords: acid mine drainage, communities, genotyping, perturbation, proteomics
Introduction
The application of ecological theory to microbial systems has demonstrated that macro-level ecological concepts can be used to describe microbial distributions and community interactions (Jessup et al., 2004; Kassen and Rainey, 2004; Martiny et al., 2006; Prosser et al., 2007; Green et al., 2008). Low complexity, extremophilic consortia that thrive in acid mine drainage (AMD) environments have been proposed as a model system for the study of natural microbial communities (Denef et al., 2010b). Ecological interactions such as competition, mutualism, predation and synergism have been described in such environments (Johnson, 1998). AMD biofilm communities from the Richmond Mine at Iron Mountain, CA, USA have been well characterized using culture independent techniques (Edwards et al., 1999; Bond et al., 2000a, 2000b; Druschel et al., 2004; Wilmes et al., 2008b), and have also been the subject of intensive genomic and proteomic studies (Tyson et al., 2004; Ram et al., 2005; Lo et al., 2007; Simmons et al., 2008; Denef et al., 2009), making them ideal for in-depth ecological analyses.
Richmond Mine communities are dominated by chemoautotrophic iron-oxidizing bacteria belonging to the Leptospirillum genus. Archaea and other low abundance bacteria colonize biofilms generated by Leptospirillum spp. to produce multi-species consortia. Characterization of AMD biofilm ultrastructure has revealed a community progression over developmental stages (Wilmes et al., 2008b). During early stages of growth, when biofilms are relatively thin, communities are dominated by Leptospirillum group II (related to L. ferriphilum). At later stages of growth, biofilms are thicker and the communities are more diverse. However, more specific details of ecological interactions and niche partitioning of low abundance bacteria and archaea are unknown.
Reconstructions of composite genomes for Leptospirillum group II have revealed two distinct genomic types (referred to as the ‘5-way CG genomic type' and the ‘UBA genomic type'), for which orthologous proteins share ∼95% identity on average (Lo et al., 2007). Analysis of environmental metagenomic data has shown that homologous recombination, as well as transposon and phage-mediated insertions or deletions are common sources of variation between the genomic types (Simmons et al., 2008). Although much of the observed variation may be adaptively neutral (Thompson et al., 2005), both gene content and sequence divergence between orthologous proteins have been proposed as mechanisms of niche differentiation (Rocap et al., 2003; Hunt et al., 2008; Wilmes et al., 2008a).
Mass spectrometry-based proteomic analyses can be used to distinguish protein sequence variants in genomically uncharacterized environmental samples, a method known as proteomics-inferred genome typing (PIGT) (Lo et al., 2007). The PIGT approach has been used to distinguish the two ‘ancestral' genotypes of Leptospirillum group II (5-way CG and UBA) in proteomic datasets, as well as four recombinant genotypes that contain distinct blocks of both the 5-way CG and UBA genomes (Lo et al., 2007; Denef et al., 2009). Detailed analysis of the distribution and protein expression of Leptospirillum group II genotypic variants has revealed differences that were correlated to environmental conditions and biofilm development, confirming the occurrence of strain-level niche partitioning within AMD communities (Denef et al., 2010a).
Laboratory growth and isolation of acidophilic organisms has been challenging, and many remain uncharacterized in terms of their functional role in community biofilms. Coculturing multi-species consortia from environmental samples has been shown as a strategy to propagate ‘unculturable' organisms for laboratory characterization (Kaeberlein et al., 2002). A laboratory community-culturing strategy has been developed for AMD biofilms (Belnap et al., 2010). Using a quantitative proteomic approach, our previous analyses have demonstrated laboratory communities to be functionally similar to a natural sample, including growth of low abundance, nonisolated bacteria and archaea. However, the distribution of specific taxa in laboratory and natural samples is not identical, and is likely influenced by fine-scale variation of community composition and geochemical conditions.
Additionally, viruses of bacteria and archaea have been predicted to be major drivers of genetic diversity (leading to niche differentiation) (Rodriguez-Valera et al., 2009) and community dynamics in prokaryotic-dominated environments (Breitbart et al., 2004; Danovaro et al., 2008). Viral genomes have been previously assembled and annotated in AMD metagenomic datasets indicating extensive viral diversity and host susceptibility (Andersson and Banfield, 2008). It is likely, then, that phage-driven mortality in AMD biofilm communities may also affect membership and function. However, distinct patterns of phage activity (for example, temporal or geospatial) are yet to be observed.
To investigate the extent to which small changes in environmental conditions influence community membership and function, the responses of laboratory-cultivated AMD biofilm communities maintained at high and low pH were compared. Utilization of community quantitative proteomic methods enabled functional characterization of dominant and low abundance taxa under the different pH conditions. Proteomic datasets were further utilized to identify the Leptospirillum group II genotype(s) present in laboratory biofilms, and to observed patterns of phage protein distribution across replicate laboratory communities.
Methods
Laboratory cultures
Biofilm communities similar to natural AMD consortia were cultured in laboratory reactors as described previously (Belnap et al., 2010). All reactors were inoculated with 50 ml from a community batch culture originating from a mixture of biofilm samples collected from several different locations within the Richmond Mine (Redding, CA, USA). Communities were grown using an acidophilic growth medium supplemented with 55.6 g l−1 FeSO4 7H20, trace elements, and acidified using H2SO4 (9K-BR medium, see Belnap et al., 2010). Unless otherwise stated, all chemicals and reagents were supplied by Sigma-Aldrich (Sigma-Aldrich Co., St Louis, MO, USA). No carbon source was provided in the growth medium. Metabolically labeled community samples were grown with 15N-ammonium sulfate (Isotec, Miamisburg, OH, USA) in place of standard ammonium sulfate. Six replicate biofilm communities, three labeled with 15N-ammonium sulfate and three unlabeled, were cultured in separate reactors. For initial growth, each reactor reservoir was supplied with pH 1.0 medium. Over a 14-day period, high cell density, multispecies biofilms formed in all reactors, and cellular metabolism gradually increased pH to 1.3–1.5. On day 14, the medium in reactor reservoirs containing 15N-ammonium sulfate was lowered to pH 0.6 (100% of reservoir volume, or 2L), which equilibrated at ∼0.85 in each reactor. Unlabeled and labeled communities were then maintained at the respective pH for a period of 19 days, and solution pH was monitored daily. In all, 50% of the total reservoir and reactor volume for each replicate was replenished with fresh medium on days 0, 5, 9, 13 and 16. On day 19, biofilms were removed from all six reactors, centrifuged for three minutes at 8500 r.p.m. and 4 °C, and cell pellets were immediately frozen at −80 °C in a dry ice/ethanol bath. A single unlabeled high-pH sample and 15N-labeled low-pH sample were mixed evenly according to biomass for each biological replicate to form three proteomic replicates subsequently used for quantitative proteomics analysis.
Microscopy
Aliquots of biofilm samples were preserved in 4% paraformaldehyde and stored at −20 °C in a 1:1 solution of ethanol and phosphate-buffered saline. Fluorescence in situ hybridization with lineage-specific probes was carried out on fixed samples as described previously (Amann, 1995; Bond and Banfield, 2001) at 630 times magnification using a Leica DMRX epifluorescence microscope (Leica, Wetzlar, Germany). Specific probes were used to target Leptospirillum groups II and III, and remaining taxa were characterized using broad-range bacterial and archaeal probes. For estimation of Leptospirillum group III abundance, representative fluorescence in situ hybridization images were collected for each of the replicate high and low pH community samples. Leptospirillum group III cells were counted in three replicate fields of view (>1000 cells counted per field of view) per image, and converted to a percentage of the total cell count found using the general nucleic acid stain 4′,6-diamidino-2-phenylindole. An average percentage of Leptospirillum group III cells for high pH and low pH treatments was calculated for the three replicate community samples and differences were tested for significance using a Standard t-test (P-value <0.05).
Protein extraction and analysis
Whole-cell protein extractions were carried out as described previously (Ram et al., 2005; Lo et al., 2007). Fractions were analyzed in duplicate by two-dimensional liquid chromatography-tandem mass spectrometry using a LTQ-Orbitrap mass spectrometer (Thermo Fisher, San Jose, CA, USA). Peptides and proteins were identified with SEQUEST (Eng et al., 1994) and DTASelect (Tabb et al., 2002) programs using an AMD genomic sequence database (Lo et al., 2007). The ProRata program was used for protein abundance ratio determination and confidence interval evaluation (Pan et al., 2006a, 2006b). Proteins were quantified on the basis of both unique peptides that can be attributed to a single protein and non-unique peptides that are shared among multiple proteins. Within each replicate dataset, proteins that were detected in only one of the growth conditions (high or low pH sample) were assigned a log2 value of±7 (that is, 128 times more abundant in the growth condition for which they were detected). For all proteins, 90% confidence intervals were calculated and used as the range of error of individual log2 abundance ratios. Nonunique peptides belonging to Leptospirillum group II proteins are assigned to both genomic types of this organism, which are referred to as the ‘5-way CG genome' and ‘UBA genome.' Genotyping analysis (PIGT, see below) indicated more than 90% of unique peptides in laboratory samples originated from the 5-way CG genome (Type I strain, see below); therefore, proteins identified as belonging to Leptospirillum group II UBA strain-types were excluded from data analyses (except those within the Type Ib recombination block).
In previous AMD quantitative proteomic experiments, protein log2 abundance ratios were normalized by shifting the median of the protein distribution for each taxa to zero, thereby removing effects caused by cell abundance and/or activity differences between samples (Belnap et al., 2010). In this study, the median of individual taxa protein distributions were shifted according to the offset from zero of Leptospirillum group II (the organism for which the most proteins are detected) within each proteomic dataset (Supplementary Table 1). Consequently, for low abundance organisms, protein distribution bias to either high or low pH, which could be caused by differences in cell numbers or activity, is evident if the median offset from zero is greater than the Leptospirillum group II correction factor. Average log2 abundance ratios were generated for proteins identified in at least two of the three biological replicates. Positive log2 ratios indicated higher abundance in pH 1.45 and negative values indicated higher abundance in pH 0.85. The 90% confidence intervals were calculated for each average log2 ratio. Note that the average dataset from the three replicates was not subjected to further normalization. Proteins with average normalized log2 ratios greater than 1 or smaller than −1 (>2 × difference) and confidence intervals excluding zero were considered significantly different between samples. All normalized proteomic results are listed in Supplementary Table 7.
For Leptospirillum group II, proteins were additionally grouped into functional categories as described previously (Denef et al., 2009), and a one-sample t-test or Wilcoxon Rank Sum test was performed using the log2-ratio values to identify categories that were significantly different in either the environmental or laboratory sample (P<0.05).
PIGT analysis of Leptospirillum group II
Genotyping of Leptospirillum group II populations by strain-resolved proteomic-based analyses have been described in detail previously (Denef et al., 2009). Briefly, the spectral counts for unique and non-unique peptides were tabulated from mass spectral data. The log2-transformed spectral counts of unique peptides were then plotted onto schematic representations of the aligned 5-way CG genome and UBA genome using Circos (http://mkweb.bcgsc.ca/circos/) to visualize points of recombination. Using this method, we defined the laboratory genotype using the Type I–VI genotype spectrum defined by Denef et al (2009), in which Type I refers to a nearly pure 5-way CG genome, Type VI refers to a nearly pure UBA genome and Types II–IV contain large genomic blocks of both types.
Results
Genotype analysis of the Leptospirillum group II population in laboratory communities
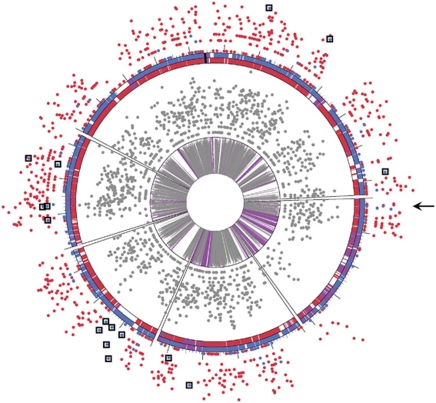
PIGT analyses for Leptospirillum group II were performed using the proteomic datasets for each of the three laboratory replicate comparisons. For all three proteomic datasets, 94–95% of unique peptides matched the 5-way CG genome, hence the genotype(s) of Leptospirillum group II present in laboratory communities is similar to the environmental Type I genotype (predominantly composed of 5-way CG genomic loci) (Figure 1 and Supplementary Figure 1A). Some peptides currently considered unique to the UBA genotype were identified at high abundance. Most of these peptides map to regions of the 5-way CG genome that correspond to sequencing gaps (see boxed proteins corresponding to gaps in red ring, Figure 1), and based on analyses across nearly 30 environmental samples likely represent currently unidentified Type I proteins in the near complete Leptospirillum group II 5-way CG population genomic dataset (Denef et al., 2009).
Figure 1.
PIGT analysis of Leptospirillum group II in laboratory proteomic datasets. Unique peptides from all three proteomic replicates were plotted onto aligned genomic representations of Leptospirillum group II 5-way CG (red ring) and UBA (blue ring) genomic variants. Purple loci represent regions of 100% amino acid identity, black represents the CRISPR repeat region, and loci for which a homolog does not exist are white. Large tick marks are spaced 100 genes apart. The inner gray ring represents percent amino acid identity between orthologs (indicated by height of tick marks, purple regions equivalent to 100% identical). Gray spots surrounding the inner ring are the total number of peptides detected at each loci. Spots on the outside of the diagram represent the number of unique peptides detected at each loci (red=Type I, blue=Type VI). Unique peptides are plotted on a log2 scale, with higher placement indicating higher detection. The arrow indicates a region of unique Type VI peptides consistently detected in the otherwise Type I dominated laboratory genotype. This region likely represents a small recombinant block. Boxes surround other abundant Type VI peptides that lack a Type I orthologue in the 5-way CG genomic dataset (and thus probably represent Type I peptides). The color reproduction of this figure is available on the html full text version of the manuscript.
In one small region of the Leptospirillum group II genome where 5-way CG genomic data is complete, several abundant unique peptides belonging to the UBA genotype were identified (Figure 1). Thus, we infer the presence of a Type VI recombination block in an otherwise Type I strain. This small, putative recombination block is a distinct feature of the laboratory strain compared with other known 5-way CG strain types (Supplementary Figure 1A). Because of its close similarity to Type I, we refer to this new genotype as Type Ib (Supplementary Figure 2). The Type Ib genotype has not been described previously in environmental samples, but has been identified in an additional laboratory sample for which PIGT analyses were carried out (Supplementary Figure 1B). Genes encoded on the Type VI block within the Type Ib genotype are listed in Supplementary Table 2. In all, 8 of the 14 proteins encoded by genes present in the recombinant block were detected in the proteomic datasets. For several proteins, spectral counts of unique Type VI peptides (UBA genomic type) were two to three times greater than unique Type I spectral counts (Supplementary Table 2). Quantitative comparisons for these proteins indicated similar abundance at high and low pH, suggesting their expression (and the selection for the Type Ib strain) was not pH dependent (Supplementary Table 2).
Unique peptides that matched the UBA genome were also present for additional genomic loci with distinct orthologs in both genomes (blue proteins, Figure 1). However, these peptides were detected at a much lower frequency, possibly indicating a low abundance of a Type VI-dominated strain.
Comparison of community function during growth at high and low pH
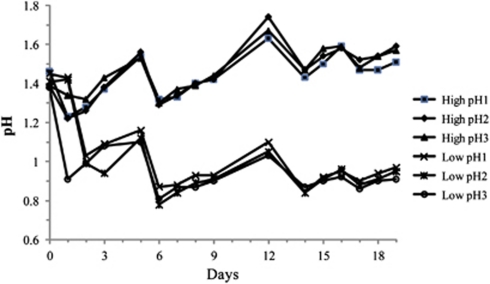
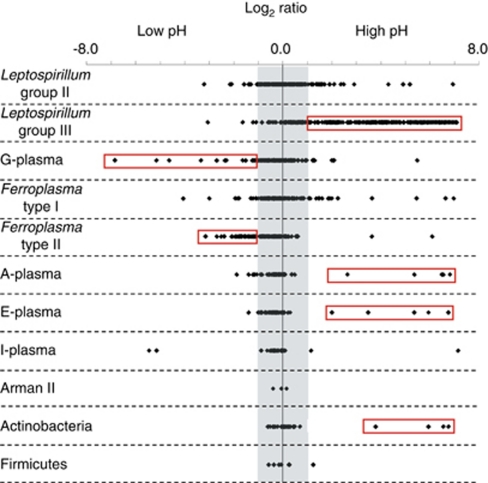
Average pH values for high-pH communities and low-pH communities were 1.45 and 0.85, respectively, over the course of the 19-day experiment (Figure 2). A total of 2112 proteins were quantified for proteomic replicate 1, 2425 for proteomic replicate 2 and 2085 for proteomic replicate 3 (Table 1). The number of proteins detected for individual taxa in each replicate dataset is tabulated in Table 1. An average log2 ratio was calculated for the 2148 proteins that were shared between at least two of the replicates. The abundance ratios for all proteins were distinguished by taxa and plotted on a log2 scale (Figure 3). For several low abundance organisms, average protein distributions were shifted to either high or low pH, suggesting a growth advantage or strong response to the respective pH condition (Figure 3).
Figure 2.
Solution pH of replicate laboratory reactors. After biofilm establishment at ∼pH 1.4, three 15N-labeled reactors were lowered to an average pH of 0.85 (day 0). pH treatments were maintained over a 19-day period, after which communities were sampled for proteomic analyses. Fresh medium was re-supplied on days 5, 9, 13 and 16.
Table 1. Summary of quantified proteins for community comparisons at high and low pH.
| Replicate 1 | Replicate 2 | Replicate 3 | Shared | |
|---|---|---|---|---|
| Leptospirillum group II | 1050 | 916 | 875 | 946 |
| Leptospirillum group III | 201 | 563 | 429 | 310 |
| G-plasma | 315 | 306 | 266 | 288 |
| Ferroplasma type II | 161 | 190 | 190 | 162 |
| Ferroplasma type I | 143 | 222 | 145 | 160 |
| A-plasma | 67 | 64 | 50 | 56 |
| E-plasma | 57 | 60 | 50 | 45 |
| I-plasma | 26 | 29 | 21 | 21 |
| Arman type II | 7 | 15 | 2 | 5 |
| Actinobacteria | 52 | 40 | 37 | 37 |
| Firmicutes | 11 | 9 | 8 | 7 |
| Plasmid | 8 | 5 | 9 | 2 |
| Virus | 14 | 6 | 3 | 1 |
| Total | 2112 | 2425 | 2085 | 2148 |
For each proteomic replicate, the number of proteins quantified for each taxa is shown. The shared column indicates the number of proteins that were detected in at least two replicates and for which average log2 ratios were calculated. Uncharacterized archaeal Thermoplasmatales lineage organisms are designated by letter abbreviations (A, E, G and I-plasma). Arman type II is an archaeal lineage distinct from the Thermoplasmatales.
Figure 3.
Distribution of average protein abundances for quantitative proteomic comparison of communities maintained at high and low pH. Log2 ratios for all proteins are plotted according to taxa. Positive log2 ratio values indicate proteins more abundant in high pH communities; negative values indicate proteins more abundant in low pH communities. The shaded area denotes values between 1 and −1 (within 2 × ), in which proteins are not considered significantly different between high and low pH comparisons. Red boxes highlight proteins from organisms that exhibited a strong response to either high or low pH. The color reproduction of this figure is available on the html full text version of the manuscript.
The strongest response to pH was associated with Leptospirillum group III. The majority of the 418 Leptospirillum group III proteins identified in at least two of the three replicates were more abundant in the high pH communities (Figure 3). Cell counts using fluorescence in situ hybridization indicated this organism made up approximately 6.6±1.0% of the total cells in high pH communities, compared with approximately 2.4±0.9% of the low pH communities. In part, the higher abundance of Leptospirillum group III proteins reflects its greater representation in pH 1.45. However, many Leptospirillum group III proteins were much more abundant, which cannot be accounted for by differences in cell numbers (that is, >3 × ). Specific proteins from all functional groups were many times more abundant in high pH samples, including proteins that carry out core metabolic processes such as energy conversion, carbohydrate metabolism, transcription, translation, and nucleotide and amino acid biosynthesis (Supplementary Table 3). Only four proteins from this organism were significantly more abundant in low pH conditions (Supplementary Table 6). In addition to greater cell abundance, the proteomic response associated with this organism suggests greater metabolic activity in the higher pH conditions.
Only 30 of 946 Leptospirillum group II proteins were significantly different between the two pH conditions. Analysis of functional categories indicated that amino acid transport and metabolism, nucleotide transport and metabolism, and cell wall/membrane/envelope biogenesis were overrepresented at high pH (Supplementary Table 4). Only the carbohydrate transport and metabolism category was overrepresented at low pH (Supplementary Table 4). Specific Leptospirillum group II proteins that were more abundant at high pH included a polysaccharide export protein, and uridine diphosphate-glucuronate 5′-epimerase (a protein putatively involved in the metabolism of sugar components of the cell wall), a periplasmic phosphate binding protein, an important energy conversion cytochrome, an NADH oxidase and two signal transduction proteins (Guanosine-5′-triphosphate pyrophosphokinase and diguanylate cyclase) (Supplementary Table 5). Proteins from this organism that were significantly more abundant at low pH included a protein that catalyzes the first step in the synthesis of the compatible solute ectoine, diaminobutyrate-2-oxoglutarate transaminase (Supplementary Table 6). Many others were classified as hypothetical or general function only (Supplementary Table 6).
Several of the low abundance organisms' log2 protein distributions were offset from zero, indicating a bias or strong response to either the low or high pH environment (Figure 3). A high number of proteins identified from the archaeal Thermoplasmatales organism G-plasma were more abundant in low pH communities. These included ribosomal proteins, enzymes involved in energy conversion reactions, as well as carbohydrate transport proteins (Supplementary Table 6). A similar trend was observed for Ferroplasma type II (Figure 3). Many of the Ferroplasma type II proteins were classified as hypothetical, however, several TCA cycle proteins and the purine biosynthesis protein adenylosuccinate synthase were several times more abundant at low pH (Supplementary Table 6). Few proteins from G-plasma or Ferroplasma type II were significantly more abundant in high pH communities (Supplementary Table 5).
In contrast to both G-plasma and Ferroplasma type II, Ferroplasma type I had a more even distribution of proteins that were significantly more abundant in either condition (7 proteins greater than 2 × more abundant in low pH vs 12 in high pH; Supplementary Tables 5 and 6). At high pH, abundant proteins included several hypothetical proteins, an amino acid and multidrug transporter, a ribosomal protein and a peroxiredoxin (Supplementary Table 5). At low pH, abundant Ferroplasma type I proteins included those belonging to energy conversion pathways (succinate dehydrogenase/fumarate reductase, aerobic-type carbon monoxide dehydrogenases, NAD-dependent aldehyde dehydrogenase), similar to the response of Ferroplasma type II (Supplementary Table 6).
For low abundance organisms for which few proteins were detected (A-plasma, E-plasma, I-plasma, Arman type II, Actinobacteria, Firmicutes; Table 1), bias to either the low or high pH condition was ambiguous, and metabolic responses could not be interpreted (Figure 3). However, several extremely abundant proteins (fold change >40 × ) in the high pH conditions were identified for both archaeal taxa A-plasma and E-plasma, as well as the Actinobacteria, and are listed in Supplementary Table 5.
Phage protein detection
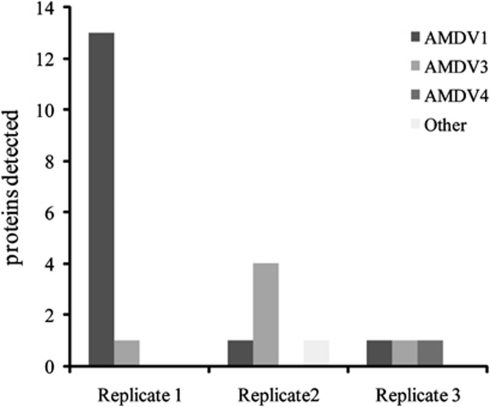
Included in the replicate proteomic datasets are 23 proteins encoded by genomes previously reconstructed for phage of bacteria and viruses of archaea. Surprisingly, the number of phage proteins identified was highly variable across the three biological replicate datasets (each comprising a mixture of labeled and unlabeled proteins). Specifically, 13 proteins from AMDV1 (a virus population that targets Leptospirillum groups II and III) were identified in replicate 1, whereas replicates 2 and 3 only had a single protein each (Figure 4).
Figure 4.
Viral proteins detected in replicate proteomic analyses used to compare communities in high and low pH solutions. The number of proteins detected for putative virus types AMDV1, AMDV3, AMDV4 and unclassified viral proteins (other) are shown for each replicate.
Discussion
Laboratory selection of near homogeneous Leptospirillum group II strain population
Cultivation of AMD biofilm communities in laboratory reactors resulted in selection for a Leptospirillum group II strain similar to that characterized as Type I (Denef et al., 2009). Low frequency detection of Type VI (UBA genomic-type) peptides identified from proteins encoded throughout the genome could signify the presence of additional strain types at low abundance (Figure 1). Alternatively, Type VI peptides may represent other small recombination points in the Type Ib genome.
Despite inoculation of laboratory reactors with a sample likely containing several Leptospirillum group II genotypes, only the Type Ib strain proliferated. Although this strain has not been detected in environmental samples to date, we hypothesize that it may have been present in environmental samples used in original reactor inoculations, but at relatively low abundance levels that prevented detection in previous PIGT analyses (Denef et al., 2009). However, an alternative explanation is that Type Ib arose by recombination between Type I and a UBA-group genotype (Types II–VI) in the laboratory communities. The existence of new recombinant types (either introduced from the environment or formed in the laboratory) is not surprising, given that Simmons et al. (2008) detected a variety of low abundance recombination points involving the 5-way CG and UBA Leptospirillum group II genomes in the original metagenomic dataset (Tyson et al., 2004).
Laboratory cultivation imposed a selective condition that allowed a normally low abundance strain type to reach high cell densities. Selection might be associated with one or a subset of proteins encoded on the recombinant Type VI block. Abundance ratios indicated approximately equal proportions of these proteins in both high and low pH datasets indicating pH did not influence their expression. It is likely that selection of the Type Ib strain was because of other factors associated with laboratory cultivation. Other proteins not identified because of the lack of the reference sequence in the proteomic database may have also been important for laboratory selection of a single strain. Regardless, selection of a single genotype in laboratory biofilms is consistent with specific functional differences between recombinant genotypes of Leptospirillum group II in environmental samples (Denef et al., 2010a).
Response of Leptospirillum spp. to high and low solution pH
Based on higher cell counts and a strong protein distribution bias to high pH for Leptospirillum group III (Figure 3), solution pH may be a significant factor that affects the distribution and metabolic activity of Leptospirillum group III in natural communities. Inhibition at low pH may contribute to the low abundance of Leptospirillum group III in environmental biofilms (Wilmes et al., 2008b; Belnap et al., 2010), where the solution pH is generally less than 1.0. As this organism has been shown to fix nitrogen and may be considered a keystone species (Tyson et al., 2005), its distribution in natural AMD communities may influence growth and metabolism of other community members.
The majority of Leptospirillum group II proteins had similar abundance between the two pH conditions (Figure 3), likely indicating comparable growth in a broad pH range. However, specific proteins and analysis of functional categories indicated Leptospirillum group II responded differently to pH 1.45 and 0.85. The functional categories distributions that were overrepresented in high pH may suggest increased metabolic activity in this condition. Increased metabolic activity is also supported by the high abundance of cytochrome 572 (Supplementary Table 5), a protein involved in iron oxidation (Jeans et al., 2008). Additionally, a periplasmic phosphate binding protein more abundant in high pH (Supplementary Table 5), possibly expressed as a response to phosphate limitation, could also be a result of increased metabolic processes (for example, energy conversion and nucleotide biosynthesis). An abundant polysaccharide export protein at high pH (Supplementary Table 5) suggests increased biopolymer production and secretion; functions that could enhance biofilm formation. Biofilm formation could have a profound impact on many other AMD taxa that colonize, and likely metabolize, exopolysaccharides generated by Leptospirillum group II.
An abundant Leptospirillum group II protein at low pH, diaminobutyrate-2-oxoglutarate transaminase (EctB) (Supplementary Table 6), is putatively involved in the first step of ectoine synthesis (Louis and Galinski, 1997). Production of ectoine, a compatible solute potentially used to balance osmotic forces in ion-rich solutions, suggests greater osmotic stress during growth in more acidic solutions.
Functional characterization of low abundance taxa
Low abundance Thermoplasmatales lineage archaea and several non-Leptospirillum bacteria have been predicted to be heterotrophs (Tyson et al., 2004). It is probable that specific organisms occupy specific niches, which can be defined by both geochemical and biological factors. Utilization of a laboratory community culturing system enabled the manipulation of one specific geochemical parameter, to which the low abundance organisms had varying response (Figure 3). Although definitive functional assessment of low abundance organisms is limited by shallow proteomic sampling, manipulation of pH has provided a glimpse of potential ecological distinctions among these organisms.
Comparison of community growth at high and low pH resulted in a strong response at low pH for both G-plasma and Ferroplasma type II. The abundance of energy conversion proteins (for both organisms) as well as ribosomal proteins, a sugar transporter, and an amino acid biosynthesis protein (for G-plasma) probably indicate a growth preference in this condition (Supplementary Table 6). Low solution pH has been suggested previously to select for higher proportions of archaea in Richmond Mine communities (Bond et al., 2000a), possibly because of their highly proton resistant tetra-ether-linked membranes (Macalady et al., 2004). Passive resistance to a highly acidic external environment may provide an energetic advantage compared with bacterial species, such as Leptospirillum group III. Interestingly, the response of Ferroplasma type I was not identical to the response of Ferroplasma type II (Figure 3), despite their close phylogenetic relationship (Eppley et al., 2007). Several Ferroplasma type I proteins abundant at high pH were indicative of growth and respiration (Supplementary Table 5). Although the laboratory isolate Ferroplasma acidarmanus, a representative of the Ferroplasma type I group, has been shown to grow in solution pH ranging from 0 to 1.5 (Edwards et al., 2000), growth of this organism in batch culture is inhibited at pH below ∼0.7 (data not shown). Within environmental biofilms, it is perhaps the case that Ferroplasma types I and II are partitioned along pH gradients, effectively minimizing competition.
Despite abundant proteins in high or low pH conditions, interpretation of the metabolism of other low abundance taxa is difficult. Extremely abundant proteins in high pH for A-plasma, E-plasma, and the Actinobacteria do not necessarily indicate a growth preference or stress response. However, an Actinobacteria cholesterol biosynthesis protein (phytoene squalene synthetase) that was extremely abundant at high pH (Supplementary Table 5) may suggest these organisms respond to pH changes by altering membrane structure.
Variation of phage protein abundance in replicate communities
Despite technical difficulties associated with identification of viral proteins (that is, incomplete viral metagenomic data and rapid mutation of viral genes), many were identified. The total number of community proteins detected in all three replicates was similar (Table 1), indicating the differences in protein counts for AMDV1 are not the result of technical variation. As reactors were given identical starting inoculum, all should have received approximately equal numbers of viral particles or cells infected with AMDV1. The high abundance of AMDV1 proteins in a single replicate is suggestive of a specific viral bloom. These results may indicate a stochastic nature of viral infection that occurs on relatively small scales (that is, within individual laboratory reactors). Increased detection of viral proteins would necessitate lytic infection of host cells. Intriguingly, approximately half the number of Leptospirillum group III proteins were detected in proteomic replicate 1 compared with both replicates 2 and 3 (Table 1). It is possible, this discrepancy is related to an AMDV1 outbreak, as AMDV1 is predicted to target Leptospirillum group III (Andersson and Banfield, 2008). However, a direct response to phage infection is not clear from Leptospirillum group III proteomic data.
Conclusion
In the current study, we utilized laboratory growth of multi-species biofilm communities to examine both membership and functional response to a specific geochemical perturbation. The findings indicate differential responses of community members to solution pH, and help to define the ecological niches of closely associated bacteria and archaea. The propagation of a single Leptospirillum group II genotype in laboratory communities supports environmental observations for functional partitioning of Leptospirillum group II strains (Denef et al., 2010a), and indicates environmental heterogeneity in natural microbial systems may allow for maintenance of variant alleles that originate through mutation or recombination and confer niche adaptations.
Acknowledgments
Mr Ted Arman (President, Iron Mountain Mines) and Dr Richard Sugarek are thanked for site access and other assistance. Dr Paul Wilmes and Dr Ryan Mueller are thanked for helpful discussions. This research was supported by the US Department of Energy, Office of Biological and Environmental Research through the Genomic Science Program (contract numbers DE-FG02-05ER64134, DE-SC0004665 and DE-SC0004918), Office of Advanced Scientific Computing Research through the SciDAC Program, the NSF Biocomplexity Program and the NASA Astrobiology Institute.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amann RI. Fluorescently labelled, rRNA-targeted oligonucleotide probes in the study of microbial ecology. Mol Ecol. 1995;4:543–554. [Google Scholar]
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Belnap CP, Pan C, VerBerkmoes NC, Power ME, Samatova NF, Carver RL, et al. Cultivation and quantitative proteomic analyses of acidophilic microbial communities. ISME J. 2010;4:520–530. doi: 10.1038/ismej.2009.139. [DOI] [PubMed] [Google Scholar]
- Bond PL, Banfield JF. Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb Ecol. 2001;2:149–161. doi: 10.1007/s002480000063. [DOI] [PubMed] [Google Scholar]
- Bond PL, Druschel GK, Banfield JF. Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl Environ Microbiol. 2000a;66:4962–4971. doi: 10.1128/aem.66.11.4962-4971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond PL, Smriga SP, Banfield JF. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl Environ Microbiol. 2000b;66:3842–3849. doi: 10.1128/aem.66.9.3842-3849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Wegley L, Leeds S, Schoenfeld T, Rohwer F. Phage community dynamics in hot springs. Appl Environ Microbiol. 2004;70:1633–1640. doi: 10.1128/AEM.70.3.1633-1640.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danovaro R, Dell'Anno A, Corinaldesi C, Magagnini M, Noble R, Tamburini C, et al. Major viral impact on the functioning of benthic deep-sea ecosystems. Nature. 2008;454:1084–1087. doi: 10.1038/nature07268. [DOI] [PubMed] [Google Scholar]
- Denef VJ, Kalnejais LH, Mueller RS, Wilmes P, Baker BJ, Thomas BC, et al. Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc Natl Acad Sci. 2010a;107:2383–2390. doi: 10.1073/pnas.0907041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef VJ, Mueller RS, Banfield JF. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 2010b;4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- Denef VJ, VerBerkmoes NC, Shah MB, Abraham P, Lefsrud M, Hettich RL, et al. Proteomics-inferred genome typing (PIGT) demonstrates inter-population recombination as a strategy for environmental adaptation. Environ Microbiol. 2009;11:313–325. doi: 10.1111/j.1462-2920.2008.01769.x. [DOI] [PubMed] [Google Scholar]
- Druschel G, Baker B, Gihring T, Banfield J. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochemical Trans. 2004;5:13. doi: 10.1186/1467-4866-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KJ, Bond PL, Gihring TM, Banfield JF. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science. 2000;287:1796–1799. doi: 10.1126/science.287.5459.1796. [DOI] [PubMed] [Google Scholar]
- Edwards KJ, Gihring TM, Banfield JF. Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl Environ Microbiol. 1999;65:3627–3632. doi: 10.1128/aem.65.8.3627-3632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Eppley JM, Tyson GW, Getz WM, Banfield JF. Genetic exchange across a species boundary in the archaeal genus Ferroplasma. Genetics. 2007;177:407–416. doi: 10.1534/genetics.107.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Bohannan BJM, Whitaker RJ. Microbial Biogeography: From Taxonomy to Traits. Science. 2008;320:1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- Jeans C, Singer SW, Chan CS, VerBerkmoes NC, Shah M, Hettich RL, et al. Cytochrome 572 is a conspicuous membrane protein with iron oxidation activity purified directly from a natural acidophilic microbial community. ISME J. 2008;2:542–550. doi: 10.1038/ismej.2008.17. [DOI] [PubMed] [Google Scholar]
- Jessup CM, Kassen R, Forde SE, Kerr B, Buckling A, Rainey PB, et al. Big questions, small worlds: microbial model systems in ecology. Trends in Ecology & Evolution. 2004;19:189–197. doi: 10.1016/j.tree.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Johnson DB. Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiology Ecology. 1998;27:307–317. [Google Scholar]
- Kaeberlein T, Lewis K, Epstein SS. Isolating ‘uncultivable' microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- Kassen R, Rainey PB. The ecology and genetics of microbial diversity. Ann Rev Microbiol. 2004;58:207. doi: 10.1146/annurev.micro.58.030603.123654. [DOI] [PubMed] [Google Scholar]
- Lo I, Denef VJ, VerBerkmoes NC, Shah MB, Goltsman D, DiBartolo G, et al. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature. 2007;446:537–541. doi: 10.1038/nature05624. [DOI] [PubMed] [Google Scholar]
- Louis P, Galinski EA. Characterization of genes for the biosynthesis of the compatible solute ectoine from Marinococcus halophilus and osmoregulated expression in Escherichia coli. Microbiology. 1997;143:1141–1149. doi: 10.1099/00221287-143-4-1141. [DOI] [PubMed] [Google Scholar]
- Macalady JL, Vestling MM, Baumler D, Boekelheide N, Kaspar CW, Banfield JF. Tetraether-linked membrane monolayers in Ferroplasma spp: a key to survival in acid. Extremophiles. 2004;8:1433–4909. doi: 10.1007/s00792-004-0404-5. [DOI] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Micro. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Pan C, Kora G, McDonald WH, Tabb DL, VerBerkmoes NC, Hurst GB, et al. ProRata: A quantitative proteomics program for accurate protein abundance ratio estimation with confidence interval evaluation. Anal Chem. 2006a;78:7121–7131. doi: 10.1021/ac060654b. [DOI] [PubMed] [Google Scholar]
- Pan C, Kora G, Tabb DL, Pelletier DA, McDonald WH, Hurst GB, et al. Robust estimation of peptide abundance ratios and rigorous scoring of their variability and bias in quantitative shotgun proteomics. Anal Chem. 2006b;78:7110–7120. doi: 10.1021/ac0606554. [DOI] [PubMed] [Google Scholar]
- Prosser JI, Bohannan BJM, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, et al. The role of ecological theory in microbial ecology. Nat Rev Micro. 2007;5:384–392. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC, II, et al. Community proteomics of a natural microbial biofilm. Science. 2005;308:1915–1920. [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado A-B, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Micro. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- Simmons SL, DiBartolo G, Denef VJ, Goltsman DSA, Thelen MP, Banfield JF. Population genomic analysis of strain variation in Leptospirillum Group II bacteria involved in acid mine drainage formation. PLoS Biology. 2008;6:e177. doi: 10.1371/journal.pbio.0060177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., III DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, et al. Genotypic diversity within a natural coastal bacterioplankton population. Science. 2005;307:1311–1313. doi: 10.1126/science.1106028. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Lo I, Baker BJ, Allen EE, Hugenholtz P, Banfield JF. Genome-directed isolation of the key nitrogen fixer Leptospirillum ferrodiazotrophum sp. nov. from an acidophilic microbial community. Appl Environ Microbiol. 2005;71:6319–6324. doi: 10.1128/AEM.71.10.6319-6324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes P, Andersson AF, Lefsrud MG, Wexler M, Shah M, Zhang B, et al. Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. ISME J. 2008a;2:853–864. doi: 10.1038/ismej.2008.38. [DOI] [PubMed] [Google Scholar]
- Wilmes P, Remis JP, Hwang M, Auer M, Thelen MP, Banfield JF. Natural acidophilic biofilm communities reflect distinct organismal and functional organization. ISME J. 2008b;3:266–270. doi: 10.1038/ismej.2008.90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.