Abstract
Background
Studying contemporary hybridization increases our understanding of introgression, adaptation and, ultimately, speciation. The sister species Ischnura elegans and I. graellsii (Odonata: Coenagrionidae) are ecologically, morphologically and genetically similar and hybridize. Recently, I. elegans has colonized northern Spain, creating a broad sympatric region with I. graellsii. Here, we review the distribution of both species in Iberia and evaluate the degree of introgression of I. graellsii into I. elegans using six microsatellite markers (442 individuals from 26 populations) and five mitochondrial genes in sympatric and allopatric localities. Furthermore, we quantify the effect of hybridization on the frequencies of the genetically controlled colour polymorphism in females of both species.
Results
In a principal component analysis of the microsatellite data, the first two principal components summarised almost half (41%) of the total genetic variation. The first axis revealed a clear separation of I. graellsii and I. elegans populations, while the second axis separated I. elegans populations. Admixture analyses showed extensive hybridization and introgression in I. elegans populations, consistent with I. elegans backcrosses and occasional F1-hybrids, suggesting hybridization is on-going. More specifically, approximately 58% of the 166 Spanish I. elegans individuals were assigned to the I. elegans backcross category, whereas not a single of those individuals was assigned to the backcross with I. graellsii. The mitochondrial genes held little genetic variation, and the most common haplotype was shared by the two species.
Conclusions
The results suggest rapid species turnover in sympatric regions in favour of I. elegans, corroborating previous findings that I. graellsii suffers a mating disadvantage in sympatry with I. elegans. Examination of morph frequency dynamics indicates that hybridization is likely to have important implications for the maintenance of multiple female morphs, in particular during the initial period of hybridization.
Background
Hybridization and introgression are increasingly recognized as important factors in the evolution of plants, animals [1,2] and prokaryotes [3], and can lead to the creation of novel genotypes and phenotypes. Thus, the study of contemporary hybridization between species and the extent of genomic introgression between them provides an excellent opportunity to examine evolutionary processes such as adaptation, gene flow and, ultimately, speciation [4-6]. Determining the degree of genetic exchange between species may be of particular interest when studying recently diverged species, since they typically show incomplete reproductive barriers.
Hybridization has an inherent spatial component, as the process requires direct contact between populations of the different species. For this reason, the spatial setting is a crucial determinant of hybridization, and, in turn, the specific conditions under which hybridization occurs can, sometimes, be inferred from the geographical distribution of hybrids. Studies of hybrid zones have indicated that natural hybridization is most likely to take place in intermediate habitats, which are often found at the ecological limits of the species' distributional ranges, and where both taxa are found in close proximity to each other [5]. When some of the interspecific matings lead to fertile first-generation (F1) hybrids, there is a possibility that these will backcross with at least one of the parental genotypes, with introgression as a consequence. If the resulting backcrossed individuals subsequently mate with the most similar parental genotype, novel genes and gene complexes can be particularly rapidly introduced into the new genetic background [7]. In some cases, stable and long-lasting hybrid zones are formed as a consequence of spatial range overlap between two species [8-10]. However, in the vast majority of cases, one of the two species, or possibly even the new hybrid cross, becomes more successful and displaces one or both of the original taxa [11].
In odonates (damselflies and dragonflies), a high level of hybridization between species is a rare phenomenon [12,13]. So far, only three molecular studies have investigated hybridization between closely related odonate species, namely between Coenagrion puella and C. pulchellum [14], Mnais costalis and M. pruinosa [15,16] and Calopteryx splendens and C. virgo [17]. All these studies failed, however, to detect extensive hybridization between the species. There was no evidence of hybridization between Coenagrion puella and C. pulchellum in any of the populations examined [14], i.e. no hybrids were found. In addition, between Mnais costalis and M. pruinosa only two F1 hybrid females were found among 900 individuals [15,16] and between Calopteryx splendens and C. virgo only seven hybrids out of 1600 putative hybrids were detected [17]. Despite the lack of evidence from molecular studies, observational and experimental studies have found evidence for putative intrageneric hybrids in some additional species [18]. For example, two of the best documented cases of hybridization among odonates are between Ischnura gemina and I. denticollis [19], and I. elegans and I. graellsii [20,21].
The latter species pair consists of two closely related damselfly species that co-occur in southern Europe [18]. Specifically, I. graellsii is a widespread species on the Iberian Peninsula (see Figure 1), while I. elegans is a species with a more northern and easterly distribution, which has recently spread into new regions within the Iberian Peninsula [20,21]. For example, the first record of I. elegans in north-west Spain was made in 1984, whereas I. graellsii is known from this area since 1917 [22]. In addition, I. elegans is the dominant species in the coastal lagoons of Galicia (north-west Spain), a region where it was a very rare species less than 30 years ago. Furthermore, I. elegans is still rapidly expanding in the area, and in several coastal populations, that are dominated by I. graellsii, immigrant individuals of I. elegans are now starting to appear [21]. In a literature revision, Monetti et al. [20] have found that at least six Spanish localities that held both species simultaneously before the 1980's, had only I. elegans in 2002. Recent observations of I. elegans individuals in south Spain (personal observation), where only I. graellsii populations have been detected until now, and in western Spain, support that this species is gradually expanding its distribution in southern latitudes and western longitudes. Ischnura elegans has also expanded elsewhere; in the UK it has expanded its northern range by approximately 168 km in the last few decades, which is more than double than the average expansion distance of other odonates [23]. Likewise, Parmesan et al. [24] showed that 22 of 35 European butterfly species have shifted their ranges over the last century. The recent change in the climate has been suggested to drive changes in both phenology [25] and distribution [23] of odonates.
Figure 1.
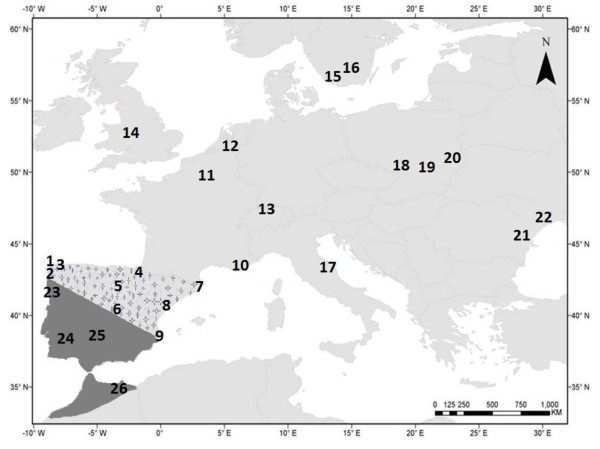
Map showing spatial distribution of I. elegans and I. graellsii in Europe and northern Africa. I. elegans (grey region) and I. graellsii (dark grey) in Europe and northern Africa, and the overlapping distribution of the two species in Spain (grey with stars). We sampled nine populations (166 samples) dominated by individuals that we phenotypically classified as I. elegans in three areas in Spain; north-west (1. Laxe, 2. Louro, 3. Doniños), north and central (4. Arreo, 5. Alfaro, 6. Baldajo) and east (Mediterranean coast: 7. Europa, 8. Amposta, 9. Marjal del Moro). Thirteen populations of allopatric I. elegans were sampled from Europe (10. Vigueirat, 11. Heuringhem, 12. Het Vinne, 13. Kaiserslautern, 14. Liverpool, 15. Värpinge, 16. Genarp, 17. Gran Sasso d'Italia, 18. Lublin-Zemborzyce, 19. Zwięczyca Reszów, 20. Breznica, 21. Suchoi Limon, 22. Enmakov Island; 220 samples). Finally, three populations of I. graellsii from the allopatric region in Iberia (23. Campus, 24. Ribeira de Cobres, 25. Córdoba) and one in northern Africa (26. Saïdia) (56 samples) were sampled.
Ischnura elegans and I. graellsii are very similar ecologically and morphologically, but they can be unambiguously identified by the morphology of prothorax and anal appendages and by the comparatively small body and short wings of I. graellsii [20]. Furthermore, genetic analyses have shown that I. elegans and I. graellsii are very similar both at allozymes [26], and at the mitochondrial Cytochrome b and Coenzyme II genes [0.2% genetic distance, 21]. Previous work has documented that the two species hybridize in the laboratory [21], and hybrids (i.e. morphologically intermediate individuals) have been detected in one sympatric locality in north-western Spain [20].
Wirtz [27] reviewed the factors promoting unidirectional or reciprocal hybridization and proposed a hypothesis based on sexual selection to explain unidirectional hybridization. He proposed that hybridization is more likely between the female of the rare species and the male of the common species. However, when the rare species is also the bigger species, hybridization can be impeded by mechanical incompatibility [28], and the outcome degree and direction of hybridization is difficult to predict. Previous findings from the field (one population) [20] and laboratory [21] indicate that I. graellsii (the common and smaller species in Iberia) suffers a mating disadvantage in sympatry with I. elegans (the less abundant and larger species in Iberia). In particular, males of I. elegans readily mate with females of I. graellsii in the laboratory but males of I. graellsii are mechanically incapable of mating with I. elegans females [21]. The resulting F1-hybrid males can only mate with I. graellsii females, whereas the F1-hybrid females are mechanically incapable of mating with males of I. graellsii but can instead mate with I. elegans males [20]. Furthermore, F1-hybrids (males and females) show a similar degree of reduced viability and fertility [Sánchez-Guillén RA, Wellenreuther M and Cordero-Rivera A: Strong asymmetry in the relative strengths of prezygotic and postzygotic barriers between two damselfly sister species, submitted]. These conditions can be hypothesized to result in a directional bias in hybridization in favour of I. elegans that could explain the recently documented range expansion of I. elegans into areas that were previously only occupied by I. graellsii [20,21]. This agrees with the colonization pattern of I. elegans in the area. Under the described scenario of introgressive hybridization, we hypothesize extensive introgression of I. graellsii genes into the Spanish I. elegans populations. Furthermore, introgression and interspecific competition are probably contributing to the fast range expansion of I. elegans and contraction of I. graellsii in Iberia.
Interestingly, several Ischnura species, including I. elegans and I. graellsii, are characterized by a conspicuous colour polymorphism that is limited to females [21,29-31]. Females exhibit three colour morphs; one androchrome 1] and two gynochromes colour morphs (the green-brown infuscans and the orange-brown infuscans-obsoleta (I. elegans) or aurantiaca (I. graellsii)) [21,29]. The colour polymorphism is controlled by a simple genetic system consisting of one gene with three alleles that are in a dominance hierarchy [21]. How the colour polymorphism of Ischnura damselflies is maintained in space and time has been a much discussed subject. Indeed, hybridization was the first mechanism proposed for maintaining the colour polymorphism in this genus [32]. For example, hybridization was hypothesized to maintain contrasting androchrome frequencies in nearby populations of I. damula and I. demorsa, and I. elegans and I. graellsii, respectively [21,32]. Under the aforementioned scenario of extensive introgressive hybridization of I. graellsii genes into the Spanish I. elegans populations, and the hypothesised role of the hybridization for the temporal maintenance of contrasting androchrome frequencies; female morph frequencies of I. elegans in nearby populations to I. graellsii must be more similar to I. graellsii frequencies, due to the absorption of the typical morph frequencies, than in populations of I. elegans located far away of I. graellsii populations. In fact, Gosden & Svensson [33] proposed that the contrasting androchrome frequencies observed in the Spanish populations of I. elegans could be a by-product of the hybridization in combination with a founder effect.
The objectives of the present study were to examine the spatial distribution of I. graellsii and I. elegans in the Iberian Peninsula, evaluate the extent of introgression of I. graellsii genes into the recently established I. elegans populations in Spain, and to understand the role of the hybridization on the temporal maintenance of female colour morph frequencies in both species. Therefore, we conducted a detailed reconstruction of the distribution of the two species in Spain to quantify the overlap in space between the species. Furthermore, we examined the degree of introgression in nine I. elegans populations within the zone of overlap using both nuclear microsatellite markers [34] and mitochondrial genes [35]. Previous studies have shown that analyses based on differentiated markers between parental species can be used to efficiently and accurately describe admixture proportions, i.e. the degree of introgression, in F1-hybrids and backcrosses see [36-39]. Furthermore, studies have also shown the usefulness of mitochondrial DNA in the study of introgressive hybridization because of their maternal inheritance [40]. Specifically, with this method F1- and F2-hybrids may be assigned to a particular maternal species based on the mtDNA haplotype they carry. However, a prerequisite is that the species do not share haplotypes, which may happen because of hybridization and ancestral polymorphisms [41]. Finally, in order to evaluate if the previously observed uncharacteristic morph frequencies of I. elegans populations in north-western Spain, where frequencies are broadly different between populations [21], extend into the sympatric area (northern, central and eastern Spain), we combined morph frequency data from previous studies [42] with new data from this study, and discuss the proposed hypothesis about the role of hybridization in the temporal maintenance of the colour polymorphism.
Results
Spatial distribution of I. graellsii and I. elegans in the Iberian Peninsula
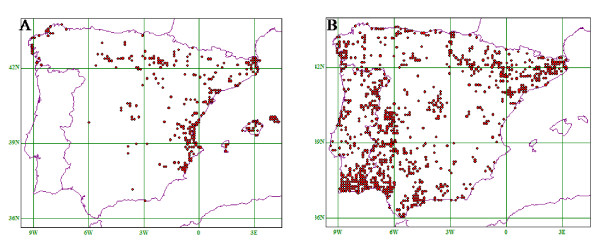
Using the available distribution data of I. elegans and I. graellsii in Spain, we constructed two geographic maps to show their overall distributional range (Figure 2A, Ischnura elegans; 2B, I. graellsii). Species overlap in north and central Spain (from west to east). In particular, I. graellsii is found all over the Iberian Peninsula, while I. elegans is very rare in southern Spain (Figure 2).
Figure 2.
Geographic distribution of I. elegans (A) and I. graellsii (B) in Spain.
Introgressive hybridization
All population groups (Spanish I. elegans populations, European I. elegans populations excluding Spain and I. graellsii populations) exhibited a high degree of molecular diversity (see Table 1). Estimates of observed and expected heterozygosity were similar and ranged from 0.56-0.70 and 0.61-0.76, respectively (Table 1). In the Spanish I. elegans populations, we detected a total of 87 alleles, 11 less than in the other European I. elegans populations. In contrast to I. elegans, we found somewhat fewer (66) alleles for I. graellsii, perhaps due to the fact that the microsatellites were specifically developed for I. elegans which could cause an ascertainment bias. Estimates of allelic richness were comparable between the Spanish and the other European I. elegans populations (6.54 and 6.03, respectively), and were considerably higher than in I. graellsii (3.41; Table 1).
Table 1.
Basic population genetic data by regions and for each population:
| Species | Populations | Region | N | HO | HE | Alleles | Richness |
|---|---|---|---|---|---|---|---|
| I. elegans | All Europe (except Spain) | 220 | 0.701 | 0.764 | 98 | 6.031 | |
| I. elegans | Spain | 166 | 0.705 | 0.767 | 87 | 6.538 | |
| I. graellsii | Iberia-Africa | 56 | 0.577 | 0.682 | 64 | 3.409 | |
| I. elegans | Doniños | Spain | 20 | 0.711 | 0.700 | 41 | 6.591 |
| I. elegans | Laxe | Spain | 14 | 0.715 | 0.805 | 32 | 6.146 |
| I. elegans | Louro | Spain | 15 | 0.712 | 0.729 | 32 | 5.792 |
| I. elegans | Arreo | Spain | 15 | 0.631 | 0.761 | 50 | 7.784 |
| I. elegans | Baldajo | Spain | 17 | 0.603 | 0.795 | 48 | 7.673 |
| I. elegans | Alfaro | Spain | 20 | 0.663 | 0.758 | 50 | 7.046 |
| I. elegans | Europa | Spain | 18 | 0.671 | 0.787 | 48 | 7.109 |
| I. elegans | Amposta | Spain | 20 | 0.691 | 0.770 | 51 | 7.156 |
| I. elegans | Marjal del Moro | Spain | 20 | 0.671 | 0.751 | 44 | 5.776 |
| I. elegans | Lublin-Zemborzyce | East Europe | 14 | 0.7505 | 0.797 | 60 | 8.081 |
| I. elegans | Vigueirat | South France | 16 | 0.733 | 0.804 | 42 | 6.252 |
| I. elegans | Gran Sasso d'Italia | Central Italy | 19 | 0.777 | 0.813 | 51 | 7.461 |
| I. elegans | Liverpool | Great Britain | 16 | 0.624 | 0.709 | 38 | 5.964 |
| I. elegans | Heuringhem | North France | 19 | 0.729 | 0.781 | 45 | 7.380 |
| I. elegans | Kaiserslautern | South Germany | 17 | 0.765 | 0.770 | 53 | 8.177 |
| I. elegans | Het Vinne | Belgium | 18 | 0.682 | 0.795 | 46 | 7.248 |
| I. elegans | Höje Å | Sweden | 20 | 0.653 | 0.717 | 43 | 7.010 |
| I. elegans | Genarp | Sweden | 20 | 0.680 | 0.753 | 44 | 7.203 |
| I. elegans | Zwięczyca Reszów | East Europe | 11 | 0.668 | 0.827 | 52 | 7.264 |
| I. elegans | Breznica | East Europe | 18 | 0.712 | 0.796 | 47 | 6.678 |
| I. elegans | Suchoi Limon | East Europe | 20 | 0.719 | 0.791 | 45 | 6.537 |
| I. elegans | Enmakov Island | East Europe | 15 | 0.713 | 0.766 | 49 | 6.811 |
| I. graellsii | Campus | Spain | 17 | 0.485 | 0.694 | 31 | 3.249 |
| I. graellsii | Córdoba | Spain | 20 | 0.647 | 0.653 | 36 | 3.466 |
| I. graellsii | Ribeira de Cobres | Portugal | 14 | 0.684 | 0.719 | 31 | 3.713 |
| I. graellsii | Saïdia | North Africa | 13 | 0.490 | 0.677 | 25 | 3.118 |
observed (HO) and expected heterozygosity (HE), number of alleles and allelic richness. N represents number of individuals genotyped per population.
Analyses of the overall genetic structure showed that I. elegans populations were significantly differentiated from one another in Europe outside Spain (FST = 0.031, P < 0.0001) as well as within Spain (FST = 0.049, P < 0.0001). Moreover, populations of I. graellsii were also significantly differentiated from one another (FST = 0.029, P < 0.0001).
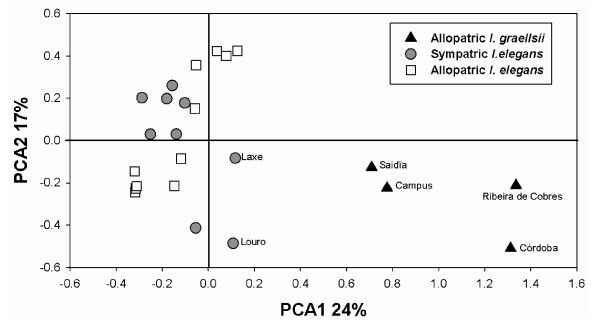
Four of 23 principal component axes accounted for a significant amount of genetic variation among samples, as indicated by a screen plot. The first axis contained 24% (FST = 0.024, P = 0.11), the second axis 17% (FST = 0.017, P = 0.08), the third axis 14% (FST = 0.014, P = 0.07) and the fourth axis 11% (FST = 0.011, P = 0.05) of the total variation (Figure 3). The first two principal components thus summarise almost half (41%) of the total variation inherent in all I. elegans and I. graellsii populations. The scores for the first principal component axis revealed a clear separation of I. graellsii and I. elegans populations, suggesting that major population differences were predominantly caused by species rather than geographic areas per se. Major population differences (nested inside species) were revealed by the second PCA axis, where two of the Spanish I. elegans populations (Louro and Laxe; Table 2 and 3) were situated in the same quadrant that was occupied by all I. graellsii populations (Figure 3).
Figure 3.
Principal component analysis of allopatric I. graellsii, and allopatric and sympatric I. elegans populations. The first (PCA 1) and second (PCA 2) axes represent the first two factorial components and sum up to 41% of explained variation.
Table 2.
Species, sampling locality, sample size for molecular analysis (N), ecology, sampling year, latitude and longitude.
| Species | Locality | Ecology | N | Year | Latitude | Longitude |
|---|---|---|---|---|---|---|
| I. graellsii | Córdoba, southern Spain | allopatric | 20 | 2008 | 37.8833 | -4.7666 |
| I. graellsii | Universitary-Campus, north-western Spain | allopatric | 20 | 1999 | 42.171 | -8.6778 |
| I. graellsii | Saïdia, northern Africa | allopatric | 20 | 2008 | 35.1217 | -2.35 |
| I. graellsii | Ribeira de Cobres, southern Portugal | allopatric | 20 | 2005 | 37.496 | -7.52 |
| Both species | Arreo, north-central Spain | sympatric | 20 | 2008 | 42.4775 | -2.5787 |
| Both species | Baldajo, central Spain | sympatric | 20 | 2008 | 40.2426 | -3.4206 |
| Both species | Alfaro, north-central Spain | sympatric | 20 | 2007 | 42.1908 | -1.7423 |
| I. elegans | Doniños, north-western Spain | sympatric | 20 | 2007 | 43.2927 | -8.1855 |
| I. elegans | Laxe, north-western Spain | sympatric | 20 | 2007 | 43.2125 | -8.9554 |
| I. elegans | Louro, north-western Spain | sympatric | 20 | 2007 | 42.758 | -9.0953 |
| I. elegans | Europa, north-eastern Spain | sympatric | 20 | 2008 | 42.2438 | 3.1028 |
| I. elegans | Amposta, central-eastern Spain | sympatric | 20 | 2008 | 40.2732 | 0.2156 |
| I. elegans | Marjal del Moro, south-eastern Spain | sympatric | 20 | 2008 | 39.0727 | -0.3135 |
| I. elegans | Liverpool, Great Britain | allopatric | 20 | 2008 | 53.2439 | -2.584 |
| I. elegans | Värpinge, southern Sweden | allopatric | 20 | 2005 | 55.7022 | 13.1437 |
| I. elegans | Genarp, southern Sweden | allopatric | 20 | 2005 | 55.6075 | 13.4042 |
| I. elegans | Het Vinne, Belgium | allopatric | 20 | 2007 | 50.833 | 5.117 |
| I. elegans | Kaiserslautern, southern Germany | allopatric | 20 | 2008 | 49.2641 | 7.4674 |
| I. elegans | Heuringhem, northern France | allopatric | 20 | 2008 | 50.4209 | 2.164 |
| I. elegans | Vigueirat, southern France | allopatric | 20 | 2008 | 43.5311 | 4.3012 |
| I. elegans | Gran Sasso, central Italy | allopatric | 20 | 2008 | 42.5015 | 13.4328 |
| I. elegans | Lublin-Zemborzyce, Poland | allopatric | 20 | 2007 | 51.15 | 22.34 |
| I. elegans | Zwięczyca Reszów, Poland | allopatric | 20 | 2007 | 50.0167 | 21.9167 |
| I. elegans | Breznica, Poland | allopatric | 20 | 2007 | 49.9696 | 19.6429 |
| I. elegans | Suchoi Limon, Ukraine | allopatric | 20 | 2006 | 46.03 | 30.047 |
| I. elegans | Enmakov Island, Ukraine | allopatric | 20 | 2006 | 45.435 | 29.525 |
Table 3.
Number of I.elegans, I. graellsii and hybrid males examined in each putative introgressed populations.
| Population | Region | Year | I. elegans | I. graellsii | Hybrids |
|---|---|---|---|---|---|
| Doniños | north-western Spain | 2007 | 80 | 0 | 0 |
| Laxe | north-western Spain | 2007 | 206 | 0 | 0 |
| Louro | north-western Spain | 2007 | 136 | 0 | 0 |
| Arreo | north-central Spain | 2008 | 160 | 2 | 0 |
| Alfaro | north-central Spain | 2007 | 38 | 63 | 4 |
| Baldajo | central Spain | 2008 | 34 | 2 | 0 |
| Europa | north-eastern Spain | 2008 | 121 | 0 | 0 |
| Amposta | central-eastern Spain | 2008 | 30 | 0 | 0 |
| Marjal del Moro | south-eastern Spain | 2008 | 25 | 0 | 0 |
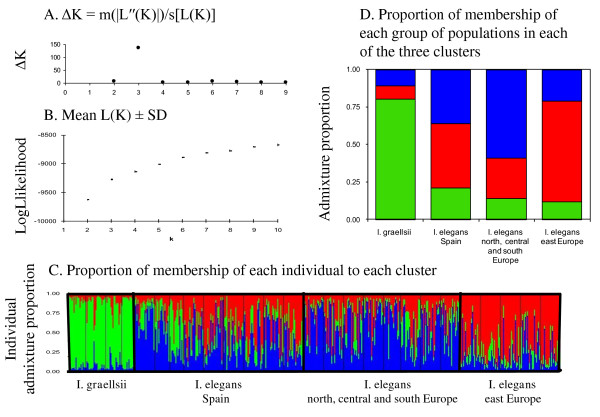
Analyses of the population structure in STRUCTURE supported the molecular differentiation between the two Ischnura species, although the ΔK-method suggested three clusters as the most likely population structure (Figure 4). More specifically, the results showed that one of the genetic clusters highly corresponded to the I. graellsii group, while the other two genetic clusters were best represented by I. elegans genotypes (one mainly corresponding to northern, central and southern European populations, and one to populations in eastern Europe), with the Spanish populations showing an intermediate assignment to each of these three genetic clusters (Figure 4).
Figure 4.
Estimated population structure of I. elegans from Bayesian structure analyses using the program STRUCTURE. 4.A. ΔK-values for different K; suggesting K = 3 as the most likely structure (ΔK = m|L"(K)|/s[L(K)]; see [67]. 4.B. Mean likelihood (± SD) of K for different numbers of clusters, K. 4.C. Individual Bayesian assignment probabilities for K = 3 for 22 populations of I. elegans and 4 I. graellsii populations. Individuals are represented by thin vertical lines, which are partitioned into K shaded segments representing each individual's estimated membership fraction. The black lines separate sampling sites. 4.D. The average Bayesian assignment probabilities for K = 3 for the four geographical areas; (1) eastern Europe, (2) northern, central and southern Europe, (3) Spanish I. elegans populations, and (4) I. graellsii populations.
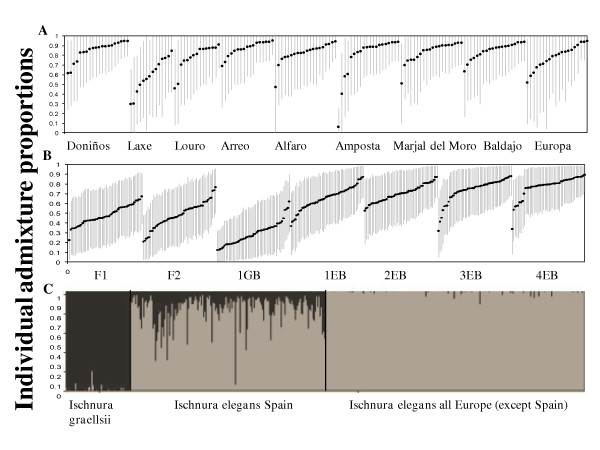
Based on these results, we included all populations of I. graellsii and I. elegans in the STRUCTURE runs (K = 2) to analyse the individual admixture proportions of I. elegans individuals in Spain (Table 4). These results showed that one genetic cluster clearly corresponded to I. graellsii, while the other cluster corresponded to I. elegans (European populations outside Spain; Table 4; Figure 5). The majority of individuals of the European I. elegans outside Spain (95%) and I. graellsii (88%) were assigned with a certainty of at least 90% to each of these clusters. However, only 27% of individuals of the Spanish I. elegans were strongly assigned to I. elegans (admixture proportions ≥ 90%) and one individual (from the sympatric population Alfaro) was assigned to I. graellsii (admixture proportion to I. elegans < 10%). The rest of the Spanish I. elegans individuals were intermediate between the two clusters (with a skew of admixture proportion towards I. elegans, see additional file 1), suggesting a significant degree of introgressed I. graellsii alleles (Table 4; Figure 5).
Table 4.
Summary of the results from the Admixture models in STRUCTURE for Ischnura populations.
| Admixture proportion to I. elegans cluster | |||||||
|---|---|---|---|---|---|---|---|
| Region/Locality | N | Species | ≥ 90 | (89-68) | (67-21) | (20-11) | ≤ 10 |
| Iberia-Africa | 56 | I. graellsii | 0 | 0 | 3 | 4 | 49 |
| All Europe (except Spain) | 220 | I. elegans | 209 | 11 | 0 | 0 | 0 |
| Spain | 166 | I. elegans | 45 | 97 | 23 | 0 | 1 |
| Spanish localities | |||||||
| Doniños | 20 | I. elegans | 8 | 10 | 2 | 0 | 0 |
| Laxe | 14 | I. elegans | 0 | 5 | 9 | 0 | 0 |
| Louro | 15 | I. elegans | 1 | 12 | 2 | 0 | 0 |
| Arreo | 17 | I. elegans | 7 | 10 | 0 | 0 | 0 |
| Baldajo | 20 | I. elegans | 4 | 15 | 1 | 0 | 0 |
| Alfaro | 20 | I. elegans | 7 | 9 | 3 | 0 | 1 |
| Europa | 20 | I. elegans | 8 | 11 | 1 | 0 | 0 |
| Amposta | 20 | I. elegans | 6 | 13 | 1 | 0 | 0 |
| Marjal del Moro | 20 | I. elegans | 4 | 12 | 4 | 0 | 0 |
Figure 5.
Admixture analysis in STRUCTURE. A and B panels show the admixture analysis in STRUCTURE of 166 genotypes of nine Spanish populations of I. elegans (upper panel) and for the artificial hybrids genotypes generated with the program HYBRID-LAB (mid panel). The estimated admixture proportion of each individual is represented by the mean assignment (± 90% credible intervals) to the I. elegans cluster. C panel shows admixture proportions of all 472 I. elegans and I. graellsii genotypes. Each individual is represented by a single vertical line broken into two segments which are proportional to the estimated membership in each of the two genetic clusters (Q1 for I. graellsii (black), and Q2 for I. elegans (grey)).
The admixture proportions of the artificial hybrids and backcrosses ranged between 11-89% (Table 5; Figure 5). The F1 and F2 showed admixture proportions to I. elegans between 67 and 21%; the 1 GB (first I. graellsii backcross) between 67 and 11%; and the four I. elegans backcrosses (1-4 EB) between 89 and 21% (Table 5; Figure 5). Based on these results, we defined three conservative assignment groups: backcrosses with I. elegans with admixture between 89-68%; backcrosses with I. graellsii ranging between 20-11%; and a mixed group of F1, F2 and backcrosses ranging between 67-21%. When the Spanish I. elegans individuals (N = 166) were allocated by their admixture proportions in one of these three groups, a total of 97 individuals (admixture proportions to I. elegans between 89-68%) were assigned to backcrosses with I. elegans (1-4 EB), whereas not a single individual was assigned to the backcross with I. graellsii (1 GB) (Table 5). Finally, the rest of the Spanish I. elegans (i.e. 23 individuals with admixture proportions to I. elegans between 67-21%), excluding the single individual from Alfaro that was assigned to I. graellsii, were assigned to the mixed-hybrid group, including F1, F2 and backcrosses with the parental species (Table 4).
Table 5.
Summary of the results from the Admixture models in STRUCTURE for artificial hybrids and backcrosses:
| Admixture proportion to I. elegans cluster | ||||||
|---|---|---|---|---|---|---|
| Type of cross | N | ≥ 90 | (90-68) | (67-21) | (20-11) | ≤ 10 |
| F1 | 50 | 0 | 0 | 50 | 0 | 0 |
| F2 | 50 | 0 | 0 | 50 | 0 | 0 |
| 1 GB | 50 | 0 | 0 | 35 | 15 | 0 |
| 1 EB | 50 | 0 | 24 | 26 | 0 | 0 |
| 2 EB | 50 | 0 | 30 | 20 | 0 | 0 |
| 3 EB | 50 | 0 | 40 | 10 | 0 | 0 |
| 4 EB | 50 | 0 | 42 | 8 | 0 | 0 |
first-generation hybrid (F1; i.e. I. graellsii × I. elegans), second-generation hybrid (F2; i.e. F1 × F1), first backcross with I. elegans (1 EB; i.e. F1 × I. elegans), first backcross with I. graellsii (1 GB; F1 × I. graellsii), second backcross with I. elegans (2 EB; 1 EB × I. elegans), third backcross with I. elegans (3 EB; 2 EB × I. elegans), and forth backcross with I. elegans (4 EB; 3 EB × I. elegans).
Low genetic variation and shared polymorphism at mitochondrial genes
The alignments for the Cytochrome C Oxidase I and II (COI-COII), Cytochrome B (CYTB), 12S rRNA (12S) and NADH Dehydrogenase 1 (ND1) fragments included 591 bp, 673 bp, 457 bp, 370 bp and 591 bp, respectively (Table 6). All new sequences were deposited in GenBank (accession numbers: HQ834794-HQ834810). COI fragment showed no polymorphic sites, revealing a unique haplotype (H1) that was shared by the two species. COII showed one polymorphic site, revealing two haplotypes (haplotype diversity, H = 0.409 ± 0.133; nucleotide diversity, π = 0.00122). Haplotype H2, the most abundant haplotype, was shared by the species in both allopatric and sympatric regions, while haplotype H3 appeared only in the three samples of I. graellsii from one allopatric region (Morocco). CYTB fragment showed two polymorphic sites revealing three haplotypes (H = 0.163 ± 0.099; π = 0.00053). Each species showed one unique haplotype (H5 and H6), in a single sample from one allopatric region (Greece and Morocco), respectively, while the rest of the samples of both species shared a common haplotype (H4). The 12S fragment showed three polymorphic sites revealing four haplotypes (H = 0.087 ± 0.047; π = 0.00024). Ischnura elegans showed three unique haplotypes (H7, H8, H9), each of which appeared in a unique sample (two haplotypes in samples from allopatric regions and one in a sample from a sympatric region), while the rest of the samples of both species shared the common haplotype H10. The last fragment, ND1, showed one polymorphic site, revealing two haplotypes (H = 0.250 ± 0.180; π = 0.00042). The most abundant haplotype (H11) was shared by both species (from allopatric and sympatric regions), while the second haplotype (H12) appeared only in one sample of I. graellsii (from an allopatric population in Portugal).
Table 6.
Number of individuals genotyped per gene (N), sequence length (bp), number of haplotypes (S), haplotype diversity (H) and nucleotide diversity (π) for each of the five analysed mitochondrial genes.
| Mitochondrial gene | Genetic measures | |||||
|---|---|---|---|---|---|---|
| N | bp | S (I. elegans) | S (I. graellsii) | H | π | |
| COI | 13 | 591 | H1 | H1 | 0.000 | 0.00000 |
| COII | 12 | 673 | H2 | H2, H3 | 0.409 ± 0.133 | 0.00122 |
| CYTB | 24 | 457 | H4, H5 | H5, H6 | 0.163 ± 0.099 | 0.00053 |
| 12S | 68 | 370 | H7, H8, H9, H10 | H10 | 0.087 ± 0.047 | 0.00024 |
| ND1 | 8 | 591 | H11 | H11, H12 | 0.250 ± 0.180 | 0.00042 |
Colour morph frequencies
All populations showed all three female morphs, with the exception of Saïdia in northern Africa (where the aurantiaca morph was missing) (Table 7). In populations dominated by I. elegans from north-central, central and eastern Spain, androchrome, infuscans and infuscans-obsoleta frequencies were highly variable between populations (range of androchrome frequencies: 3.3-42.9%; infuscans: 6.7-68.8%; infuscans-obsoleta: 11.8-76.7%) which is consistent with what has previously been observed for the I. elegans populations from north-western Spain [21]. In allopatric populations of I. graellsii, androchrome frequencies were always below 20% (3.0-18.8%); with the infuscans morph being the most abundant morph type (64.9-89.7%). The sympatric populations, O Vilar and Xuño, which have previously shown very high frequencies of androchromes for both species [21], also showed in our study high levels of androchrome frequencies (12.5-15.7% in I. graellsii and 40.0-47.6% in I. elegans). In addition, the sympatric populations Las Cañas and Alfaro from central Spain further showed high androchrome frequencies (13.0-17.1% in I. graellsii and 69.7-70.8% in I. elegans).
Table 7.
Frequencies of female colour morphs of Ischnura elegans and I. graellsii.
| Species | Ecology | Locality | Date | N | A | O | I | Source |
|---|---|---|---|---|---|---|---|---|
| I. graellsii | Allopatric | Saïdia, northern Africa | Jun-09 | 29 | 10.3 | 89.7 | 0 | 39 |
| I. graellsii | Allopatric | Puente de los Arenales, southern Spain | Sep-08 | 33 | 3 | 78.8 | 18.2 | 39 |
| I. graellsii | Allopatric | Doñana, southern Spain | Jun-03 | 77 | 10.4 | 76.6 | 13 | 21 |
| I. graellsii | Allopatric | Granjuela, southern Spain | Sep-08 | 35 | 11.4 | 77.1 | 11.4 | 39 |
| I. graellsii | Allopatric | Jaraiz de la Vera, central Spain | Jun-07 | 67 | 13.5 | 64.9 | 21.6 | 39 |
| I. graellsii | Allopatric | Ribeira de Cobres, southern Portugal | Apr-2003 | 48 | 18.8 | 72.9 | 8.3 | 21 |
| I. graellsii | Sympatric | Troi, northern Spain | Jul-08 | 54 | 14.8 | 72.2 | 13 | 39 |
| Sympatric-I. graellsii | Sympatric | O Vilar, north-western Spain | May-Jul 2006 | 121 | 15.7 | 33.06 | 51.24 | This study |
| Sympatric-I. elegans | Sympatric | O Vilar, north-western Spain | May-Jul 2007 | 21 | 47.62 | 47.62 | 4.76 | This study |
| Sympatric-I. graellsii | Sympatric | Xuño, north-western Spain | Jun-Sept 2006 | 48 | 12.5 | 33.33 | 54.17 | This study |
| Sympatric-I. elegans | Sympatric | Xuño, north-western Spain | Jun-Sept 2007 | 15 | 40 | 40 | 20 | This study |
| Sympatric-I. graellsii | Sympatric | Las Cañas, north-central Spain | Jul-07 | 23 | 17.4 | 69.6 | 13 | 39 |
| Sympatric-I. elegans | Sympatric | Las Cañas, north-central Spain | Aug-07 | 24 | 70.83 | 25 | 4.17 | This study |
| Sympatric-I. graellsii | Sympatric | Alfaro, north-central Spain | Jul-07 | 23 | 13 | 60.9 | 26.1 | 39 |
| Sympatric-I. elegans | Sympatric | Alfaro, north-central Spain | Aug-07 | 33 | 69.7 | 27.3 | 3 | This study |
| I. elegans | Sympatric | Arreo, north Spain | Jul-08 | 30 | 6.3 | 68.8 | 25 | 39 |
| I. elegans | Sympatric | Almoquera, central Spain | Aug-2008 | 28 | 42.9 | 42.9 | 14.3 | 39 |
| I. elegans | Sympatric | Baldajo, central Spain | Aug-2008 | 34 | 29.4 | 58.8 | 11.8 | 39 |
| I. elegans | Sympatric | Europa, north-eastern Spain | Jul-08 | 30 | 16.7 | 6.7 | 76.7 | 39 |
| I. elegans | Sympatric | Amposta, north-eastern Spain | Jul-08 | 30 | 3.3 | 33.3 | 63.3 | 39 |
| I. elegans | Sympatric | Barranco de Caixanet, south-eastern Spain | Sep-08 | 27 | 25.9 | 25.9 | 48.1 | 39 |
| I. elegans | Sympatric | Marjal del Moro, south-eastern Spain | Sep-08 | 25 | 36 | 20 | 44 | 39 |
The number of mature female examined in each locality is indicated (N). A: androchrome, I: infuscans and O: infuscans-obsoleta (I. elegans) or aurantiaca (I. graellsii).
Discussion
Hybridization and genomic introgression are repeatedly suggested to be important elements in evolutionary processes such as maintenance of genetic variation, adaptation and speciation [4-6]. Determining the degree of genetic exchange between recently diverged species may be of particular interest in this sense since they typically show incomplete reproductive barriers. Moreover, the pattern and extent of hybridization and introgression can have important conservation implications because it may lead to the replacement of one of the hybridizing taxa [10,43-45].
In the present study, we have revealed extensive hybridization and introgression in I. elegans populations in Iberia where it co-occurs sympatrically with its closely related sister species I. graellsii. Distribution maps for both species show an extended area of overlap in northern and central Spain. Ischnura graellsii is generally very abundant all along the Iberian Peninsula, while I. elegans has a patchy distribution. For example, I. elegans is very rare in southern Spain. In the sympatric regions, I. elegans is also less abundant than I. graellsii (with the exception of the area around Valencia) and is not present at all in some provinces (Figure 2). Consequently, I. graellsii occurs allopatrically in southern Iberia (Figure 2), whereas I. elegans is exclusively found in areas that are also occupied by I. graellsii. Our admixture analyses in STRUCTURE revealed clear evidence of past and present hybridization and introgression between I. elegans and I. graellsii over a large geographic area in northern Spain. The degree of introgression in the populations is consistent with I. elegans backcrosses and occasional F1-hybrids. Thus, hybridization between these two damselfly species is a relatively recent and widespread phenomenon. Interestingly, the studied populations have been going through a recent species turn-over and are now dominated by I. elegans individuals that appear to carry introgressed I. graellsii genetic material. These kinds of dramatic demographic and genetic effects have not previously been documented in odonates, although it is known from other taxa. For instance, over the past century, the blue-winged (Vermivora pinus) warbler has rapidly replaced the golden-winged warbler (V. chrysoptera) over an extensive part of their hybrid zone in eastern north America. Marker-based analyses show asymmetric and rapid introgression from blue-winged warbler into golden-winged warbler in some areas and bidirectional maternal gene flow in others [46,47]. Rapid introgression has also been detected in other taxa, e.g. in pocket gophers (Geomys bursarius major and G. knoxjonesi) [48].
Extent of hybridization and direction of introgression
In vertebrates, hybridization is particularly common in fish, where several hundred interspecific and intergeneric crosses have been reported, and in birds, with roughly 10% of all species known to have bred in nature with another species e.g. [5,27,46-50]. In comparison, detailed genetic studies of hybridization and introgression, and thus the knowledge about these phenomena, are lacking for most odonates [12,13]. As mentioned above, three previous genetic studies failed in detecting extensive hybridization between different pairs of odonate species [14-17]. Our admixture analysis in STRUCTURE showed that the I. elegans populations Louro and Laxe in north-western Spain had the highest degree of introgression of I. graellsii alleles. This is not surprising, given that north-western Spain is the region where hybridization between I. elegans and I. graellsii was most recently detected [20]. In 1990, I. elegans, I. graellsii and hybrids were found in Foz (north-western Spain), which is geographically close to Louro and Laxe. However, after only ten years, the population was dominated by individuals that were phenotypically classified as I. elegans, although morphological intermediate and I. graellsii males were occasionally detected [21]. The population Laxe was visited for the first time in June 2001 (two visits) and only I. elegans were detected, although at a low densities (between 0.2 to 1.3 captured males/minute). At a visit in June 2002, I. elegans were found at a similar density (0.5 males/minute). Five years later (in 2007), we revisited the population and the density had reached the highest value in the region. A total of seven visits were conducted in 2007, between June and August, and the density ranged between 3.3 and 14.7 males/minute; and neither I. graellsii nor putative hybrids were detected. Nevertheless, the admixture analyses showed that none of the 14 examined individuals at Laxe could be genetically assigned to pure I. elegans status. Nine individuals were assigned to F1, F2, or backcrosses with I. elegans (admixture proportion between 67-21%), and five individuals were assigned to backcrosses with I. elegans (admixture proportion between 89-68%). In 1980, Louro was visited for first time by Ocharan [22] and mainly I. graellsii individuals were observed. However, Torralba and Ocharan [51] reviewed Ocharan's samples from Louro (sampled in 1980) and have now identified both species in the sample; nevertheless, I. graellsii species still remains the dominant species. However, on our first visit in 2000 and on subsequent visits, I. elegans individuals completely dominated in numbers (a couple of I. graellsii individuals were detected among hundreds of I. elegans). Only one out of the 15 Louro individuals that were molecularly examined in the present study was assigned to be pure I. elegans. In addition, the vast majority of the individuals (93%) showed an assignment proportion between 89-68%, suggesting that these individuals were backcrosses, and not F1 or F2 hybrids.
These results are corroborated by the PCA-analysis. Both Laxe and Louro were placed within the I. graellsii quadrant, indicating a significant degree of molecular similarity between these I. elegans populations and I. graellsii. Analyses of the remaining Spanish I. elegans populations showed that only 31% of the individuals were pure I. elegans, and that the remaining 69% showed admixture proportions expected for hybrids and backcrosses with I. elegans. In the Alfaro population in the north-central Spain where both species co-occur in equal numbers, one individual classified as I. elegans had a very high proportion of I. graellsii alleles. This either suggest that this individual was misclassified despite the fact that we only collected males to minimise misidentifications [20], or that it is a backcross that has inherited a very high proportion of I. graellsii alleles at the few markers analysed.
Our suggest that hybridization between I. elegans and I. graellsii is asymmetric and largely unidirectional. This corroborates previous findings in the field [20] and laboratory [21] showing that I. graellsii has a mating disadvantage in sympatry with I. elegans. Heterospecific matings, and matings between the F1-hybrids and the parental species, rarely take place with I. elegans females, but occur more frequently among I. graellsii females, I. elegans males and hybrids [Sánchez-Guillén RA, Wellenreuther M and Cordero-Rivera A: Strong asymmetry in the relative strengths of prezygotic and postzygotic barriers between two damselfly sister species, submitted]. The reason for the almost complete lack of hybridization between I. graellsii males and I. elegans females is that males cannot grasp the female by their protothorax [20], a mechanical handicap that appears to be a very efficient prezygotic isolation mechanism. Previous work on plants [52,53] and animals [27] has suggested that unidirectional hybridization usually occurs between the females of the rare species and the males of a common species, but not vice versa. Wirtz [27] reviewed the factors promoting unidirectional or reciprocal hybridization and proposed a sexual selection hypothesis for unidirectional hybridization based on the fact that females normally invest more in offspring and, therefore, are more discriminating than males. When heterospecific males are less abundant than conspecific males, females rarely mate with heterospecific males. Consequently, under such a condition, the rare species is usually the maternal parent of the hybrids. However, this is not the case in our study where the more abundant species is initially I. graellsii. Ischnura elegans, on the other hand, appears to be the intruding species and is hence initially the rare species, which has been expanding its range in Spain and is now displacing I. graellsii from some populations and regions. Thus, the direction of hybridization between these two Ischnura species is opposite that expected based on the rare female hybridization hypothesis just outlined e.g. [27], but follows the prediction proposed by Grant and Grant [28], namely that when the rare species is also the bigger species (in our study I. elegans), hybridization can be impeded by mechanical incompatibility. Laboratory tests have detected that I. elegans females and I. graellsii males are mechanically impaired to form a tandem, preventing over 93% of all matings, while only 13% of the matings between I. graellsii females and I. elegans males are mechanically prevented [Sánchez-Guillén RA, Wellenreuther M and Cordero-Rivera A: Strong asymmetry in the relative strengths of prezygotic and postzygotic barriers between two damselfly sister species, submitted].
The role of hybridization in the maintenance of colour polymorphisms
Johnson [33] proposed that the colour polymorphisms in odonates could be maintained due to hybridization between closely related species and balanced by differential predation pressure. According to this hypothesis, androchrome females benefit from avoiding matings with heterospecific males, while gynochromes females are involved in heterospecific matings (usually sterile); the relative fitness of the colour morphs is balanced by a higher probability of predation of the androchrome females. In a recent study on I. elegans and I. graellsii [21], a role of hybridization for the temporal maintenance of contrasting androchrome frequencies in nearby populations in north-western Spain was suggested. The relatively low androchrome frequencies in I. elegans populations located close to I. graellsii populations, and the relatively high androchrome frequencies in I. graellsii populations adjacent to I. elegans populations, were hypothesized to be caused by the absorption of the, in other circumstances, typical morph frequency [21]. Ischnura elegans data from other sympatric regions in the Iberian Peninsula now shows that the substantial variation in female morph frequencies between different populations in north-western Spain is not unique to this region; also in other parts of Iberia morph frequencies are drastically variable between populations (androchrome: 3.3-70.8%; infuscans: 6.7-72.9%; infuscans-obsoleta: 3.0-76.7%). Furthermore, other recent studies have revealed that disparate morph frequencies are not restricted to Iberian populations [53,54], although androchrome frequencies are typically more abundant in northern latitudes and show less variation between populations than in Iberia [with the exception of Iberia, 53]. In our study, the highest frequencies of androchromes were found in the four populations with both species present (Xuño, O Vilar, Alfaro and Las Cañas; Table 7); in I. graellsii androchrome frequencies ranged between 12.5 and 17.4% and in I. elegans between 40.0 and 70.8%. Nevertheless, the I. graellsii population Ribeira de Cobres had androchrome frequencies of 18.8%, for this species an atypically high level. However, this comparatively high androchrome frequency cannot be explained by the influence of nearby I. elegans populations, because this population is located in the allopatric region (southern Portugal). Furthermore, in I. elegans the lowest androchrome frequencies were found in north-central Spain (population named Arreo) where the nearest I. graellsii population (named Troi) had higher androchrome frequencies (14.8%) than the I. elegans population (3.3%), and in a coastal population near the Mediterranean coast (eastern Spain) (Amposta; 3.3% androchromes), but here we lack data of I. graellsii morph frequencies in nearby populations.
In conclusion, hybridization is likely to have important implications for the maintenance of multiple female morphs, but only during the short period when two species with contrasting morph frequencies start to hybridize.
The evolution, maintenance and adaptive function of genetic colour polymorphism have received very much attention in a broad range of organisms, e.g. in birds [55], reptiles [56] and insects [57]. However, the role of hybridization in this context has received little attention, which may simply reflect the fact that only a few suitable study systems are available to study; one being the presently described Ischnura damselfly complex in Spain. Another case is the hybrid zone of the land snails Mandarina mandarina and M. chichijimana in the oceanic Bonin Islands [58]. In that system, the variability of the colour polymorphism in the hybrid population was substantially higher than that in the pure populations, suggesting that morphological variation was maintained by hybridization [58]. These results highlight the importance of hybridization as a source of morphological variation, diversity and evolutionary novelty.
Conclusions
When hybrids are fertile and backcross with one of the parental species, hybridization will inevitably result in introgression, thereby increasing the genetic variability of the introgressed species [59]. Even at low levels, introgression of novel genetic material could be an important factor as a source of new genetic and phenotypic variability, and subsequent evolution. Thus a certain degree of hybridization could create favourable conditions for new adaptations [4-6]. In contrast, extensive hybridization and introgression can, as mentioned above, sometimes have important conservation implications when leading to the replacement of one of the hybridizing taxa [10,42-44]. We have documented a unique case of hybridization and unidirectional introgression in polymorphic Ischnura damselflies, where hybridization is likely to having important implications on the temporal maintenance of multiple female morphs. The potential adaptive significance of introgression in the system, and whether this per se has contributed to the rapid species turn-over in sympatric populations in the region, remains to be evaluated.
Methods
Spatial distribution of I. graellsii and I. elegans in the Iberian Peninsula
We conducted a revision of the distribution data of the two species along the Iberian Peninsula from 1866-2008 using data from Baixeras et al. [60], Jödicke [61] and Ocharan [22,62], in the region around la Rioja using data from Tomás Latasa (personal communication) and along the Iberia using data from Jean Pierre Boudot (personal communication). Using DMAP (Distribution mapping software, Version 7.0) we constructed two geographic maps showing the distribution of both species in Iberia (Figure 2).
Study populations and sample collection
Samples of I. elegans and I. graellsii were collected from 26 populations from Europe and northern Africa (see Table 2 for details of sampling locations). In particular, all European populations, except the populations from Spain, were classified as allopatric I. elegans populations and included a total of 220 individuals from 13 I. elegans populations. In depth sampling of both I. elegans and I. graellsii was carried out in the sympatric region in Spain; in the north-western corner, the central parts and along the east coast (Figure 1). In these areas, nine sympatric populations (166 individuals) were sampled, and these were, with the exception of Alfaro, dominated by individuals that were phenotypically classified as I. elegans (see Table 3 for details of species proportions in the sampled populations). In addition, four allopatric I. graellsii populations (56 I. graellsii individuals) from the north, central and south Iberia, and northern Africa were sampled.
At each allopatric and sympatric I. elegans population (see Table 2), and at the four allopatric I. graellsii populations, a minimum of 20 adult males were collected during the flight season between 1999-2008 using hand nets. Captured individuals were stored in 100% ethanol until DNA extraction. Only males were sampled because the identification of male I. elegans, I. graellsii and hybrids is more reliable than that of females. Note that the few individuals classified as I. graellsii and hybrids in the Spanish I. elegans populations, mainly found in Alfaro in north-central Spain (Table 3), were not included in the genetic analyses because the aim of this study was to test for introgression in I. elegans populations.
DNA extraction and microsatellite genotyping
To extract DNA from the samples, the head of each damselfly was removed, dried and then homogenized using TissueLyser (Qiagen). DNA was extracted by proteinase K digestion followed by a standard phenol/chloroform-isoamylalcohol extraction [63]. The purified DNA was re-suspended in 50-100 μl of sterile water. The genotypes of all damselflies were assayed at six microsatellite loci previously isolated for this species [I-002, I-015, I-041, I-053, I-095, I-134, for details see 33]. These loci did not deviate statistically from Hardy-Weinberg expectations and linkage equilibrium, and showed no evidence for presence of common null-alleles (using Micro-Checker; [64], within populations of both species [34]. One primer of each pair was 5'-labelled with 6-FAM, HEX or NED florescent dyes. Polymerase chain reactions (PCRs) were carried out in 10 μL volumes on a GeneAmp PCR System 9700 (Applied Biosystems) and contained 4 pmol of each primer, 15 nmol MgCl2, 1.25 nmol dNTP, 0.5 U Ampli-taq polymerase and 10-20 ng template. The conditions were: denaturation step of 94°C for 2 minutes, then 35 cycles at 94°C for 30 s, touch-down from 62-58°C for 30 s, 72°C for 30 s followed by 72°C for 10 minutes. Multiplex primer reactions were performed for combinations of primers with matching annealing temperatures but differing size ranges and dye labels, then mixed with a labelled size standard and electrophoresis was conducted on an ABI PRISM 3730 Genetic Analyzer (Applied Biosystems). GeneMapper 3.0 (Applied Biosystems) was used for fragment size determination and allelic designations.
Microsatellite DNA Analyses
The program FSTAT [65] was used to calculate several basic population genetic measures, namely, the expected heterozygosity (HE), observed heterozygosity (Ho), number of alleles, and the allelic richness for each population. These aforementioned measures as well as the genetic differentiation between populations (FST) were also calculated for three regions, namely the Spanish populations of I. elegans, the European populations of I. elegans (excluding Spain), and the entire sample of I. graellsii.
Principal component analysis (PCA) was used to reduce the variation in the multivariate data set (consisting of 117 alleles at six loci) to two linear combinations. The analysis was done using PCA-GEN [66]. The significance of each principal component was assessed from 5000 randomisations of genotypes. The allocation of each species across the two principal components provides a quantitative measure of the degree of the genetic dissimilarity among the populations/species [18].
The Bayesian statistical framework provided by the program STRUCTURE [version 2.2.3, 67] was used to understand the genetic structure among populations and to determine which individuals from the allopatric and sympatric populations of I. elegans and I. graellsii can be classified to a high degree as pure species. STRUCTURE applies a Bayesian Markov chain Monte Carlo (MCMC) approach that uses model-based clustering to partition individuals into groups. The model accounts for the presence of Hardy-Weinberg and linkage disequilibrium by introducing group structure and attempts to find groupings that (as far as possible) are in equilibrium [68]. We applied the 'admixture model' with 'correlated allele frequencies' for more details [69]. For the model, a 'burn-in' period of 20,000 MCMC replicates and a sampling period of 100,000 replicates were used. We performed runs for a number of genetic clusters (K), ranging from one to ten; and for each K, 20 iterations were run. In this way, multiple posterior probability values (log likelihood (lnL) values) for each K were generated, and the most likely K was evaluated by the ΔK-method following Evanno et al. [66].
Admixture analyses in STRUCTURE were also used to assign all individuals of the Spanish Ischnura populations into each of two genetic clusters, one representing I. graellsii genotypes and one I. elegans. We used the 'prior population information' option in the models to (i) facilitate the clustering process of the reference individuals (i.e. pure I. elegans from central and eastern Europe, and I. graellsii, respectively), and (ii) to calculate the admixture proportions (and ± 90% credible regions) of each individual in the Spanish I. elegans populations. This approach was hence used to measure of the degree of introgression of I. graellsii genetic material into the genome of I. elegans in Spain. The model was run for K = 2, where one cluster corresponded to I. graellsii and the other to I. elegans. We used the 'population flag' option to exclude Spanish I. elegans as reference individuals, which implied that the clustering process was based on only I. graellsii samples and I. elegans samples collected outside of Spain. The model was run for 100,000 MCMC replicates, after an initial burn-in period of 20,000 replicates, using the admixture model and correlated frequencies for five iterations [32-34,63]. To generate simulated genotypes of hybrids and backcrosses, we applied the program HYBRID-LAB [70] using the genotypes of 66 individuals of I. graellsii and 240 genotypes of I. elegans collected outside of Spain as initial genotypes. We generated 50 genotypes of each of the following crosses: first-generation hybrid (F1; i.e. I. graellsii × I. elegans), second-generation hybrid (F2; i.e. F1 × F1), first backcross with I. elegans (1 EB; i.e. F1 × I. elegans), first backcross with I. graellsii (1 GB; F1 × I. graellsii), second backcross with I. elegans (2 EB; 1 EB × I. elegans), third backcross with I. elegans (3 EB; 2 EB × I. elegans), and forth backcross with I. elegans (4 EB; 3 EB × I. elegans). We then evaluated the admixture proportions (± 90% credible intervals) of these artificial crosses with STRUCTURE in the same way as done for the I. elegans samples from Spain (above). To determine the level of introgression of I. graellsii into the Spanish I. elegans populations, the individual admixture proportions of the I. elegans samples from Spain were compared to the admixture proportion for the artificial hybrids and backcrosses.
Mitochondrial sequencing
Three to six damselflies from sympatric and allopatric localities were amplified by polymerase chain reaction (PCR) for part of the mitochondrial Cytochrome C Oxidase I and II genes (COI-COII), part of the mitochondrial Cytochrome B (CYTB) gene and part of the mitochondrial 12S rRNA (12S) and NADH Dehydrogenase 1 (ND1). The amplification was done using universal primers: 591 bp of the COI with the primers COI-H (5'-TCAGGGTGACCAAAAAATCA-3') and COI-L (5'-GGTCAACAAATC ATAAAGATATTGG-3') (Marina Magaña Ramos, personal communication), 673 pb of the COII with the primers TL2-J-3037 (5'- ATGGCAGATTAGTGCAATGG-3') and C2-N-3494 (5'-GGTAAAACTACTCGATTATCAAC-3') and C2-J-3400 (5'-ATTGGACATCAATGATATTGA-3') and TK-N-3785 (5'-GTTTAAGAGACCAGTACTTG-3') [34], 457 pb of the CYTB with the primers CB-J-10933 (5'-TATGTACTACCATGAGGACAAATATC-3') and TS1-N-11683 (5'-TATTTCTTTATTATGTTTTCAAAAC-3') [34], 370 bp of the 12S with the primers SR-J-14233 (5'-AAGAGCGACGGGCGATGTGT-3') and SR-N-14588 (5'-AAACTAGGATTAGATACCCTATTAT-3') [30], and 591 bp of the ND1 gene with CB-J-11545 (5'ACATGAATTGGAGCTCGACCAGT-3') and N1-N-12051 (5'-GATTTTGCTGAAGGTGAATCAGA-3') [34]. DNA amplification was done in a total reaction volume of 20 μl. The amplification conditions were as follows: 50 ng of DNA (1 μL), 1 unit (0.2 μL) of Taq DNA polymerase (Ecogen), 2 μL 10x of reaction buffer (Ecogen), 0.5 μL of MgCl2 (50 mM) (Ecogen), 0.5 μL of dNTPs Mix Sigma (200 μM), and 1 μL of each primer (10 pmol). All PCR reactions were completed in a "GeneAmp PCR system 2700" thermocycler (Applied Biosystems). The PCR program had an initial cycle of 95°C for 3 min, the annealing temperature for 1 min, and an elongation period at 72°C for 45 s, followed by 34 cycles at 95°C for 30 s, with annealing for 45 s, and an elongation phase at 72°C for 45 s, and a final extension phase at 72°C for 10 min. PCR products were sent to an external sequencing service (University of Valencia) where bidirectional sequencing reactions were conducted using Bigdye™terminator cycle sequencing kit (Applied Biosystems) using automatic sequencer ABI3100 (Applied Biosystems). Forward and reverse sequences were edited in Codon Code Aligned (CodonCode, Dedham, MA, USA) and consensus sequences were aligned with Clustal X [71] implemented in MEGA v. 4.0 [72,73]. Variable positions were revised by eye, and only high quality sequences were considered.
Mitochondrial data analyses
Samples of both species were pooled in two groups, depending on the sympatric or allopatric origin. For each gene, genetic diversity was assessed in terms of number of haplotypes (S), and haplotype diversity (H) and nucleotide diversity (π) according to Nei [74] with DNASP V4.10 [74]. Further analyses could not be done because overall low genetic diversity and shared haplotype frequencies.
Colour morph frequencies
We reviewed female morph frequencies from the literature in seven I. graellsii populations from Iberia and northern Africa and seven I. elegans populations from Iberia [41]. In addition, we estimated female colour morph frequencies in four sympatric populations; Xuño and O Vilar from north-western Spain, and Alfaro and Las Cañas from north-central Spain. Populations were visited between June and September during sunny days, in two different years (2006 and 2007) and the sampling was done with entomological nets. Only single, solitary and mature females were used, and colour morph frequencies were estimated from the number of each morph divided by the total number of females.
Authors' contributions
RSG designed the study, collected genetic samples, did laboratory work, analysed data and wrote the paper. MW designed the study, did laboratory work, analysed data and wrote the paper. ACR designed the study and wrote the paper. BH designed the study, analysed data and wrote the paper. All authors approved the final version of the manuscript.
Supplementary Material
Bayesian assignment probabilities. The average Bayesian assignment probabilities in each of the two genetic clusters (Q1 for I. graellsii (grey), and Q2 for I. elegans (grey dark)), for the nine Spanish populations of I. elegans.
Contributor Information
Rosa A Sánchez-Guillén, Email: rguillen@uvigo.es.
Maren Wellenreuther, Email: maren.wellenreuther@zooekol.lu.se.
Adolfo Cordero-Rivera, Email: adolfo.cordero@uvigo.es.
Bengt Hansson, Email: bengt.hansson@zooekol.lu.se.
Acknowledgements
We would like to thank for help with fieldwork Iñaki Mezquita, Tomás Latasa, Mario García-París, Bernat Garrigós, Pere Luque, Xoaquín Baixeras, Francisco Cano, Jean Pierre Boudot, Jürgen Ott, Cedrick Vanappelghem, Philippe Lambret, and Phill Watts. We would like to thank Geert de Knijf, Tom Kompier, Martin Waldhauser, Xoaquín Baixeras, Tomás Latasa and Jean Pierre Boudot for help with the distribution maps. This research was funded by the Spanish Ministry of Science and Innovation (grant CGL2008-02799 and CGL2008-03197-E to ARC) and the Swedish Research Council (grant 621-2007-5381 and 621-2009-4945 to BHN). RSG is supported by a grant (Formación de Personal Investigador) from Spanish Ministry of Science and Innovation, and MW by postdoctoral scholarships from the Wenner-Gren Foundation, Tryggers Foundation and the Swedish Research Council. Permits to capture damselflies in Spain were issued by each Regional Government to RSG.
References
- Grant PR, Grant BR, Markert JA, Keller L, Petren K. Convergent evolution of Darwin's finches caused by introgressive hybridization and selection. Evolution. 2004;58:1588–1599. doi: 10.1111/j.0014-3820.2004.tb01738.x. [DOI] [PubMed] [Google Scholar]
- Kronforst MR. Gene flow persists millions of years after speciation in Heliconius butterflies. BMC Evol Biol. 2008;8:98. doi: 10.1186/1471-2148-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RM, Rivera C, Moore JE, Lake JA. Horizontal gene transfer in microbial genome evolution. Theor Popul Biol. 2002;61:489–495. doi: 10.1006/tpbi.2002.1596. [DOI] [PubMed] [Google Scholar]
- Anderson E. Introgressive hybridization. Chapman & Hall: London; 1949. [Google Scholar]
- Dowling TE, Secor CL. The role of hybridisation and introgression in the diversification of animals. Annu Rev Ecol Syst. 1997;28:593–619. doi: 10.1146/annurev.ecolsys.28.1.593. [DOI] [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol Evolut. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Barton NH. Speciation Trends Ecol Evol. 2001. pp. 325–413. [DOI] [PubMed]
- Jiggins CD, Mallet J. Bimodal hybrid zones and speciation. Trends Ecol Evol. 2000;15:250–255. doi: 10.1016/S0169-5347(00)01873-5. [DOI] [PubMed] [Google Scholar]
- Redenbach Z, Taylor EB. Evidence for bimodal hybrid zones between two species of char (Pisces Salvelinus) in north western North America. J Evol Biol. 2003;16:1135–1140. doi: 10.1046/j.1420-9101.2003.00619.x. [DOI] [PubMed] [Google Scholar]
- Secondi JB, Faivre B, Bensch S. Spreading introgression in the wake of a moving contact zone. Mol Ecol. 2006;15:2463–2475. doi: 10.1111/j.1365-294X.2006.02948.x. [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- Tennessen KJ. Review of reproductive isolating barriers in Odonata. Adv Odonat. 1982;1:251–265. [Google Scholar]
- Battin TJ. The odonate mating system, communication, and sexual selection: A review. Ital J Zool. 1993;60:353–360. doi: 10.1080/11250009309355839. [DOI] [Google Scholar]
- Lowe CD, Harvey IF, Thompson DJ, Watts PC. Strong genetic divergence indicates that congeneric damselflies Coenagrion puella and C. pulchellum (Odonata: Zygoptera: Coenagrionidae) do not hybridise. Hydrobiologia. 2008;605:55–63. doi: 10.1007/s10750-008-9300-9. [DOI] [Google Scholar]
- Hayashi F, Dobata S, Futahashi R. Macro- and microscale distribution patterns of two closely related Japanese Mnais species inferred from nuclear ribosomal DNA, its sequences and morphology (Zygoptera: Calopterygidae) Odonatologica. 2004;33:399–412. [Google Scholar]
- Hayashi F, Dobata S, Futahashi R. Disturbed population genetics; Suspected introgressive hybridization between two Mnais damselfly species (Odonata) Zool Sci. 2005;22:869–881. doi: 10.2108/zsj.22.869. [DOI] [PubMed] [Google Scholar]
- Tynkkynen K, Grapputo A, Kotiaho JS, Rantala MJ, Vaananen S, Suhonen J. Hybridization in Calopteryx damselflies: the role of males. Anim Behav. 2008;75:1431–1439. doi: 10.1016/j.anbehav.2007.09.017. [DOI] [Google Scholar]
- Corbet PS. Dragonflies: behavior and ecology of Odonata. Harley Books, Essex; 1999. [Google Scholar]
- Leong JM, Hafernik JE. Hybridization between two damselfly species (Odonata: Coenagrionidae): Morphometric and genitalic differentiation. Ann Entomol Soc Am. 1992;85:662–670. [Google Scholar]
- Monetti L, Sánchez-Guillén RA, Cordero-Rivera A. Hybridization between Ischnura graellsii (Vander Linder) and I. elegans (Rambur) (Odonata: Coenagrionidae): are they different species? Bio J Linn Soc. 2002;76:225–235. doi: 10.1046/j.1095-8312.2002.00060.x. [DOI] [Google Scholar]
- Sánchez-Guillén RA, Van Gossum H, Cordero Rivera A. Hybridization and the inheritance of female colour polymorphism in two Ischnurid damselflies (Odonata: Coenagrionidae) Bio J Linn Soc. 2005;85:471–481. doi: 10.1111/j.1095-8312.2005.00506.x. [DOI] [Google Scholar]
- Ocharan F. PhD thesis. University of Oviedo; 1987. Los Odonatos de Asturias y de España, aspectos sistemáticos y faunísticos. [Google Scholar]
- Hickling RD, Roy B, Hill JK, Thomas CD. A northward shift of range margins in British Odonata. Glob Chan Biol. 2005;11:502–506. doi: 10.1111/j.1365-2486.2005.00904.x. [DOI] [Google Scholar]
- Parmesan CN, Ryrholm C, Steganescu C, Hill JK, Thomas CD, Descimon B, Huntley B, Kaila L, Kullberg J, Tammaru T, Tennent WJ, Thomas JA, Warren M. Poleward shifts in geographical ranges of butterfly species associated with regional warming. Nature. 1999;399:579–583. doi: 10.1038/21181. [DOI] [Google Scholar]
- Hassall CD, Thompson J, French GC. Historical changes in the phenology of British Odonata are related to climate. Glob Chan Biol. 2007;13:933–941. doi: 10.1111/j.1365-2486.2007.01318.x. [DOI] [Google Scholar]
- Carchini G, Cobolli M, De Mathaesis E, Utzeri C. A study on genetic differentiation in the Mediterranean Ischnura Charpentier (Zygoptera: Coenagrionidae) Adv Odonat. 1994;6:11–20. [Google Scholar]
- Wirtz P. Mother species-father species: unidirectional hybridisation in animals with female choice. Anim Behav. 1999;58:1–12. doi: 10.1006/anbe.1999.1144. [DOI] [PubMed] [Google Scholar]
- Grant PR, Grant BR. Mating patterns of Darwin's Finch hybrids determined by song and morphology. Bio J Linn Soc. 1997;60:317–343. doi: 10.1111/j.1095-8312.1997.tb01499.x. [DOI] [Google Scholar]
- Hinnekint BON. Population dynamics of Ischnura e. elegans (Vander Linden) (Insecta: Odonata) with special reference to morphological colour changes, female polymorphism, multiannual cycles and their influence on behaviour. Hydrobiologia. 1987;146:3–31. doi: 10.1007/BF00007574. [DOI] [Google Scholar]
- Cordero A. The inheritance of female polymorphism in the damselfly Ischnura graellsii (Rambur) (Odonata: Coenagrionidae) Heredity. 1990;64:341–346. doi: 10.1038/hdy.1990.42. [DOI] [Google Scholar]
- Cordero A, Andrés JA. Colour polymorphism in Odonates: females that mimic males? J British Dragonfly Soc. 1996;12:50–60. [Google Scholar]
- Johnson C. Polymorphism and natural selection in Ischnuran damselflies. Evol Theory. 1975;1:811–90. [Google Scholar]
- Gosden T. PhD thesis. Lund University; 2008. [Google Scholar]
- Wellenreuther M, Sánchez-Guillén RA, Cordero-Rivera A, Hansson B. Development of 12 polymorphic microsatellite loci in Ischnura elegans (Odonata: Coenagrionidae) Mol Ecol Res. 2010;10:576–579. [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Amer. 1994;87:651–701. [Google Scholar]
- Beaumont M, Barratt EM, Gottelli D, Kitchener AC, Daniels MJ, Pritchard JK, Bruford MW. Genetic diversity and introgression in the Scottish wildcat. Mol Ecol. 2001;10:319–336. doi: 10.1046/j.1365-294x.2001.01196.x. [DOI] [PubMed] [Google Scholar]
- Vähä JP, Primmer CR. The efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- Hansen MM, Mensberg KLD. Admixture analysis of stocked brown trout populations using mapped microsatellite DNA markers: indigenous trout persist in introgressed populations. Bio Letters. 2009;5:656–659. doi: 10.1098/rsbl.2009.0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz N, Araguas RM, Fernández R, Vera M, García-Marín JL. Efficiency of markers and methods for detecting hybrids and introgression in stocked populations. Conserv Genet. 2009;10:225–236. doi: 10.1007/s10592-008-9550-0. [DOI] [Google Scholar]
- Funk DJ, Omland KE. Species-Level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 2003;34:397–423. doi: 10.1146/annurev.ecolsys.34.011802.132421. [DOI] [Google Scholar]
- Good JM, Demboski JR, Nagorsen DW, Sullivan J. Phylogeography and introgressive hybridization: Chipmunks (Genus Tamias) in the Northern Rocky Mountains. Evolution. 2003;57:1900–1916. doi: 10.1111/j.0014-3820.2003.tb00597.x. [DOI] [PubMed] [Google Scholar]
- Sánchez-Guillén RA, Hansson B, Wellenreuther M, Svensson EI, Cordero-Rivera A. The influence of stochastic and selective forces in the population divergence of female colour polymorphism in damselflies of the genus Ischnura. Heredity. [DOI] [PMC free article] [PubMed]
- Rhymer JM, Simberloff D. Extinction by hybridization and introgression. Annu Rev Ecol Syst. 1996;27:83–109. doi: 10.1146/annurev.ecolsys.27.1.83. [DOI] [Google Scholar]
- Randler C. Avian hybridization, mixed pairing and female choice. Anim Behav. 2002;63:103–119. doi: 10.1006/anbe.2001.1884. [DOI] [Google Scholar]
- Seehausen O. Conservation: losing biodiversity by reverse speciation. Current Biology. 2006;16:R334–R337. doi: 10.1016/j.cub.2006.03.080. [DOI] [PubMed] [Google Scholar]
- Gill FB. Historical aspects of hybridization between bluewinged and golden-winged warblers. Auk. 1980;97:1–18. [Google Scholar]
- Bradley RD, Davis SK, Baker RJ. Genetic control of premating isolating behavior: Kaneshiro's hypothesis and asymmetrical sexual selection in pocket gophers. Heredity. 1991;82:192–196. [Google Scholar]
- Vallender R, Friesen VL, Lovette J. Complex hybridization dynamics between golden-winged and blue-winged warblers (Vermivora chrysoptera and Vermivora pinus) revealed by AFLP, microsatellite, intron and mtDNA markers. Mol Ecol. 2007;16:2017–2029. doi: 10.1111/j.1365-294X.2007.03282.x. [DOI] [PubMed] [Google Scholar]
- Stone G. Phylogeography, hybridisation and speciation. Trends Ecol Evol. 2000;15:354–355. doi: 10.1016/S0169-5347(00)01947-9. [DOI] [Google Scholar]
- Price T. Speciation in birds. Roberts & Co. Publishers, CO, USA; 2007. [Google Scholar]
- Torralba A Burrial, Ocharan FJ Larrondo. Presencia de Ischnura elegans (Vander Linden, 1829) (Odonata: Coenagrionidae) en 1980 en Louro (Galicia, noroeste de España) Boletin de la Sociedad Entomológica Aragonesa. 2010;46:466–468. [Google Scholar]
- Rieseberg LH, Gerber D. Hybridization in the Catalina Island Mountain Mahogany (Cercocarpus traskiae): RAPD Evidence. Conserv Biol. 1995;9:199–203. doi: 10.1046/j.1523-1739.1995.09010199.x. [DOI] [Google Scholar]
- Rieseberg L. Hybrid origins of plant species. Annu Rev Ecol Syst. 1997;28:359–389. doi: 10.1146/annurev.ecolsys.28.1.359. [DOI] [Google Scholar]
- Hammers M, Van Gossum H. Variation in female morph frequencies and mating frequencies: random, frequency-dependent harassment or male mimicry? Anim Behav. 2008;76:1403–1410. doi: 10.1016/j.anbehav.2008.06.021. [DOI] [Google Scholar]
- Roulin A. The evolution, maintenance and adaptive function of genetic colour polymorphism in birds. Biol Rev Camb Phil Soc. 2004;79:815–848. doi: 10.1017/S1464793104006487. [DOI] [PubMed] [Google Scholar]
- Sinervo B, Lively CM. The rock-paper-scissors game and the evolution of alternative male strategies. Nature. 1996;380:240–243. doi: 10.1038/380240a0. [DOI] [Google Scholar]
- Chamberlain NL, Hill RI, Kapan DD, Gilbert LE, Kronforst MR. Polymorphic Butterfly Reveals the Missing Link in Ecological Speciation. Science. 2009;326:847–850. doi: 10.1126/science.1179141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S. Novel colour polymorphisms in a hybrid zone of Mandarina (Gastropoda: Pulmonata) Bio J Linn Soc. 1997;61:369–384. [Google Scholar]
- Grant PR, Grant BR. Hybridization of bird species. Science. 1992;256:193–197. doi: 10.1126/science.256.5054.193. [DOI] [PubMed] [Google Scholar]
- Baixeras J, Michelena JM, González P, Ocharan FJ, Quirce C, Marcos MA, Soler E, Domingo J, Montagud G, Guitérrez A, Arles M. Les Libèl. Lules De La Comunitat Valenciana. Generalitat Valenciana, conselleria de Territori i habitatge, Generalitat Valenciana. 2006.
- Jödicke R. In: Studies on Iberian Dragonflies. Jödicke R, editor. Adv. Odonat. Ursus Scientific Publishers, Bilthoven, Netherlands; 1996. [Google Scholar]
- Ocharan F. Composición de la Odonatofauna Ibérica. Rev Biol Univ Oviedo. 1988;6:83–93. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York; 1989. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Resources. 2004;4:535–538. [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices, version 2.9.3. 2001. http://www.unil.ch/popgen/softwares/fstat.htm
- Goudet J. PCAGEN. http://www2.unil.ch/popgen/softwares/pcagen.htm
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol Notes. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen EEG, Bach LA, Kotlicki P. Hybridlab (version 1.0): a program for generating simulated hybrids from population samples. Mol Ecol Notes. 2006;6:971–973. doi: 10.1111/j.1471-8286.2006.01433.x. [DOI] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak FDR, Jeanmougin FO, Higgins DG. The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press, New York; 1987. [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian assignment probabilities. The average Bayesian assignment probabilities in each of the two genetic clusters (Q1 for I. graellsii (grey), and Q2 for I. elegans (grey dark)), for the nine Spanish populations of I. elegans.