Abstract
Ostreococcus is a marine picophytoeukaryote for which culture studies indicate there are ‘high-light' and ‘low-light' adapted ecotypes. Representatives of these ecotypes fall within two to three 18S ribosomal DNA (rDNA) clades for the former and one for the latter. However, clade distributions and relationships to this form of niche partitioning are unknown in nature. We developed two quantitative PCR primer-probe sets and enumerated the proposed ecotypes in the Pacific Ocean as well as the subtropical and tropical North Atlantic. Statistical differences in factors such as salinity, temperature and NO3 indicated the ecophysiological parameters behind clade distributions are more complex than irradiance alone. Clade OII, containing the putatively low-light adapted strains, was detected at warm oligotrophic sites. In contrast, Clade OI, containing high-light adapted strains, was present in cooler mesotrophic and coastal waters. Maximal OI abundance (19 555±37 18S rDNA copies per ml) was detected in mesotrophic waters at 40 m depth, approaching the nutricline. OII was often more abundant at the deep chlorophyll maximum, when nutrient concentrations were significantly higher than at the surface (stratified euphotic zone waters). However, in mixed euphotic-zone water columns, relatively high numbers (for example, 891±107 18S rDNA copies per ml, Sargasso Sea, springtime) were detected at the surface. Both Clades OI and OII were found at multiple euphotic zone depths, but co-occurrence at the same geographical location appeared rare and was detected only in continental slope waters. In situ growth rate estimates using these primer-probes and better comprehension of physiology will enhance ecological understanding of Ostreococcus Clades OII and OI which appear to be oceanic and coastal clades, respectively.
Keywords: picoeukaryotes, Ostreococcus, quantitative PCR, mamiellales, prasinophytes, niche differentiation
Introduction
Over the past two decades, the diversity of small eukaryotic phytoplankton has been investigated with increasing intensity (for example, Rappé et al., 1998; Diez et al., 2001; Not et al., 2007; Shi et al., 2009). Picoeukaryotes have also been shown to contribute significantly to picophytoplankton biomass and primary production (for example, Li, 1994; Worden et al., 2004; Grob et al., 2007; Cuvelier et al., 2010; Jardillier et al., 2010). Several ‘new' picoeukaryotic (⩽2–3 μm diameter) groups have been reported (Vaulot et al., 2008; Worden and Not, 2008), some having been successfully cultured and characterized, like Ostreococcus (Courties et al., 1994). This photosynthetic picoprasinophyte falls in the order Mamiellales with Micromonas and Bathycoccus (Worden and Not, 2008).
Ostreococcus is the smallest known free-living eukaryote (∼0.8–1.2 μm) and several studies have explored its diversity using environmental clone libraries (Guillou et al., 2004; Worden, 2006; Viprey et al., 2008; Worden and Not, 2008). Thus, far Ostreococcus diversity has been best resolved using the internal transcribed spacer (ITS), which is more diverged than the 18S ribosomal RNA (rRNA) gene (Rodriguez et al., 2005). Laboratory photophysiology studies indicate that two differently photoadapted ecotypes exist and are represented by four distinct clades defined using ITS phylogeny, ITS Clades A, C and D being high-light adapted and ITS Clade B being low-light adapted (Rodriguez et al., 2005). The same clade designations have been used for 18S ribosomal DNA (rDNA) trees (Guillou et al., 2004; Viprey et al., 2008), although not necessarily retaining bootstrap support in analyses of either marker (see also Rational section of Results and Discussion for details). These clades have also been termed Clades OI and OII based on 18S rDNA bootstrap support, with OI representing ITS/18S rDNA Clades A and C, OII representing ITS/18S rDNA Clade B and ITS/18S rDNA Clade D excluded because only one published sequence from a cultured strain was available at the time of analysis (Worden and Not, 2008; Worden et al., 2009). Among isolates, Ostreococcus tauri as well as strains RCC356, RCC344 and RCC501 are regarded as high-light ecotypes (falling within Clade OI) whereas Ostreococcus RCC393, RCC143 and RCC809 (formerly RCC141) are considered low-light ecotypes (falling within Clade OII) (Rodriguez et al., 2005; Six et al., 2009). This photophysiology-based ecotypic differentiation has been likened to niche partitioning reported for strains of the cyanobacterium Prochlorococcus (Moore et al., 1998; Moore and Chisholm, 1999; Six et al., 2008).
Ostreococcus abundance has been explored in several studies using fluorescence in situ hybridization (FISH) (Not et al., 2004, 2005, 2008) or quantitative PCR (qPCR) (Zhu et al., 2005; Countway and Caron, 2006), in both cases with genus-level probes. However, ecotype distributions have not been explored systematically or quantitatively in nature. Moreover, comparison of clone library and metagenomic data have led to the proposal that the ‘deep-adapted' clade is not low-light adapted per se but rather better adapted to life in open-ocean conditions, which include periods of time deep in the euphotic zone (Worden, 2006).
We explored Ostreococcus clade niche-partitioning in nature in relation to the laboratory-based hypotheses on irradiance and photophysiology. To this end, qPCR primer-probe sets were developed that target the proposed ecotypes independently. We quantified Clades OI and OII in marine samples and discuss how the data reshapes concepts of niche partitioning.
Materials and methods
Sampling
Twelve cruises were performed between 2001 and 2007 (Table 1). Samples were also utilized from the Scripps Pier (Worden et al., 2004; Worden, 2006), the Monterey Bay Time Series (MBTS, http://www.mbari.org/bog/Projects/CentralCal/summary/ts_summary.htm) and the Bermuda Atlantic Time-Series Study (BATS) station program. Water was typically collected in Niskin bottles mounted on a rosette along with a Conductivity, Temperature Depth sensor.
Table 1. Cruises on which the environmental sample set was collected.
| Location | Cruise | Station/number |
∼Start or station |
∼Site farthest from start |
Dates | ||
|---|---|---|---|---|---|---|---|
| Lat | Lon | Lat | Lon | (Date Month Year) | |||
| South Pacific | KM0703 | Aust—S. Pac./26 | 18° 10′12″S | 147° 25′12″E | 15° 0′0″S | 170° 0′0″W | Mar–Apr 2007 |
| North Pacific | KM0715 | ALOHA | 22° 45′0″N | 158° 0′0″W | NA | NA | Aug 2007 |
| CN107 | CalCOFI line 67/8 | 36° 27′36″N | 122° 25′8″W | 33° 17′9.6″N | 129° 25′41″W | Jul 2007 | |
| CN207a | CalCOFI line 67/7 | 36° 17′35″N | 123° 8′2″W | 33° 10′16″N | 129° 15′25″W | Oct 2007 | |
| MBTS | Sts C1, M1, M2 | 36° 47′60″N | 121° 50′60″W | 36° 42′0″N | 122° 23′24″W | Jan–Dec 2007 | |
| NA | Scripps Pier | 32° 31′48″N | 117° 9′0″W | NA | NA | Jan–Dec 2001 | |
| Trop. Atlantic | SJ0609 | FL-E. Atl./19 | 18° 12′37″N | 67° 28′27″W | 12° 24′49″N | 35° 16′14″W | Jun–Jul 2006 |
| Florida Straits | WS0503a | Sts 01, 04, 14 | 25° 30′07″N | 80° 04′04″W | 25° 29′59″N | 79° 20′58″W | Mar 2005 |
| WS0518a | Sts 01, 04, 14 | 25° 30′04″N | 80° 03′59″W | 25 29′55″N | 79° 20′54″W | Aug 2005 | |
| WS0705 | Sts 01, 04, 08 | 25° 30′ 0″N | 80° 3′ 58″W | 25° 30′ 0″N | 79° 42′ 36″W | 27 Feb 2007 | |
| North Atlantic | EN351 | Shelf—BATS/5 | 31° 49′44″N | 64° 10′30″W | 40° 15′7″N | 70° 25′23″W | Mar–Apr 2001 |
| EN360 | Shelf—W. Sarg/5 | 39° 59′38″N | 71° 48′1″W | 34° 25′34″N | 72° 3′28″W | Sep 2001 | |
| OC374 | Shelf—BATS/5 | 31° 38′55″N | 64° 12′17″W | 40° 12′57″ | 70° 6′19″W | Mar 2002 | |
| OC413a | N. BATS, BATS/2 | 35° 09′24″N | 66° 33′46″W | 31° 39′20″ | 64° 37′21″W | May–Jun 2005 | |
| Sargasso Sea | 138b | BATS | 31° 34′59″ | 64° 8′ 2.4″ | NA | NA | 14 Mar 2000 |
| 173b | BATS | 31° 42′36″N | 64° 13′16″W | NA | NA | 14 Feb 2003 | |
| 174b | BATS | 31° 42′36″N | 64° 13′16″W | NA | NA | 04 Apr 2003 | |
| 179b | BATS | 31° 42′36″N | 64° 13′16″W | NA | NA | 13 Aug 2003 | |
Abbreviations: Aust, Australia; BATS, Bermuda Atlantic Time-series Study; MBTS, Monterey Bay Time Series; NA, not applicable; Sts, stations.
Clone libraries sequenced.
Samples were provided by the BATS program (see acknowledgements).
Samples were collected on multiple dates from each MBTS stations (Supplementary Figure 2) and from the Scripps Pier (Worden, 2006).
Station name is provided for known time-series sites, and station number is provided for transect cruises, indicating the total number of stations from which samples were evaluated.
Euphotic zone DNA samples were collected by filtering 500–2000 ml seawater through a 0.2- or 0.45-μm pore size Supor filter (Pall Gelman, East Hills, NY, USA). In all, 24 of 301 samples were first gravity filtered through a 2-μm pore size polycarbonate filter (Osmonics, Trevose, PA, USA) and subsequently onto a 0.2 μm Supor filter. Ostreococcus is between ∼0.8 and 1.2 μm in diameter (Worden and Not, 2008), hence pre-filtration is not expected to significantly reduce counts. Filters were placed into sterile cryovials and frozen in liquid nitrogen before storage at −80 °C. BATS euphotic zone samples were filtered (∼75–100 l) as described in Treusch et al. (2009). For cruises with nutrient and Chl a data many of the data have been reported previously (see below).
DNA extraction, clone library and plasmid preparation
All samples, except those from BATS in 2000 and 2003, were extracted using the DNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions with filters being subjected to additional freeze fracture and bead-beating steps according to (Moisander et al., 2008) or crushed using sterile pestles (63 CN cruise samples) at the start of extraction procedure. DNA was eluted using either one or two sequential elutions, resulting in a final volume of 50 or 100 μl. Extracts were aliquoted and stored at −80 °C. For 2000 and 2003 BATS cruises a sucrose-based extraction was used, as in Treusch et al. (2009). 18S rRNA gene clone libraries were constructed from some samples (Table 1) as described previously (Cuvelier et al., 2008) and sequences deposited under accession numbers HQ847732–HQ847735.
To generate 18S rDNA insert-bearing plasmid standards for qPCR, the 18S rRNA gene was amplified from culture DNA (O. tauri OTH95 and RCC809, extracted as above) using the eukaryotic primers 5′-ACCTGGTTGATCCTGCCAG-3′ and 5′-TGATCCTTCYGCAGGTTCAC-3′ (Moon-van der Staay et al., 2000). PCR was performed using HotStar Taq polymerase (Qiagen) and cycling conditions were 95 °C (15 min), proceeded by 32 cycles at 94 °C (30 s), 55 °C (30 s) and 72 °C (1 min), and final extension at 72 °C (10–15 min). PCR products were cloned (TOPO-TA, Invitrogen, Carlsbad, CA, USA) and plasmids purified (QIAprep kit, Qiagen) according to the manufacturer's procedures. Sequencing used Big Dye Terminator chemistry (Applied Biosystems, Foster City, CA, USA). Clone UEPACAAp1, taken from a Pacific Ocean environmental clone archive (Worden, 2006), was also used. OTH95 and UEPACAAp1 have 98 and 99% identity to O. lucimarinus CCE9901 over 1722 positions, respectively, and represent Clade OI while RCC809 represents Clade OII. DNA from RCC789, a clonal ITS Clade D strain derived from RCC501, as well as DNA or plasmids from Bathycoccus and several Micromonas were also used (see below, Table 3, Supplementary Tables 2 and 3).
qPCR, FISH, primer-probe design and qPCR analysis
Initially, published Ostreococcus genus-level SYBR-green qPCR primers (Zhu et al., 2005) and a Tyramide signal Amplification FISH (TSA–FISH) probe (Not et al., 2004; with details in Cuvelier et al., 2010) were used. To develop primer-probe sets that could discriminate between different clades, 18S rRNA gene sequences for cultured prasinophytes, other organisms, and environmental sequences were retrieved from GenBank (last retrieval December 2008). Primer-probe sets for Clade OI and OII (Table 2), as well as Bathycoccus and Micromonas (Supplementary Table 1), were designed manually using Seqman (DNASTAR Inc., Madison, WI, USA). Melting temperature and secondary structures were checked using Primer Express (AB), Beacon Designer 7.0 (PREMIER Biosoft International, Palo Alto, CA, USA) and an online Tm calculator (IDT, San Diego, CA, USA). The reporter dye at the probe 5′ terminus was 6-FAM (fluorescein) and a minor-groove binding non-fluorescent quencher or a Black-Hole quencher was used at the 3′ terminus. DNA extracts and plasmid standard curves from cultured relatives and more distant phytoplankton were used to test specificity of the Ostreococcus Clade OI and OII (Table 2), Bathycoccus (Supplementary Table 2) and Micromonas (Supplementary Table 3) primer-probe sets. The reaction efficiency, calculated as (10(−1/m)−1) where m is the slope of a linear regression against standard curve cycle threshold (CT) values, was 90–101% (n=14, OI primer-probe) and 91–99% (n=13, OII primer-probe). For Micromonas and Bathycoccus primer-probe sets it was 98 and 95–103%, respectively.
Table 2. Nucleotide sequences of the clade-specific Ostreococcus primer-probe sets.
| Targeted clade | Full name | Sequence (5′–3′) |
|---|---|---|
| OI.08F | GGATTTTGGCTGAGAACGGTC | |
| OI | OI.08R | CGATGAAGCACACCTCCTCAC |
| OI.08P | 6-FAM TGCACTGACTGGTCTC MGBNFQ | |
| OII.08F | GGATTTTGGCTGAGAACGAA | |
| OII | OII.08R | AAAGTAACCACGGTGACTAAGTGGC |
| OII.08P | 6-FAM TGCACTGTTTGGTCTCA MGBNFQ |
Abbreviation: MGBNFQ, minor-groove binding non fluorescent quencher.
Before qPCR, plasmids were quantified using the NanoDrop system (Thermo Scientific, Waltham, MA, USA) and diluted to 0.5 × 109 gene copies per μl, followed by 10-fold serial dilutions, creating a standard curve. Reaction volumes were 25 μl, being composed of 12.5 μl TaqMan Universal PCR Master Mix (AB), 2.5 μl (each) of probe (250 n final concentration), forward and reverse primers (900 n final concentration), 3 μl H2O and 2 μl template (DNA extract or plasmid). For environmental samples, DNA was diluted according to inhibition test results (see below). qPCR cycling conditions were 10 min at 95 °C (initial denaturation) followed by 45 cycles at 95 °C (15 s) and 60 °C (1 min) using an AB7500. Data were collected during the annealing phase. Of 271 global environmental samples (not including MBTS), 81 were run in triplicate, and the rest in duplicate to conserve DNA. The 29 MBTS samples were run in triplicate (17 samples, which required dilution based on inhibition results) or duplicate (12 samples). Each run (96 well-plate) included triplicated no-template controls and plasmid standard curves ranging from 108 to 100 copies per rxn, although dilutions from 108 to 101 were used for a small subset. 100 rDNA copies per rxn were detected 39±19% of the time among triplicates. The number of standard copies detected consistently, where the s.d. was smaller than the mean for triplicates, was 101 rDNA copies per rxn.
The possibility that environmental DNA extracts were inhibited was tested by spiking samples with 2 μl of plasmid (104 or 105 gene copies per rxn). CT values from standard curve points with the same concentration were compared with plasmid spiked samples to ensure values were equivalent, or assess the extent of inhibition (Short and Zehr, 2005). Inhibition tests were performed on each sample using one of the four primer-probe sets developed herein. Although some samples were not diluted, typically dilutions ranged between 1:4 and 1:100 to avoid inhibition, or in some cases to conserve DNA. The resulting theoretical detection limits, based on the volume of sample filtered, the elution volume, the dilution used for the final run and consistent detection (101 rDNA copies per rxn) was 0.1–12.5 rDNA copies per ml for 264 of 271 discrete samples (detection down to 0.1–<3 copies per ml, 151 samples; 3–13 copies per ml, 113 samples), between 16 and 20 rDNA copies per ml for six samples and for one sample the minimum detectable was 130 rDNA copies per ml. Owing to limited extract, 12 Florida Straits and 4 Sargasso Sea samples were not tested for inhibition; inhibition was not seen in other samples from these regions using the same extraction and dilution methods. Scripps Institution of Oceanography pier samples (five total), most of which contained high Ostreococcus counts, were not checked for inhibition. For each run, threshold and baseline values were calculated treating each measurement as a unique run (AB7500 software package, Applied Biosystems, Foster City, CA, USA). rDNA copies per ml in seawater samples were determined according to CT values fitted on linear regression of CT versus copy numbers (in log scale) of the standard curve, also taking into account sample volume filtered, elution volume, dilution and template volume. If the mean rDNA copies per ml in the environmental sample was at least twice the detection limit, the data was considered quantitative and in this case typically at least 10 copies per rxn were detected. The clade was considered detected but not quantifiable for ratios of sample copies to detection limit <2 and ⩾1, and undetected for ratios <1.
Interrelating gene copies per ml and cells per ml
The genome sequence of O. tauri OTH95 (Derelle et al., 2006) shows one rRNA gene cluster while O. lucimarinus CCE9901 (Palenik et al., 2007) and RCC809 (a representative of the putatively deep-adapted Clade OII), have two copies. Given the identical nature of this gene cluster, genome assemblies sometimes do not render the correct copy number. However, based on these genome sequences, qPCR rDNA copies per ml likely correspond to either an equivalent number of cells per ml, or two times the cell count, assuming G1 phase of the cell cycle and that the amplification efficiency of the plasmid standards matches that of cells.
Statistics and climatology
Environmental data for samples containing the respective Ostreococcus clades were tested for normality using SigmaStat (Systat software, Inc., Chicago, IL, USA), which typically failed. T-tests were performed using Mann–Whitney rank sum when environmental parameters did not have normal distributions and medians, rather than means and deviations, then reported. In addition, for the MBTS, which had a consistent set of environmental metadata, PRIMER-E v6 (PRIMER-E Ltd., Plymouth Marine Laboratory, Plymouth, UK) was used to investigate the relationship between gene copies per ml and physico–chemical data. An environmental matrix was assembled that contained the following variables: salinity, temperature, Chl a, NO3, NO2 and PO4 concentrations, and NO3 to PO4 ratios. The environmental matrix was normalized by subtracting the mean of each variable from every value and dividing by the s.d. Sea surface temperature climatology for 2009 (as in Figure 3) was determined using the daily 9 km blended microwave and infrared optimum interpolated sea surface temperature from remote sensing systems.
Ostreococcus growth curve and field growth estimate
OTH95 (axenic) and RCC809 (not axenic) were grown in K-media made with artificial (http://www.mbari.org/phyto-genome/Resources.html) or Sargasso seawater bases, respectively, at 21.5 °C on a 14:10 light:dark cycle. Triplicate OTH95 cultures were grown in semi-continuous batch mode and monitored daily by fluorometry for each light-level investigated. Cells were acclimated to light levels, maintained for 10 generations of acclimated mid-exponential growth, and rates then calculated from four successive transfers (after the 10 generations). A previously described Sargasso Sea dilution experiment (Cuvelier et al., 2010) was also further analyzed using flow cytometry (FCM) and qPCR results from herein. Average growth rate of all non-prymnesiophyte phytoplankton in the 70 m dilution experiment, including OII cells, was estimated as:
abbreviations as follows: Aph, FCM abundance of photosynthetic eukaryotes; Apr, FISH abundance of pico-prymnesiophytes (from Cuvelier et al., 2010); subscripts indicate time point and values are for the 20% raw seawater treatment (80% filtered seawater), representing a conservative growth estimate (see Worden and Binder, 2003), or the Y-intercept of a regression (FCM-based only).
Results and discussion
Rationale, development and verification of qPCR primer-probe sets
Ostreococcus abundance in Sargasso Sea samples determined by TSA–FISH were not above the nonspecific negative control probe (∼30 cells ml−1). Surprisingly, in one such sample, 32 of 36 18S rDNA sequences recovered from known photosynthetic taxa belonged to Clade OII and qPCR showed 2514±401 genome copies per ml using genus-level Ostreococcus primers (Zhu et al., 2005; calculated using a standard curve based on genome copies and assuming 100% extraction efficiency for the genome copy-based standard curve). Anomalously low Ostreococcus counts by TSA–FISH have been seen in another open-ocean study (Jardillier personal communication). Small cell size or other factors may make TSA–FISH enumeration more challenging for Ostreococcus in certain samples than for other taxa. Inability to confidently enumerate Ostreococcus by TSA–FISH in our open-ocean samples and desire to enumerate the clades independently prompted us to design, test and implement Clade OI- and OII-specific qPCR primer-probe sets.
To verify specificity, primer-probe sets were tested against several species and strains (Figure 1, Table 2, Supplementary Table 1). No non-target amplification was detected using either Ostreococcus primer-probe set (Table 3). The Micromonas and Bathycoccus primer-probe sets were used for a subset of inhibition tests only. The former showed no cross-reactivity with non-target standards, while a low level of non-linear cross-reactivity by the latter was detected at CT values of 38–39 against an OI standard curve (Supplementary Tables 2 and 3). Given the high plasmid copy number (106–108 per rxn) where Ostreococcus amplification was detected with the Bathycoccus primer-probe set, and the dramatically higher amplification efficiency (several orders of magnitude) against its target taxon versus Ostreococcus, this primer-probe set should still be Bathycoccus-specific in environmental samples. Eukaryotic picophytoplankton concentrations (and hence gene copies per ml) at these plasmid levels (106–108) are rarely reported (see, for example, Li, 1994; Worden and Not, 2008; Cuvelier et al., 2010).
Figure 1.
Regions of the 18S rRNA gene targeted by the Ostreococcus qPCR primer-probe sets. Three sequences (highlighted in grey) are from Ostreococcus Clade OII that were characterized as deep-adapted in Rodriguez et al. (2005), as well as two environmental clones. Five sequences are shown for Clade OI (highlighted in yellow), composed of O. tauri and the four other sequences, two from cultures and two from environmental samples (including sequences from two of the clades identified in Rodriguez et al., 2005 as being more high-light adapted). RCC501 represents the third putatively high-light adapted clade and is not targeted by either primer-probe set. Where highlighted sequences have perfect identity to respective primers and probes.
Table 3. Verification of Ostreococcus clade-specific primer-probe sets against target and non-target DNA samples.
|
Organism source material |
Primer-probe set |
|||||||
|---|---|---|---|---|---|---|---|---|
|
OI.08 |
OII.08 |
|||||||
| Class | Genus | Strain | Clade | DNA | Detect. | Mismatch | Detect. | Mismatch |
| Prasinophytae | Ostreococcus | OTH95 | OI | C, PSTD | Y | 1, 0, 2 | UD | 3, 2, 0 |
| Prasinophytae | Ostreococcus | UEPACAAp1 | OI | EPSTD | Y | 0, 0, 0 | ND | 2, 2, 0 |
| Prasinophytae | Ostreococcus | RCC809 | OII | C, PSTD | UD | 2, 2, 2 | Y | 0, 0, 0 |
| Prasinophytae | Ostreococcus | RCC789 | ITS D | C | UD | 1, 0, 7 | UD | 3, 3, 5 |
| Prasinophytae | Bathycoccus | BBAN 7 | NA | C, PSTD | UD | 7, 16, 5 | UD | 6, 6,13 |
| Prasinophytae | Micromonas | RCC472 | MII | C | UD | 6, 4, 19 | UD | 5, 4, 6 |
| Prasinophytae | Micromonas | RCC451 | MI | C | UD | 6, 4, 19 | UD | 5, 4, 6 |
| Prasinophytae | Micromonas | UEPACOp3 | MIV | EP | ND | 6, 4, 22 | ND | 4, 4, 7 |
| Prasinophytae | Micromonas | CCMP1545 | MV | C | UD | 5, 2, 20 | UD | 5, 4, 7 |
| Prasinophytae | Micromonas | CCMP1646 | MIII | C, PSTD | UD | 4, 2, 19 | UD | 4, 4, 6 |
| Prasinophytae | Micromonas | RCC299 | MII | C | UD | 6, 4, 19 | UD | 5, 4, 6 |
| Cryptophyceae | Rhodomonas | CCMP1319 | NA | C | UD | ND | UD | ND |
| Bolidophyceae | Bolidomonas | CCMP1866 | NA | C | UD | ND | UD | ND |
| Prymnesiophyceae | Isochrysis | CCMP1323 | NA | C | UD | ND | UD | ND |
| Dictyochophyceae | Rhizochromulina | CCMP2174 | NA | C | UD | ND | UD | ND |
| Pelagophyceae | Pelagomonas | CCMP1756 | NA | C | UD | ND | UD | ND |
Abbreviations: C, culture DNA; Detect., indicates whether the primer-probe set rendered a detectable signal; EP, environmentally derived plasmid; EPSTD, environmentally derived plasmid standard curve; Mismatch, provides the number of mismatches between the forward primer, probe and reverse primer to the source material indicated to the left; NA, not applicable; ND, not determined; PSTD, plasmid standard curve (101 to 108); UD, undetected (CT>40, or no value detected); Y, yes detected; RCC472 is the same as CCMP492, RCC451 is the same as CCMP1764.
With regard to published phylogenetic studies the OII primer-probe set amplifies cultured strains formerly considered ‘deep-adapted', all of which belong to ITS Clade B (Rodriguez et al., 2005), designated Clade OII based on 18S rDNA analyses (Worden and Not, 2008; Worden et al., 2009). The OI primer-probe set amplifies a combination of ITS/18S Clade A (for example, clone UEPACAAp1, and many other environmental clones) and Clade C (for example, O. tauri), both of which are considered to be more high-light adapted (Rodriguez et al., 2005). Ostreococcus RCC789 (the clonal version of RCC501), which belongs to ITS/18S Clade D was not amplified by either primer-probe set because of mismatches (Figure 1, Table 3). We scanned for similar sequences in GenBank and found that, apart from RCC501, isolated from Barcelona Harbor, Spain, only three other sequences representing this clade have been deposited (⩾99–100% 18S rDNA identity to each other; 97–98% identity to O. tauri and O. lucimarinus). All are from estuarine or brackish settings, specifically the Baltic Sea (FN690726, Gulf of Finland and FN263267, Gulf of Gdansk, Poland) and Lake Pontchartrain (FJ350825, LA, USA). Five unpublished Roscoff Culture Collection (RCC) cultures from estuarine/lagoon or bay settings also appear to belong to Clade D (http://www.sb-roscoff.fr/Phyto/RCC/), suggesting this clade generally resides in brackish, not coastal or open-ocean, environments. Clade C also appears to be lagoonal based on isolation locations. Given the environmental samples investigated here, the primer-probe sets are likely amplifying cells from OI belonging to ITS/18S Clade A and OII (ITS/18S Clade B).
Characteristics of the sample set
The environments investigated (Table 1) represent a range of euphotic zone conditions. Environmental and other data have been published elsewhere for most cruises (Cuvelier et al., 2008, 2010; Treusch et al., 2009; Moisander et al., 2010; Santoro et al., 2010). Briefly, all Atlantic cruise samples were from relatively oligotrophic waters (see, for example, Steinberg et al., 2001; Lomas et al., 2010), with the exception of some stations on transit to the Sargasso Sea, which passed through continental shelf and slope waters, before crossing into the oligotrophic Gulf Stream and beyond. EN351, OC374 and EN360 traversed these settings (Table 1). The former two cruises also sampled stations north of BATS and BATS, while EN360 sampled more western Sargasso Sea sites. BATS and Sargasso Sea stations north of BATS undergo winter deep mixing (Steinberg et al., 2001; Treusch et al., 2009; Lomas et al., 2010). The BATS euphotic zone was well mixed during OC374 (down to ∼120 m with no apparent deep chlorophyll maximum (DCM); 5 March), with stratification developing during EN351 (DCM apparent at 60 m, 31 March) and OC413 (DCM developing at ∼100 m, May–June). Sargasso sites were stratified during EN360 (DCM at 95 m; 22 September). The South Pacific transect was also oligotrophic (Moisander et al., 2010) as were samples from the North Pacific Gyre (Station ALOHA). The eastern North Pacific transects (CN107 and CN207) originated in Monterey Bay and encompassed more varied conditions than other Pacific cruises, with eight stations spread over 800 km, ranging from coastal, to mesotrophic and relatively oligotrophic waters. Finally, 2007 MBTS samples were analyzed separately from the global set (see below).
Relationship of Ostreococcus Clades OI and OII to environmental parameters
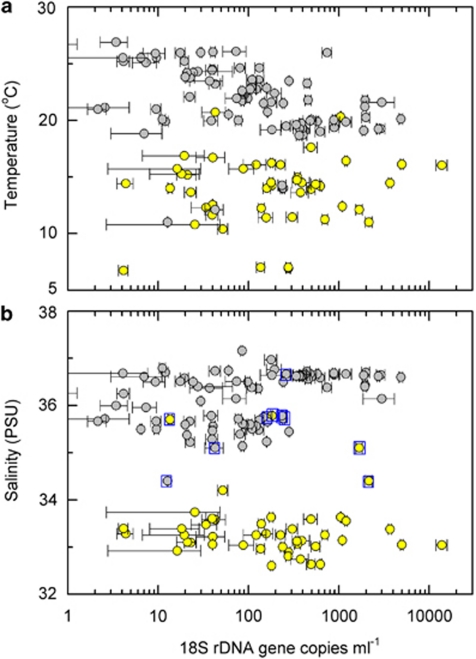
Most environmental parameters were strongly delineated within the global sample set between those samples containing Clade OI and those containing Clade OII (Supplementary Table 4). Of the 271 global samples evaluated, 64 contained OI and 99 contained OII. OI was detected largely in colder, lower salinity coastal waters while OII was detected in gyre-like conditions with relatively warm waters and high salinities (Figures 2 and 3). The mean temperatures (Figure 2a) and s.d. for the two groups were significantly different, 14±3 °C versus 22±3 °C, for OI and OII, respectively. Salinity (Figure 2b) and depth were also significantly different (P<0.001), with median values of 33 and 36 PSU (Practical Salinity Unit), and 30 and 75 m for OI and OII, respectively. Fewer measurements were available for NO3, PO4 and Chl a and data availability was uneven for oligotrophic versus coastal and mesotrophic sites. Still, NO3 (Supplementary Figure 1) was significantly different (P<0.02) for samples containing OI (median 0.980 μ, n=41) versus OII (median 0.545 μ, n=18), as was PO4 (P<0.001) for OI (median 0.686 μ, n=41) versus OII (median 0.002 μ, n=23) containing samples. Chl a concentrations at sites with OI were also higher than those with OII, with median values of 0.74 μg l−1 (n=35) and 0.31 μg l−1 (n=30), respectively.
Figure 2.
Environmental parameters (a) temperature and (b) salinity as a function of Ostreococcus Clade OI (yellow) and Clade OII (grey) rDNA copies per ml. Only data considered quantifiable are shown. In (a), the three yellow symbols above 17 °C are from the Scripps Pier, which can warm significantly during day time hours at ∼0.5 m where the samples were collected, while grey symbols below 15 °C are from OC374 and EN351 continental slope samples. In (b), data points from OC374 and EN351 slope samples are indicated by blue boxes, although, for some, only one clade was found at the particular depth; for data from all other samples shown only one clade was quantifiable. Error bars represent the s.d. of averaged technical replicates, where not visible they are within the symbol.
Figure 3.
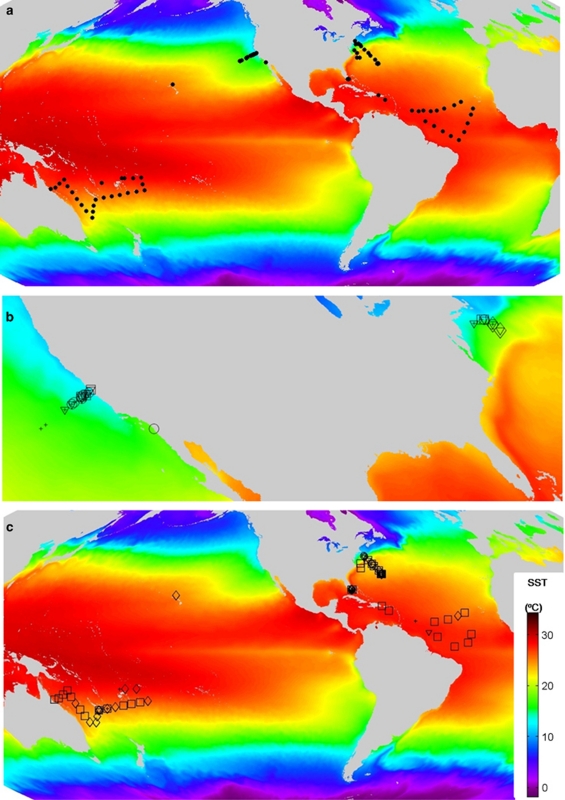
Distribution of Ostreococcus Clades OI and OII. (a) Global sample sites. (b) Sites at which Clade OI 18S rDNA copies per ml were quantitated (open symbols) or detected but not quantifiable (plus sign) and (c) the same for Clade OII. Depth ranges are 0–6 m (open circle), 10–59 m (open triangle), 60–99 m (open square) and ⩾100 m (open diamond). Several measurements can be contained within a single depth range, but are represented by a single symbol for simplicity. As OI were not detected outside of the geographical range shown in (b) a high-resolution zoom to this area is shown. Note that Clade OI members have been detected in clone libraries in coastal European regions, for example, near Roscoff, FR.
In oligotrophic waters, that is, subtropical and tropical Atlantic, Gulf Stream, South Pacific and ALOHA, seawater temperatures were significantly higher where Ostreococcus was undetected (26±3 °C) than where detected (only OII at these sites; 22±3 °C), The median sample depth for oligotrophic samples (from 200 m and up), was also significantly different (P<0.001) for samples with no Ostreococcus detected (6 m, n=88) versus OII containing samples (83 m, n=85). Nitrate was significantly lower (P<0.001), with median concentrations of 0.004 and 0.545 μ and PO4 lower (0.000 versus 0.002 μ, but not significantly different), where Ostreococcus was not detected versus samples containing OII, respectively. The latter two analyses compare measurements generated by different methodologies, however, in the case of NO3 only one sample was below detection using the less sensitive method and hence comparisons should be valid. Finally, Chl a concentrations were significantly (P<0.02) lower for samples with no Ostreococcus detected (median 0.17 μg l−1, n=44) versus those containing OII (median 0.31 μg l−1, n=30).
The MBTS covers a productive coastal/bay area (Monterey Bay) and these samples were analyzed separately to avoid weighting global results with data from a single region and year. OI, and only OI, was detected in 27 of 29 samples spanning February to December (Supplementary Figure 2). Average temperature for OI containing samples, or even those with ⩾1000 rDNA copies per ml, was within the overall mean (12±1 °C). MBTS environmental metadata was measured using a consistent set of protocols and a Relate test (Clarke, 1993) was performed to correlate qPCR data with the entire environmental matrix (depth, temperature, NO3, NO2, PO4, Chl a and salinity) and each variable independently. A Euclidean distance matrix of environmental data was also compared with a Bray–Curtis dissimilarity matrix of gene copies per ml for each sample. However, there was no significant correlation for either the entire environmental matrix or the individual variables, nor did Euclidean distance provide significant results. For example, although the mean NO3 concentration was lower (9.3±5.7 μ) for samples with ⩾1000 rDNA copies per ml than for samples with fewer or none (13.6±7.6 μ), the differences were insignificant. Nitrate ranged from 4.0 to 26.3 μ and was 19.7 μ and 24.0 μ for the two samples in which OI was undetected. Furthermore, no statistical differences were detected between NO3, PO4, Chl a data from samples containing ⩾100, 1000 or 5000 copies per ml versus those containing fewer than each of these abundance categories. Only differences presumably related to other aspects of seasonal variability (based on date of collection) seemed apparent—with generally lower OI rDNA copies per ml from April to August than other months (Supplementary Figure 2).
Co-occurrence and overall trends
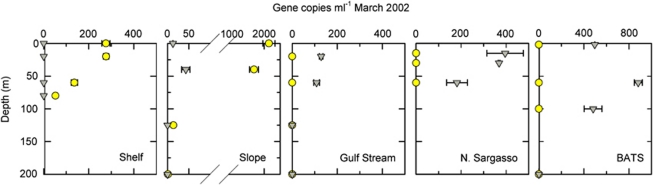
The two Ostreococcus clades were co-localized in continental slope samples from transects from coastal New England, USA to the Sargasso Sea. Specifically, OI and OII were quantifiable in the same slope water samples in early March (OC374, Figure 4) and late March/early April (EN351, Supplementary Figure 3a). Notably, a differential in abundance related to depth was not apparent and co-localization may have resulted from physical mixing of different water masses. In a September transect, OI was close to the detection limit at the Narragansett Pier and in low numbers in shelf waters, but not found elsewhere, while OII was detected in the Gulf Stream DCM and one of two Sargasso Sea Stations (EN360, Supplementary Figure 3b).
Figure 4.
18S rDNA copies per ml of Ostreococcus Clade OI (yellow) and Clade OII (grey) along a transect from the coastal Northwest Atlantic to BATS for all samples evaluated. Similar trends in Clade distribution were seen in EN351 in March–April 2001, although in EN360 in September 2001 few Ostreococcus were detected (Supplementary Figure 3). Error bars represent the s.d. of averaged technical replicates, where not visible they are within the symbol.
These data indicate that factors beyond light, such as temperature, salinity, nutrient availability or related variables, have a role in clade dynamics. With respect to depth, OI was detected as deep as 173 m (160±16 copies per ml) at a slope station (Supplementary Figure 3a) with maximum abundance at 40 m at the base of the MBTS Station M2 mixed layer (Supplementary Figure 2). Across the global data set, the median depth for OI was 30 m, both for locations where it was detected and for samples with ⩾1000 OI rDNA copies per ml. Notable OII counts (for example, 2914±316 copies per ml, March 2000, 40 m) were detected close to the surface at BATS. The influence of irradiance was not explored directly because sampling was performed throughout the day–night period and hence irradiance data were not comparable. However, even using depth as an irradiance proxy, establishing it as a driver for distributions was confounded by the fact that nutrient concentrations increased with depth (except under deep-mixing conditions) and could also serve as a driver. Parameters such as NO3 were significantly different among those samples containing one versus the other clade or open-ocean samples in which OII was present versus not present. Furthermore, OII was detected at relatively high abundances in BATS surface samples in March 2002 (Figure 4) and April 2003 (385±19 rDNA copies per ml, 4 m). In Northern Sargasso Sea Stations, equivalent concentrations were found at 15 and 30 m in early March 2002 (33° 13′ 31″ N, 64° 57′ 8″ W) with 395±81 and 369±4 rDNA copies per ml, respectively and at 15 and 150 m in March/April 2001 (33° 14′ 7″ N, 64° 53′ 19″ W) with 402±40 and 365±25 rDNA copies per ml, respectively. Nutrient samples were not taken at these sites, however, the euphotic zone was well mixed during the sampling period. In addition to similar cell concentrations at the surface and DCM, temperature, salinity and in vivo fluorescence (and presumably nutrients by proxy) were more similar through the euphotic zone than in periods after stratification set in. Thus, nutrient availability appears to have a role in the vertical distributions of OII through the euphotic zone.
From a broader geographical perspective, the two clades appear to differentiate along lines of coastal versus oligotrophic water masses (Figure 3). Samples from mesotrophic locations close to the Pacific coast, near-shore samples and the Scripps Institution of Oceanography Pier contained OI, as did Atlantic Narragansett Pier, Shelf and Slope samples. In contrast, apart from Atlantic slope sites, OII was not detected at these locations but present in the North Pacific Gyre, the South Pacific Ocean, the North Atlantic Gyre, subtropics and tropics. Similarly, only OII was detected in the Florida Straits, which are physically close to land but composed of oligotrophic Gulf Stream forming waters.
Correspondence between clone library and qPCR results
Evaluation of clone libraries and published sequences (for example, Worden, 2006; Viprey et al., 2008; Worden and Not, 2008) support the qPCR findings (Figure 3). First, when the qPCR primer-probe sets developed here were applied to environmental samples from which Ostreococcus sequences were recovered, 18S rDNA clade sequence affiliations were consistent with clades detected by qPCR. In the Florida Straits and Sargasso, only OII 18S rDNA sequences were recovered (for example, clone FS_St01Mar05_5mB060T1F from 5 m station 01, 30 March 2005 has 100% identity to RCC143 over 707 positions). At the Scripps Pier (Worden, 2006) and in the eastern North Pacific (CN207), only OI 18S rDNA sequences were recovered (for example, clone CN207H3_5m, 5 m station H3, 10 October 2007 has 99% identity to O. lucimarinus over 1765 positions) and only this clade was detected by qPCR. The high representation of deposited OI environmental sequences and strains likely indicates a sampling bias towards coastal settings, which are investigated more frequently. For example, the high-light adapted representative O. tauri was isolated from a shallow lagoon (Six et al., 2008) and O. lucimarinus from ∼0.5 m in coastal California waters (Worden et al., 2004). From a broader perspective, in addition to the proposal that the ‘deep-adapted' clade (OII) are not strictly low-light adapted but rather better adapted to life in open-ocean conditions, including periods of time in deep euphotic-zone waters (Worden and Not, 2008), a Mediterranean Sea clone library study (Viprey et al., 2008) suggested that Ostreococcus Clade A (within OI herein) prefers surface waters influenced by low temperature, low salinity Atlantic waters, whereas Clade B (OII) was present throughout euphotic zone waters including surface waters, for example, 5 and 15 m. Clade OII (but not OI) sequences have also been recovered from the Indian Ocean (Not et al., 2008).
Light intensity and clade distributions
In our study, Clade OII was relatively abundant in Sargasso Sea surface waters. Without further research, factors such as physical mixing, potentially displacing OII to the surface, even if not growing there, or the possibility of differently adapted ecotypes within the 18S OII rDNA grouping cannot be ruled out as contributing factors. Still, the discrepancy between environmental results and culture-based inferences are not surprising given the light intensities explored in the laboratory studies.
In oceanic systems light levels of 800 μmol photons m−2 s−1, where either the onset of photoinhibition (OI representatives) or lack of growth (OII representatives) has been observed for cultures (Rodriguez et al., 2005), are probably rarely encountered. Irradiance quickly decreases with depth. We recorded 21 and 6% (Photosynthetically active radiation) relative to levels in ‘air' (detector out of the water) at 2 m (590 μmol photon m−2 s−1) and 15 m (175 μmol photon m−2 s−1), respectively, on a sunny day in oligotrophic waters (33° 10′ 16″ N, 129° 15′ 25″ W). At 76 and 93 m, PAR was 12 and 5 μmol photon m−2 s−1 or 0.4 and 0.2% of surface (air) irradiance, respectively. Although PAR detector calibration could have a role (air measurement 2863 μmol photon m−2 s−1, a relatively high measurement) the percentages should reflect light attenuation. Thus, the same percentages applied to what is typically considered full sun irradiance (2000 μmol photon m−2 s−1) render PAR of 412 μmol photon m−2 s−1 at 2 m, 122 μmol photon m−2 s−1 at 15 m, 8 μmol photon m−2 s−1 at 76 m, 2 μmol photon m−2 s−1 at 106 m (the 0.1% irradiance level). The published μmax values for RCC141 and RCC143, belonging to OII, appear to occur between 85 and 180 μmol photon m−2 s−1 and photoinhibition was only apparent at ⩾400 μmol photon m−2 s−1 under the experimental conditions used. Here, we observed a higher μmax for O. tauri (1.6–1.8 per day), occurring between 150 and 250 μmol photon m−2 s−1, and growth rates as high as 0.3 per day at 4 μmol photon m−2 s−1 (Supplementary Figure 4). These values are higher than those published previously for O. tauri or even those for OII representatives at similarly low irradiances (Rodriguez et al., 2005). However, the results cannot be compared directly because of potential differences in culturing conditions, for example, a different K-media seawater base. Moreover, RCC809 (representing OII) growth rates were not attained because we could not maintain this strain in acclimated, mid-exponential growth at any light level (because of it ‘crashing', despite daily attention), which we felt necessary for establishing the relationship between irradiance and growth rate (as done for the O. tauri measurements). Still, these comparisons, and reflection on natural irradiance levels, emphasize difficulties with relating culture-based experiments to field conditions. Only the highest irradiances encountered in the field, occurring above ∼10 m seem similar to photoinhibition levels seen for cultures. Furthermore, water column stability influences the extent and duration that cells are exposed to high surface irradiance. Given that such exposure may be rare in a well-stratified column, and of limited duration in less well-stratified waters, it seems unlikely that high culture irradiances relate directly to natural conditions.
These observations do not negate reported photophysiology-based differences. For example, maximal growth rates were reported to be ∼1.1 per day at 180 μmol photon m−2 s−1 (OTH95, RCC356 and RCC420), or for RCC501, at 400 μmol photon m−2 s−1. In contrast, OII isolates grew at 0.2±∼0.2 per day to ∼0.6 per day at 400 μmol photons m−2 s−1 for RCC143 and RCC141, respectively, with maximal growth rates at 85 μmol photon m−2 s−1 (∼0.65 per day) and 180 μmol photon m−2 s−1 (∼0.9 per day), respectively (Rodriguez et al., 2005). Six et al. (2008) saw significant differences between the growth optima of a clonal version of RCC141 (that is, RCC809) compared with the original strain used by Rodriguez et al. (2005), and showed that below 15 μmol photon m−2 s−1 both RCC809 and OTH95 had growth rates of ∼0.4 per day. More recently, differences in photoprotection capacity (greater in O. tauri, representing OI) and light-harvesting capacity (greater in RCC809, representing OII), which correspond with conditions at the strain isolation site (Cardol et al., 2008) have been reported. Responses to high-light exposure indicate these two strains differ in terms of recovery from photoinactivation (Six et al., 2009). The extent to which these differences extend to other isolates, or natural populations, and how they respond under optimal growth conditions (for example, temperature) for each respective isolate, requires further investigation. Experiments show that O. tauri and RCC809 have the most disparate responses and photosystem characteristics, with O. lucimarinus (Clade A/OI) often appearing to form a more intermediate response (Six et al., 2008, 2009). Differences in O. tauri, relative to O. lucimarinus and RCC809 may also reflect its unique isolation environment (a shallow lagoon).
Results from a single dilution experiment rendered a picoeukaryote growth rate of 0.5 per day at 70 m in the Sargasso Sea. This regression-based rate represents the average for all small eukaryotic phytoplankton by FCM. The average minimum growth rate for all non-prymnesiophyte eukaryotic phytoplankton, with OII included, was 0.4 per day based on the 20% treatment (see also Cuvelier et al., 2010). OII sequences were the most abundant in an associated clone library, having 100% identity to RCC143 (32 of 36 clones from photosynthetic taxa; also for 15 m clone library, e.g. OC413_NSSJun05_15mQ004T1F), although Clade MI/MII Micromonas, Bathycoccus, a likely photosynthetic stramenopile (96% identity to Pinguiochrysis pyriformis, AB058926) were also present as were sequences likely from non-photosynthetic taxa (for example, novel alveolates). qPCR showed high OII abundance at To (4864±224 rDNA copies per ml; 6441 FCM eukaryotic phytoplankton cells per ml, with FISH prymnesiophytes excluded). Depending on how OII copy numbers relate to cellular abundance and assuming 100% recovery during extraction, either 38% (2 copies per genome, the more likely number) or 76% (1 copy per genome) of the eukaryotic phytoplankton belonged to OII, a significant portion of the picophytoeukaryote community present during this experiment.
Conclusions
Ecotypic differentiation based on photophysiology and other characteristics has been demonstrated for the cyanobacterium Prochlorococcus (Moore et al., 1998, 2002; Moore and Chisholm, 1999). The offset between irradiances where cultured Prochlorococcus strains grow or are inhibited appears to be greater over a narrower range of irradiances (than for Ostreococcus). Furthermore, these irradiances (used in Prochlorococcus studies) are more similar to those at different depths in situ and Prochlorococcus niche partitioning occurs along depth gradients at the same geographic location. Clear shifts can be seen from dominance by high-light adapted ecotypes in surface waters to low-light adapted ecotypes at depth although some clades tolerate a greater irradiance range than others (Johnson et al., 2006; Zinser et al., 2007).
These observations for Prochlorococcus are quite different than those for Ostreococcus here. Although photophysiology may have a role in allowing OII to survive in low-irradiance waters, it seems unlikely that it is the primary environmental driver for Ostreococcus clade distributions. One caveat is that our sampling was not exhaustive. For example, we had no open-ocean ‘winter' samples (for example, January, Sargasso Sea). If OI and OII thrive together at this time, or partition the euphotic zone vertically to strata where one dominates the other, we would not have detected this due to the composition of our global sample set.
In light of our current knowledge, it seems premature to classify the clades as either depth-specialists or generalists. Comparative genomic analyses and physiology studies should facilitate identification of specific niche defining factors. Based on this study, we conclude that Ostreococcus Clade OI thrives in cooler, more nutrient rich waters found in more coastal environments, including the base of the mixed layer in such settings. In contrast, Clade OII thrives in warmer, higher salinity open-ocean settings and its success in such environments may involve the capacity to grow at deeper nutricline depths in gyre euphotic zones.
Acknowledgments
We thank the captains and crews of the research vessels employed for this research. We are grateful to B Binder for berthing for AZW on EN351, EN360 and OC374 and for MLC on OC413, F Chavez and K Johnson for berthing for ED on CN107 and CN207, J Montoya for KM0703 cruise orchestration as well as F Not, RM Welsh, A Engman, MP Simmons and SM Dostal for cruise participation. In addition, K Vergin, R Parsons, SJ Giovannoni and C Carlson provided 2000 and 2003 BATS DNA samples, K Turk provided invaluable assistance, R Foster provided significant input on qPCR primer-probe set development, and P Moisander helped with nutrient data. H Moreau kindly provided Ostreococcus RCC809 and RCC789 DNA or cultures. R Welsh and H Wilcox assisted with clone library construction while A Ortiz, D McRose and H Alexander assisted with initial library analyses. We also thank T Campbell for performing multivariate statistical analyses on MBTS. F Chavez and K Johnson provided nutrient data for CN and MBTS cruises, most having been published elsewhere. Nutrient analyses for Florida Straits were kindly run by L Zamora. Finally, we thank anonymous reviewers for suggestions and comments. This research was funded by a Gordon and Betty Moore Foundation (GBMF) Young Investigator Award to AZW as well as grants from the Lucille and David Packard Foundation and NSF OCE-0623928/OCE-0836721 and GBMF grant 1668 to AZW.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Author contributions
JPZ and AZW orchestrated cruises. ED, MLC, JPZ and AZW performed cruise sampling. ED, MLC, SS, JPZ and AZW performed extractions/provided DNA. MLC performed O. tauri physiology studies, FISH, initial qPCR and the dilution experiment. ED designed, tested and implemented Ostreococcus primer-probe sets, as well as Micromonas and Bathycoccus primer-probe sets. MLC and AZW constructed clone libraries. ED compiled qPCR data. ED, SS and AZW compiled environmental data. AZW performed statistical analyses and global data synthesis. ED synthesized MBTS data. SS further verified Micromonas and Bathycoccus primer-probe and performed a subset of inhibition analysis. CLG processed and integrated sea surface temperature data, created projections and mapped Ostreococcus data. AZW wrote the paper and ED provided written contributions. SS and JPZ provided edits. All authors read and commented on the paper.
Supplementary Material
References
- Cardol P, Bailleul B, Rappaport F, Derelle E, Beal D, Breyton C, et al. An original adaptation of photosynthesis in the marine green alga Ostreococcus. Proc Natl Acad Sci USA. 2008;105:7881–7886. doi: 10.1073/pnas.0802762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
- Countway PD, Caron DA. Abundance and distribution of Ostreococcus sp. in the San Pedro Channel, California, as revealed by quantitative PCR. Appl Environ Microbiol. 2006;72:2496–2506. doi: 10.1128/AEM.72.4.2496-2506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courties C, Vaquer A, Troussellier M, Lautier J, Chretiennot-Dinet MJ, Neveux J, et al. Smallest eukaryotic organism. Nature. 1994;370:255. [Google Scholar]
- Cuvelier ML, Allen AE, Monier A, McCrow JP, Messié M, Tringe SG, et al. Targeted metagenomics and ecology of globally important uncultured eukaryotic phytoplankton. Proc Natl Acad Sci USA. 2010;107:14679–14684. doi: 10.1073/pnas.1001665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvelier ML, Ortiz A, Kim E, Moehlig H, Richardson DE, Heidelberg JF, et al. Widespread distribution of a unique marine protistan lineage. Environ Microbiol. 2008;10:1621–1634. doi: 10.1111/j.1462-2920.2008.01580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouze P, Worden AZ, Robbens S, et al. From the cover: genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez B, Pedros-Alio C, Massana R. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol. 2001;67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob C, Ulloa O, Claustre H, Huot Y, Alarcon G, Marie D. Contribution of picoplankton to the total particulate organic carbon concentration in the eastern South Pacific. Biogeosciences. 2007;4:837–852. [Google Scholar]
- Guillou L, Eikrem W, Chretiennot-Dinet M, Le Gall F, Massana R, Romari K, et al. Diversity of picoplanktonic prasinophytes assessed by direct nuclear SSU rDNA sequencing of environmental samples and novel isolates retrieved from oceanic and coastal marine ecosystems. Protist. 2004;155:193–214. doi: 10.1078/143446104774199592. [DOI] [PubMed] [Google Scholar]
- Jardillier L, Zubkov MV, Pearman J, Scanlan DJ. Significant CO2 fixation by small prymnesiophytes in the subtropical and tropical northeast Atlantic Ocean. ISME J. 2010;4:1180–1192. doi: 10.1038/ismej.2010.36. [DOI] [PubMed] [Google Scholar]
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EM, Chisholm SW. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- Li WKW. Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraplankton: measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–175. [Google Scholar]
- Lomas MW, Steinberg DK, Dickey T, Carlson CA, Nelson NB, Condon RH, et al. Increased ocean carbon export in the Sargasso Sea linked to climate variability is countered by its enhanced mesopelagic attenuation. Biogeosciences. 2010;7:57–70. [Google Scholar]
- Moisander PH, Beinart RA, Hewson I, White AE, Johnson KS, Carlson CA, et al. Unicellular cyanobacterial distributions broaden the oceanic N2 fixation domain. Science. 2010;327:1512–1514. doi: 10.1126/science.1185468. [DOI] [PubMed] [Google Scholar]
- Moisander PH, Beinart RA, Voss M, Zehr JP. Diversity and abundance of diazotrophic microorganisms in the South China Sea during intermonsoon. ISME J. 2008;2:954–967. doi: 10.1038/ismej.2008.51. [DOI] [PubMed] [Google Scholar]
- Moon-van der Staay S, van der Staay G, Guillou L, Vaulot D, Claustre H, Medlin L. Abundance and diversity of prymnesiophytes in the picoplankton community from the equatorial Pacific Ocean inferred from 18S rDNA sequences. Limnol Oceanogr. 2000;45:98–109. [Google Scholar]
- Moore L, Post A, Rocap G, Chisholm S. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol Oceanogr. 2002;47:989–996. [Google Scholar]
- Moore L, Rocap G, Chisholm S. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- Moore LR, Chisholm SW. Photophysiology of the marine cyanobacterium Prochlorococcus: Ecotypic differences among cultured isolates. Limnol Oceanogr. 1999;44:628–638. [Google Scholar]
- Not F, Gausling R, Azam F, Heidelberg JF, Worden AZ. Vertical distribution of picoeukaryotic diversity in the open ocean. Environ Microbiol. 2007;9:1233–1252. doi: 10.1111/j.1462-2920.2007.01247.x. [DOI] [PubMed] [Google Scholar]
- Not F, Latasa M, Marie D, Cariou T, Vaulot D, Simon N. A single species, Micromonas pusilla (Prasinophyceae), dominates the eukaryotic picoplankton in the Western English Channel. Appl Environ Microbiol. 2004;70:4064–4072. doi: 10.1128/AEM.70.7.4064-4072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Not F, Latasa M, Scharek R, Viprey M, Karleskind P, Balague V, et al. Protistan assemblages across the Indian Ocean, with a specific emphasis on the picoeukaryotes. Deep-Sea Res Part I-Oceanographic Res Papers. 2008;55:1456–1473. [Google Scholar]
- Not F, Massana R, Latasa M, Marie D, Colson C, Eikrem W, et al. Late summer community composition and abundance of photosynthetic picoeukaryotes in Norwegian and Barents Seas. Limnol Oceanogr. 2005;50:1677–1686. [Google Scholar]
- Palenik B, Grimwood J, Aerts A, Rouze P, Salamov A, Putnam N, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé MS, Suzuki MT, Vergin KL, Giovannoni SJ. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States. Appl Environ Microbiol. 1998;64:294–303. doi: 10.1128/aem.64.1.294-303.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F, Derelle E, Guillou L, Le Gall F, Vaulot D, Moreau H. Ecotype diversity in the marine picoeukaryote Ostreococcus (Chlorophyta, Prasinophyceae) Environ Microbiol. 2005;7:853–859. doi: 10.1111/j.1462-2920.2005.00758.x. [DOI] [PubMed] [Google Scholar]
- Santoro A, Casciotti KL, Francis CA. Activity, abundance, and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol. 2010;12:1989–2006. doi: 10.1111/j.1462-2920.2010.02205.x. [DOI] [PubMed] [Google Scholar]
- Shi XL, Marie D, Jardillier L, Scanlan DJ, Vaulot D. Groups without cultured representatives dominate eukaryotic picophytoplankton in the oligotrophic South East Pacific Ocean. PLoS ONE. 2009;4:e7657. doi: 10.1371/journal.pone.0007657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SM, Zehr JP. Quantitative analysis of nifH genes and transcripts from aquatic environments. Environ Microbiol. 2005;397:380–394. doi: 10.1016/S0076-6879(05)97023-7. [DOI] [PubMed] [Google Scholar]
- Six C, Finkel ZV, Rodriguez F, Marie D, Partensky F, Campbell DA. Contrasting photoacclimation costs in ecotypes of the marine eukaryotic picoplankter Ostreococcus. Limnol Oceanogr. 2008;53:255–265. [Google Scholar]
- Six C, Sherrard R, Lionard M, Roy S, Campbell DA. Photosystem II and pigment dynamics among ecotypes of the green alga Ostreococcus. Plant Physiol. 2009;151:379–390. doi: 10.1104/pp.109.140566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg DK, Carlson CA, Bates NR, Johnson RJ, Michaels AF, Knap AH. Overview of the US JGOFS Bermuda Atlantic Time-series Study (BATS): a decade-scale look at ocean biology and biogeochemistry. Deep-Sea Res Part II-Topical Stud Oceanography. 2001;48:1405–1447. [Google Scholar]
- Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3:1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- Vaulot D, Eikrem W, Viprey M, Moreau H. The diversity of small eukaryotic phytoplankton (⩽3 μm) in marine ecosystems. FEMS Microbiol Rev. 2008;32:795–820. doi: 10.1111/j.1574-6976.2008.00121.x. [DOI] [PubMed] [Google Scholar]
- Viprey M, Guillou L, Ferreol M, Vaulot D. Wide genetic diversity of picoplanktonic green algae (Chloroplastida) in the Mediterranean Sea uncovered by a phylum-biased PCR approach. Environ Microbiol. 2008;10:1804–1822. doi: 10.1111/j.1462-2920.2008.01602.x. [DOI] [PubMed] [Google Scholar]
- Worden AZ. Picoeukaryote diversity in coastal waters of the Pacific Ocean. Aquat Microb Ecol. 2006;43:165–175. [Google Scholar]
- Worden AZ, Binder BJ. Application of dilution experiments for measuring growth and mortality rates among Prochlorococcus and Synechococcus populations in oligotrophic environments. Aquat Microb Ecol. 2003;30:159–174. [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouze P, Simmons MP, Aerts AL, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- Worden AZ, Nolan JK, Palenik B. Assessing the dynamics and ecology of marine picophytoplankton: The importance of the eukaryotic component. Limnol Oceanogr. 2004;49:168–179. [Google Scholar]
- Worden AZ, Not F.2008Ecology and diversity of picoeukaryotesIn: Kirchman DL (ed).Microbial Ecology of the Oceans2nd edn.Wiley: New York; p594 [Google Scholar]
- Zhu F, Massana R, Not F, Marie D, Vaulot D. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol. 2005;52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Zinser ER, Johnson ZI, Coe A, Karaca E, Veneziano D, Chisholm SW. Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol Oceanogr. 2007;52:2205–2220. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.