Abstract
Admixture mapping is a rapidly developing method to map susceptibility alleles in complex genetic disease associated with continental ancestry. Theoretically, when admixture between continental populations has occurred relatively recently, the chromosomal segments derived from the parental populations can be deduced from the differences in genotype allele frequencies. Progress in computational algorithms, in identification of ancestry informative single nucleotide polymorphisms, and in recent studies applying these tools suggests that this approach will complement other strategies for identifying the variation that underlies many complex diseases.
Introduction
The approach of using admixture for disease studies was first advanced to examine linkage disequilibrium (LD) between susceptibility genes and markers in a process termed ‘mapping by admixture disequilibrium’ (MALD) [1,2]. In brief, the LD created by admixture between alleles with large frequency variations in the different populations can facilitate the mapping of traits in admixed populations, if any disease susceptibility alleles or disease protective alleles are present in a sufficiently different frequency distribution in the parental populations.
Alternatively, admixture mapping can be viewed and implemented by examining the linkage between a trait and ancestry segments from one or another founding population. Instead of testing for allelic association, markers provide ancestry information and this information is used to examine the linkage with the trait. This concept, originally advanced by McKeigue [3], underlies the approach that my co-workers and I, and others have taken to address the etiopathogenesis of complex genetic disease [4–7]. Here, I consider the situation where admixture has occurred between two continental populations. Although it is theoretically possible to perform these studies in more complex admixed populations, achieving statistical confidence in ancestry assignment becomes more difficult.
General parameters and considerations
Modeling studies suggest that between 2000 and 5000 well-distributed ancestry informative markers (AIMs) distinguishing parental origins are sufficient for admixture mapping when two continental populations have admixed within the last 15 generations [4–6,8••,9,10]. This number contrasts with the 250 000 or more markers suggested to be necessary for association studies. Admixture mapping suffers, however, from the disadvantages that it can map only disease-associated alleles that are present in different frequencies in the parental populations, and increased regions of LD can hinder fine-scale mapping in the latter stages of identifying the causative genetic variation underlying disease susceptibility. Admixture mapping has an advantage over general association studies in that it is not deterred by multiple independent mutational events, because only the ancestral identity of an allele is used in computations. General association studies have been criticized because of their decreased power in the presence of allelic heterogeneity, particularly because allelic heterogeneity is likely to be very common in complex genetic diseases [11].
Admixture mapping also has the potential to map genes that are not sufficiently polymorphic within a non-admixed population to be detected by either association or linkage studies. Some functional polymorphisms might be fixed or nearly fixed in one of the parental populations. For the cell-surface glycoprotein Duffy, for example, the null allele is fixed in the African population and a functional allele is fixed in the European population. This particular example is a function of positive selection and is thought to be due to Duffy’s role as a receptor for Plasmodium vivax [12]. Furthermore, selection has been suggested to have been an important factor in shaping the differences between the major ancestral groups; in other words, many AIMs might have acquired their ethnic differences in allele frequencies owing to selection [13]. This possibility is supported by recent studies [14,15••,16] and might enhance the possibility of successful admixture mapping for some diseases.
Although definitive studies demonstrating that admixture mapping is an effective strategy for identifying genetic factors in complex disease are still lacking, studies in African Americans for skin pigmentation, hypertension and multiple sclerosis have provided data suggesting the applicability of these methods [17–19,20•]. More recent studies have provided further enthusiasm for admixture mapping. Reich’s group [21••] has reported mapping a strong susceptibility locus for early onset prostate cancer in African Americans. In addition, studies led by Ziv report the ability of the admixture mapping approach to identify an amino acid variant linked to African ancestry that determines the levels of soluble interleukin-6 receptor. This work, presented at the 2006 annual meeting of the American Society of Human Genetics, might provide the best proof in concept for the admixture mapping approach (http://www.ashg.org).
Application to specific admixed populations and diseases
In the American continents, admixture has occurred between different continental groups from European, Amerindian and African ancestry. Different ethnic groups – for example, Mestizo Mexicans, African Americans and Puerto Ricans – have different allelic contributions from the ‘parental’ populations [22•]. In addition, the admixture characteristics differ with respect to the number of generations and probably the history of gene flow. These differences have an impact on power characteristics and on the number of AIMs needed for defining chromosomal segment ancestry.
Table 1 provides a partial list of diseases that might be particularly amenable to study in specific admixed populations on the basis of the prevalence in founding populations.
Table 1.
Diseases with epidemiological differences in prevalence on the basis of continental ancestry.
| Disease | Admixed population | Higher risk continental population | Reference |
|---|---|---|---|
| Multiple sclerosis | African American | European | [30] |
| Osteoporosis | African American | European | [31] |
| Prostate cancer | African American | African | [32] |
| Rheumatoid arthritis | Mexican American | Amerindian | [33] |
| Type 2 diabetes | Mexican American | Amerindian | [34] |
| Systemic lupus erythematosus | African American | African | [35] |
Ancestry informative markers
Over the past several years, many groups have developed and characterized sets of markers that can distinguish among major ethnic groups in their genetic ancestry (i.e. AIMs) [8••,23–26]. Using HapMap data, for example, my co-workers and I [8••] have recently identified and validated a set of 4222 single nucleotide polymorphisms (SNPs) distributed throughout the genome that have very large differences in allele frequencies between African and European continental populations. These SNPs have been selected to show little difference between disparate African populations and also show very few differences in allele frequencies between different European subpopulations.
Current efforts are nearing completion with respect to a similar set of AIMs to distinguish European and Amerindian ancestry throughout the genome. Our recent studies and those of other groups indicate that the differences among Amerindian groups are usually larger than those found in other subpopulations. Our studies suggest that using a single specific Amerindian group to represent one of the parental populations contributing to the Mexican American admixed population can be problematic. By screening multiple Amerindian populations, however, it is possible to identify sufficient numbers of SNPs that have large differences between Amerindian and European populations and yet show little variation among different Amerindian groups. These studies should enable the development of sets of AIMs for admixture mapping in the Mexican, Mexican American and other ‘Latino’ populations.
Algorithms
Admixture mapping is based on the assumption that some susceptibility variants will be associated with continental ancestry and that this association can be discerned in admixed populations by examining linkage to ancestry. Several different algorithms and computational programs have been developed to facilitate admixture mapping [3–7]. Each of these relies on using a hidden Markov model (HMM) to determine ancestral states along the chromosome (transition probabilities) on the basis of the typing results of markers that are informative for ancestry. The model formulation relies on the prior probability that any given locus in the current generation is derived from one of the founder populations, and depends on the occurrence of different ancestral states along a chromosome that are a result of recombinant events in previous generations since admixture. This HMM approach is designed to infer the unobserved local ancestries for each individual and uses multipoint information from linked markers. Thus, the transition probabilities in HMM, simulated by Poisson arrivals, provide an approximation to the correlations in ancestry between linked markers.
Although the actual underlying model is unknown, simulation studies have shown the ability of these methods to discern ancestry linkage in various admixture models and conditions. These approaches use either case–control analyses or both case–control and case-only analyses. For case-only algorithms, the detection of linkage to ancestry is based on the difference in distribution of the ancestry of chromosomal segments for the loci associated with disease as compared with those in which there is no association. We note that appropriate genome-wide α levels (i.e. the meaningful significance level) should be less when both case-only and case–control algorithms are used to analyze the same set of probands; however, extensive simulations will be necessary to establish this relationship.
Computational programs that are readily available include AncestryMap [5], Structure/MALDsoft [9] and Admix-Map [4]. In our application of these programs to simulated data based on real genotypes, my colleagues and I [8••] have found that, although each algorithm can yield appropriate results in many models, the AdmixMap program performs the best when the admixture model is more complex (more generations and different gene flow schemes). Unlike the other methods, AdmixMap estimates both the admixture proportion and the number of generations for each gamete in each individual. We have also noted that the potential issue of LD within parental populations does not seen to be a problem with our current AIM sets using this algorithm [8••].
A new Markov-HMM (MHHM) algorithm has been recently developed that explicitly accounts for LD in parental populations [27•]. The power in real or simulated data sets using real genotyping data has not, however, been robustly examined; thus, the efficacy of this algorithm in admixture mapping is not yet clear. This study has also suggested that the MHHM algorithm will enable admixture mapping without the use of AIMs. In practice, this approach might be problematic because some or many SNPs might not have the appropriate characteristics – namely, little variation in allele frequencies within multiple parental populations that could have contributed to one continental founder population (see ‘Ancestry informative markers’ above).
Power, sample size and marker considerations
The power of admixture mapping is determined by several factors: (i) the sample size; (ii) the risk, here termed the ‘ethnicity risk ratio’ (ERR), conferred by one parental ancestry as compared with the other parental ancestry in the admixed population; (iii) the admixture characteristics (e.g. continuous gene flow and number of generations since admixture); and (iv) the ability to define the ancestry of each chromosomal segment derived from both parental gametes in the study subjects. Importantly, modeling studies suggest that multiple waves of parental contribution to the admixed population might enhance rather than diminish the ability of admixture mapping to identify chromosomal regions of interest [5,6,28]. The reader should note that ERR equals the genotypic risk ratio (GRR) in the admixed population if the responsible allelic variant is fixed in opposite directions in the founding populations. This method is generally limited to detecting genes with a GRR of more than 1.5 in the admixed population in studies using ~1000 cases.
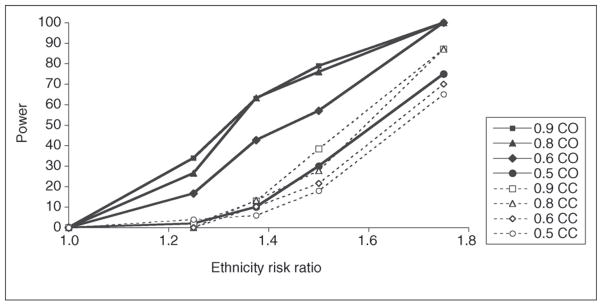
For admixture mapping in African Americans, my coworkers and I [8••] have recently examined both the admixture mapping information of AIM sets and the power as a function of admixture mapping information. A set of 4222 AIMs extracts ~70% admixture mapping information genome-wide and provides good power. A subset of 2000 AIMs selected from this set provides only marginally less coverage. Figure 1 shows the power relationship using real marker sets and using case-only and case–control algorithms under an admixture model (six generations, continuous gene flow, 80:20 admixture ratio) consistent with both our own [29] and other assessments of African American admixture characteristics. Although the case-only algorithms provides greater power, it is not yet certain whether segregation distortion or other factors unrelated to phenotype selection can result in false positives. For Mexican American subjects these relationships are different. Our current studies suggest that a total of ~15 generations fits the admixture characteristics of this population and that the admixture proportion is close to 50% European and 50% Amerindian. Under these conditions, substantially more markers (~5000) are necessary for adequate admixture mapping extraction. Simulations under these conditions, however, show that the power is greater and that the critical region defined by ancestry segments is substantially smaller. In addition, for Mexican Americans the power for case–control algorithms is nearly equal to that for case-only algorithms.
Figure 1.
Power of admixture mapping as a function of admixture mapping information. The power was determined from simulations using 700 cases and 700 controls and SNP sets with admixture information corresponding to the key for the SNP set used [8••]. The power curves shown were determined by using the AdmixMap program and deCODE genetic map for either case-only (CO) or case–control (CC) analyses. The appropriate α level for these analyses, based on extensive simulations, was a normalized score of 4.0. The results were obtained from a minimum of 50 separate simulations and analysis of each measurement. Similar results were obtained with AncestryMap and MALDsoft algorithms in more limited analyses (data not shown). Figure reproduced, with permission, from study by Tian et al. [8••].
From chromosomal segments to disease alleles
The above sections have focused on identification of the chromosomal region containing susceptibility loci linked to continental ancestry. The size of the critical region identified by these studies depends on the ERR, the sample size and the admixture characteristics that resulted in the population being studied [4]. Identification of the actual susceptibility gene within the critical region requires the same approach used in general association tests or to identify candidate genes; in other words, further narrowing depends on LD within the ancestry-associated segment that is not due to continental ancestry or identification of particular nucleotide variations that can be shown to have a physiological effect.
Conclusion
This brief review has highlighted an emerging methodology – admixture mapping – that is a promising approach in many complex common diseases. Advances in high-throughput genotyping might facilitate the extension of this methodology to admixed populations with several rather than two founding populations. Many ongoing studies using admixture mapping will indicate whether differences between continental populations are particularly useful in identifying variations that result in disease, response to pharmacological therapy and toxicity.
Acknowledgments
This work was supported by grants from the National Institutes of Health (AR050267 and DK071185).
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Rife DC. Populations of hybrid origin as source material for the detection of linkage. Am J Hum Genet. 1954;6:26–33. [PMC free article] [PubMed] [Google Scholar]
- 2.Stephens JC, Briscoe D, O’Brien SJ. Mapping by admixture linkage disequilibrium in human populations: limits and guidelines. Am J Hum Genet. 1994;55:809–824. [PMC free article] [PubMed] [Google Scholar]
- 3.McKeigue PM. Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations, by conditioning on parental admixture. Am J Hum Genet. 1998;63:241–251. doi: 10.1086/301908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O’Brien SJ, Altshuler D, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C, Chen K, Seldin MF, Li H. A hidden Markov modeling approach for admixture mapping based on case–control data. Genet Epidemiol. 2004;27:225–239. doi: 10.1002/gepi.20021. [DOI] [PubMed] [Google Scholar]
- 7.Zhu X, Cooper RS, Elston RC. Linkage analysis of a complex disease through use of admixed populations. Am J Hum Genet. 2004;74:1136–1153. doi: 10.1086/421329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Tian C, Hinds DA, Shigeta R, Kittles R, Ballinger DG, Seldin MF. A genomewide single-nucleotide-polymorphism panel with high ancestry information for African American admixture mapping. Am J Hum Genet. 2006;79:640–649. doi: 10.1086/507954. This study takes advantage of the HapMap studies to develop markers for a comprehensive genome-wide admixture mapping analysis in African American populations and to provide realistic simulations and modeling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montana G, Pritchard JK. Statistical tests for admixture mapping with case-control and cases-only data. Am J Hum Genet. 2004;75:771–789. doi: 10.1086/425281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKeigue PM. Prospects for admixture mapping of complex traits. Am J Hum Genet. 2005;76:1–7. doi: 10.1086/426949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terwilliger JD, Weiss KM. Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr Opin Biotechnol. 1998;9:578–594. doi: 10.1016/s0958-1669(98)80135-3. [DOI] [PubMed] [Google Scholar]
- 12.Hamblin MT, Thompson EE, Di Rienzo A. Complex signatures of natural selection at the Duffy blood group locus. Am J Hum Genet. 2002;70:369–383. doi: 10.1086/338628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12:1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 15••.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. This study uses novel methods and emerging genomic resources, including primate sequences, to provide lists of putative chromosomal positions and genes for which there is evidence of positive selection. The study emphasizes the potential of using such information to link biology including disease genes with population events. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson CS, Thomas DJ, Eberle MA, Swanson JE, Livingston RJ, Rieder MJ, Nickerson DA. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005;15:1553–1565. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, et al. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat Genet. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 18.Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003;112:387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- 19.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, et al. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet. 2005;37:177–181. doi: 10.1038/ng1510. One of the first studies to provide evidence that examining continental ancestry can be useful in identifying putative susceptibility genes. [DOI] [PubMed] [Google Scholar]
- 21••.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, Penney K, Steen RG, Ardlie K, John EM, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. This study uses an admixture mapping approach to provide strong evidence for a prostate cancer susceptibility gene originating in Africa that might underlie the increased frequency of this disease in African Americans. This study might have broad implications for both prostate cancer and admixture mapping studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Yang N, Li H, Criswell LA, Gregersen PK, Alarcon-Riquelme ME, Kittles R, Shigeta R, Silva G, Patel PI, Belmont JW, et al. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Hum Genet. 2005;118:382–392. doi: 10.1007/s00439-005-0012-1. This study demonstrates the ability of ancestry informative markers to identify continental and admixed individuals using only genomic DNA. It provides the first evidence that population substructure can also be examined using specific sets of SNP markers. [DOI] [PubMed] [Google Scholar]
- 23.Smith MW, Lautenberger JA, Shin HD, Chretien JP, Shrestha S, Gilbert DA, O’Brien SJ. Markers for mapping by admixture linkage disequilibrium in African American and Hispanic populations. Am J Hum Genet. 2001;69:1080–1094. doi: 10.1086/323922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins-Schramm HE, Phillips CM, Operario DJ, Lee JS, Weber JL, Hanson RL, Knowler WC, Cooper R, Li H, Seldin MF. Ethnic-difference markers for use in mapping by admixture linkage disequilibrium. Am J Hum Genet. 2002;70:737–750. doi: 10.1086/339368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins-Schramm HE, Chima B, Morii T, Wah K, Figueroa Y, Criswell LA, Hanson RL, Knowler WC, Silva G, Belmont JW, et al. Mexican American ancestry-informative markers: examination of population structure and marker characteristics in European Americans, Mexican Americans, Amerindians and Asians. Hum Genet. 2004;114:263–271. doi: 10.1007/s00439-003-1058-6. [DOI] [PubMed] [Google Scholar]
- 27•.Tang H, Coram M, Wang P, Zhu X, Risch N. Reconstructing genetic ancestry blocks in admixed individuals. Am J Hum Genet. 2006;79:1–12. doi: 10.1086/504302. This study develops a new algorithm for admixture mapping that addresses potential problems with linkage disequilibrium that might be present in parental or founding populations in an admixed population. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaff CL, Parra EJ, Bonilla C, Hiester K, McKeigue PM, Kamboh MI, Hutchinson RG, Ferrell RE, Boerwinkle E, Shriver MD. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet. 2001;68:198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seldin MF, Morii T, Collins-Schramm HE, Chima B, Kittles R, Criswell LA, Li H. Putative ancestral origins of chromosomal segments in individual African Americans: implications for admixture mapping. Genome Res. 2004;14:1076–1084. doi: 10.1101/gr.2165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallin MT, Page WF, Kurtzke JF. Multiple sclerosis in US veterans of the Vietnam era and later military service: race, sex, and geography. Ann Neurol. 2004;55:65–71. doi: 10.1002/ana.10788. [DOI] [PubMed] [Google Scholar]
- 31.Looker AC, Wahner HW, Dunn WL, Calvo MS, Harris TB, Heyse SP, Johnston CC, Jr, Lindsay R. Updated data on proximal femur bone mineral levels of US adults. Osteoporos Int. 1998;8:468–489. doi: 10.1007/s001980050093. [DOI] [PubMed] [Google Scholar]
- 32.Freedland SJ, Isaacs WB. Explaining racial differences in prostate cancer in the United States: sociology or biology? Prostate. 2005;62:243–252. doi: 10.1002/pros.20052. [DOI] [PubMed] [Google Scholar]
- 33.Del Puente A, Knowler WC, Pettitt DJ, Bennett PH. High incidence and prevalence of rheumatoid arthritis in Pima Indians. Am J Epidemiol. 1989;129:1170–1178. doi: 10.1093/oxfordjournals.aje.a115238. [DOI] [PubMed] [Google Scholar]
- 34.West KM. Diabetes in American Indians and other native populations of the New World. Diabetes. 1974;23:841–855. doi: 10.2337/diab.23.10.841. [DOI] [PubMed] [Google Scholar]
- 35.Molokhia M, Hoggart C, Patrick AL, Shriver M, Parra E, Ye J, Silman AJ, McKeigue PM. Relation of risk of systemic lupus erythematosus to West African admixture in a Caribbean population. Hum Genet. 2003;112:310–318. doi: 10.1007/s00439-002-0883-3. [DOI] [PubMed] [Google Scholar]