Abstract
A key step during the synthesis of collagen constructs is the incubation of monomeric collagen in phosphate buffer saline (PBS) to promote fibrillogenesis in the collagen network. Optimal PBS treatment conditions for monomeric collagen solutions to induce gelation are well established in the literature. Recently, a report in the literature[1] showed a novel method to fabricate highly oriented electrochemically aligned collagen (ELAC) threads which have orders of magnitude greater packing density than collagen gels. The optimal PBS treatment conditions for induction of D-banding pattern in such dense and anisotropic collagen network are unknown. This study aimed to optimize PBS treatment of ELAC threads by investigating the effect of phosphate ion concentration (0.5×, 1×, 5× or 10×) and incubation time (3, 12 or 96 hours) on the mechanical strength and ultrastructural organization by monotonic mechanical testing, small angle X-ray scattering and transmission electron microscopy. ELAC threads incubated in water (No PBS) served as the control. ELAC threads incubated in 1× PBS showed significantly higher extensibility compared to 0.5× or 10× PBS along with the presence of D-banded patterns with a periodicity of 63.83 nm. Incubation of ELAC threads in 1× PBS for 96 hours resulted in significantly higher ultimate stress compared to 3 or 12 hours. However, these threads lacked D-banding pattern. TEM showed no significant differences in the microfibril diameter distribution of ELAC threads treated with or without PBS. This indicates that microfibrils lacked D-banding following electrochemical alignment and the subsequent PBS treatment induced D-banding by reorganization within microfibrils. It was concluded that incubation of aligned collagen in 1× PBS for 12 hours results in mechanically competent, D-banded ELAC threads which can be used for the regeneration of load bearing tissues such as tendons and ligaments.
Keywords: collagen type-I, biomaterial, aligned, PBS, D-banding, microfibril
Introduction
Type-I collagen is the most abundant fibrous protein in load-bearing tissues like ligaments or tendons [2, 3]. The typical structure of type-I collagen in tendons has a unique hierarchical structure that is the key to resist mechanical forces parallel to the fiber direction [4-6]. Therefore, tendon properties are anisotropic. In an effort to mimic the hierarchical and mechanical characteristics of tendons, our laboratory has previously developed a novel method to produce densely packed electrochemically aligned collagen (ELAC) threads [1]. These ELAC threads are mechanically stronger [1] and more receptive to cell migration and population [7] compared to the traditional randomly oriented collagen constructs. A key step during the synthesis of ELAC is the incubation of aligned collagen post electrochemical manipulation in a phosphate buffer saline (PBS) solution to promote fibrillogenesis. Traditionally, collagen gels are synthesized by adding 10× PBS to monomeric collagen solution, neutralizing the pH, and facilitating fibrillogenesis by incubation at 37 °C for 1 – 2 hours. However, unlike collagen gels, the packing density of the collagen molecules within dehydrated ELAC (1030 mg/mL) is one of the highest that has been reported to date [8, 9]. The optimal PBS treatment conditions which apply to loose and isotropic networks of collagen gels may not necessarily apply to ELAC threads. Therefore, PBS concentration and incubation time needs to be optimized to allow fibrillogenesis to take place efficiently in such dense tissue-mimicking collagen constructs. Well-orchestrated fibrillogenesis is critical to synthesize robust tendon-like ELAC threads with the desired mechanical strength and hierarchical organization for tissue engineering applications.
Collagen fibrillogenesis in vitro can be defined as the process in which successive steps of lateral as well as longitudinal aggregation occur along a collagen molecule long axis [10, 11]. This process is known to be temperature-dependent, as well as entropy-driven by the displacement of water-bound molecules around the collagen surface [12]. In addition, collagen fibrillogenesis can also be attributed to hydrophilic interactions via hydrogen bonding between polar residues when measured by osmotic-stress techniques [13, 14]. The growth of fibrillar collagen aggregates has been studied in vitro under various conditions of buffer composition and concentration [15-18], ionic strength [15, 19, 20], pH [15, 17, 18, 20, 21], temperature [15, 16, 20], and stock collagen concentration [15, 19, 20]. These studies have been successful to produce well-organized collagen fibrils in the nano-, and micron-scale. However, there is a lack of studies that analyze the optimal fibrillogenesis conditions to induce fibrillogenesis in dense collagen constructs such as ELAC threads.
The current study focused on elucidating the optimal PBS concentration and incubation duration to obtain ELAC threads that are both mechanically competent and hierarchically ordered. At first, ELAC threads post electrochemical manipulation were incubated with different concentrations of PBS (0.5, 1, 5 or 10×) for 12 hours and the mechanical properties and structural organization of the resultant ELAC threads were determined by monotonic mechanical testing, small angle X-ray scattering (SAXS) and transmission electron microscopy (TEM). Once the optimal concentration was identified, aligned collagen was incubated in PBS for different times (3, 12 or 96 hours) and similar analyses were performed. Monotonic tensile testing revealed the ultimate stress, ultimate strain and Young's Modulus of ELAC threads. SAXS and TEM are two complementary techniques used to characterize the hierarchical structure of ELAC bioscaffolds [19]. SAXS provided global structural order about the packing of collagen molecules within ELAC threads, and TEM provided collagen microfibril diameter information of cross sections of ELAC threads.
Materials and Methods
ELAC fabrication
The methodology employed to fabricate ELAC threads was similar to that reported in Cheng et. al [1]. Briefly, a dialyzed solution of monomeric type-I collagen (Nutragen, Advanced Biomatrix, San Diego, CA) was subjected to isoelectric focusing in a 10-cm long electrochemical cell. The electrochemical cell was placed in a humidity-controlled chamber at 60% relative humidity. Collagen alignment along the isoelectric point occurred after supplying 20 VDC to the electrochemical cell. Immediately after alignment, aligned collagen was incubated at 37 °C in 50 mL of PBS (Sigma-Aldrich, St. Louis, MO) at different concentrations (0.5×, 1×, 5× or 10×) or incubation time (3, 12 or 96 hours). The mechanical properties, D-banding pattern and fibril diameter of the resultant ELAC threads post PBS treatment were analyzed by monotonic testing, SAXS and TEM. ELAC threads incubated in a PBS-free solution (DNAse, RNAse free water –Invitrogen, Carlsbad, CA-) under the same conditions as the PBS-treated groups served as negative control. All PBS solutions were diluted in DNAse, RNAse-free water (Invitrogen, Carlsbad, CA).
Monotonic Mechanical Testing
Cross-sectional area of ELAC threads was reconstructed digitally using a confocal laser scanning microscope (Olympus Fluoview FV1000, Melville, NY). The cross-sectional area was calculated as the average of six measurements along the 2 cm long ELAC thread to be mechanically tested. Measurements of ELAC cross-sectional areas were done using ImageJ (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, MD). ELAC threads were tested under monotonic tension until failure in strain-control (ARES, TA Instruments, New Castle, DE). End regions of ELAC segments were fixed to plastic frames using medical grade adhesive (Loctite 4851). All ELAC threads were hydrated in DNAse, RNAse-free water (Invitrogen, Carlsbad, CA) for 30-45 minutes prior to testing. Plastic frames were mounted on aluminum fixtures which were controlled to maintain a nominal gage length of 4-5 mm. Immediately before testing the edges of the plastic frames were cut and samples were pre-loaded to 0.05 g to provide them with baseline tautness. ELAC threads were tested using a strain rate of 10 mm/min. Load was measured with a 250-gram load cell, and load values were normalized with the cross-sectional area to obtain stress. Gage extension was normalized by initial gage-length at pre-load to obtain the strain. Ultimate tensile stress (UTS), ultimate failure strains (UFS), and Young's moduli (E) were calculated from strain-stress curves. For each group 8 to 12 ELAC threads were tested.
Small Angle X-ray Scattering (SAXS)
ELAC threads for SAXS were dehydrated in a vacuum chamber overnight. ELAC threads were sectioned into 0.5 cm segments and placed perpendicular to the x-ray beam. To compare the SAXS pattern of a sample having a well-defined D-banding configuration to SAXS patterns of ELAC threads, bleached rabbit tendon was used as the positive control. Rabbit tendon samples were harvested by firmly pulling individual bundles from the Achilles tendon using forceps and scalpel. The bundles were bleached following the procedure from Habelitz et al. [22] with slight modifications. Achilles tendon bundles were bleached in 1% NaOCl solution for 2 minutes to expose the collagenous matrix. SAXS patterns were collected using a three-pinhole SAXS camera (Molecular Metrology, The Woodlands, Texas) with a microfocus X-ray source, an Osmic MaxFlux confocal X-ray optic, and a 2D Fujifilm image plate detector. The length between SAXS camera and detector was 1647 mm. All SAXS patterns were collected during a 24-hour period, and all samples were kept in a desiccator prior SAXS experiments. The 2D SAXS scattering data was processed to calculate the D-spacing as described in Cheng et al. [1].
Transmission Electron Microscopy (TEM)
For TEM analysis, freshly harvested ELAC threads were cut into small pieces (1 mm length) using a scalpel blade and fixed in 2% glutaraldehyde (in PBS) for 1 hour. After washing with buffer, the samples were post-fixed in 1% OsO4 (in PBS) for 1 hour. The samples were then washed and dehydrated in a graded series of ethanol, infiltrated with Spurr's epoxy resin and flat embedded in a mold by polymerization at 60 °C for 48 hours. Ultrathin sections were cut in the transverse direction and collected on grids. The sections were stained with 2% uranyl acetate in 70% methanol for 5 minutes and lead citrate for 3 minutes, dried and examined with TEM (FEI/Philips CM-10, FEI Company, Hillsboro, OR). TEM images in the transverse region were analyzed for fibril diameter distribution using ImageJ (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, MD). Images were normalized using quantile-based normalization algorithms to obtain similar histograms for all images. Thresholding and size analysis algorithms were performed to localize particles with sphericity larger than 0.75, and area larger than 20 μm2. The sphericity discrimination prevented the quantification of microfibrils that were 1) off-axis from the plane perpendicular to the image plane, and 2) clumped as aggregates with irregular perimeters. That is, only semi-spherical and spherical microfibrils were considered and quantified.
Statistics
Significant differences between groups were tested with a non-parametric one-way ANOVA (Kruskal-Wallis) test. If the difference between groups was significant, then differences between paired groups were assessed using a non-parametric Mann-Whitney U-test. For all statistical tests, the level of significance was set at p < 0.05, and tests were performed using Minitab-16 (College Station, PA). Significant differences between fibril diameter distributions were analyzed following the protocol developed by Alaseirlis et al. [23]. A Kolmogorov-Smirnov test was used to compare microfibril diameters for ELAC threads incubated in a PBS-free solution, 1× for 12 and 96 hours, and 10× PBS for 12 hours. A level of significance set at p < 0.05, and the test was evaluated using Minitab-16.
Results
Monotonic Mechanical Testing
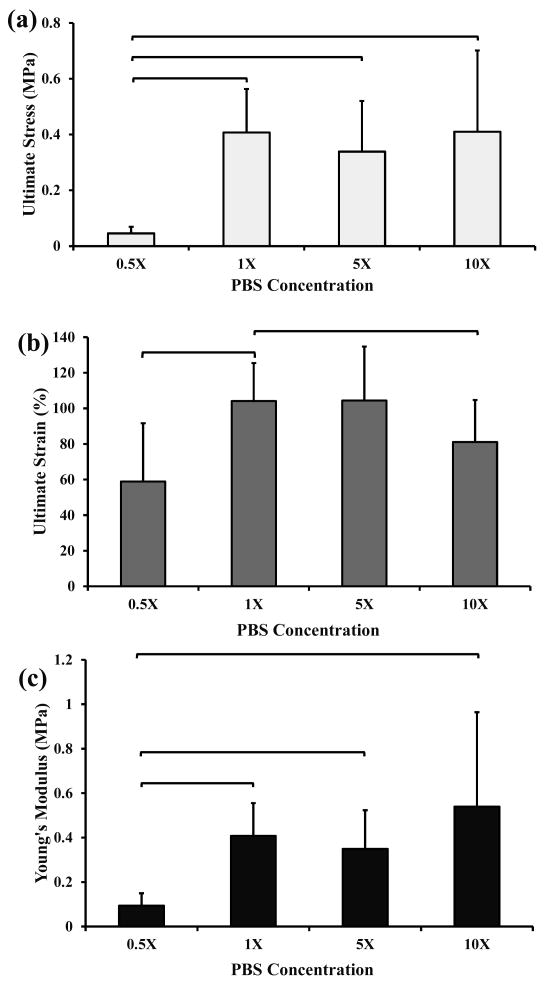
The ultimate stress of ELAC threads incubated in 1, 5, and 10× PBS was significantly higher than that obtained when ELAC threads were incubated in 0.5× PBS (figure 1a). Typical stress values for ELAC threads incubated in 1, 5, and 10× PBS was on average 0.5 MPa. ELAC threads without PBS treatment (negative control) were not tested because they were extremely weak and did not allow physical manipulation and handling. The ultimate strain of ELAC threads incubated in 1× PBS was significantly higher than those incubated at 0.5× and 10× PBS (figure 1b). The Young's modulus of ELAC threads incubated in 1, 5, and 10× PBS was significantly higher than that found for ELAC threads incubated in 0.5× PBS (figure 1c). These results indicate that the mechanical strength of ELAC threads improve significantly when threads are incubated in a PBS concentration of 1× and higher. Also, ELAC threads exhibited significantly higher strain (>100%) when they were incubated in 1× and 5× PBS solutions compared to 0.5× and 10× (figure 1b).
Figure 1.
Mechanical test results of ELAC threads incubated at different PBS concentrations. Mean and standard deviation values were provided in the graphs. Any two groups connected via brackets were significantly different from each other (n=8-12, Mann-Whitney U-test at p<0.05). All the ELAC threads were incubated in PBS for 12 hours.
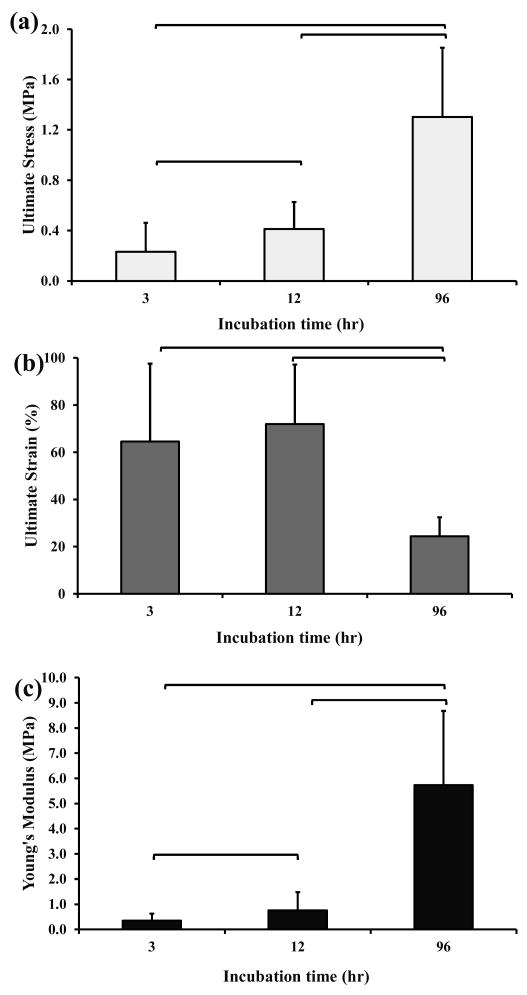
The ultimate stress of ELAC threads incubated for 96 hours (1.2 MPa) was three times higher than that found for ELAC threads incubated for 12 hours (0.4 MPa), and six times higher for ELAC threads incubated for 3 hours (0.2 MPa) (figure 2a).
Figure 2.
Mechanical test results of ELAC threads incubated at different PBS incubation times. Mean and standard deviation values were provided in the graphs. Any two groups connected via brackets were significantly different from each other (n=8-12, Mann-Whitney U-test at p<0.05).
The ultimate strain of ELAC threads incubated for 3 and 12 hours was comparable (> 60%), but it was significantly greater than the strain found in ELAC threads incubated for 96 hours (figure 2b). Additionally, the threads incubated for 96 hours were stiffer than any other ELAC threads, showing Young's moduli values as high as 6 MPa (figure 2c).
Small Angle X-ray Scattering (SAXS)
Bleached rabbit Achilles tendon served as a positive control to confirm the presence of a distinguishable pattern under the operational conditions of the SAXS instrument. ELAC threads incubated in PBS-free solution did not exhibit a periodic SAXS pattern as demonstrated in an earlier study [1]. Additionally, ELAC threads incubated in 10× PBS did not show any distinguishable periodic arc pattern (figure 3a). The periodicity in the SAXS pattern appeared only when ELAC threads were incubated in 1× PBS. Comparison of the location of periodic peaks of native rabbit Achilles tendon with those of ELAC threads incubated in 1× PBS indicated that the peaks from ELAC threads had nearly the same q-value, shifted slightly to larger spacing (Figure 3b and Table 1). The positions of all the diffraction peaks observed from each sample can be explained by the periodic d-banding (D), given by D = n2π/qpeak, where n is the order of the peak 1, 2, 3, etc. Seven peaks of higher order were observed for the rabbit tendon sample yielding a periodic distance of 63.60 nm (Table 1). The seven peaks observed were the n = 8, 13, 16, 18, 21, 24, and 29. Lower order peaks hit the beam stop or were at too small of an angle to be observed by the apparatus. The position of the two peaks at n = 16, and 24 for dehydrated ELAC threads incubated in 1× PBS showed a slightly larger d-banding pattern of 63.83 nm. This is in the lower range of characteristic D-banding pattern (60-70 nm) found in type-I collagenous materials [24-28].
Figure 3.
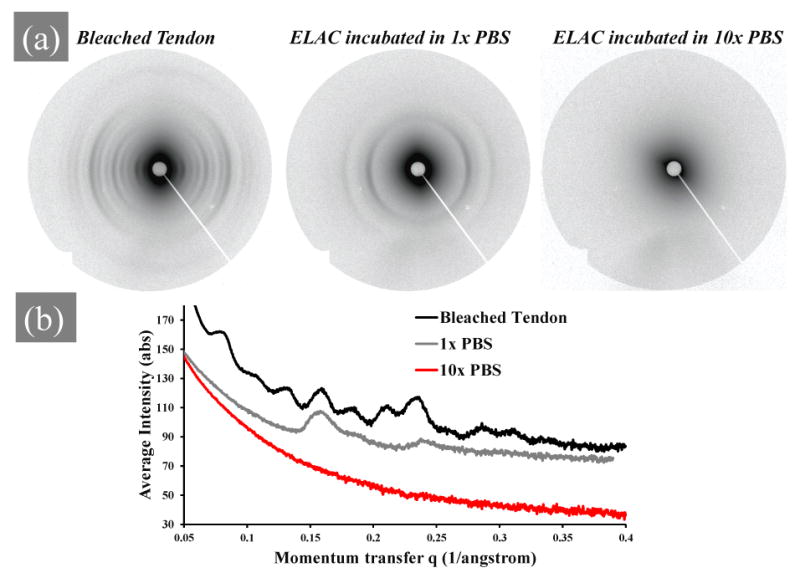
Small angle X-ray scattering (SAXS) results. a) SAXS patterns of dehydrated bleached rabbit Achilles tendon, ELAC thread incubated in 1× PBS, and ELAC thread incubated in 10× PBS. b) Intensity vs. peak position of X-ray scattering data of the patterns in panel A. ELAC threads were incubated in the PBS solution for 12 hours.
Table 1.
Structural parameters obtained from meridional reflections obtained from SAXS diffraction patterns of bleached rabbit Achilles tendon as a reference and ELAC threads incubated in 1× PBS.
| Order (n) | Bleached Tendon | 1× PBS | ||
|---|---|---|---|---|
| q(1/A) | d(nm) | q(1/A) | d(nm) | |
| 1 | - | 63.60 | - | 63.83 |
| 2 | - | 31.80 | - | 31.91 |
| 3 | - | 21.20 | - | 21.28 |
| 4 | - | 15.90 | - | 15.96 |
| 5 | - | 12.72 | - | 12.77 |
| 6 | - | 10.60 | - | 10.64 |
| 7 | - | 9.09 | - | 9.12 |
| 8 | 0.08 | 7.95 | - | 7.98 |
| 9 | - | 7.07 | - | 7.09 |
| 10 | - | 6.36 | - | 6.38 |
| 11 | - | 5.78 | - | 5.80 |
| 12 | - | 5.30 | - | 5.32 |
| 13 | 0.13 | 4.78 | - | 4.91 |
| 14 | - | 4.43 | - | 4.56 |
| 15 | - | 4.14 | - | 4.26 |
| 16 | 0.16 | 3.97 | 0.16 | 3.99 |
| 17 | - | 3.74 | - | 3.75 |
| 18 | 0.18 | 3.45 | - | 3.51 |
| 19 | - | 3.27 | - | 3.33 |
| 20 | - | 3.10 | - | 3.16 |
| 21 | 0.21 | 2.98 | - | 3.01 |
| 22 | - | 2.84 | - | 2.87 |
| 23 | - | 2.72 | - | 2.75 |
| 24 | 0.23 | 2.68 | 0.24 | 2.63 |
| 25 | - | 2.57 | - | |
| 26 | - | 2.47 | - | |
| 27 | - | 2.38 | - | |
| 28 | - | 2.29 | - | |
| 29 | 0.29 | 2.17 | - | |
ELAC threads incubated in 1× PBS for 3 hours show little periodic D-banding pattern (figure 4), a pattern that emerges only after 12 hours of 1× PBS incubation. Ninety-six hours incubation of ELAC threads in 1× PBS was detrimental to the D-banding pattern of collagen molecules since the peaks at n = 16 and 24 disappeared. The color of ELAC threads incubated in 1× PBS for 96 hours tinted to yellow unlike shorter treatment durations which were transparent.
Figure 4.
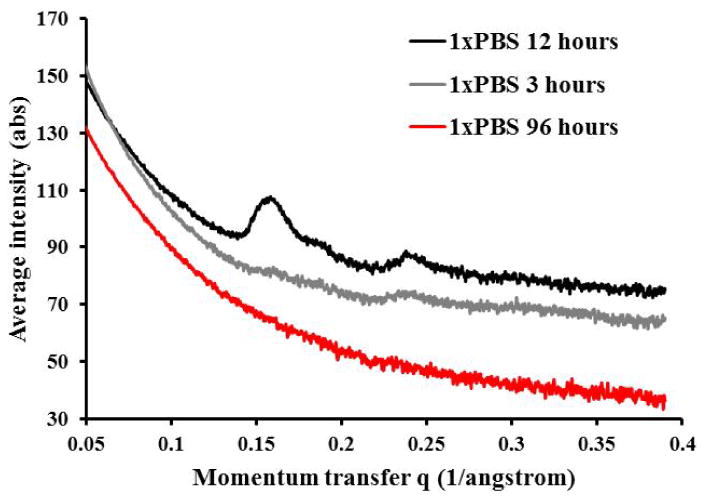
SAXS patterns of ELAC threads incubated in 1× PBS for 3, 12, and 96 hours.
Transmission Electron Microscopy (TEM)
The distribution of the fraction of microfibrils was presented by normalizing the number of microfibrils having a particular diameter to the total number of microfibrils in a field of view of approximately 1.4 × 1.6 μm2 (figure 5a). The distributions of collagen microfibrils from ELAC threads incubated with and without PBS was comparable (p>0.05). For all groups, the diameter of ≈80% of the microfibrils ranged from 5-7 nm. This diameter corresponds well to that of a collagen microfibril, which was the smallest supramolecular unit that could demonstrate axial periodicity or D-banding pattern [29]. The appearance of collagen microfibrils in the transverse plane to longer axis of ELAC threads that was typical of all groups is shown in figure 5b.
Figure 5.
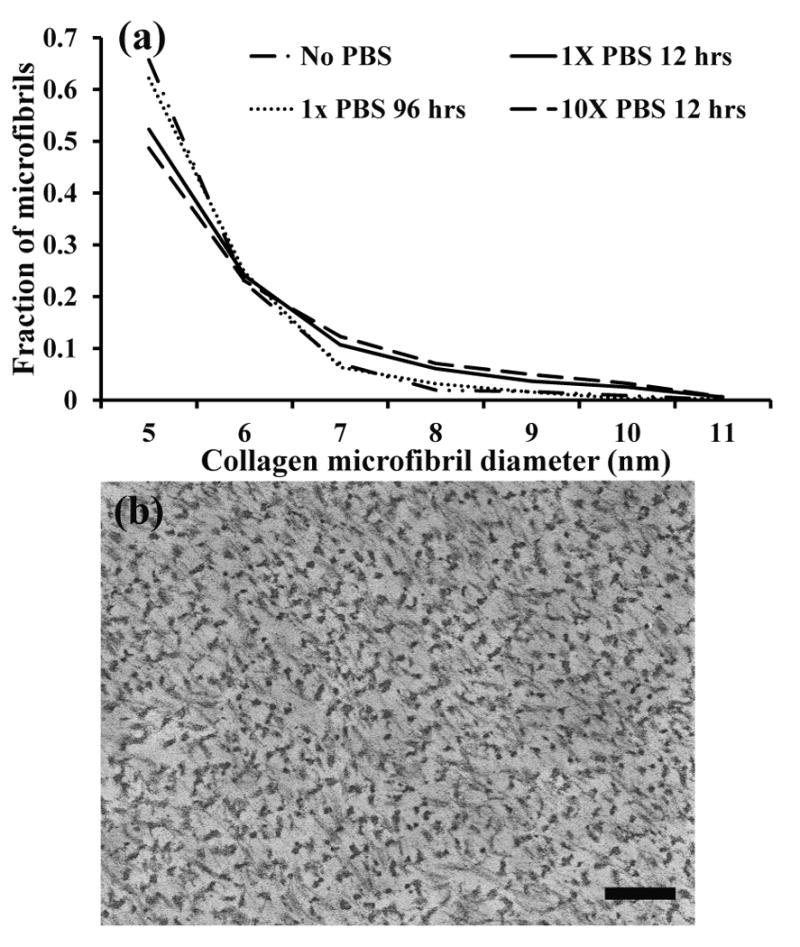
a) Distribution of collagen microfibril diameters of ELAC threads incubated in different PBS-treatment combinations. A Kolmogorov-Smirnov test indicates that all distributions are statistically similar (p>0.05). b) Representative TEM image of the cross sections of ELAC threads incubated at different PBS-treatment combinations. Throughout the groups, the microfibril distribution was not statistically different (p>0.05). Scale bar represents 200 nm.
Discussion
The current study aimed to optimize the strength and D-banding pattern formation in ELAC threads by modulating the phosphate ion concentration and incubation time. The key findings of the study were as follows: a) ELAC threads incubated in 1× PBS exhibited higher strain compared to 0.5× or 10× PBS, b) Incubation of aligned collagen in 1× PBS promoted the formation of D-banding pattern with a periodicity of 63-64 nm, c) the D-banding arc pattern was at its maximum after 12 hours of incubation in 1× PBS as per SAXS measurements, d) extended periods of 1× PBS incubation was detrimental for the D-banding pattern of ELAC threads, e) ELAC threads were composed of tightly packed microfibrils with a mean diameter between 5-7 nm, and, f) fibrillogenesis occured within existing microfibrils as of electrochemical alignment step without any lateral aggregation of microfibrils.
This work focused on a system that has been widely studied over the past fifty years: in vitro fibrillogenesis of collagen type-I molecules. However, the unique feature of this study relies on the fact that fibrillogenesis was being promoted in already formed densely packed collagen assemblies (ELAC threads). ELAC biomaterial was 25-50 times denser than collagen matrices found in the literature [8, 9]. Most of the studies about in vitro collagen fibrillogenesis focused on the polymerization and self-assembly of collagen molecules dissolved in solution [10, 17, 30]. Current results indicate that a limited amount of D-banding formation is possible in densely packed ELAC threads (figure 3). The absence of D-banding in the microfibrils within ELACs incubated at PBS concentrations lower or higher than 1× suggested that the surface interaction between adjacent microfibrils was interrupted. The interruption may be due to the absence or saturation of PBS in the incubation solution. Mechanical and structural observations demonstrated that the optimal PBS concentration was at about 1×. This concentration was in agreement with that observed for fibrillogenesis in collagen solutions [15, 30-32]. It has been shown that mechanics and hierarchical arrangement of type-I collagen molecules in ELAC threads depends closely on the PBS concentration and incubation time (figures 1 to 4).
A typical strength of 0.5 to 0.7 MPa was observed for ELAC threads incubated in PBS at 1× or higher concentrations. These results were comparable to a previous study which reported that the ultimate tensile stress (UTS) of un-crosslinked extruded collagen fibers was below 2 MPa [33]. It was observed that collagen treated in 1× PBS for 96 hours were stronger, stiffer and less extensible than threads subjected to shorter treatment durations (figure 2). However, the D-banding which was present by 12 hours of treatment disappeared after 96 hours (figure 4). Furthermore, the 96 hours samples displayed yellow discoloration. In the face of these observations, the increase in the strength and stiffness of 96 hours treated samples was not due to fibrillogenesis, rather, due to some crosslinking of unknown nature.
Extrusion is an alternative method to the electrochemical route for making dense and aligned collagen fibers [33-37]. In un-crosslinked ELAC threads, as well as in extruded un-crosslinked collagen fibers, hydrogen and electrostatic bonding between molecules are the main interactions. The stiffness of un-crosslinked ELAC threads incubated in PBS concentrations above 1× was about 0.4 MPa. Un-crosslinked extruded collagen fibers incubated in PBS for one hour have shown stiffness below 0.2 MPa [34]. Cornwell et al. [35] have used un-crosslinked extruded collagen fibers to study cell-matrix interactions. The mechanical properties of these extruded fibers are comparable to un-crosslinked ELAC threads in terms of ultimate tensile stress and Young's modulus. However, reported ultimate tensile strains (42 ± 12 %) are well below the ultimate tensile strains reported here for ELAC. ELAC threads, regardless of the PBS concentration or the incubation time they were subjected to, showed strains at failure between 60 and 120%. The reason for such high extensibility may be the packing and hierarchical arrangement of collagen molecules within ELAC threads. ELAC threads incubated in 1× PBS for 12 hours showed D-banding pattern with a periodicity of ≈ 64 nm. Pins et al. [36] performed TEM on self-assembled collagen fibers and reported the presence of D-banding period. Caves et al. [37] have developed a modified extrusion process which includes a fiber extrusion tube and a fiber rinsing bath. This continuous spinning system is able to produce D-banded collagen fibrils when the fibers are incubated in phosphate buffer solutions.
Phosphate ions can accumulate inside collagen fibrils either as bound to collagen or as dissolved ions in interstitial water [38]. Only dibasic phosphate (HPO42-) exhibit binding to collagen molecules via direct hydrogen bonding between positively charged amino acids, but the main contribution of the binding energy comes from electrostatic interactions. Mertz et al. [38] have proposed that divalent phosphate anions bind at collagen sites with excess positive charge. These divalent phosphate ions form salt bridges between two positively charged amino acid residues not involved in ion pairs with negatively charged residues. Thus the role of phosphate anion binding in fibrillogenesis is dependent on the phosphate concentration. That is, diluted phosphate solutions have not shown to be detrimental in the collagen fibrillogenesis kinetics [15, 39]. However, if collagen molecules are incubated in solutions of high phosphate concentrations (10× PBS), phosphate binding becomes more significant and can affect collagen-to-collagen interactions. In support of this, we have observed diminishment of D-banding in 10× PBS concentrations (figure 3b). The evidence suggests that the distance between nearest molecules remains unchanged, but the distance between microfibrils varies [38] disrupting the hierarchical arrangement. Finally, the influence of phosphate concentration on in vitro collagen fibril formation has been evaluated on pepsin-digested collagen from calf skin [40]. High concentration of various divalent anions such as phosphate, sulfate, thiosulfate, citrate, malonate, and bicarbonate decreased the rate of collagen fibril formation. This effect was possible due to the ability of divalent anions to change the nature of collagen molecular interactions[41].
It is important to emphasize that 50-200 nm diameter fibers can be obtained from sea urchin ligament and sea cucumber [30, 42], and 33 ± 4 nm diameter fibrils can be obtained using a continuous spinning system [37], whereas fibrillar assembly in ELAC threads were limited to microfibrils within 10 nm in diameter. The collagen microfibril diameters reported here were mainly between 5-7 nm (figures 5a and 5b). These diameters are significantly less than those found for self-assembled collagen fibers produced via the extrusion method (20 to 26 nm) [36], and the continuous spinning method [37]. On the other hand, the microfibril diameter for ELACs found in this study is similar to the building block of immature tissues of 18 days old fetal rats (8 nm) [43].
The synthesis of ELAC threads involves two stages: 1) electrochemical alignment to obtain collagen threads, and 2) PBS treatment to induce D-banding to the electrochemically aligned threads. Electrochemical focusing and alignment occurs by electrophoretic motion of molecules towards the isoelectric point [1]. In this process, molecules are packed and aligned at the isoelectic line by fluid convection forces. Therefore, the lateral aggregation during alignment (but before PBS treatment) is induced by physical forces. This lateral aggregation occurs randomly without any evidence of D-banding. The D-banding emerges following the PBS treatment by staggering of collagen molecules in existing microfibrils. TEM results indicated that the fibril size before and after PBS treatment did not change, indicating that there were no further lateral accretion of the collagen fibrils during PBS treatment.
In conclusion, this study demonstrates that incubation of aligned collagen in 1× PBS for 12 hours results in mechanically competent ELAC threads with D-banding period similar to native tendons. These ELAC threads are composed of tightly packed fibrils with diameters comparable to those of microfibrils in native tendon. Overall, ELAC as a biomimetic material presents the ideal characteristics of a unit that can be used in the synthesis of tissue-engineered tendons.
Acknowledgments
This study was funded by the National Science Foundation (CBET-0754442) and the National Institute of Health (NIH 1R21AR056060). The authors would like to thank Steve Gaik at the Purdue University X-Ray Scattering Facility for his help in SAXS data analysis and processing.
References
- 1.Cheng X, Gurkan UA, Dehen CJ, Tate MP, Hillhouse HW, Simpson GJ, Akkus O. Biomaterials. 2008;29:3278–3288. doi: 10.1016/j.biomaterials.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 2.Butler DL, Juncosa N, Dressler MR. Annu Rev Biomed Eng. 2004;6:303–329. doi: 10.1146/annurev.bioeng.6.040803.140240. [DOI] [PubMed] [Google Scholar]
- 3.Cen L, Liu W, Cui L, Zhang W, Cao Y. Pediatr Res. 2008;63:492–496. doi: 10.1203/PDR.0b013e31816c5bc3. [DOI] [PubMed] [Google Scholar]
- 4.Jozsa L, Kannus P, Balint JB, Reffy A. Acta Anat (Basel) 1991;142:306–312. doi: 10.1159/000147207. [DOI] [PubMed] [Google Scholar]
- 5.Magnusson SP, Hansen P, Kjaer M. Scand J Med Sci Sports. 2003;13:211–223. doi: 10.1034/j.1600-0838.2003.00308.x. [DOI] [PubMed] [Google Scholar]
- 6.Ottani V, Martini D, Franchi M, Ruggeri A, Raspanti M. Micron. 2002;33:587–596. doi: 10.1016/s0968-4328(02)00033-1. [DOI] [PubMed] [Google Scholar]
- 7.Gurkan UA, Cheng X, Kishore V, Uquillas JA, Akkus O. J Biomed Mater Res A. 2010;94:1070–1079. doi: 10.1002/jbm.a.32783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross VL, Zheng Y, Won Choi N, Verbridge SS, Sutermaster BA, Bonassar LJ, Fischbach C, Stroock AD. Biomaterials. 2010;31:8596–8607. doi: 10.1016/j.biomaterials.2010.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guille MMG, Helary C, Vigier S, Nassif N. Soft Matter. 2010;6:4963–4967. doi: 10.1039/c0sm00260g. [DOI] [PubMed] [Google Scholar]
- 10.Trelstad RL, Hayashi K, Gross J. Proc Natl Acad Sci U S A. 1976;73:4027–4031. doi: 10.1073/pnas.73.11.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cisneros DA, Hung C, Franz CM, Muller DJ. J Struct Biol. 2006;154:232–245. doi: 10.1016/j.jsb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Cooper A. Biochem J. 1970;118:355–365. doi: 10.1042/bj1180355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cassel JM. Biopolymers. 1966;4:989–997. [Google Scholar]
- 14.Leikin S, Rau DC, Parsegian VA. Nat Struct Biol. 1995;2:205–210. doi: 10.1038/nsb0395-205. [DOI] [PubMed] [Google Scholar]
- 15.Williams BR, Gelman RA, Poppke DC, Piez KA. J Biol Chem. 1978;253:6578–6585. [PubMed] [Google Scholar]
- 16.Liu MY, Yeh ML, Luo ZP. Biomed Mater Eng. 2005;15:413–420. [PubMed] [Google Scholar]
- 17.Harris JR, Reiber A. Micron. 2007;38:513–521. doi: 10.1016/j.micron.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 18.Jiang FZ, Horber H, Howard J, Muller DJ. Journal of Structural Biology. 2004;148:268–278. doi: 10.1016/j.jsb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Gobeaux F, Mosser G, Anglo A, Panine P, Davidson P, Giraud-Guille MM, Belamie E. J Mol Biol. 2008;376:1509–1522. doi: 10.1016/j.jmb.2007.12.047. [DOI] [PubMed] [Google Scholar]
- 20.Wood GC, Keech MK. Biochemical Journal. 1960;75:588–598. doi: 10.1042/bj0750588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li YP, Asadi A, Monroe MR, Douglas EP. Mat Sci Eng C-Bio S. 2009;29:1643–1649. [Google Scholar]
- 22.Habelitz S, Balooch M, Marshall SJ, Balooch G, Marshall GW., Jr J Struct Biol. 2002;138:227–236. doi: 10.1016/s1047-8477(02)00029-1. [DOI] [PubMed] [Google Scholar]
- 23.Alaseirlis DA, Li Y, Cilli F, Fu FH, Wang JH. Connect Tissue Res. 2005;46:12–17. doi: 10.1080/03008200590935501. [DOI] [PubMed] [Google Scholar]
- 24.Baselt DR, Revel JP, Baldeschwieler JD. Biophys J. 1993;65:2644–2655. doi: 10.1016/S0006-3495(93)81329-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belton JC, Michaeli D, Fudenberg HH. Arthritis Rheum. 1975;18:443–450. doi: 10.1002/art.1780180503. [DOI] [PubMed] [Google Scholar]
- 26.Hosoyamada Y, Kurihara H, Sakai T. J Anat. 2000;196(Pt 3):327–340. doi: 10.1046/j.1469-7580.2000.19630327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto S, Hashizume H, Hitomi J, Shigeno M, Sawaguchi S, Abe H, Ushiki T. Arch Histol Cytol. 2000;63:127–135. doi: 10.1679/aohc.63.127. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto S, Hitomi J, Sawaguchi S, Abe H, Shigeno M, Ushiki T. Jpn J Ophthalmol. 2000;44:318. doi: 10.1016/s0021-5155(00)00156-8. [DOI] [PubMed] [Google Scholar]
- 29.Veis A, Miller A, Leibovich SJ, Traub W. Biochim Biophys Acta. 1979;576:88–98. doi: 10.1016/0005-2795(79)90487-2. [DOI] [PubMed] [Google Scholar]
- 30.Kadler KE, Holmes DF, Trotter JA, Chapman JA. Biochem J. 1996;316(Pt 1):1–11. doi: 10.1042/bj3160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham HK, Holmes DF, Watson RB, Kadler KE. J Mol Biol. 2000;295:891–902. doi: 10.1006/jmbi.1999.3384. [DOI] [PubMed] [Google Scholar]
- 32.Holmes DF, Chapman JA, Prockop DJ, Kadler KE. Proc Natl Acad Sci U S A. 1992;89:9855–9859. doi: 10.1073/pnas.89.20.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang MC, Pins GD, Silver FH. Biomaterials. 1994;15:507–512. doi: 10.1016/0142-9612(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 34.Caruso AB, Dunn MG. Journal of Biomedical Materials Research Part A. 2004;69A:164–171. doi: 10.1002/jbm.a.20136. [DOI] [PubMed] [Google Scholar]
- 35.Cornwell KG, Lei P, Andreadis ST, Pins GD. Journal of Biomedical Materials Research Part A. 2007;80A:362–371. doi: 10.1002/jbm.a.30893. [DOI] [PubMed] [Google Scholar]
- 36.Pins GD, Christiansen DL, Patel R, Silver FH. Biophysical Journal. 1997;73:2164–2172. doi: 10.1016/S0006-3495(97)78247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caves JM, Kumar VA, Wen J, Cui WX, Martinez A, Apkarian R, Coats JE, Berland K, Chaikof EL. J Biomed Mater Res B. 2010;93B:24–38. doi: 10.1002/jbm.b.31555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mertz EL, Leikin S. Biochemistry. 2004;43:14901–14912. doi: 10.1021/bi048788b. [DOI] [PubMed] [Google Scholar]
- 39.Kuznetsova N, Chi SL, Leikin S. Biochemistry. 1998;37:11888–11895. doi: 10.1021/bi980089+. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi T, Nagai Y. J Biochem. 1973;74:253–262. [PubMed] [Google Scholar]
- 41.Pogany G, Hernandez DJ, Vogel KG. Arch Biochem Biophys. 1994;313:102–111. doi: 10.1006/abbi.1994.1365. [DOI] [PubMed] [Google Scholar]
- 42.Thurmond FA, Trotter JA. Journal of Molecular Biology. 1994;235:73–79. doi: 10.1016/s0022-2836(05)80015-4. [DOI] [PubMed] [Google Scholar]
- 43.Craig AS, Parry DAD. P Roy Soc Lond B Bio. 1981;212:85–92. doi: 10.1098/rspb.1981.0026. [DOI] [PubMed] [Google Scholar]