Legionella pneumophila actively modulates host vesicle trafficking pathways to facilitate its intracellular replication with effectors translocated by the Dot/Icm type IV secretion system (T4SS)1. The SidM/DrrA protein functions by locking the small GTPase Rab1 into an active form by its guanine nucleotide exchange factor (GEF) and AMPylation activity2-4. Here we demonstrate that the L. pneumophila protein SidD preferably deAMPylates Rab1. We found that the deAMPylation activity of SidD could suppress the toxicity of SidM to yeast and is required to efficiently release Rab1 from bacterial phagosomes. A molecular mechanism for the temporal control of Rab1 activity in different phases of L. pneumophila infection is thus established. These observations indicate that AMPylation-mediated signal transduction is a reversible process regulated by specific enzymes.
L. pneumophila, the etiological agent of Legionnaires’ disease is capable of colonizing a wide range of eukaryotic cells. Successful infection by this pathogen requires the Dot/Icm T4SS, which translocates numerous protein substrates into host cells1. These proteins modulate various host cellular pathways, such as vesicle trafficking, cell death, lipid metabolism and protein synthesis to construct a phagosome permissive for intracellular bacterial replication5. Accumulating evidence suggests that a unique repertoire of effectors is required for each phase of the infection. There is a need for the bacterium to regulate the activity of its virulence factor, because some effectors are potentially detrimental to host cells. Such regulation can be achieved by various mechanisms, including the control of the amount of protein delivered into host cells, the stability of the protein or susceptibility of the protein to host degradation machinery. L. pneumophila has evolved unique mechanisms to neutralize the activity of effectors whose functions presumably have become detrimental to or no longer important for the development of the bacterial phagosome. For example, several hours after infection, the bacterial E3 ubiquitin ligase, LubX, targets the effector SidH for proteasomal degradation6. Similarly, within several hours after uptake, LepB, a GTPase activation protein (GAP) for Rab1, antagonizes the effects of SidM (also known as DrrA), which recruits the small GTPase to the Legionella containing vacuole (LCV) and converts it into the active GTP-bound form via its guanine nucleotide exchange factor (GEF) activity2, 3, 7. SidM also catalyzes an adenosine monophosphate modification (AMPylation) on the tyrosine 77 of Rab1 to lock it into the active form 4. Posttranslational modification by AMPylation has recently emerged as a novel cellular signaling mechanism utilized by all domains of organisms8, 9. However, little is known about the regulation of this signaling mechanism and naturally occurring enzymes involved in the reversal of the modification remain elusive.
In an earlier study we isolated a number of Dot/Icm substrates toxic to yeast, such as SidI, Lgts, SidM and AnkX10-12. To determine whether the activity of any of these proteins is under direct regulation of bacterial factors, we initiated screenings to identify L. pneumophila proteins capable of suppressing the toxicity to yeast. A plasmid-borne L. pneumophila genomic library was introduced into yeast strains expressing toxic effectors from a galactose-inducible promoter, leading to the identification of a number of clones that efficiently suppress the toxicity of SidM. Sequencing revealed that all of these clones harbored sidD (lpg2465), which encodes a Dot/Icm substrate of 507 amino acids13. In the L. pneumophila genome, sidD is localized next to sidM and these two genes are transcribed in divergent orientations2, 13, 14.
Co-expression of sidD completely rescued the growth of the SidM-producing yeast strain on inducing media (Fig. 1a). SidD was unable to suppress the toxicity of AnkX, which is believed to interfere with host vesicle trafficking by AMPylating yet unidentified substrate(s) in a Fic domain-dependent manner15 (Fig. S1), suggesting that the suppressor activity of SidD is specific for SidM.
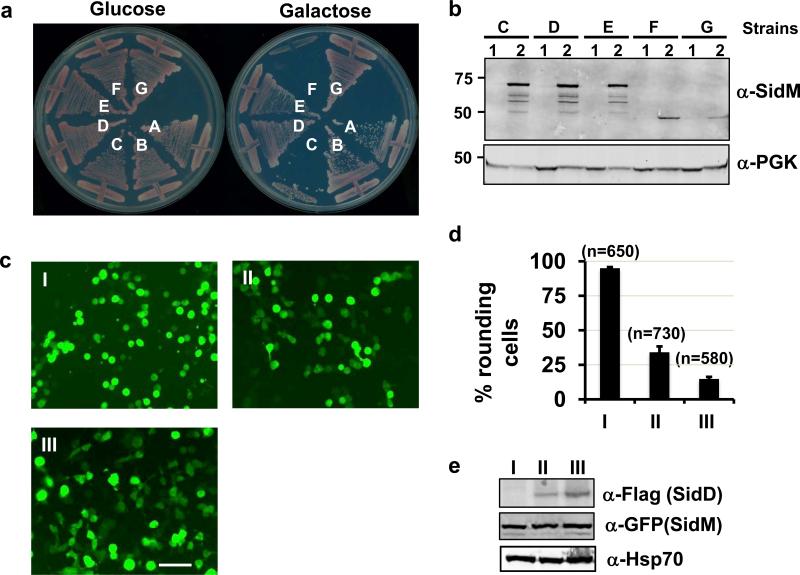
Fig. 1. Suppression of the cytotoxicity of SidM by SidD.
a. Suppression of yeast toxicity of SidM. Yeast strains expressing SidM or SidM1-339 from a galactose-inducible promoter was transformed with various plasmids harboring sidD and the cells were streaked onto plates containing glucose or galactose. Plates were incubated at 30 °C for 3 days before acquiring the images. Yeast strains: A, vector/vector; B, vector/pSidD; C, pSidM/vector; D, pSidM/pSidD (original clone #1); E, pSidM/pSidD; F, pSidM1-339/vector; and G, pSidM1-339/pSidD. b. SidD did not affect the protein level of SidM or SidM1-339 in yeast cells. Subcultures of relevant yeast strains were grown in raffinose (1) or in galactose (2) medium. Crude lysates resolved by SDS-PAGE were probed with SidM-specific antibody. The 3-phosphoglycerate kinase (PGK) was used as a loading control (lower). c-e. Co-expression of SidD rescued the cell-rounding phenotypes caused by SidM1-339. 293T cells were transfected to express SidM1-339. A SidD plasmid was not included (I) or was used at 1:3 (II), or 1:5 (III) molar ratio, respectively. 24 hrs after transfection, samples were analyzed by acquiring images (c), by enumerating green cells exhibiting the rounding phenotype (d) or by immunoblotting to examine the protein levels of SidM1-339 and SidD (e). Experiments were repeated at three times and similar results were obtained. Error bars indicate s.d. Hsp70 was probed as a loading control. Bar, 50 μm.
SidM is a protein of multiple functions, which by binding to phosphatidylinositol 4′-monophosphate, anchors on the Legionella vacuole, recruits and activates the small GTPase Rab12, 3, 16. In particular, its N-terminal domain (SidM1-339) possesses an adenosine monophosphorylation (AMPylation) activity, which covalently modifies Rab1 at tyrosine 77 in a process that requires the G98X(11)D110XD112 motif conserved between SidM and the glutamine synthetase adenylyl transferase 4. We thus examined whether SidD is able to suppress the toxicity induced by SidM1-339. Expression of SidM1-339 strongly inhibited yeast growth (Fig. 1a, strain F) and such inhibition can be suppressed by SidD (Fig. 1a, strain G), suggesting that SidD interferes with the activity conferred by the AMPylation function. We also examined the ability of SidD to suppress the AMPylation-dependent toxicity of SidM1-339 in mammalian 293T cells4. Transfection of these cells with GFP-SidM1-339 led to extensive cell rounding. Co-transfection of the cells to express Flag-SidD suppressed this toxicity (Fig. 1c-e). When 5 times more DNA of the SidD expressing plasmid was used, at least 85 % of the cells were protected (Fig. 1c-e). Together, these data suggest that SidD is able to neutralize the effect of SidM in eukaryotic cells either by inhibiting its AMPylation activity or by reversing its effects on the target molecule Rab1.
To determine the mechanism of action of SidD, we tested its effect on the AMPylation activity of SidM with purified recombinant proteins. Incubation of GSTSidM with GST-Rab1 in the presence of 32P-α-ATP for 30 min led to robust production of radiolabeled Rab1 (Fig. 2a, lane I). Consistent with the genetic data, inclusion of His6-SidD in the AMPylation reaction abolished the formation of 32P-α-AMP-Rab1 (Fig. 2a, lane II). The lack of AMPylation can be a result of SidD-mediated inhibition of SidM activity or of a deAMPylation function of SidD. Because full-length SidM has a high binding affinity for Rab12-4, its presence may interfere with SidD activity. We distinguished between these two possibilities by producing 32P-α-AMP-Rab1 using His6-SidM1-339, which catalyzes the reaction but did not detectably bind GST-Rab1 in vitro 2-4 (Fig. S2). After removing His6-SidM1-339, AMPylated Rab1 was incubated with various amounts of His6-SidD for 5 min. Under these conditions, the amount of radiolabeled GSTRab1 significantly decreased when as little as 0.6 μg His6-SidD was included in the reaction, and 1.2 μg His6-SidD can almost completely deAMPylate 32P-α-AMP-Rab1 (Fig. 2b). In a time course analysis, incubation of 1.0 μg SidD with 10 μg modified Rab1 for 1 min led to more than a 50% reduction in radiolabeled substrate (Fig. 2c). SidD is also able to remove the AMP moiety from other Rab proteins modified by SidM 4 (Fig. S3).
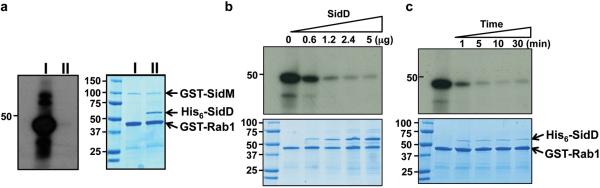
Fig. 2. SidD is a deAMPylase that targets SidM-modified Rab1.
a. SidD prevented SidM-mediated AMPylation of Rab1. Shown were AMPylation reactions containing GSTSidM and GST-Rab1without (II) or with (I) His6-SidD. After SDS-PAGE, 32P-α-AMPRab1 was detected by autoradiography (left panel) and proteins were detected by Coomassie bright blue staining (right panel). b. Dose-dependent deAMPylation by SidD. His6-SidD was added to identical samples containing AMPylated GST-Rab1 to establish reactions in which the molar ratio of Rab1 and SidD is 8, 4, 2 and 1, respectively; reactions were terminated after 5 min of incubation. AMPylated GST-Rab1 and proteins in reactions were detected as described in a. c. Time course of SidD activity. AMPylated GST-Rab1 was mixed with His6-SidD at a molar ratio of 10:1, reactions were terminated at the indicated time intervals. AMPylated GST-Rab1 (upper panel) and proteins (lower) were similarly detected. Markers for protein size (kDa) are indicated.
To verify the SidD-mediated removal of the AMP moiety from modified Rab1, AMPylated Rab1 treated with His6-SidD completely digested with trypsin was subjected to mass spectrometric analysis. The relative intensity ratio data revealed that approximately 50% of the Rab1 was AMPylated when 10 μg Rab1 was incubated with 1.5 μg SidM for 30 min (Fig. S4a). The ratio of unmodified Rab1 increased about 10-fold when 4 μg His6-SidD was added to the reaction for 10 min and further increased to 20-fold when the incubation was extended for 30 min (Fig. S4 b-c). Furthermore, we did not detect any mass loss of the Rab1 peptide containing tyrosine 77, suggesting that deAMPylation by SidD was not caused by other hydrolytic reaction. Taken together, these results indicate that SidD is a deAMPylase that functions to reverse the SidM-mediated posttranslational modification of Rab1.
Sequence analysis with HHPred17 revealed that the predicted secondary structure of SidD N-terminal portion is detectably similar to some protein phosphatases (Fig. S5). In particular, the Asp residues at positions 92 and 110 of SidD are conserved with residues critical for the biochemical activity of SaSTP and tPphA, phosphatases from Streptococcus agalactiae and Thermosynechococcus elongates, respectively18, 19 (Fig. S5). Substitution of D92 or D110 with alanine completely abolished the ability of SidD to suppress the toxicity for SidM to yeast whereas a mutation in D60 did not detectably affect its activity (Fig. (Fig. 3 a-b). Consistently, SidDD92A and SidDD110A both had lost the activity to remove the AMP moiety from AMPylated Rab1 (Fig. 3c). These results indicate that residues D92 and D110 participate in the formation of the catalytic pocket or structural features important for its enzymatic function.
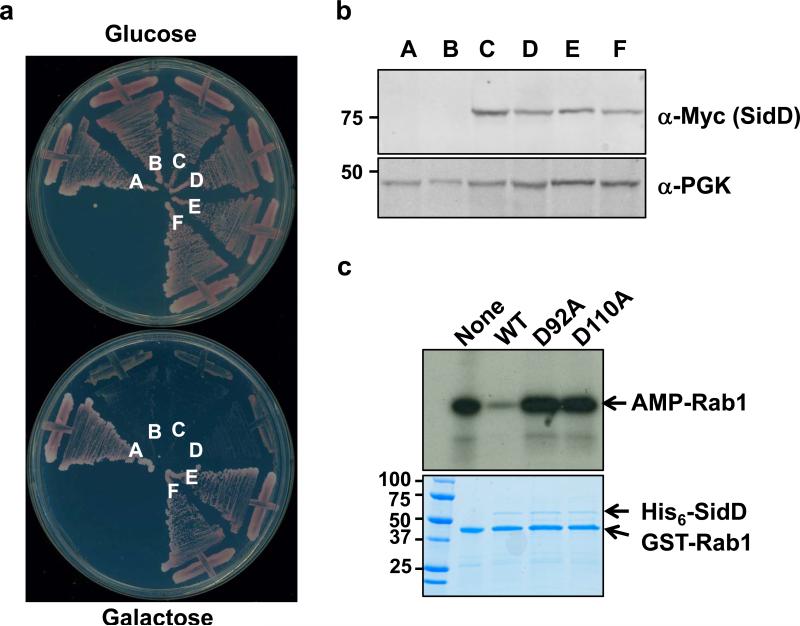
Fig. 3. The Asp residue at position 92 or 110 is important for SidD activity.
a. Mutations of D92 and D110 abolished the ability of SidD to suppress the yeast cytotoxicity of SidM. Plasmids harboring sidD or its mutants were transformed into the SidM-expression yeast strain; cells were streaked onto plates containing glucose or galactose. Yeast strains: A, vector/vector; B, pSidM/vector; C, pSidM/pSidDD92A; D, pSidM/pSidDD110A; E, pSidM/pSidDD60A and F, pSidM/pSidD. b. Expression of the SidD mutants in yeast, samples prepared as described in Figure 1 and were probed for SidD (Myc-tagged) and for PGK. c. SidDD92A and SidDD110A have lost the deAMPylation activity. 1.5 micrograms purified proteins were added to reactions containing AMPylated Rab1. After 30 min incubation, reactions were terminated by SDS sample buffer. 32P-α-AMP-GST-Rab1 was detected by autoradiography and the proteins were detected by Coomassie bright blue staining (lower panel). Protein size (kDa) references are indicated on the left lane of the gel.
In bone marrow derived mouse macrophages, the association of Rab1 with LCVs peaks at 2 hrs after bacterial uptake and begins to disappear when infection has proceeded for 4 hrs7. The removal of Rab1 from LCVs is presumably due to extraction by RabGDI, which only interacts with the inactive GDP-bound form20. LepB is unable to induce GTP hydrolysis of AMPylated Rab1, suggesting that a deAMPylation factor is necessary for the production of the GDP-bound, inactive Rab14. Thus, we examined whether SidD is required for efficient removal of Rab1 from the LCVs. In infections with wild type bacteria, the percentages of vacuoles staining positively for Rab1 began to diminish at 2 hrs (Fig. 4a). On the other hand, in infections with the sidD deletion mutant, the rates of Rab1 positive vacuoles remained at the maximal level (about 60%) 4 hrs after infection (Fig. 4). The kinetics of Rab1-positive LCVs was restored to the pattern of wild type bacteria when SidD was expressed in the mutant (Fig. 4a-b). Interestingly, the association of Rab1 with the LCV did not persist after 4 hrs of infection with the sidD mutant (Fig. 4a), suggesting the contribution of host deAMPylases or additional bacterial proteins for reversal of the modification. These results indicate that SidD plays an important role in the efficient removal of Rab1 from the Legionella phagosome.
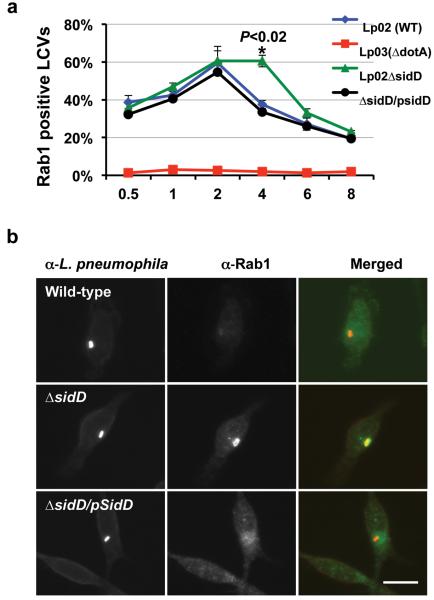
Fig. 4. SidD is required for efficient removal of Rab1 from L. pneumophila phagosome.
a. Mouse macrophages were infected with relevant L. pneumophila strains. At the indicated time points, fixed samples were probed for L. pneumophila and Rab1 with specific antibodies followed by Texas red and FITC-conjugated secondary antibodies, respectively. Processed samples were scored for co-localization of Rab1 with the bacterial phagosomes. Data shown are from two independent experiments performed in triplicate in which at least 100 phagosomes were scored per sample. b. Association of Rab1 with L. pneumophila phagosome 4 hrs after infection. Shown are images of wild type (Lp02, dot/icm+), the sidD deletion mutant (Lp02ΔsidD, dot/icm+) and the complementation strain (Lp02ΔsidD/Flag-SidD) residing in macrophages 4 hrs after infection. L. pneumophila and Rab1 are labeled as described in a, with bacteria marked in red and Rab1 marked in green. Bar, 10 μm. Note the difference in the intensity of Rab1 staining signals among the three bacterial strains. At least 150 vacuoles were scored each sample and error bars indicate s.d. Similar results were obtained in at least three independent experiments.
AMPylation of proteins often leads to alteration of their enzymatic activity or the ability to interact with target molecules 4, 8, 21, 22. Proteins with AMPylation activity dependent upon the Fic domain are present in all kingdoms and at least one has been characterized in humans8, 9. Although the importance of signaling pathways regulated by AMPylation has not yet been fully recognized, the fact that this reaction can be catalyzed by proteins lacking a Fic domain such as SidM suggests extensive involvement of this posttranslational modification in cellular signaling processes. The identification of a deAMPylase revealed that like other post-translational modifications involved in signal transduction, AMPylation is a reversible process regulated by specific enzymes. Similar to sidM2, 3and lepB 7, sidD is not required for bacterial intracellular growth 13, probably due to functional redundancy among the several hundreds Dot/Icm substrates 23 or the limitation of the experimental systems. Nevertheless, further study on the structure and function of SidD will advance not only our understanding of its roles in bacterial pathogenesis but also the involvement of such enzymes in other cellular processes.
Methods Summary
Bacterial, yeast strains and plasmid construction
All L. pneumophila strains used in this study were derivatives of the Philadelphia 1 strain Lp02 24. E. coli strains were grown and maintained on LB agar or LB broth. When necessary antibiotics were included as described 25. Strains of L. pneumophila were grown and maintained on CYE medium or in AYE broth as previously described 25. The sidD in-frame deletion mutant was constructed in an earlier study 13. In this mutant, the open reading frame of sidD was replaced by a 32-amino acid (aa) polypeptide consisting of the first and the last 15 aa and two aa encoded by the sequence of the BamHI restriction enzyme 13. For complementation experiments, a Flag-tagged sidD gene was inserted into the sidD locus of the deletion mutant using a two-step site-specific recombination with the π protein dependent plasmid pSR47s 24 with an established procedure 26. Successful insertion of the gene into the bacterial chromosome was determined by PCR reactions with flag-tag specific primers and by the expression of Flag-tagged SidD. All infections were performed with bacterial cultures grown to the post-exponential phase as judged by optical density of the cultures (OD600=3.3-3.8) as well as increase of bacterial motility. For expression in mammalian cells, genes were cloned into pEGFPC1 (Clontech) or a 4×Flag vector (Sigma). The integrity of all constructs was verified by sequencing analysis. The sequences of all primers used in this study are listed in Table S1.
All yeast strains used were derived from W303 27; yeast was grown at 30 °C in YPD medium or in appropriate amino acid dropout synthetic media with glucose or galactose at a final concentration of 2% as the sole carbon source. Yeast transformation was performed according to a standard procedure 28.
Supplementary Material
Acknowledgments
We thank Dr. Ralph Isberg (Tufts Medical School, Boston, USA) for the antibody against SidM and Drs. Art Aronson and Andy Tao (Purdue University, West Lafayette, USA) for critical reading of the manuscript and for helpful discussion. This work was supported by NIH-NIAID grants R01AI069344, K02AI085403 and R21AI092043 (Z.-Q.L).
Methods
Construction of a L. pneumophila genomic library on a yeast expression vector
L. pneumophila genomic DNA partially digested with the restriction Sau3AI was separated by agarose gel electrophoresis. DNA fragments of 1-8 kbp were recovered from the gels and ligated to BamHI digested yeast vector pGBKT7 (Clontech). Ligated plasmid DNA was introduced into the E. coli strain DH5α by electroporation; approximately 4X105 independent colonies were pooled and used to extract total plasmid DNA.
Identification of L. pneumophila genes capable of suppressing yeast toxicity of SidM
The open reading frame of sidM was inserted into pSB157 12 to generate pSB157::SidM, which was digested with StuI and transformed into the yeast strain W303 27. The yeast strain W303 (pSB157::SidM), which consistently exhibits galactose-dependent SidM toxicity was used for the subsequent screenings. Plasmid DNA of the L. pneumophila genomic library was transformed into the yeast strain and the transformants were plated onto the selective medium with galactose as the sole carbon source. From about 2X106 potential transformants, we obtained a total of 25 colonies that harbor potential suppressor genes. Plasmids carried the potential suppressing genes were rescued and re-introduced into the original yeast strain. Inserts of 16 plasmids that reproducibly suppress the SidM toxicity were sequenced.
Protein purification
To express recombinant proteins, the orfs of sidM, sidD, rab1 and other rab genes were amplified with specific primer pairs (Table S1) and were inserted into appropriately digested pGEX6-P-1 or pQE30 (Qiagen), respectively to produce GST-tagged or His6-tagged proteins. The cDNA of other Rab proteins were purchased from Open Biosystems (Huntsville, AL). The integrity of each gene was verified by double strand sequencing analysis. For protein production, E. coli strains harboring the appropriate expression vector were grown at 37°C in LB medium (100 μg/ml Ampicillin) to an OD600 of 0.5. After adding isopropylthio-d-galactopyranoside (IPTG) to a final concentration of 0.2 mM, the cultures were incubated at 18°C in a shaker for 18 hrs. Harvested cells were suspended in PBS buffer and were lysed in a French press at 1,500 psi. The soluble fraction obtained by centrifugation at 6,000 g for 10 min at 4°C was incubated with glutathione Sepharose resin or Ni2+ resin (Qiagen) equilibrated with PBS. The proteins were purified as described 29. Protein concentrations were determined by the Bradford assay; the purity of all proteins was more than 95% as assessed by SDS-PAGE followed by Coomassie bright blue staining.
Cell culture, infection and transfection
Mouse macrophages were prepared from bone marrow of female A/J mice of 6-10 weeks of age following published protocols 13. 293T cells were cultured in Dulbecco's modified minimum Eagle's medium (DMEM) supplemented with 10% FBS. Established protocols 29 were used for transfection and infection.
Cytotoxicity Assay
Yeast strains harboring the Pgal::sidM constructs co-integrated into the chromosome grown on glucose medium were streaked onto glucose and galactose media, respectively. The growth the cells was assessed after 3-day incubation at 30°C. To examine the effects of SidD on the cytotoxicity of SidM1-339 on mammalian cells, we cotransfected 293T cells with plasmids coding for SidD and SidM1-339 at molar ratios of 3 and 5, respectively. Twenty-four hours after transfection, cells expressing the GFP protein were inspected for the cell-rounding phenotype.
In vitro AMPylation and deAMPylation assays
For AMPylation assays using purified recombinant proteins, 1.5 μg GST-SidM was incubated with 10 μg of GST-Rab1 for 30 minutes at 35°C in an AMPylation buffer containing 25 mM Tris-HCl, pH 7.5, 50 mM NaCl, 3 mM MgCl2, 0.5 mM EDTA and 32P-α-ATP (5μCi) (Perkin Elmer). The AMPylation reaction was stopped by the addition of SDS sample buffer. Samples were boiled for 5 minutes, separated by SDS-PAGE, and detected by autoradiography. The same gels were stained by Coomassie brilliant blue to assess the levels of each protein in the reaction.
To assay the time course of the SidD deAMPylation activity, a 240-μl master reaction containing 9 μg His6-SidM1-339, 60 μg GST-Rab1 bound on glutathione beads and 30 μCi 32P-α-ATP was first set up and was allowed to proceed for 30 min at 35°C. We then washed the beads in spin columns (Sigma) with 10X bed volumes of the ampylation buffer containing 1% Triton X-100 for 5 times to remove His6-SidM1-339 followed by 5X washes with the ampylation buffer. This washing regime was effective as no His6-SidM1-339 was detected by Commassie brilliant blue staining (Fig. S2). Sub-reactions were then established by splitting the master reactions into 6 different test tubes and 1 μg His6-SidD was added to 5 of these reactions. The reaction that never received His6-SidD was terminated with SDS sample buffer and used as a control. After incubation at 35°C for the indicated time duration, the deAMPylation reaction was terminated with SDS loading buffer. To test dose-dependent deAMPylation of Rab1, a master reaction was similarly set up, and after AMPylation, different amounts of SidD (0.6, 1.2, 2.4, 5μg) were added into 4 of the 5 sub-reactions. The 5th sub-reaction terminated after AMPylation was again used as a control. After incubation at 35°C for 5 min, the reactions were stopped with SDS loading buffer. Reaction products were separated on 4-20% SDS-PAGE gels (Bio-Rad) and AMPylated GST-Rab1 was detected by autoradiography. Proteins in the gels were detected by Coomassie brilliant blue staining.
Mass spectrometric analysis
A reaction containing 1.5 μg GST-SidM, 10 μg GST-Rab1 and 1 mM ATP was allowed to proceed for 30 min at 37°C. One third of the reaction was withdrawn and terminated with SDS sample buffer to serve as the modified sample. Identical samples were withdrawn at 10 min and 30 min after the addition of 4 μg of His6-SidD to the remaining reaction. Proteins were separated by SDS-PAGE and the protein bands corresponding to GST-Rab1 were excised. After complete trypsin digestion, the levels of AMPylation of the samples were analyzed by mass spectrometric by the Taplin Mass Spectrometry Facility at Harvard Medical School.
Antibodies, immunostaining and Western Blot
Antibodies against Legionella and GFP were described elsewhere 29. The antibody against Flag (F1804), Rab1 (sc-599) or Myc (sc-40) was purchased from Sigma and Santa cruz Biotechnology, respectively. The antibody against the yeast metabolic 3-phosphoglycerate kinase (PGK) (a6457) was from Invitrogen. The SidM-specific antibody 2 was a kind gift from Dr. Ralph Isberg at Tufts Medical School. Cell fixation, permeabilization and immunostaining were performed as described 25. The concentrations of antibodies and procedure for Western blots were followed established protocols 29.
Footnotes
The authors declare no competing financial interests.
Reference
- 1.Ensminger AW, Isberg RR. Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr Opin Microbiol. 2009;12:67–73. doi: 10.1016/j.mib.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Murata T, et al. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 4.Muller MP, et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 5.Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubori T, Shinzawa N, Kanuka H, Nagai H. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 2010;6:e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 8.Worby CA, et al. The fic domain: regulation of cell signaling by adenylylation. Mol Cell. 2009;34:93–103. doi: 10.1016/j.molcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinch LN, Yarbrough ML, Orth K, Grishin NV. Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS One. 2009;4:e5818. doi: 10.1371/journal.pone.0005818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belyi Y, Tabakova I, Stahl M, Aktories K. Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J Bacteriol. 2008;190:3026–3035. doi: 10.1128/JB.01798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen X, et al. Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 2009;11:911–926. doi: 10.1111/j.1462-5822.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien M, et al. The genomic sequence of the accidental pathogen Legionella pneumophila. Science. 2004;305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 15.Roy CR, Mukherjee S. Bacterial FIC Proteins AMP Up Infection. Sci Signal. 2009;2:e14. doi: 10.1126/scisignal.262pe14. [DOI] [PubMed] [Google Scholar]
- 16.Brombacher E, et al. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem. 2009;284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 18.Rantanen MK, Lehtio L, Rajagopal L, Rubens CE, Goldman A. Structure of Streptococcus agalactiae serine/threonine phosphatase. The subdomain conformation is coupled to the binding of a third metal ion. FEBS J. 2007;274:3128–3137. doi: 10.1111/j.1742-4658.2007.05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlicker C, et al. Structural analysis of the PP2C phosphatase tPphA from Thermosynechococcus elongatus: a flexible flap subdomain controls access to the catalytic site. J Mol Biol. 2008;376:570–581. doi: 10.1016/j.jmb.2007.11.097. [DOI] [PubMed] [Google Scholar]
- 20.Seabra MC, Wasmeier C. Controlling the location and activation of Rab GTPases. Curr Opin Cell Biol. 2004;16:451–457. doi: 10.1016/j.ceb.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Woolery AR, Luong P, Broberg CA, Orth K. AMPylation: Something Old is New Again. Front Microbiol. 2010;1:113. doi: 10.3389/fmicb.2010.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yarbrough ML, et al. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- 23.Zhu W, et al. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One. 2011;6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 25.Conover GM, Derre I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Gao P, Banga S, Luo ZQ. An in vivo gene deletion system for determining temporal requirement of bacterial virulence factors. Proc Natl Acad Sci U S A. 2008;105:9385–9390. doi: 10.1073/pnas.0801055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan HY, Cheng KK, Klein HL. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 delta of Saccharomyces cerevisiae. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 29.Xu L, et al. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 2010;6:e1000822. doi: 10.1371/journal.ppat.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.