Abstract
Lactobacillus reuteri strain 100-23 together with a Lactobacillus-free mouse model, provides a system with which the molecular traits underpinning bacterial commensalism in vertebrates can be studied. A polysaccharide was extracted from sucrose-containing liquid cultures of strain 100-23. Chemical analysis showed that this exopolysaccharide was a levan (β-2, 6-linked fructan). Mutation of the fructosyl transferase (ftf) gene resulted in loss of exopolysaccharide production. The ftf mutant was able to colonise the murine gastrointestinal tract in the absence of competition, but colonisation was impaired in competition with the wild type. Biofilm formation by the mutant on the forestomach epithelial surface was not impaired and the matrix between cells was indistinguishable from that of the wild type in electron micrographs. Colonisation of the mouse gut by the wild-type strain led to increased proportions of regulatory T cells (Foxp3+) in the spleen, whereas colonisation by the ftf mutant did not. Survival of the mutant in sucrose-containing medium was markedly reduced relative to the wild type. Comparison of the genomic ftf loci of strain 100-23 with other L. reuteri strains suggested that the ftf gene was acquired by lateral gene transfer early in the evolution of the species and subsequently diversified at accelerated rates. Levan production by L. reuteri 100-23 may represent a function acquired by the bacterial species for life in moderate to high-sucrose extra-gastrointestinal environments that has subsequently been diverted to novel uses, including immunomodulation, that aid in colonisation of the murine gut.
Keywords: Lactobacillus reuteri, exopolysaccharide, survival, regulatory T cells, evolution
Introduction
Vertebrates are colonised by complex microbial communities that influence host resistance to infections, as well as host nutrition and immunological development (Backhed et al., 2005). The phylogeny of bacterial communities of vertebrates has been studied extensively (Ley et al., 2008) but autecological studies of commensals to reveal the molecular traits that are necessary for autochthony are less advanced.
Lactobacillus reuteri 100-23 populates the surface of the forestomach epithelium of mice and is present in the digesta throughout the remainder of the digestive tract. This strain, together with Lactobacillus-free mice, provides a model system by which molecular aspects of bacterial commensalism can be studied (Tannock, 2004). A high molecular weight surface-protein adhesin, acid-resistance through D-alanylation of teichoic acids and enzymic reversal of methionine oxidation in proteins have already been revealed as facilitators of colonisation for strain 100-23 (Walter et al., 2005, 2007). Mice previously naïve with respect to exposure to lactobacilli develop innate immune tolerance to the presence of strain 100-23 in the bowel (Hoffman et al., 2008). This tolerance is associated with increased proportions of regulatory T cells (Foxp3+) in the mesenteric lymph nodes and spleens of mice colonised with L reuteri 100-23 relative to noncolonised animals (Livingston et al., 2009). The molecular mechanisms by which this tolerance is achieved are unknown.
Extracellular polysaccharide (exopolysaccharide) production by lactic acid bacteria such as lactobacilli affects the rheological and textural properties of fermented foods (Korakli et al., 2001; Tieking et al., 2003). Exopolysaccharides may also be important components of ‘functional' foods because they might interact with the immune system of consumers (Vinderola et al., 2006), or act as prebiotics in the bowel (Bello et al., 2001). Some lactobacilli produce exopolysaccharides in the form of glucans or fructans from sucrose through the action of glycosyltransferases (Tieking et al., 2005). Exopolysaccharides are important in the colonisation of dental surfaces by streptococci (Banas and Vickerman, 2003) but their relevance to the ecology of lactobacilli that inhabit the gastrointestinal biome has received little attention.
Exopolysaccharide produced by L. reuteri 100-23 could influence colonisation of the murine gut in several ways. The results of experiments to characterize the exopolysaccharide, and to test the possible effects of exopolysaccharide production on the ability of L. reuteri 100-23 to inhabit the gut of mice, are reported in this article.
Materials and methods
Extraction of exopolysaccharide from cultures
L. reuteri 100-23 (DSM 17509) was cultured using Lactobacilli MRS medium (Difco, Becton Dickinson Co., Sparks, MD, USA), which was modified to contain 5% sucrose (w/v), whereas glucose was omitted. Cultures were incubated anaerobically at 37 °C. Genetic manipulations described in this report utilised a plasmid-free derivative, 100-23C (Heng et al., 1999). Exopolysaccharide was extracted from supernatants obtained by centrifugation of liquid MRS cultures that had been incubated for 48 h at 37 °C. Yeast extract, which contains mannans that coprecipitate with polysaccharide, was omitted from the MRS medium. An equal volume of cold ethanol (−20 °C) was added to the supernatant and held at 4 °C for 48 h. The precipitate was harvested by centrifugation (12 000 × g, 30 min, 5 °C) and dissolved in water. The solution was centrifuged again and the supernatant retained. The polysaccharide was re-precipitated with an equal volume of ethanol at 4 °C during a further 24 h. The harvested precipitate (centrifugation as above) was dissolved in water, re-precipitated, dissolved in water, and then dialysed (12 000–14 000 mw cutoff) against water for 72 h with twice daily changes of water. Proteins were removed by addition of proteinase K (6 mg per ml) to exopolysaccharide solution and incubation for 24 h at 37 °C. Peptides were precipitated using 20% trichloroacetic acid for 1 h on ice, followed by centrifugation at 14 365 × g for 10 min, 5 °C. The supernatant was then dialysed against water at 4 °C for 72 h. The dialysate was lyophilised and stored at 4 °C. Protein content was measured using the Bradford assay according to the manufacturer's instructions (Bio-Rad, Hercules, CA, USA).
Measuring the yield of exopolysaccharide
The yield of exopolysaccharide was obtained by culture of strain 100-23 in 500 ml of Lactobacilli MRS-sucrose medium for 48 h, after which bacterial cells and supernatant were collected after centrifugation of the culture. Exopolysaccharide was obtained as described above and the bacterial cells were lyophilised. The weights of dried polysaccharide and cells were recorded and the yield (grams of exopolysaccharide per gram of cells) was calculated.
Chemical analysis of exopolysaccharide
The procedures used to analyse exopolysaccharide included colorimetry, fructanase digestion, size exclusion high performance liquid chromatography, high performance anion exchange chromatography (HPAEC), gas chromatography, mass spectrometry and nuclear magnetic resonance spectrometry. The materials and methods used in these analyses are described in the Supplementary Material.
Characterisation and mutagenesis of the genomic fructosyltransferase (ftf) locus
The Integrated Microbial Genomes genome browsing system (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi) was used to characterize the ftf locus in L. reuteri 100-23 and perform genomic comparisons. Tests of selection (Z-test: Nei-Gojobori method) were performed using the MEGA4 software after generating ClustalW alignments of conserved regions of ftf and inu genes of L. reuteri. dN/dS ratios (the ratios of nonsynonymous to synonymous polymorphisms) were calculated using the Nei and Gojobori method using START2 software (Jolley et al., 2001). Ftf gene sequences used in these comparisons had been obtained from L. reuteri strains by PCR, genomic alignments and sequencing of specific loci (van Hiijum et al., 2002; Tieking et al., 2003, 2005).
To inactivate ftf of strain 100-23C, an internal region of the gene was amplified by PCR with primers 71010 ins for (5′-TCACGCGGATCCTAACCAAGTTCATCGGCTTCTTC-3′) and 71010 ins rev (5′-TCACCGGAATTCCTCGTGTATATGCTGCATCC-3′) and cloned into pORI28 by directional cloning as described by Russell and Klaenhammer (2001), resulting in plasmid pORIdftf. Insertional mutagenesis of ftf was achieved by site-specific integration of pORIdftf into the chromosome of L. reuteri 100-23C using the temperature-sensitive plasmid pVE6007 as a helper plasmid as described previously (Walter et al., 2005). The correct integration of pORI28 was confirmed by PCR using primers 71010 test for (5′-CTCTACTGCTGCGCTGGTA-3′) and 71010 test rev (5′-CATGGTGTGGTCGTCCCA-3′) flanking the target region. In vitro growth curves of Lactobacilli MRS cultures of the wild type and mutant did not differ (data not shown).
Colony morphologies of the wild type (100-23, 100-23C) and the ftf mutant were compared after 48 h incubation on Lactobacilli MRS agar plates in which glucose was replaced with sucrose (2% w/v).
Lactobacillus-free mice
Lactobacillus-free mice aged 6-weeks old were used in animal experiments. The colony of BALB/c mice were maintained in gnotobiotic isolators as described previously (Tannock et al., 1988). Lactobacilli are absent from the gut of the animals but a complex microbiota consisting of about 600 bacterial phylotypes is present in the caecum (Tannock, 2009). The mice therefore provide animal hosts that are naïve with respect to lactobacilli but whose tissues have been conditioned by exposure to a complex microbiota. The constituents of the sterile diet fed to the animals are given in Supplementary Table S1.
Analysis of mouse stomach contents
Sucrose concentrations in stomach contents were measured using high performance liquid chromatography as described in the Supplementary Material. Detection of exopolysaccharide in stomach contents was by HPAEC (Supplementary Material).
Experiments with Lactobacillus-free mice
Colonisation of the gut of 6-week old Lactobacillus-free mice was accomplished as described previously (Walter et al., 2005). Briefly, mice were inoculated by gavage on a single occasion with about 106 lactobacilli per dose. To measure in vivo competition between wild-type and mutant strains, mice were inoculated with a 1:1 mixture of the ftf mutant and wild-type (100-23C) cultures. The animals were killed and samples were taken for culture 7 days after inoculation. To determine colonisation ability, mice inoculated with a pure culture of the ftf mutant were examined 14 days after inoculation. The Lactobacillus populations present in the forestomach and cecum were quantified by plating dilutions of organ homogenates on Rogosa SL agar plates (Difco) that were then incubated at 37 °C for 48 h. Differential counts on Rogosa SL agar with or without erythromycin (5 μg ml−1) were used to determine the proportion formed by the ftf mutant of the total Lactobacillus population in the competition experiment, and to measure in vivo stability of the mutation.
Biofilm structure was examined in sections prepared from forestomachs fixed in ruthenium red-glutaraldehyde solution and observed by transmission electron microscopy, as described previously (Walter et al., 2007). The forestomachs were examined 7 days after inoculation with either 100-23 (two mice) or the ftf mutant (three mice).
Proportions of Foxp3-positive cells in spleens
Lactobacillus-free mice were inoculated (∼106 Lactobacillus cells) by gavage on a single occasion at 6 weeks of age with L. reuteri 100-23C or the ftf mutant. Inoculated mice and matched noncolonised animals were killed 3 weeks later. Colonisation by L. reuteri 100-23 was confirmed by culture of caecal contents at necropsy. Spleens were removed at necropsy and used for Foxp3 measurements. Single-cell suspensions were prepared from spleens and labelled with antibodies to CD4, CD25 and Foxp3 using a kit supplied by eBioScience (San Diego, CA, USA) following the manufacturer's instructions. Briefly, aliquots of cells (∼1 × 106) were incubated with FITC-labeled CD4 antibody together with PE-labeled CD25 antibody or corresponding isotype controls. The cells were washed and permeabolised, washed again, then incubated with Fc-blocking antibody. PE-labeled Foxp3 antibody or PE-labeled isotype antibody was added. Following incubation, the cells were washed and analysed by flow cytometry (FACSCalibur, BD Biosciences, San Jose, CA, USA).
Survival experiments in high-sucrose medium
A volume of 10 ml of MRS medium containing 5% sucrose or 5% glucose, in triplicate, were inoculated with 100 μl of overnight culture of wild-type 100-23C or isogenic ftf mutant and incubated for 48 h, anaerobically at 37 °C. The cultures were then placed in an incubator at 25 °C and held for a total period of 8 days during which time CFU per ml measurements were made in duplicate for each culture every 2 days using diluted culture aliquots and Lactobacilli MRS agar plates. Turbidities of wild-type and mutant cultures were similar (A600 2.7 and 2.9, respectively), as were the pH of culture superanatants (both 5.1).
Determination of proton motive force (ΔpH)
For the determination of ΔpH in cultures during survival experiments (see above at pH 5.1), cells (3 × 1 ml samples) were taken directly from the culture and added to glass tubes containing one of the following: benzoate (11 μ, pKa=4.2), 3H-taurine (50 μ; 5–30 Ci mmol−1) and 3H-water (1 m; 25 mCi g−1). Taurine was not metabolised by L. reuteri 100-23 and was used to determine the intracellular volume. After incubation for 10 min at 37 °C, the cultures were centrifuged in 1.5 ml microcentrifuge tubes (13 000 × g, 5 min, 22 °C) containing 350 μl of a 40% (v/v) mixture of phthalic acid bis (2-ethyl-hexyl ester) and 60% silicone oil (40% part mixture of DC200/200 silicone oil and 60% DC 550), which had been left overnight in an anaerobic chamber at room temperature to equilibrate), then 20 μl samples of supernatant were removed. The tubes and contents were frozen (−70 °C), and cell pellets were retained. Supernatant and cell pellets were added to scintillation fluid, and the radioactivity was measured by using a scintillation counter (LKB Wallac 1214 Rackbeta). In a second set of experiments, the ΔpH was determined on overnight cultures that were harvested by centrifugation (8000 × g, 20 min, 4 °C) and washed twice in 100 m potassium citrate–phosphate buffer. Cells were suspended to a final A600 of 1.0 at the external pH being studied (pH 3.0–7.0) and a volume of 5 ml, energised for 20 min with 15 mM arginine (final concentration), and the ΔpH determined as above. The intracellular volume (3.1±0.4 μl mg−1 of protein) was estimated from the difference between the partitioning of 3H-water and 3H-taurine. The ΔpH was determined from the distribution of benzoate using the Henderson–Hasselbalch equation (Riebeling et al., 1975), and ZΔpH was calculated as 62 mV × ΔpH. Protein from NaOH-hydrolyzed cells (0.2 M NaOH, 100 °C, 20 min) was assayed by the method of Markwell et al., 1978.
Statistical comparisons and ethical approvals
The Mann–Whitney nonparametric test was used to make statistical comparisons. Animal experiments were conducted with approval from the University of Otago Animal Ethics Committee (approval numbers 32/07and 2/09).
Results
Yield of exopolysaccharide from growth in sucrose-containing medium
Strain 100-23 produced a measure of 1.038 g of exopolysaccharide in culture supernatant per gram of bacterial cells (dry weights).
Chemical analysis of exopolysaccharide
Total sugar analysis showed that the putative exopolysaccharide contained 96.8% w/w carbohydrate. The fructose content was 98.7% w/w, indicating that the exopolysaccharide was comprised predominantly of fructose. Protein content was less than 1% w/w.
Size exclusion chromatography (SEC) (Supplementary Figure S1A) showed a peak eluting between 25 and 29 ml that represented approximately 85% of the eluted material as judged by refractive index. This peak was absent in fructanase-treated samples and the polymer was thus identified as a fructan. The weight-average molecular weight of this fructan was 5.7 kDa by comparison with pullulan standards and 7.7 kDa by light scattering. HPAEC showed a series of peaks eluting from 4 to >40 ml, indicating the presence of one major and one minor series of oligosaccharides ranging in size from degree of polymerisation about 3 to >40 (Supplementary Figure S1C). Again, the absence of peaks when fructanase digested samples were chromatographed showed that the observed peaks represented fructan. The broad peak at about 33 min corresponds to a degree of polymerisation of about 35, giving an estimated molecular weight of 5.6 kDA in good agreement with SEC data. For comparison, separation of chicory inulin (Cichorium intybus), that contains predominantly a linear series β-2,1-linked fructofuranosyl residues (degree of polymerisation about 3–50), is shown (Supplementary Fig. S1B).
Glycosyl linkage analysis with acid hydrolysis at 50 °C gave mostly 2,6-linked Fruf together with smaller amounts of terminal and 2,1,6-linked Fruf. This identified the fructan as a levan (Table 1).
Table 1. Glycosyl linkage composition of Lactobacillus reuteri exopolysaccharide.
Terminal Fruf deduced from 2,5-di-O-acetyl-(2-deuterio)-1,3,4,6-tetra-O-methylhexitol and so on.
Average of duplicate determinations.
The 13C-nuclear magnetic resonance spectrum of the exopolysaccaharide showed six sharp resonances that were deduced to correspond to the C-1 to C-6 of the → 6)-D-Fruf-β-(2 → residues observed in the glycosyl linkage analysis (Supplementary Figure S1F). Similarly, the 1H-nuclear magnetic resonance spectrum showed resonances that were deduced to correspond to H-1–H-6 of the same residues (Supplementary Figure S1E) The anomeric region of the 1H-nuclear magnetic resonance spectrum did not show any major resonances as expected for the presence of a ketose sugar (that is, fructose). A minor resonance at 5.23 p.p.m. was deduced to correspond to H-1 of an α-D-Glcp that are terminal residues of fructans. Analysis by 2D HSQC-COSY (data not shown) showed cross-peaks that enabled complete assignment of the fructose residues of the exopolysaccharide (Table 2).
Table 2. Summary of 13C and 1H NMR chemical shifts (δ, p.p.m.) for Lactobacillus reuteri exopolysaccharide.
| Residue |
Chemical shifts (p.p.m.) |
|||||
|---|---|---|---|---|---|---|
| C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | |
| H-1 | H-2 | H-3 | H-4 | H-5 | H-6/6′ | |
| → 6)-D-Fruf-β-(2 → | 61.4 | 105.6 | 77.8 | 76.7 | 81.7 | 64.8 |
| 3.71/3.61 | — | 4.13 | 4.03 | 3.89 | 3.83/3.50 | |
Abbreviation: p.p.m., parts per million.
Annotation and analysis of the ftf genomic locus
Homologues of Ftf occur in strains of L. reuteri isolated from various vertebrate species (rodent, pigs and ducks) as well as from sourdough. The L. reuteri 100-23 ftf gene is 2391 bp in size and encodes a protein of 796 amino acids. Ftf contains a leader peptide and a gram-positive cell wall anchor (LPXTG motif), indicating that it is a surface protein attached to the peptidoglycan of the cell wall. The protein consists to a large extent of a pfam02435 domain (glycosyl hydrolase 68 family), responsible for levansucrase and/or invertase activity. As shown in Figure 1a, the ftf gene is preceded by a gene predicted to encode a peptide (29 amino acids: LLYQRLFSVNVRLFYQKSLTDFKIMQKSE) of unknown function. This peptide is 96% identical to a peptide (27 amino acids) in the same position upstream of inu of L. reuteri strain 121. These small peptides might therefore have a role in the function or expression of the fructosyltransferases.
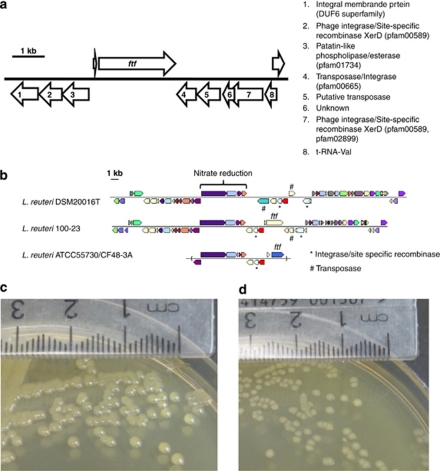
Figure 1.
(a) Genome region in L. reuteri 100-23 containing the ftf gene. (b) Comparison of the loci containing ftf in L. reuteri 100-23, ATCC55730 (which is identical to strain CF48-3A) and DSM20016T. The IMG platform provided by the Joint Genome Institute was used to generate these comparisons. (c) L. reuteri wild type colonies (convex, shiny) on Lactobacilli MRS-sucrose agar medium. (d) L. reuteri ftf mutant colonies (flat, matt) on Lactobacilli MRS-sucrose agar medium.
The closest known homologues of Ftf of strain 100-23 are proteins found in other strains of L. reuteri, namely FTFA from L. reuteri LTH5448 (77% identity over 811 amino acids), Inu from L. reuteri strain 121 (67% identity over 806 amino acids), Inu from L. reuteri TMW1.106 (67% identity over 806 amino acids) and Ftf from L. reuteri strains CF48-3A and ATCC55730 (82% identity over 604 amino acids) and Ftf from L. reuteri MLC1 (78% identity over 597 amino acids). Ftf homologues of L. reuteri are present in the genomes of L. gasseri, L. johnsonii and Streptococcus mutans (>54% identity) but the absence of close orthologues of Ftf in the more closely related species L. fermentum (none) and L. vaginalis (49% amino acid identity), suggests that ftf was acquired horizontally by L. reuteri after division of the lineage. A lateral acquisition of ftf is supported by the presence of mobile DNA elements that flank the locus in the genome of 100-23 (annotated as transposases, site specific recombinases, and integrases) and the location of the locus adjacent to a tRNA gene (Figure 1a).
To gain a more detailed insight into the evolution of Ftf in L. reuteri, we analysed both its genomic loci and its diversification. As shown in Figure 1b, ftf is located in the same genomic cluster in L. reuteri strains 100-23 and ATCC 55730/CF48-3A. It is likely that ftf was acquired early in the evolution of the species L. reuteri because these strains belong to subpopulations that are very distinct genetically. It appears that Ftf diversified much faster than proteins encoded within the same genomic region (for examples nitrate reductases, putative esterase) or by house keeping genes (Ftf <84% amino acid similarity compared with >95% amino acid similarity for other genes) after acquisition. The codon-based Z-test (Nei–Gojobori method) suggested stabilisation (purifying) selection for Ftf (average dN/dS=0.158) in L. reuteri using analysis of the ftf genes listed above. However, the analysis of the loci in different strains also revealed that selection pressure was not homogenous in all strains and might be influenced by animal host. Genomic comparisons indicated that the ftf loci is in a process of erosion in human isolates, as the gene is absent in the genome of L. reuteri DSM20016T, probably deleted through the action of transposons present at the site (Figure 1b). In addition, ftf of strains ATCC55730 and CF48-3A appear to be pseudogenes because they contain a stop codon that truncates the proteins. This has led to the formation of genes that encode proteins that contain the active domain but lack signal peptides, indicating that these Ftf are likely to be nonfunctional.
The strains 100-23 and 100-23C produced mucoid, glistening colonies characteristic of exopolysaccharide production, when cultured on sucrose-containing solid medium (Figure 1c). Mutation of the ftf gene resulted in matt, nonmucoid colonies when grown on sucrose-containing medium (Figure 1d). This indicated that levan production was catalysed by Ftf.
Analysis of mouse stomach contents
Although there was variation between mice, the concentration of sucrose in stomach contents tended to be greater in the case of Lactobacillus-free mice (mean 2.65 μg ml−1 (dry weight); s.e.m. 1.71; n=13) relative to that of animals colonised with strain 100-23 (1.31; 0.49; 13). This indicated that strain 100-23 utilised sucrose in the stomach habitat. HPAEC of the dialysed extract of stomach contents of mice colonised with L. reuteri is shown in Supplementary Figure S1D. There were several minor peaks detected at about 7, 14, 21 and 24 min, but there was no indication of the presence of a series of oligosaccharides similar to that of the exopolysaccharide produced by cultures in vitro. The chromatogram of the extract from colonised mice was similar to that of the stomach contents from Lactobacillus-free mice (data not shown). Levan, although, therefore, probably produced by lactobacilli from sucrose in the stomach, was below the detection limit of HPAEC analysis.
Colonisation of the murine gut
The ftf mutant strain colonised the gut of Lactobacillus-free mice when administered in pure culture, attaining mean log10 population values of 8.5 (s.e.m. 0.1) in the forestomach and 7.4 (0.1) in the caecum, similar to those of the wild-type strain. CFU per gram did not differ when counts were made from agar plates containing erythromycin or in the absence of erythromycin, indicating that the insertional mutation was stable in vivo. When administered to mice with equal numbers of 100-23C, however, the ftf mutant formed on average only 4.2% (s.e.m. 2.3; n=5) of the total Lactobacillus population of the forestomach and 2.2% (s.e.m. 1.4; n=5) of that of the caecum. Therefore the ability to produce exopolysaccharide enhanced the ability of strain 100-23C to colonise the gut.
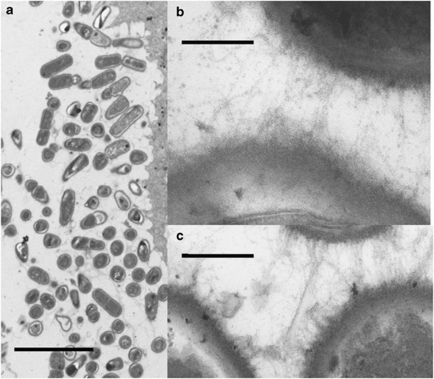
The ftf mutant associated with the forestomach epithelium, and the matrix surrounding and linking the bacterial cells was indistinguishable from that of strain 100-23C (Figure 2).
Figure 2.
Transmission electron micrographs of bacterial cells associated with the surface of the forestomach epithelium of mice. (a) Layer of bacterial cells (biofilm) association with the forestomach epithelium of mouse colonised by the ftf mutant, bar=five micrometres. (b and c) Strands of amorphous material between bacterial cells in biofilm preparations from mice colonised with wild-type strain (b) or ftf mutant (c), bars=0.2 micrometre.
Induction of T regulatory cell populations in spleens
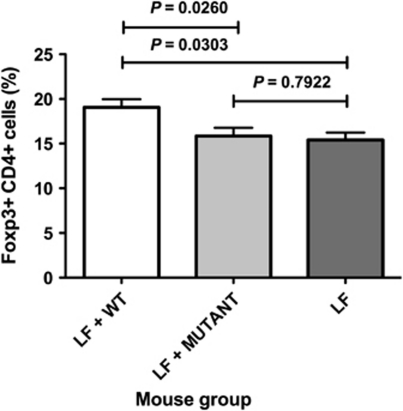
Increased proportions of Foxp3+ T cells in spleens were observed in mice colonised by the wild-type strain relative to Lactobacillus-free animals (Figure 3). In contrast, the proportions of Foxp3+ T cells in mice colonised by the ftf mutant were not different from those of Lactobacillus-free mice (Figure 3). Therefore, in vivo levan production by L. reuteri 100-23 enhanced the induction of regulatory T cells in the systemic immune system.
Figure 3.
Proportions of CD4-positive T cells that were Foxp3 positive in the spleens of mice. LF=Lactobacillus-free mice. Means and s.e.m. are shown; n=6 animals per group.
Survival in a high-sucrose environment
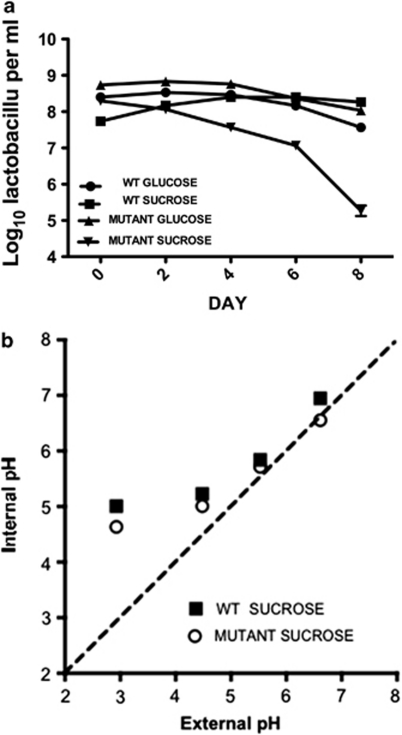
Growth of the wild-type strain was slower in high-sucrose medium compared with the ftf mutant strain. Hence, CFU values did not match at the first sampling time (Figure 4a). It was clear, however, that the CFU per ml of the ftf mutant declined markedly, relative to the wild type, when the cultures containing sucrose were held at 25 °C. The viability of the ftf mutant and wild type was unaffected when held in glucose-containing medium (Figure 4a). The external pH of either sucrose- or glucose-containing medium was 5.1 and this raised the possibility that the decline in viability was because of a defect in pH homeostasis of the ftf mutant in sucrose-containing medium. At day 8 (Figure 4a), the internal pH of cells in the stationary phase cultures was measured. The internal pH was: pH 5.53±0.04 glucose-grown wild-type; pH 5.79±0.03 sucrose-grown wild-type; pH 5.73±0.05 glucose-grown ftf mutant; pH 5.53 sucrose-grown ftf mutant. Further experimentation using washed cell suspensions of wild type versus ftf mutant demonstrated no significant difference in pH homeostasis over the pH range 3–7 (Figure 4b). Based on these data, the internal pH of wild-type and mutant cells held in glucose- or sucrose-containing media was not different (Figure 4b). Therefore, the ability to produce exopolysaccharide in a high sucrose environment was an important survival attribute for strain 100-23, but did not involve the maintenance of intracellular pH.
Figure 4.
(a) Viability of L. reuteri wild type and ftf mutant in media containing glucose or sucrose. Means and s.e.m. are shown; three replicate cultures per sampling time. (b) The effect of extracellular pH on the internal pH of L. reuteri wild-type and the ftf mutant. The cells (energised with 15 m arginine) were suspended in potassium citrate-phosphate buffer at the pH indicated. The ΔpH values are the means of three technical replicates and the standard error associated with these determinations was less than 15%.
Discussion
Sucrose is abundant in many fruits and cereal grains (Schwab et al., 2007). Some bacteria produce exopolysaccharides when they encounter moderate-to-high sucrose concentrations such as in the human oral cavity (Banas and Vickerman, 2003), or on fermenting plant- or animal-derived materials (Smitinont et al., 1999). The structural diversity of bacterial polysaccharides is created by several features including sugar composition, variations in glycosidic linkages between sugars, enantiomeric form of the sugars (D- or L-form), number of carbon molecules that form a ring structure of the sugar (furanose or pyranose) and configuration of the anomeric centre of each sugar. These features are important beyond biochemistry because they contribute to differences in immunological epitopes of polysaccharides. The variations in these features result in the generation of a diversity of structures that interact specifically with the immune system (Mazmanian and Kasper, 2006). Our experiments showed that L. reuteri 100-23 synthesised a levan (β--2, 6-linked fructan) that was produced through the action of a Ftf.
Exopolysaccharides, especially glucans, are important mediators of biofilm formation (dental plaque) in the human oral cavity. S. mutans, for example, produces insoluble glucan (mutan) that contributes to the matrix in which the plaque bacteria accumulate on dental surfaces (Parisotto et al., 2010). The colonisation of the forestomach of mice by a sourdough strain of L. reuteri (TMW1.106) was impaired when the glucosyltransferase (gtf) gene was mutated but was unaffected when the ftf gene was disrupted (Walter et al., 2008). Therefore the ability of the ftf mutant of strain 100-23 to form a biofilm similar to that of the wild type in the mouse forestomach is consistent with the previous observation. This indicates that levan does not form the matrix seen in electron micrographs of forestomach biofilms.
Accumulating evidence suggests that exopolysaccharides have a role in bacterial survival under conditions of stress. L. reuteri strains produce exopolysaccharides from sucrose during type II sourdough fermentation (Tieking et al., 2003). The importance of exopolysaccharide production to the lactobacilli in sourdough is unknown but might be related to the enhanced survival of L. reuteri when stressed by chemical and physical factors affecting membrane lateral pressure (Schwab and Ganzle, 2006). Heterologous expression of the glucosyltransferase (gtf) gene of Pediococcus parvulus 2.6 in L. paracasei NFBC 338 was recently shown to result in increased tolerance of the lactobacilli to heat and acid stress (Stack et al., 2010). Donoghue and Newman (1976) reported that an exopolysaccharide-producing strain of S. mutans survived better in media with high glucose (5%) or high sucrose (5%) content relative to a strain that did not produce exopolysaccharides. The authors concluded that exopolysaccharides affected susceptibility of the cells to acidic pH by virtue of anions associated with the exopolysaccharides. These anions could restrict access of acids to bacterial cells. We found that the survival of the ftf mutant was impaired in high-sucrose (5%) medium but internal pH of the mutant and wild-type cells did not differ. Viability of the mutant was not affected in high-glucose medium. It can be concluded from these results that the detrimental effects of a high sucrose environment, the specifics of which are currently unknown, were negated by fructan production. It is unlikely that the levan is a stored nutritional resource because L. reuteri, including strain 100-23, is unable to ferment fructose. Strain 100-23 in the mouse stomach reduced the sucrose concentration of the habitat by about 50%. This must therefore have been because of the formation of levan. Although the stomach digesta is not a high-sucrose environment, utilisation of sucrose in the stomach habitat by conversion to levan was important because inability to produce the polysaccharide resulted in impaired colonisation in a competitive environment. Overall, levan production seems likely to have importance in resistance to stresses encountered in the gut as well as survival of the bacteria ex vivo. Speculatively, this latter aspect might assist the horizontal transmission of the commensal among murine populations.
Recent studies have provided evidence that there are regulatory T cell (Treg) populations that are induced in tissues rather than emerging from the thymus (Round and Mazmanian, 2010). These Treg subsets, marked by expression of the transcription factor Foxp3, suppress proinflammatory T cell responses to bacteria. Polysaccharide A of Bacteroides fragilis mediates the conversion of CD4+ T cells into Foxp3+ Treg cells. It appears, therefore, that B. fragilis co-opts the Treg lineage differentiation pathway to induce mucosal tolerance (Round and Mazmanian, 2010). Larger Foxp3+ T cell populations have been detected in mice colonised by L. reuteri 100-23 relative to mice naïve to lactobacilli (Livingston et al., 2009). The presence of larger Treg populations in the spleen coincide with downregulation of the mild inflammatory response that occurs in the intestinal mucosa during the initial colonisation period after strain 100-23 has been introduced into the gastrointestinal tract of Lactobacillus-free mice (Hoffman et al., 2008). Data reported here confirm this previous observation concerning induction of Foxp3+ T cells by colonisation of the gut by strain 100-23. Additionally, the data now show that levan produced by L. reuteri 100-23 has a role in mediating this immunoregulatory phenomenon. Foxp3+ T cell populations were not different in size from those of Lactobacillus-free mice when animals were colonised by the ftf mutant, which does not make the levan.
It seems likely that the ftf gene was acquired by lateral gene transfer early in the evolution of L. reuteri and then underwent diversification. As indicated by its population genetic structure, the evolution of L. reuteri resulted in host-confined phylogenetic lineages with a specialization to particular hosts (Oh et al., 2010). Lactobacilli are ubiquitous in environments with high-carbohydrate content such as decaying fruits, cereals, vegetation, meats and seafoods (Smitinont et al., 1999; Hammes and Hertel, 2006). It seems likely that these carbohydrate-rich habitats were the evolutionary source of lactobacilli that subsequently adapted to life in the gut of vertebrates that consumed vegetation and cereals. Exopolysaccharide production by L. reuteri strains may reflect the evolutionary origins of the species where survival in environments of moderate-to-high osmolarity, as shown by in vitro experimentation, would have been advantageous. Later, after adopting a commensal relationship with vertebrate hosts, our data indicate that levan production has assumed other functions that enhanced colonisation potential. The most notable of these is the induction of regulatory T cells that would generate immunological tolerance towards the commensal. This comprehensive, autecological study of a commensal has therefore provided new insights into evolutionary adaptations that underpin microbe-host relationships, specifically those that enable life in the gut.
Acknowledgments
IMS thanks the Ministry of Research, Science and Technology for financial support. Transmission electron micrographs were prepared by Richard Easingwood, Otago Centre for Electron Microscopy, University of Otago.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Banas JA, Vickerman MM. Glucan-binding proteins of the oral streptococci. Crit Rev Oral Biol Med. 2003;14:89–99. doi: 10.1177/154411130301400203. [DOI] [PubMed] [Google Scholar]
- Bello FD, Walter J, Hertel C, Hammes WP. In vitro study of prebiotic properties of levan-type exopolysaccharides from lactobacilli and non-digestible carbohydrates using denaturing gradient gel electrophoresis. Syst Appl Microbiol. 2001;24:232–237. doi: 10.1078/0723-2020-00033. [DOI] [PubMed] [Google Scholar]
- Donoghue HD, Newman HN. Effect of glucose and sucrose on survival in batch culture of Streptococcus mutans C67-1 and a noncariogenic mutant, C67-25. Infect Immun. 1976;13:16–21. doi: 10.1128/iai.13.1.16-21.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes WP, Hertel C.2006The genera Lactobacillus and CarnobacteriumIn: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds). The Prokayotes3rd edn., volume 4, pp320–403.
- Heng NC, Bateup JM, Loach DM, Wu X, Jenkinson HF, Morrison M, et al. Influence of different functional elements of plasmid pGT232 on maintenance of recombinant plasmids in Lactobacillus reuteri populations in vitro and in vivo. Appl Environ Microbiol. 1999;65:5378–5385. doi: 10.1128/aem.65.12.5378-5385.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M, Rath E, Holzlwimmer G, Quintanilla-Martinez L, Loach D, Tannock G, et al. Lactobacillus reuteri 100-23 transiently activates intestinal epithelial cells of mice that have a complex microbiota during the early stages of colonisation. J Nutr. 2008;138:1684–1691. doi: 10.1093/jn/138.9.1684. [DOI] [PubMed] [Google Scholar]
- Jolley KA, Feil EJ, Chan MS, Maiden MC. Sequence type analysis and recombinational tests (START) Bioinformatics. 2001;17:1230–1231. doi: 10.1093/bioinformatics/17.12.1230. [DOI] [PubMed] [Google Scholar]
- Korakli M, Rossmann A, Ganzle MG, Vogel RF. Sucrose metabolism and exopolysaccharide production in wheat and rye sourdoughs by Lactobacillus sanfranciscensis. J Agric Food Chem. 2001;49:5194–5200. doi: 10.1021/jf0102517. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston M, Loach D, Wilson M, Tannock GW, Baird M. Gut commensal Lactobacillus reuteri 100-23 stimulates an immunoregulatory response. Immunol Cell Biol. 2009;88:99–102. doi: 10.1038/icb.2009.71. [DOI] [PubMed] [Google Scholar]
- Markwell MA, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nature Rev. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, et al. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 2010;4:377–387. doi: 10.1038/ismej.2009.123. [DOI] [PubMed] [Google Scholar]
- Parisotto TM, Steiner-Oliveira C, Duque C, Peres RC, Rodrgues LK, Nobre-dos-Santos M. Relationship among microbiological composition and presence of dental plaque, sugar exposure, social factors and different stages of early childhood caries. Arch Oral Biol. 2010;55:365–373. doi: 10.1016/j.archoralbio.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Riebeling V, Thauer RK, Jungermann K. The internal-alkaline pH gradient, sensitive to uncoupler and ATPase inhibitor, in growing Clostridium pasteurianum. Eur J Biochem. 1975;55:445–453. doi: 10.1111/j.1432-1033.1975.tb02181.x. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. PNAS USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WM, Klaenhammer TR. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl Environ Microbiol. 2001;67:4361–4364. doi: 10.1128/AEM.67.9.4361-4364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab C, Ganzle MG. Effect of membrane lateral pressure on the expression of fructosyltransferases in Lactobacillus reuteri. Syst Appl Microbiol. 2006;29:89–99. doi: 10.1016/j.syapm.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Schwab C, Walter J, Tannock GW, Vogel RF, Ganzle MG. Sucrose utilization and impact of sucrose on glycosyltransferase expression in Lactobacillus reuteri. Syst Appl Microbiol. 2007;30:433–443. doi: 10.1016/j.syapm.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Smitinont T, Tasakul C, Tanasupawat S, Keeratipibul S, Navarini L, Bosco M, et al. Exopolysaccharide-producing lactic acid bacteria strains from traditional thai fermented foods: isolation, identification and exopolysaccharide characterization. Int J Food Microbiol. 1999;51:105–111. doi: 10.1016/s0168-1605(99)00094-x. [DOI] [PubMed] [Google Scholar]
- Stack HM, Kearney N, Stanton C, Fitzgerald GF, Ross RP. Association of beta-glucan endogenous production with increased stress tolerance of intestinal lactobacilli. Appl Environ Microbiol. 2010;76:500–507. doi: 10.1128/AEM.01524-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW. A special fondness for lactobacilli. Appl Environ Microbiol. 2004;70:3189–3194. doi: 10.1128/AEM.70.6.3189-3194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW. Research for the 21st century: can we draw a blueprint of the bowel ecosystem. Bioscience Microflora. 2009;28:75–80. [Google Scholar]
- Tannock GW, Crichton C, Welling GW, Koopman JP, Midtvedt T. Reconstitution of the gastrointestinal microflora of lactobacillus-free mice. Appl Environ Microbiol. 1988;54:2971–2975. doi: 10.1128/aem.54.12.2971-2975.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieking M, Kaditzky S, Valcheva R, Korakli M, Vogel RF, Ganzle MG. Extracellular homopolysaccharides and oligosaccharides from intestinal lactobacilli. J Appl Microbiol. 2005;99:692–702. doi: 10.1111/j.1365-2672.2005.02638.x. [DOI] [PubMed] [Google Scholar]
- Tieking M, Korakli M, Ehrmann MA, Ganzle MG, Vogel RF. In situ production of exopolysaccharides during sourdough fermentation by cereal and intestinal isolates of lactic acid bacteria. Appl Environ Microbiol. 2003;69:945–952. doi: 10.1128/AEM.69.2.945-952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hiijum SA, van Geel-Schutten GH, Rahaoui H, van der Maarel MJ, Dijkhuizen L. Characterization of a novel fructosyltransferase from Lactobacillus reuteri that synthesizes high-moleulcar-weight inulin and inulin oligosaccharides. Appl Environ Microbiol. 2002;68:4390–4398. doi: 10.1128/AEM.68.9.4390-4398.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinderola G, Perdigon G, Duarte J, Famworth E, Matar C. Effects of the oral administration of the exopolysaccharide produced by Lactobacillus kefiranofaciens on the gut mucosal immunity. Cytokine. 2006;36:254–260. doi: 10.1016/j.cyto.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Walter J, Chagnaud P, Tannock GW, Loach D, Bello FD, Jenkinson HF, et al. A high-molecular mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl Environ Microbiol. 2005;71:979–986. doi: 10.1128/AEM.71.2.979-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Loach DM, Alqumber M, Rocket C, Hermann C, Pfitzenmaier M, et al. D-alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100-23 results in impaired colonization of the mouse gastrointestinal tract. Environ Microbiol. 2007;9:1750–1760. doi: 10.1111/j.1462-2920.2007.01292.x. [DOI] [PubMed] [Google Scholar]
- Walter J, Schwab C, Loach DM, Ganzle MG, Tannock GW. Glucosyltransferase A (GtfA) and inulosucrase (Inu) of Lactobacillus reuteri TMW1.106 contribute to cell aggregation, in vitro biofilm formation, and colonization of the mouse gastrointestinal tract. Microbiol. 2008;154:72–80. doi: 10.1099/mic.0.2007/010637-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.