Abstract
Background
Vinyl chloride is a widespread groundwater pollutant and Group 1 carcinogen. A previous comparative genomic analysis revealed that the vinyl chloride reductase operon, vcrABC, of Dehalococcoides sp. strain VS is embedded in a horizontally-acquired genomic island that integrated at the single-copy tmRNA gene, ssrA.
Results
We targeted conserved positions in available genomic islands to amplify and sequence four additional vcrABC -containing genomic islands from previously-unsequenced vinyl chloride respiring Dehalococcoides enrichments. We identified a total of 31 ssrA-specific genomic islands from Dehalococcoides genomic data, accounting for 47 reductive dehalogenase homologous genes and many other non-core genes. Sixteen of these genomic islands contain a syntenic module of integration-associated genes located adjacent to the predicted site of integration, and among these islands, eight contain vcrABC as genetic 'cargo'. These eight vcrABC -containing genomic islands are syntenic across their ~12 kbp length, but have two phylogenetically discordant segments that unambiguously differentiate the integration module from the vcrABC cargo. Using available Dehalococcoides phylogenomic data we estimate that these ssrA-specific genomic islands are at least as old as the Dehalococcoides group itself, which in turn is much older than human civilization.
Conclusions
The vcrABC -containing genomic islands are a recently-acquired subset of a diverse collection of ssrA-specific mobile elements that are a major contributor to strain-level diversity in Dehalococcoides, and may have been throughout its evolution. The high similarity between vcrABC sequences is quantitatively consistent with recent horizontal acquisition driven by ~100 years of industrial pollution with chlorinated ethenes.
Background
Chlorinated ethene congeners ("chloroethenes") are among the most frequently detected groundwater contaminants in the United States of America and other industrialized countries [1]. Chloroethenes are often incompletely dechlorinated by bacteria in these anoxic environments, leading to an accumulation of vinyl chloride, a Group 1 human carcinogen [2,3]. Growth-linked reductive dechlorination of vinyl chloride is critical to avoid its accumulation and achieve in situ remediation of chloroethenes [1], but vinyl chloride respiration has only been observed in certain strains of Dehalococcoides [4,5]. Dehalococcoides is a genus-level phylogenetic group within the Chlorofiexi phylum [6]. Dehalococcoides are strictly anaerobic bacteria that gain metabolic energy exclusively via the oxidation of H2 coupled to the reduction of organohalide compounds [7-9]. This catabolic reductive dehalogenation of organohalide compounds ("organohalide respiration") is catalyzed in Dehalococcoides by heterodimeric, membrane-bound enzymes called "reductive dehalogenases" [10]. Reductive dehalogenases typically contain corrinoid and iron-sulfur clusters as cofactors, and have varied substrate ranges that do not necessarily overlap [10,11].
The catalytic subunit of reductive dehalogenases is encoded in Dehalococcoides by reductive dehalogenase homologous genes (rdhA). Dehalococcoides possess as many as 36 rdhA per genome [9], but few of the encoded enzymes, RdhA, have been purified and characterized in vitro. Many rdhA are co-expressed [12-16], further confounding a determination of the RdhA responsible for catalysis of an observed reductive dehalogenation activity. The only reductive dehalogenase shown to catabolically reduce vinyl chloride, VcrA, was purified from a highly-enriched vinyl chloride respiring culture dominated by Dehalococcoides strain VS [5]. The operon encoding VcrA, vcrABC, was identified by reverse genetics, and highly-similar vcrA were detected in other vinyl chloride respiring Dehalococcoides cultures [5,17,18]. Primers targeting vcrA are now commonly used as an indicator of attenuation potential at vinyl chloride contaminated sites ([5], U.S. Patent Application 20090176210). A putative VC reductase operon, bvcAB, shares only limited similarity with vcrAB and is present in a different VC respiring Dehalococcoides strain, BAV1, which does not contain vcrABC [19].
Although Dehalococcoides are the only known microorganisms capable of vinyl chloride respiration, both vcrA and bvcA appear to be horizontally acquired [9]. Both vcrA and bvcA have a highly unusual, low %(G+C) codon bias that appears maladapted to Dehalococcoides genomes [20], and both are found within a low %(G+C) "genomic island" (GI) [21] that interrupts local gene synteny relative to other Dehalococcoides strains. In strain VS, this vcrABC -containing genomic island (vcr-GI) integrated at the ssrA locus, and as a result is flanked by ssrA and a 20 bp direct repeat of the ssrA 3' end [9]. ssrA is a single-copy gene essential in bacteria [22] encoding transfer messenger RNA (tmRNA), which plays a key role in maintaining the fidelity of protein synthesis [23]. Specific integration of genetic elements at ssrA is also common across many bacterial phyla, and often results in a direct repeat at the genomic island boundary opposite the site of integration [24]. In addition to the vcr-GI, over a dozen ssrA direct repeats were previously detected downstream of ssrA in Dehalococcoides, collocated with many strain-specific rdhA in a region of high genomic variability between Dehalococcoides strains [9]. To further understand the acquisition and dissemination of vcrABC, as well as the impact of ssrA-specific integration on Dehalococcoides genome dynamics, we determined the conserved features of Dehalococcoides ssrA-specific genomic islands (ssrA-GIs) from all publicly available genomes and metagenomes of Dehalococcoides cultures, including the recently-sequenced Dehalococcoides strain GT [17] and the metagenome sequences of the vinyl chloride respiring Dehalococcoides enrichment cultures KB-1 [25,26] and ANAS [27]. We also amplified and sequenced ssrA-GIs from the vinyl chloride respiring Dehalococcoides enrichment cultures Evanite (EV) [28], PM [28], WBC-2 [29], and WL [30] using primers designed to target either vcr-GIs specifically, or conserved features present in all available Dehalococcoides ssrA-GIs. Previous studies have implicated a subset of rdhAB with horizontal gene acquisition, but evidence for the method of integration, mobilization, replication, and transfer is limited [31,32]. We describe here a family of putative ssrA-specific integrative and mobilizable elements [33] that share a conserved 'integration module' while also encoding a broad variety of putative and unknown functions, including reductive dehalogenation. The key conserved integrase encoded on these elements is a homolog of the CcrB family of site-specific serine recombinases that specifically integrate/excise the methicillin-resistance element "SCCmec" in Staphylococcus aureus [34]. Using a robust whole-genome phylogeny and several estimates for mutation rate, we estimate the age of the most recent common ancestor of contemporary Dehalococcoides strains, as well as the age of divergence for Dehalococcoides ssrA-GI integration module components.
Results
ssrA Genomic Islands in Dehalococcoides
The region downstream of ssrA in available Dehalococcoides (meta)genome sequences contains multiple tandem genomic islands that are primarily distinguished by their boundaries - ssrA or its 20 bp direct repeat - as well as disruption to local gene synteny and in many cases the presence of a characteristic cluster of integration-associated genes adjacent to the left edge (Figure 1). All direct repeats are located within 100 kbp downstream of ssrA, with varying numbers per strain and no duplicate genomic islands within any strain. These findings are consistent with ssrA-specific integration described for other bacteria [21,24], as well as a class of integrating and mobilizing elements that encode their own specific integration but do not replicate independently from the chromosome nor encode for conjugation [33]. From available Dehalococcoides genomic data (including this study) we have detected a total of 31 ssrA-GIs containing 47 rdhA, 75 hypothetical protein encoding genes, 2 putative complete CRISPR modules and arrays [35], as well as other genes; most of which are not believed to encode a core function and are present in only a subset of Dehalococcoides strains.
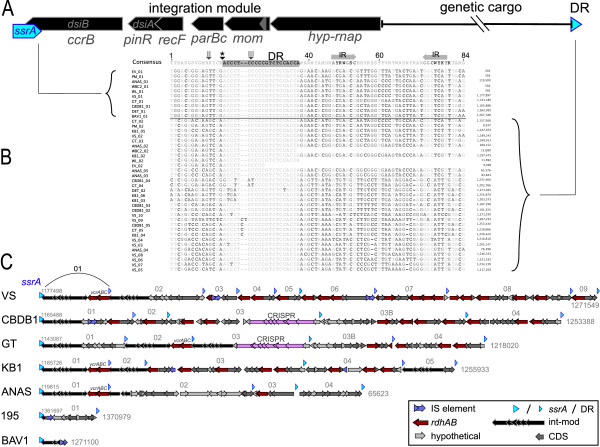
Figure 1.
General structure of ssrA genomic islands (ssrA-GIs). (A) Generalized structure of Dehalococcoides ssrA-GIs, oriented according to published Dehalococcoides complete genomes. Labels below genes in the integration module indicate the most informative homolog of the protein encoding gene. (B) Alignment of the 84 bp region surrounding the 3' end of ssrA or its direct repeat (DR) fragments (shaded black in the consensus) from 40 such positions in available Dehalococcoides genomes. Each sequence is labeled by its strain or enrichment name, underscore, and the order in which it occurs, beginning with the 3' end of ssrA. Positions in the alignment that disagree with the 75% consensus sequence are shaded in darker grey. The alignment is ordered such that sequences corresponding to Dehalococcoides ssrA ("_01") are the top 10 sequences, emphasizing a conserved position of disagreement between ssrA sequences and the direct repeat regions, position 15 in the alignment, 333 in ssrA. The two bases flanking the inferred integration site are marked with a hash. (C) To-scale genomic maps of region downstream of Dehalococcoides ssrA in (meta)genomic datasets. Orientation of genes is indicated with arrows. Key genes are shaded according to the provided legend.
Sixteen of the identified Dehalococcoides ssrA-GIs contain an integration module comprised of 6 syntenic protein encoding genes oriented on the reverse strand and located adjacent to attL (in this context, attL and attR are the ssrA direct repeat sequence at the left or right boundary, respectively; Figure 1A).
Beginning from attL, the integration module contains genes that appear to encode (1) a 540 residue serine recombinase family putative site-specific integrase we call Dehalococcoides ssrA-specific integrase, DsiB (Figure 2); (2) a smaller (200aa) PinR (COG1961) homolog that also contains a serine recombinase catalytic domain (cd00338), DsiA; (3) a small (150aa) RecF homolog likely involved in DNA recombination or repair [36], (4) a 210 residue protein with ParBc domain, possibly catalyzing single-stranded DNA cleavage, circular element nicking, element segregation ([37,38], PF02195); (5) a Mom [39] homolog (270 aa), predicted to play a role in restriction endonuclease resistance via methylation [40,41]; and (6) a large (700 aa) protein containing a DNA-directed RNA polymerase domain in the first 85 residues (GO:0003899). These integration modules also contain a 76 bp conserved tRNA-like locus embedded within the first 150 bp of the fifth protein encoding gene, approximately 4400 bp from attL (Figure 1A). It is usually labeled as 'pseudo-tRNA' by automated annotation pipelines, but alignment-based RNA folding analysis predicts a complete tRNA-Gly-like structure (Additional file 1 Figure S1). The elevated sequence conservation at its 3' end provides an effective target for primers, as do the regions surrounding ssrA direct repeats and a site of locally high nucleotide conservation within dsiB (Figures 1B, Additional file 1 Figure S2).
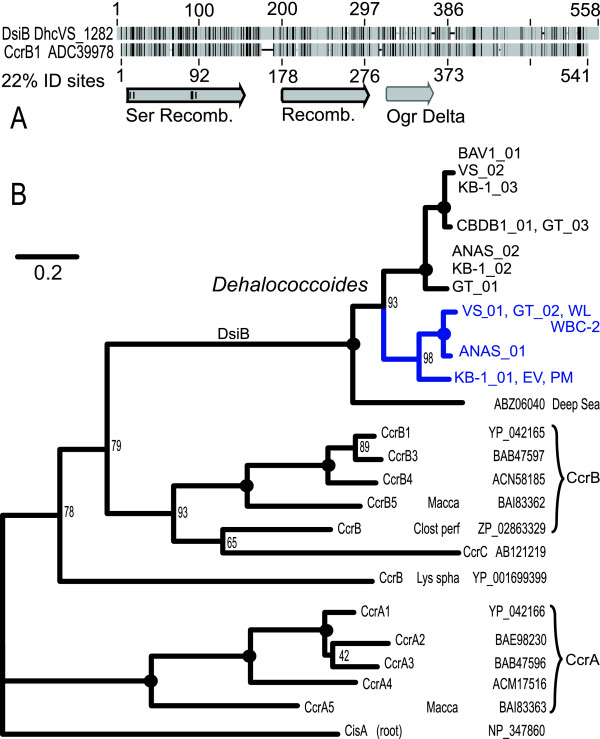
Figure 2.
Phylogeny of ssrA-GI integrase, DsiB. (A) Grey-scale similarity 'barcode' representation (black is identical sites) of pairwise global alignment (Needleman-Wunsch, free end-gaps) between a representative DsiB [DhcVS_1282, Genbank: ACZ62382], and CcrB1 of Staphylococcus aureus [Genbank: ADC39978]. Key domains of CcrB1 are annotated below the alignment, and traced in black if they are also detected in DhcVS_1282 by the conserved domain database search [84] incorporated in PSI-BLAST [85]. (B) Maximum Likelihood tree of the putative integrases encoded on Dehalococcoides ssrA-GIs, DsiB, as well as key integrases involved in mobility of SCCmec in Staphylococcus aureus [64] (unless otherwise noted). The clade of integrase sequences found on vcr-GIs are shaded in blue. Nodes with 100% bootstrap support are bolded with a filled circle. CisA of Clostridium acetobutylicum ATCC 824 is rooted as an outgroup, as in [86]. The following abbreviations are used to label CcrA, CcrB, or CcrC from bacteria other than S. aureus: 'Lys spha' - Lysinibacillus sphaericus C3-41; 'Clost perf' - Clostridium perfringens C str. JGS1495; 'Macca' - Macrococcus caseolyticus [86].
Alignment of the ~85 bp surrounding each of the 28 ssrA direct repeats reveals additional nucleotide conservation and allows identification of the site of insertion in ssrA (Figure 1B). A 15 bp motif (TTCAGRSMGMRKCCA) occurs adjacent upstream of the direct repeat and does not align well with the corresponding positions in Dehalococcoides ssrA (318-333), indicating that insertion likely occurs between 333 and 334 in ssrA. This location corresponds to the middle of the T-loop of the encoded tmRNA, between the canonical insertion positions called 'Sublocations II and III' [24] (Figure 1B).
Specific features of vcr-GIs
vcr-GIs are a distinct subset of Dehalococcoides ssrA-specific genomic islands, present in two Dehalococcoides genomes (VS [GenBank:CP001827], GT [GenBank:NC_013890]) and two metagenomes (KB-1 [JGI:4083612], ANAS [JGI:4085297]). Using primers that target conserved features of all ssrA-GIs or specific features of vcrABC, we amplified and sequenced 4 additional vcr-GIs from independently derived vinyl chloride respiring Dehalococcoides enrichment cultures (WBC-2, PM, EV, WL [GenBank:JN034252-JN034255] see Methods). In all instances the vcr-GI is located immediately adjacent to ssrA, except strain GT where it is the second genomic island downstream of ssrA. Because ssrA is an essential single-copy gene encoding a structural RNA [42], its sequence provides a coarse phylogenetic identity of the chromosome from which it was amplified [43]. This allowed confirmation that the vcr-GIs acquired via metagenomic and targeted sequencing are Dehalococcoides chromosomal segments, even though the source genomic DNA was from a mixed culture (Figure 3, Additional file 1 Figure S3).
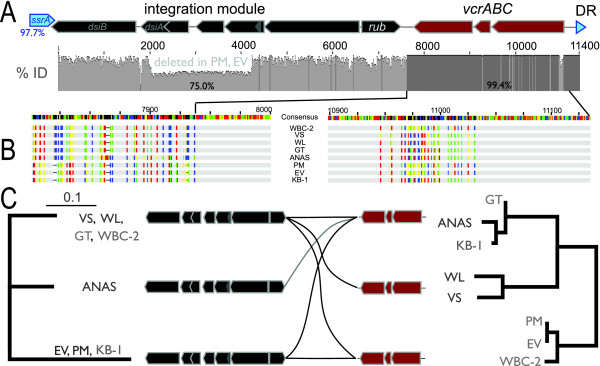
Figure 3.
Organization, alignment, and phylogenetic comparison of 8 vinyl chloride reductase genomic islands (vcr-GIs). (A) To scale summary plot (0 - 100% ID. 14 bp window) of a multiple alignment of all 8 vcr-GIs. Horizontal axis numbers indicate the distance downstream of ssrA, in nucleotides. Bar heights are shaded darker grey when their value is 100%. Position and orientation of genes are annotated above the plot, shaded according to Figure 1. Regions of categorically different similarity correspond to the integration and vcrABC cargo modules, with region-wide average % IDs of 75.0 and 99.4, respectively. (B) Enlarged view of the multiple alignment at key positions. (Left) The presumed boundary between integration and cargo modules. (Right) The region of atypically high substitutions occurring in the leader sequence of vcrA. Identical sequence is shaded light-grey, nucleotides that disagree with the consensus are indicated with tick marks shaded red, green, yellow or blue representing nucleotides A, T, G, C, respectively. (C) Phylogenetic discontinuity between integration modules (left) and their attached vcrABC cargo (right), represented by separately calculated Maximum Likelihood trees. Middle cartoon summarizes the major phylogenetic separations of the trees, with curves connecting modules if one of the 8 vcr-GIs contains the corresponding combination of module types. vcr-GIs sequences are from Dehalococcoides strain VS [GenBank:CP001827], strain GT [GenBank:NC_013890]) and two metagenomes (KB-1 [JGI:4083612], ANAS [JGI:4085297]), as well as targeted sequencing (this study) from the vinyl chloride-respiring Dehalococcoides enrichment cultures WBC-2, PM, EV, and WL [GenBank:JN034252-JN034255].
These vcr-GIs contain integration and cargo (vcrABC ) modules with discordant evolutionary histories. The boundary between integration module and vcrABC is delineated by an unambiguous difference in nucleotide identity, 75.0 and 99.4%, respectively (Figure 3). This boundary reveals that vcr-GI integration modules contain a seventh protein encoding gene oriented in the opposite (forward) direction encoding a rubredoxin domain protein, in addition to the 6 integration module genes described previously (Figure 3). These integration modules (9164 - 11361 bp) are related as three distinct branches with nearly-identical leaves (masking a ~2200 bp deletion in PM, EV), grouped in a topology that is discordant with the corresponding tree of the 3784 bp vcrABC cargo (Figure 3). Relatedness of vcrABC -cargo sequences was estimated based on the 44 variant positions in their alignment, appearing mostly (66%) in the form of substitutions in the leader sequence of vcrA [5] (Figure 3). Ka/Ks ratios for the vcrA leader sequence (first 129 bp) ranged between approximately 0.05 and 0.2 for different pairwise combinations [44] and phylogenetic nodes [45], suggesting this region is under purifying selection. By contrast, the Ka/Ks ratio for the remainder of vcrA was incalculable because all 15 variant positions (out of 1431 bp) were non-synonymous substitutions, suggesting recent positive selection on the mature VcrA enzyme. This latter conclusion must be tempered by the limited information available in just 15 variant sites, the complete lack of indels detected in any vcrA (suggesting some purifying selection in the leader sequence), as well as the clear influences of recent horizontal gene transfer and recombination on these vcr-GIs. For example, vcrC is identical across all strains, within a 1650 bp region of perfect identity.
Age of Dehalococcoides
A core-gene phylogenetic tree was constructed to support age estimates based on evolutionary models. The core-gene tree was built from 432 core orthologous protein encoding genes shared between available Dehalococcoides (meta)genomes and Dehalogenimonas lykanthroporepellens BL-DC-9, a Chlorofiexi strain that is a phylogenetic outgroup to Dehalococcoides and its closest completely-sequenced relative [46]. Age estimates depend heavily on the assumed rate of mutation. We iterated our calculations on multiple published mutation rates (see Methods), as well as an empirical observation for mutation rate derived from the known divergence time (16 years, S. Zinder, pers. comm.) between the isolation of Dehalococcoides ethenogenes strain 195 [6] and the generation of a metagenome of its parent culture, DONNA2 (R. E. Richardson, pers. comm.). The latter empirical rate is substantially faster than the published values of faster-growing microbes (Additional file 4 Table S1), possibly because it includes mutations that already existed between strain variants within the DONNA2 culture prior to isolation of strain 195. Although we expect a long-term average mutation rate in the natural environment to be slower, and hence ages based on this rate to be an underestimate, it remains useful as a conservative bound on the 'recentness' of the events in question. Similarly, we used a range of growth rates to estimate the age of Dehalococcoides. For a recent bound we used the fastest reported Dehalococcoides doubling time (0.8 days [6]), as well as a range of slower reported growth rates from anaerobic environmental systems for more realistic estimates (11-14 days [47-49]). The corresponding estimates and lower (recent) bounds are presented in Table 1.
Table 1.
Divergence Time Estimates Under Different Rates of Evolution.
| Divergence of interest | Tree Calculation Method |
Divergence time estimates from different proposed rates | |||||
|---|---|---|---|---|---|---|---|
|
Universal bacterial rate in nature |
Empirical E. coli rates in culture |
DONNA2/ strain 195 divergence |
16S clock | ||||
| Dehalogenimonas/Dehalococcoides MRCA | Splitstree | 5 | (1.9/28/33/34) | 0.9 | (0.4/5.3/6.2/6.3) | 0.5 | 200-600 |
| ML | 3 | (1.2/18/21/21) | 0.5 | (0.2/3.2/3.8/3.9) | 0.3 | ||
| Dehalococcoides MRCA | Splitstree | 0.3 | (0.14/2/2.4/2.4) | 0.06 | (0.03/0.37/0.44/0.44) | 0.04 | 30-60 |
| ML | 0.4 | (0.17/2.5/3/3) | 0.08 | (0.03/0.47/0.55/0.56) | 0.04 | ||
| ssrA-GI integration modules MRCA | ML | 3 | (1.1/16/19/19) | 0.5 | (0.2/3/3.5/3.6) | 0.2 | |
| integration modules MRCA, vcr-GIs only | ML | 1 | (0.4/5.5/6.5/6.6) | 0.2 | (0.07/1/1.2/1.2) | 0.08 | |
| vcrAB MRCA | ML | 0.05 | (0.02/0.27/0.31/0.32) | 0.008 | (0.03/0.47/0.55/0.56) | 0.004 | |
| vcrAB, leader masked | ML | 0.01 | (0.004/0.057/0.067/0.068) | 0.002 | (0.001/0.010/0.012/0.013) | 0.0009 | |
Reported age estimates (left, no parenthesis) are based on the rate of evolution listed in the column header as well as the average Dehalococcoides doubling time from published values (2 days). All estimates are reported in units of 1 million years. Age estimates of the Dehalococcoides clade from the Splitstree consensus network are based on branch lengths of strains to the common network node. '16S clock' is based on 1-2% 16S rRNA gene divergence per 50 million years [83]. Values in parenthesis are results from different doubling time estimates: 0.8 days - the fastest reported doubling time for Dehalococcoides [6]; 11.71 days - anaerobic benthic sediment community doubling time [49]; 13.76 days - bacterial doubling time in anaerobic seawater [48]; 14 days - approximate doubling time of the strictly anaerobic annamox bacteria [47]. For the secondary calculations of age estimates, an extra significant digit has been reported to allow distinguishing of estimates.
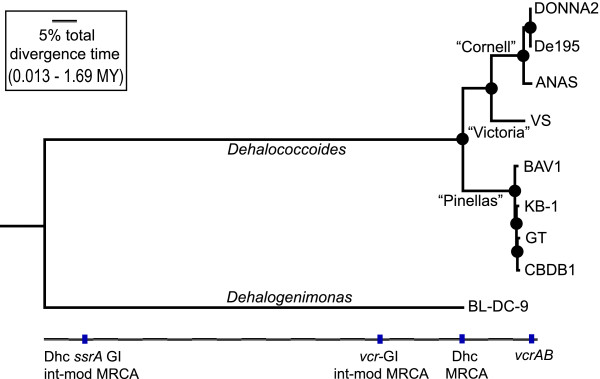
In relative terms, the divergence of Dehalococcoides and Dehalogenimonas are comparable to the predicted most recent common ancestor (MRCA) of available integration modules, approximately an order of magnitude earlier than the MRCA of Dehalococcoides strains. The MRCA of vcr-GI integration modules also significantly precedes the divergence of contemporary Dehalococcoides strains. In contrast, the high similarity among vcrABC sequences results in an estimated age that is at least an order of magnitude younger than Dehalococcoides speciation (Figure 4, Table 1).
Figure 4.
Date Estimates of Key Events in Dehalococcoides Evolution. Maximum likelihood phylogeny of 432 'core' orthologs. Timing of key evolutionary events are mapped onto the tree. The horizontal line below the tree represents the divergence time to the Dehalococcoides (Dhc) and Dehalogenimonas (Dehly) MRCA, while vertical hash marks indicate the relative divergence times of Dehalococcoides ssrA genomic island (GI) components. These include Dehalococcoides ssrA integration modules, vcrABC-attached integration modules, and vcrAB. Relative divergence times are based on the estimated age of the MRCA of Dehalococcoides and Dehalogenimonas (set to 1). The scale bar represents 5% of the total divergence time. Absolute time scales are from published mutation rate estimates and rate estimates based on the Dehalococcoides strains DONNA2/strain 195 divergence. Black points on the tree are nodes with 100% bootstrap support.
Discussion
Age and specific features of vcrABC acquisition
Nucleotide similarity is significantly higher between vcrABC cargo modules than can be expected if it was an orthologous locus present in the Dehalococcoides common ancestor (Figures 3, 4). In all cases vcrABC is located within a syntenic putatively-mobile element, vcr-GI, that is part of a broader class of ssrA-specific mobile elements that appear to be common among Dehalococcoides. In all vcrABC -containing strains except GT, the vcr-GI is located adjacent to the primary site of integration, ssrA, structural evidence that vcr-GIs are among the most recently integrated of the available Dehalococcoides ssrA-GIs. Within phylogenetic branches, integration modules are perfectly identical, except for a large identical deletion in the EV and PM vcr-GIs. The significantly unusual nucleotide signature of vcr-GIs [9,20], as well as the discordance between the vcrA tree and the corresponding Dehalococcoides strain phylogeny, indicate that vcrABC has not been stably maintained in Dehalococcoides genomes since their divergence. Taken together, these observations suggest recent horizontal acquisition and dissemination of vcrABC across all Dehalococcoides ecotypes by way of a ssrA-specific mobile element with conserved attachment site and integration module.
Because anthropogenic release of chloroethenes into the environment is a relatively recent phenomenon (~100 years [3]), we are particularly interested in the recent bounds for estimates of the age of the MRCA of these vcrABC sequences as a proxy for their horizontal acquisition by Dehalococcoides. Using our highest estimated rates of mutation and chromosomal replication, the divergence of these vcrABC sequences appears to have occurred 4000 years ago. This value is in flated by the inexplicably high variation within the leader sequence of vcrA. If we remove the vcrA leader sequence from the calculation, the age of divergence decreases to 900 years. However, there is clear signal for positive selection in the remaining vcrAB sequence alignment: all 16 variant positions (15 in vcrA and 1 in vcrB) are predicted to result in amino acid substitutions. If positively selected, these mutations may have accumulated faster than the background rates assumed in our molecular dating calculations. Because the relative increase in substitution rate is unclear and the total information represented by just 16 variant positions is low, we cannot confidently distinguish the divergence of these vcrABC from the first industrial production of chloroethenes. By contrast, our most conservative estimate for the MRCA of contemporary Dehalococcoides strains is 40,000 years ago (ranging as high as 3 Mya, Table 1), long before industrial civilization had a chance to influence the evolution of Dehalococcoides and their streamlined genomes specialized for organohalide respiration.
It is important to note that these molecular dating estimates use the available vcrABC sequences to predict the first horizontal acquisition of vcrABC by Dehalococcoides. This analysis is not meant to predict the age of genesis of the first vinyl chloride reductase. We did not detect partial homology with other rdhA that would suggest vcrA is a chimera resulting from a recent homologous recombination event. Moreover, the existence of an alternate vinyl chloride reductase from strain BAV1, BvcA [19], that shares deeply branching ancestry with VcrA on a tree of available RdhA [9], suggests that vinyl chloride reductases have existed for a considerable period of time, just not within strains of Dehalococcoides for which sequence data is currently available. In fact, naturally occurring vinyl chloride has been detected in soils [50], providing a plausible source of selective pressure to explain the existence of vinyl chloride reductases in nature prior to human pollution. However, we have not identified any candidate lineages as the possible progenitor of vinyl chloride reductases, and we have no way of knowing whether the primary substrate for the ancestral VcrA or BvcA was consistently vinyl chloride, leaving their ancestral history unclear.
The phylogenetic discord between integration modules and their attached vcrABC indicates that homologous recombination - or perhaps a more directed form of 'module swapping' - has recently occurred between vcr-GIs (Figure 3). This additional inter-element recombination may be independent of ssrA-specific integration, but it would still require horizontal transfer so that 2 or more vcr-GIs are collocated within the same cell. Multiple vcr-GI variants have not been detected in the same complete genome. However, we did detect a low-coverage variant in the KB-1 metagenome assembly with 3 corroborating reads that perfectly match a different vcr-GI integration module found in VS, WL, GT, and WBC-2 cultures, providing preliminary evidence of the physical collocation of two vcr-GIs within the KB-1 culture (Additional file 5 Figure S4).
ssrA-GIs appear to be integrative and mobilizable elements
A subset of Dehalococcoides rdhAB were previously implicated in horizontal transfer [31,32], including the trichloroethene reductase gene, tceAB [32]. Although the selective conditions in chloroethene-contaminated environments favors maintenance of tceAB and vcrABC, the genes implicated in tceAB transfer [32] share no detectable homology with the ssrA-specific system described in detail here. We hypothesize that these Dehalococcoides ssrA-GIs behave as integrative and mobilizable elements ("IMEs") because they do not appear to encode conjugation, although they share many other features of the broadly defined class of integrative and conjugative elements ("ICEs") [33]. It may be possible that conjugation is encoded by a surprisingly minimal gene set within the integration modules [33], similar to the small (10.9 kbp) integrating and conjugating element 'pSAM2' of Streptomyces ambofaciens, which requires only a single gene, traSA, for inter-mycelial (conjugal) transfer [51]. Dehalococcoides core genes do include putative pil genes, the functions of which are unclear but may play a role in conjugation. Some strains of Dehalococcoides contain unambiguous prophages, providing an alternative hypothesis for the mechanism of ssrA-GI transfer, via illegitimate packaging of the excised ssrA-GI into a phage capsule. The length of Dehalococcoides ssrA-GIs is within the range of typical phage genomes. However, evidence for a complete prophage is not as ubiquitous among Dehalococcoides as the presence of ssrA-GIs, and there have been no descriptions to date of Dehalococcoides phage that also encode an rdhA, leaving the influence of phage on rdhA evolution unclear. Based on currently available evidence, we hypothesize that Dehalococcoides ssrA-GIs are mobilizable but not conjugating elements that sometimes mobilize adjacent tandem islands but in all cases rely on a host- or phage-encoded system for cell-cell transfer of a transient, presumably circular, intermediate.
Dehalococcoides also contains comEA, and it is unknown if Dehalococcoides is transiently competent for uptake of exogenous DNA. However, transfer via stochastic competence is an unsatisfying explanation, mainly because Dehalococcoides ssrA-GIs appear to lack genes encoding independent replication, and stable non-phage extrachromosomal elements have not been observed in Dehalococcoides [7-9].
Occasionally integrating and conjugating elements do have replicative forms [33], as in the case of rolling circle replication of pSAM2 in the donor cell [52]. Maphosa et al. recently described a field site in which there were 1 to 2 orders of magnitude more vcrA copies detected than copies of tceA, bvcA, or Dehalococcoides 16S rRNA genes [53]. vcrA was also more abundant than Dehalococcoides 16S rRNA genes in a dechlorinating bioreactor inoculated from the site [53], suggesting either (1) there exists a vcr-IME that can replicate independently or has integrated within an element that can replicate independently, or (2) they detected a non-Dehalococcoides population that also possesses vcrA, coexisting with a Dehalococcoides population.
It is important to note that, while a conspicuous and common feature, not all Dehalococcoides ssrA-GIs contain an integration module. We identified 15 ssrA-GIs without integration modules, containing a total of 38 rdhA as well as other genes. These might be 'cis-mobilizable elements' that encode neither integration nor transfer, but retain functional attL/attR sites [33] and are occasionally or constitutively mobilized with adjacent genomic islands through a process known as accretion [54]. In some cases these tandem ssrA-GIs may have been previously mobile but are now fixed in the chromosome. For example, there is a region immediately downstream of the direct repeats furthest from ssrA that is similarly dense in rdhA while also syntenic across Dehalococcoides strains, phylogenetically coherent with whole genome estimates, and devoid of ssrA-GI signatures (Additional file 6 Figure S5); suggesting this region was present in the MRCA of available Dehalococcoides [9]. Some or all of this region may have been acquired originally as an ssrA-GI, but deletion and amelioration has erased evidence of horizontal gene transfer.
Likely Roles within ssrA-GI Integration Modules
The first identified Dehalococcoides ssrA-specific integrase gene (dsiB) (DhcVS_1292) was sequenced following the original identification and characterization of VcrA, and noted for its proximity to vcrA on the chromosome [5]. It is now clear that DhcVS_1292 is part of an integration module in an adjacent downstream ssrA-GI (GI 02 in VS, Figure 1), one of 16 dsiB homologs detected in Dehalococcoides genome sequences. The closest relative to dsiB in the public database is present on a fully-sequenced metagenomic fosmid from a deep (4000 m) ocean subsurface sample (EU016565, Figure 2), within an apparent integration module that also includes homologs to dsiA, parB, mom, and a putative tRNA embedded in mom, as well as an unambiguous ssrA-direct repeat at the homologous attL position embedded in dsiB (Additional file 7 Figure S6). This is especially intriguing in light of the recent sequencing of 32 novel rdhA amplified from various marine subsurface sediments [55], many of which appear phylogenetically within a major rdhA branch (Cluster I [9]) that is otherwise populated only by rdhA from Dehalococcoides or Dehalogenimonas. Given this indirect evidence and the large diversity of organohalogens detected in marine systems [56], it is tempting to speculate that Dehalococcoides plays a role in these settings. However, in the absence of direct observation of Dehalococcoides-like microorganisms in marine (subsurface) settings, this role remains unclear.
A more sensitive database search indicated that DsiB is a structurally similar homolog of CcrB, containing the serine-recombinase-catalytic domain at the N terminus, as well as similar motifs along its ~500 residue length (mean 22% ID, Figure 2A). CcrB specifically integrates/excises the so-called 'Staphylococcus Cassette Chromosome' (SCC [57]) family of mobile elements that are a vector of antimicrobial resistance (among other phenotypes [58,59]), with major consequences for hospitals and the greater community [60-63]. CcrB was shown to have DNA-binding and recombination activity for attS of SCC [64], but SCC integration [57] and attB-specific excision both required the product of a smaller, co-transcribed serine recombinase gene, ccrA, that does not encode a DNA-binding domain [64]. Similarly, Dehalococcoides integration modules encode on a putative operon a second, smaller serine recombinase, DsiA, that also lacks a detectable DNA-binding motif. Dehalococcoides ssrA-GIs and SCC also share overlapping size ranges and specifically integrate at a non-tRNA, single-copy essential gene. We hypothesize that integration/excision of Dehalococcoides ssrA-GIs occurs in a homologous mechanism to SCC, via DsiB in concert with DsiA, with other integration module elements likely playing a role in regulation of integrase/excisionase activity or modification of the excised element to facilitate transfer or maintenance. Unfortunately, the mode of SCC transfer among Staphylococcus is unclear [65], and so does not provide additional clues regarding a likely transfer mechanism.
Interestingly, dsiB is always found overlapping attL at its 3' end. A stop codon occurs only upstream of the genomic island, even if that means overlapping substantially with an adjacent genomic island or ssrA itself. Complimentary overlap of ssrA with small open reading frames has been detected in some bacteria with ambiguous implications [66]. It seems unlikely in this instance that the 3' terminal ~70 bp of ssrA also encode a functional region of dsiB on its complementary strand. Accordingly, alignments of DsiB are divergent at this portion of their sequence, both in length and amino acid similarity. The majority of dsiB is upstream of ssrA or its direct repeat, and already comprises the expected length for homologs of ccrB (1600 bp). In addition to a trivial explanation in which dsiB undergoes low-efficiency translation that is variable at the C-terminus, it may be that dsiB is only fully functional when encoded on the circularized element, or alternatively when encoded on the chromosome downstream of an adjacent genomic island containing the requisite 3' gene fragment. In any case, the overlap of dsiB with attP/attL leaves the stop codon of dsiB unclear, and may have functional relevance or affect regulation of dsiB.
Conclusions
Structural comparison of new (meta)genomic data, as well as targeted sequencing from unsequenced vinyl chloride respiring enrichment cultures, resulted in identification of 8 homologous mobile elements containing the vinyl chloride reductase operon, vcrABC. These vcr-GIs are a subset of mobile genetic elements in Dehalococcoides that specifically integrate at the single-copy gene, ssrA. A detailed comparison of these ssrA-GIs allowed identification of the precise position of insertion, the direct repeat created by the insertion event, as well as a conserved module of syntenic integration-associated genes that includes the likely ssrA-specific integrase, which we named dsiB. ssrA-GIs are most likely 'integrating and mobilizable elements' (IMEs) that do not encode their own mechanism of cellular transfer. Core (meta)genome phylogenetic analysis allowed an estimation of timing of divergence of Dehalococcoides strains, between 40,000 and 400,000 years ago, suggesting that the specialization by Dehalococcoides for respiration of organohalide compounds far precedes industrial synthesis by humans. By contrast, time estimates for the first horizontal acquisition of vcrABC sequences by Dehalococcoides are not confidently distinguishable from the first industrial synthesis of chloroethenes ~100 years ago. Vinyl chloride reductases may be ancient, nevertheless, and the donor(s) of recent genetic diversity to Dehalococcoides remain undetermined.
Methods
Identification of Dehalococcoides sequences in metagenome data
For ANAS and KB-1 enrichment cultures, complete genomes have not been assembled. However, identification of Dehalococcoides contigs was performed by comparison with closely-related complete genomes of strains 195 and CBDB1, respectively. For KB-1, gap closure was performed to create a closed draft genome of the dominant Dehalococcoides strain in the metagenome, including primer-walking of gap-spanning fosmid inserts, as well as gap-spanning PCR amplification from an aliquot of the originally-submitted KB-1 genomic DNA. For ANAS, structural genomic information comes from a single contig (contig 2014738012; 119815 bp) that contains a Dehalococcoides ssrA on one end and a series of tandem ssrA-GIs downstream (Figure 1). A second contig containing a Dehalococcoides ssrA was also detected (2014739670), but it contained no detectable ssrA-GIs. Dehalococcoides orthologs present in the ANAS dataset were detected using reciprocal best-hit BLASTp criteria described previously [8,9], treating the collection of all Dehalococcoides protein encoding genes in ANAS as if it were one genome. Although ANAS contains more than one Dehalococcoides strain, this approach had little effect on the number of orthologous groups considered 'core' between all Dehalococcoides, mainly because sequencing was sufficiently deep and because the Dehalococcoides strains in ANAS are closely-related to the Cornell lineage from which ethenogenes 195 is the only completely-sequenced representative [67].
DNA Source, Primer Design, Amplification Optimization
Enrichment cultures were derived from samples from geographically distinct contaminated sites (Additional file 8 Figure S7): WL from Western Louisiana [30]; EV from the Evanite site in Corvallis, Oregon [28]; PM from the Point Mugu Naval Weapon Facility, California [28]; and WBC-2 from the West Branch Canal Creek, Aberdeen Proving Ground, Maryland [29]. Culture, culture pellets, or genomic DNA were provided by L. Semprini (EV, PM) or E. Edwards (WL, WBC-2).
Nucleotide positions strongly conserved at ssrA, its direct repeat, and a few locations within integration modules were used for primer design (Additional file 2 Figure S2). Amplification was successful with a variety of combinations of predicted melting temperature and degeneracy. We described only the best-performing primer pairs, especially those that contributed to vcr-GI amplification and sequencing. All PCR amplifications from mixed cultures were performed using Phusion polymerase under standard conditions using 'HF' buffer. Primer pairs were optimized toward amplification of regions of ssrA-GIs in mixed genomic DNA template by DMSO and annealing temperature gradients. For most target amplicons the optimal DMSO concentration was at or near 4%, with optimal annealing temperature depending on the primer, and summarized in Additional file 2 Figure S2. In particular, a 20 bp forward primer beginning at Dehalococcoides ssrA position 8 (CGTGGTTTCGACAGGGAAGG - 'ssrA_03F'), successfully amplified ~90% of all 4 novel vcr-GIs when paired with a reverse primer upstream of vcrA (GTTCCTGACCATGCCGTACC - 'vcrA_05R'). The resulting (8.3 - 10.5 kbp) amplicons were purified in agarose gel electrophoresis and sequenced directly by the Sanger method (MCLAB, ELIM) and primer walking. No single primer-pair was determined that could amplify a complete ssrA-GI in one reaction from attL to attR, as these would be reverse complements of one another and produce primer dimers. Instead, combinations of PCR reactions were amplified and sequenced separately, and their resulting sequence data was assembled in silico and verified manually. For example, forward primers targeting a conserved position in the integration module (TGGAGCGCCGCCGTNGG - 'REC_003eF') amplify a portion of the integration module and all of the genetic cargo (~7 kbp) when coupled with a reverse primer that targets the ssrA-direct repeat (TGGTGGAGACGGGGGAGGG - 'REC_001eR'). Two-fold or greater coverage and perfect agreement between different amplicons from the same sample was required in assembly. In some instances ssrA-GI-derived amplicons were cloned in Escherichia coli following agarose gel purification. Efficient ligation to a vector was achieved with Enzymatic Assembly [68] and pSMART-LC-Kan (AF532106; Lucigen Corp.) or by blunt ligation into the pJAZZ-OK linear vector (FJ160465; Lucigen). Transformation was achieved chemically in E. coli DH5α or electrically in E. coli BigEasy-TSA (Lucigen) cells, respectively.
Core Genome and Genomic Island Phylogenies
The reciprocal BLASTp procedure for identifying orthologous groups among Dehalococcoides was also applied to Dehalogenimonas lykanthroporepellens BL-DC-9 in comparison to Dehalococcoides ANAS, KB-1, 195, DONNA2, CBDB1, BAV1, GT, and VS; resulting in 432 core orthologous groups that were also free of paralogs. Global alignments of each orthologous group were performed by Muscle (version 3.8.31) [69]. Single gene trees were generated using RAxMLHPC (version 7.0.3) [70] under the GTR + γ model [71] with Dehalogenimonas lykanthroporepellens BL-DC-9 constrained as the outgroup to prevent long-branch artifacts. The resulting trees were entered into Splitstree4 [72] and a consensus network was generated. The single gene global alignments were concatenated to generate a single large alignment for the 9 organisms. A core-genome phylogeny was generated using RAxMLHPC as described above, with 10 initial random starting tree iterations and 100 bootstrap replications. The tree with the highest likelihood is presented in Figure 4 and used for evolutionary analysis. Alignments for components of genomic islands were generated using Muscle and refined with hmmer (version 2.3.2) [73], then masked manually. Phylogenies were generated in RAxMLHPC under the GTR + γ model with 10 random starting trees and 100 bootstrap replications. In each case, the appropriate sequence was constrained as an outgroup. The trees with the best likelihood were identified and used for further age estimate calculations.
Date Estimations
Estimates of the age of the Dehalococcoides/Dehalogenimonas divergence, the Dehalococcoides clade, as well as the various components of the genomic islands were determined under three different estimates for the rate of Dehalococcoides evolution. Two mutation rates from published values were used: one from a universal estimate of bacterial mutation rates in natural environments [74], and one from an empirical analysis of E. coli in lab cultures [75] (Additional file 3 Table S1). A third rate was based on a known divergence time of approximately 16 years between the separation of Dehalococcoides ethenogenes strain 195 ("strain 195") [6] from its mother culture - the "TCE/MeOH" culture (Prof. S. Zinder, pers. comm.) - in 1992, and the 2008 metagenome sequencing of the "DONNA2" enrichment culture. DONNA2 was also derived from the TCE/MeOH culture and maintained in parallel from strain 195 until its subsequent metagenome sequencing (R. E. Richardson, pers. comm., see DONNA2 Mutation Detection, below). Branch lengths between strain 195/DONNA2 were calculated from single-gene trees of the 387 core protein encoding genes, after excluding 45 trees that did not have strain 195 and DONNA2 as a monophyletic group, most likely resulting from frame-shift mutations. The mean branch length of the 387 protein encoding gene trees, the core-gene concatenated ML tree, as well as the splitstree [72] network average branch length were all approximately 3(10) -5. For a minimum separation of 16 years, this corresponds to 2(10) -6 branch length per year of Dehalococcoides divergence. It is important to note that some unknown fraction of the observed mutations could have already existed within the mother culture prior to isolation of strain 195 if parents of the two contemporary strain variants coexisted at that time. Combined with the imposed pressures for rapid growth inherent to a laboratory culture, we expect that the Dehalococcoides mutation rates observed by this approach represent an unrealistic upper bound to what is likely to occur in Dehalococcoides in nature. This value is still useful, however, for creating lower bounds in molecular dating estimates that are compared with relatively recent events (e.g. human civilization, anthropogenic chloroethene pollution, etc.).
DONNA2 Dehalococcoides Mutation Detection
Because the dominant bacterium in the DONNA2 culture was our target variant of strain 195, the DONNA2 metagenome data included a high coverage of this variant. A comparative assembly of the DONNA2 shotgun reads on the strain 195 genome allowed identification of reliable mutations between these two strains, using the Variant Ascertainment Algorithm (VAAL) under default settings [76]). The DONNA2 metagenome project has gone through successive rounds of sequencing, and the mutation analysis described here is based on the raw 454 GS FLX Titanium shotgun reads available on 06 November 2009, which were subsequently filtered by alignment to the genome sequence of Dehalococcoides strain 195. The resulting 455,062 Dehalococcoides-derived reads had a mean length of 365 ± 142 nucleotides, and %(G+C) of 48.8. Our version of VAAL did not produce assembly statistics, but a separate comparative assembly using Geneious Pro v5.4 (medium-sensitivity default parameters) successfully aligned 454,342 reads to the strain 195 genome, for a coverage of 115.2 ± 41.2. The consensus sequence of the comparative assembly produced by VAAL formed the basis for the subsequent strain-level mutation analysis. Gene annotations from strain 195 were mapped onto the DONNA2-variant genome sequence and the protein-encoding genes among these were extracted and included as a separate whole-genome collection in the genome-wide core gene phylogenetic analysis (above). The cumulative length of the protein encoding genes shared between 195 and DONNA2 was 1,301,665 bp; and among these genes we detected a total of 192 mutations, with adjacent SNPs considered part of a single mutation. Of these 192 mutations, 39% were deletions, 28% were insertions, 28% were transitions, and 4% were transversions. With respect to the predicted effects relative to the encoded protein in strain 195, 40% were frame-shift mutations, 40% were synonymous (probably no change), 19% were non-synonymous substitutions, and 1% were predicted to cause a truncation due to an early stop codon. It should be noted that frame-shift and truncation mutations would probably not directly affect our subsequent tree calculations because those genes would likely fail our orthology criteria (above) and thus would not be included in the set of "core" genes.
Ka/Ks ratios
Ka/Ks ratios are an intrinsically pairwise calculation that was performed on a subset of the most different pairs of vcrA (full-length, and leader sequence only) using the 'kaks' function in the SeqinR package [44] of R [77]. Ka/Ks ratios were also calculated for all adjacent branches in a phylogenetic tree of the 8 sequences, using the Ka/Ks Calculation tool [45].
Integration Module tRNA Secondary Structure
The putative tRNA sequence was originally detected by ARAGORN [78] and annotated previously in publicly available annotations of Dehalococcoides strains CBDB1 and VS. Secondary structure was predicted from the alignment of all 16 detected tRNAs in available ssrA-GI integration modules, submitted to the RNAalifold [79,80], Pfold [81], and PETfold [82] web servers for independent calculations. The resulting structures were compared manually, including a comparison to classical tRNA secondary structure for identification of the conserved "DCC" anti-codon within a 5 nt anti-codon loop (Additional file 1 Figure S1).
Abbreviations
vcrABC: the vinyl chloride reductase operon; ssrA: the tmRNA encoding gene; ssrA-GI: ssrA-specific genomic island; vcr-GI: a vcrABC -containing ssrA-specific genomic island, a subclass of ssrA-GI; DsiB: predicted Dehalococcoides ssrA-specific integrase; dsiB: gene encoding DsiB; MRCA: most recent common ancestor; ML: Maximum Likelihood; DMSO: dimethyl sulfoxide; CRISPR: clustered regularly interspaced short palindromic repeats; SCC: Staphylococcus Cassette Chromosome; attL/attR: the extreme left or right edge, respectively, of the genomic island that likely participates in site-specific recombination; attB/attP: the DNA motif on the bacterial chromosome or mobile element, respectively, that likely participates in site-specific recombination; IME: integrative and mobilizable elements; ICE: integrative and conjugative elements; pSAM2: an integrative and conjugative plasmid found in many Streptomyces; EV, PM, WBC2, WL, ANAS, KB1, DONNA2: Dehalococcoides enrichment culture names; CBDB1, GT, VS, BAV1: Dehalococcoides strain names.
Authors' contributions
PJM conceived of the experiments, carried out the molecular experiments, performed the comparative analyses, and drafted the manuscript. LAH maintained the WBC-2 and WL cultures, assisted with the molecular experiments, performed the molecular dating analyses and helped to draft the manuscript. EAE participated in the design of the study and helped to draft the manuscript. SH consulted on the molecular dating analyses and helped to draft the manuscript. AMS participated in the design of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Description of additional data files
Additional data file 1 is a PDF format file containing the supplemental figures and associated legends. Additional data file 2 is a Microsoft excel (.xls) file containing tables of growth rates and rates of evolution, as well as other parameters and example calculations used in the molecular dating analyses.
Supplementary Material
Figure S1: Alignment and Predicted Secondary Structure of Putative tRNA-gly. These tRNA-gly are strongly conserved in 16 Dehalococcoides ssrA-GI integration modules. Bases are shaded according to the Vienna RNA conservation coloring schema in both the alignment (A) and secondary structure cartoon indicating the majority consensus with degeneracy (B). Secondary structure prediction was unanimous from three independent secondary structure prediction servers [80-82]. Free energy of the thermodynamic ensemble is -54.26 kcal/mol [80]. Substructure labels correspond to classical tRNA, including the apparent anti-codon 'DCC'.
Figure S2: Primers Mapped onto an Alignment of 16 ssrA Integration Modules. (A) Annotated alignment of the 16 integration modules discussed in this study. Individual sequences are shown as a thick black line, with gaps indicated by a thin horizontal line. Plot of average nucleotide identity (14 bp window) for all 16 sequences is shown along the top of the alignment. Three main target locations for primer design are indicated with downward-pointing black triangles, numbered beginning at ssrA (left). (B) Zoomed-in view of the alignment at the three target locations for primer binding. The 75% Consensus sequence is depressed slightly at the region targeted by primers, which are annotated along the top. Exact position of putative tRNA-gly is also shown.
Figure S3: Phylogenetic Tree of ssrA Versus 16S rRNA gene. The most likely of 100 bootstrap Maximum Likelihood trees with bootstrap support shown at nodes. Support not shown at nodes with poor or ambiguous support. (A) Phylogenetic tree of ssrA, the ~350 bp gene encoding tmRNA. (B) Similarly calculated tree based on the 16S rRNA gene (~1500 bp), reflected relative to typical tree orientation to emphasize topological similarity with (A). Other Chlorofiexi are included, with Staphylococcus aureus as an outgroup. Full name and accession number correspond to the following abbreviations: Dehalococcoides - Dhc; CBDB1 - Dhc CBDB1 NC_007356; GT - Dhc GT NC_013890; BAV1 - Dhc BAV1 NC_009455; 195 - Dhc ethenogenes 195 NC_002936; VS - Dhc VS NC_013552; Deha lyk - Dehalogenimonas lykanthroporepellens BL-DC-9 NC_014314; Staph aur - Staphylococcus aureus NC_002952; Rose cast - Ro-seiflexus castenholzii DSM 13941 NC_009767; Rose RS-1 - Roseiflexus sp. RS-1 NC_009523; Chlo aur - Chloroflexus aurantiacus J-10-fl NC_010175; Chlo agg - Chloroflexus aggregans DSM 9485 NC_011831.
Table S1: Parameters and example calculations utilized in divergence age estimates. (Top table) Summary of age estimates for Dehalococcoides-related genetic divergence utilizing four different models for rate of evolution: (1) estimated universal bacterial rate of evolution in nature [74], (2) in vitro E. coli empirically derived rate of evolution [75], (3) empirical Dehalococcoides rate based on observed mutations in the whole genomes of strain 195 and its resequenced variant in the DONNA2 sister culture (see Methods), and (4) the 16S rRNA gene clock model. For ages based on the first two rates of evolution, we further considered six different values for doubling time that span a range relevant to Dehalococcoides, including four published values for Dehalococcoides growth in laboratory culture [4,6,88,96], other anaerobic bacterial growth rates [47], and values derived from environmental anaerobic systems [48,49], as well as one arbitrarily large value (130 days) intended to represent general substrate-limited conditions. The left two columns indicate the divergence being considered and the tree calculation method, respectively. Ages are presented in units of 1 million years. (Middle Two Tables) Referenced summary of growth rates utilized for the age estimate calculations. (Bottom Table) Sample calculation for length of time to a single mutation, given rates of evolution taken from literature and the averaged Dehalococcoides growth rate.
Figure S4: KB-1 variant at vcr-GI module transition. (A) Cartoon representation of the vcr-GI observed in all 8 versions, as shown in Figure 3. (B) Alignment of the region at the transition between integration and vcrABC cargo modules, including reads in the KB-1 metagenome dataset that disagree with the main consensus at this location. All 3 of these variant reads are perfectly identical to the VS, GT, WL, and WBC-2 vcr-GIs at this position.
Figure S5: Genetic Map of Putative Fixed rdhA Region Downstream of Direct Repeats. (Top) Genetic map output from a Mauve alignment of the portion of High Plasticity Region 2 (HPR2) downstream of any ssrA direct repeats in the Dehalococcoides genomes. Each sequence was first aligned at tRNA-Ala-3 previously defining the boundary of HPR2 closest to the Ori [9], with local collinear blocks (LCBs) indicating large collinear homologous region that are free from rearrangements, but not necessarily indels. Large gaps were manually inserted such that vertical positions also containing the identity graph indicate aligned positions within the LCB. The darker grey LCB is the putative 'fixed' region of HPR2 downstream of any ssrA direct repeats. The lighter grey LCB is a portion of the Dehalococcoides core genome that surrounds the Ori. Annotated genes are shown beneath each LCB, with genes on the forward and reverse strands drawn as rectangles above or below the midline, respectively. rdhA are shaded red for emphasis. Scale bar shown in top left corner. Note that two different contigs from the ANAS genome are included. (Bottom) Phylogenetic trees of three semi-core (missing strain BAV1) rdhA that share a syntenic neighborhood within the putative fixed region. Each orthologous rdhA group recapitulates the topology and approximate genetic distances of the whole-genome tree (Figure 4). HPR2 was deleted in strain BAV1 [9], save for a ~600 bp rdhA fragment (DehaBAV1_1302) that is the basis for the tree on the right-hand side.
Figure S6: Genetic Map of a dsiB-Containing Deep-Sea Environmental Fosmid. The fosmid, EU016565, contains the most similar non-Dehalococcoides integration module(s) detected in the public database. EU016565 is part of an environmental shotgun sequencing dataset of genomic DNA obtained from a 4000 m sub-seafloor sediment [87]. Two partial Dehalococcoides ssrA integration modules are detectable, one of which contains an ssrA direct repeat at the expected location within a dsiB homolog. It also contains 4 of the 6 protein encoding genes typically found in integration modules as well as the putative tRNA embedded within mom homolog. The reverse-complement of EU016565 is displayed for consistent orientation with other figures. Light grey, dark grey, and black indicate protein encoding genes for which the annotation is hypothetical, identifiable, or part of the integration module, respectively.
Figure S7: Geographic locations of Dehalococcoides strains and cultures mentioned in this article. The underlying map was created using Google Earth. Labels have a dark red border if they are cultures/strains for which high throughput sequencing data is available and vinyl chloride respiration is reported. Blue borders indicate the vinyl chloride respiring cultures for which genomic island data was obtained during this study. White stars indicate cultures/strains for which no high throughput sequencing data was available at the time of this publication. The origin of the Dehalococcoides isolate FL2 [88] and the Dehalococcoides enrichment culture 'Pinellas' [89] are also shown. The following isolated bacterial strains were discussed in the manuscript: Dehalococcoides ethenogenes 195 - Ithaca Wastewater Treatment Plant, Ithaca, NY, USA [6,90]; CBDB1 - Saale River, Jena, Germany [91-93]; BAV1 - Bachman Road Site, Oscada, MI, USA [94]; VS - Contaminated Site, Victoria, Texas, USA [95]; GT - Hydrite Chemical Co., Cottage Grove, WI, USA [17]; Dehalogenimonas lykanthroporepellens BL-DC-9 [46]. The following Dehalococcoides enrichments were discussed. An asterisk indicates that no high-throughput sequence data is currently available: KB-1 - Southern Ontario, Canada [25]; ANAS - Alameda Naval Air Station, CA, USA [27] *PM - Point Mugu Naval Weapon Facility, CA, USA [28]; *EV - Evanite contaminated site, Corvallis, Oregon, USA [28]; *WBC-2 - West Branch Canal Creek, Aberdeen Proving Ground, MD [29] *WL - contaminated site, Western Louisiana, USA [30].
Contributor Information
Paul J McMurdie, Email: mcmurdie@stanford.edu.
Laura A Hug, Email: laura.hug@utoronto.ca.
Elizabeth A Edwards, Email: elizabeth.edwards@utoronto.ca.
Susan Holmes, Email: susan@stat.Stanford.EDU.
Alfred M Spormann, Email: spormann@stanford.edu.
Acknowledgements
The authors would like to thank Ruth Richardson (Cornell University) for advance access to the DONNA2 metagenome raw sequence data, Steve Zinder (Cornell University) for sharing insight into the minimum timing of culture divergence, Lew Semprini (Oregon State) for providing samples from the Evanite and Point Mugu enrichment cultures, Michelle Lorah for providing the WBC-2 enrichment culture, and John Roth for helpful comments on the manuscript, as well as Koshlan Mayer-Blackwell and Daniel M. Callahan for assistance with cloning and sequencing. This work was supported by the Strategic Environmental Research Defense Project (SERDP) to AMS by grant ER-1588. SH was partially funded by NIH grant R01GM086884, PJM was partially funded by fellowship grant FP-91671901 from the U.S. Environmental Protection Agency's Science to Achieve Results (STAR) program. (Meta)Genome sequencing and assembly was made available under the auspices of the U.S. Department of Energy Joint Genome Institute, supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
References
- McCarty PL. Breathing with chlorinated solvents. Science. 1997;276(5318):1521–1522. doi: 10.1126/science.276.5318.1521. [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (IARC) Monographs on the evaluation of carcinogenic risks to humans. IARC; 1987. Vinyl Chloride; pp. 311–443. [Google Scholar]
- Kielhorn J, Melber C, Wahnschaffe U, Aitio A, Mangelsdorf I. Vinyl chloride: still a cause for concern. Environ Health Perspect. 2000;108(7):579–88. doi: 10.1289/ehp.00108579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Ritalahti KM, Yang KL, Koenigsberg SS, Löffler FE. Detoxiflcation of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature. 2003;424(6944):62–5. doi: 10.1038/nature01717. [DOI] [PubMed] [Google Scholar]
- Muller JA, Rosner BM, Von Abendroth G, Meshulam-Simon G, McCarty PL, Spormann AM. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl Environ Microbiol. 2004;70(8):4880–4888. doi: 10.1128/AEM.70.8.4880-4888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maymó-Gatell X, Chien Y, Gossett JM, Zinder SH. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276(5318):1568–71. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- Seshadri R, Adrian L, Fouts DE, Eisen JA, Phillippy AM, Methe BA, Ward NL, Nelson WC, Deboy RT, Khouri HM, Kolonay JF, Dodson RJ, Daugherty SC, Brinkac LM, Sullivan SA, Madupu R, Nelson KE, Kang KH, Impraim M, Tran K, Robinson JM, Forberger HA, Fraser CM, Zinder SH, Heidelberg JF. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science. 2005;307(5706):105–108. doi: 10.1126/science.1102226. [DOI] [PubMed] [Google Scholar]
- Kube M, Beck A, Zinder SH, Kuhl H, Reinhardt R, Adrian L. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat Biotechnol. 2005;23(10):1269–1273. doi: 10.1038/nbt1131. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Behrens SF, Müller JA, Göke J, Ritalahti KM, Wagner R, Goltsman E, Lapidus A, Holmes S, Löffler FE, Spormann AM. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 2009;5(11):e1000714. doi: 10.1371/journal.pgen.1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson JK, Stern RV, Gossett JM, Zinder SH, Burris DR. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl Environ Microbiol. 1998;64(4):1270–5. doi: 10.1128/aem.64.4.1270-1275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliger C, Wohlfarth G, Diekert G. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev. 1998;22(5):383–398. doi: 10.1111/j.1574-6976.1998.tb00377.x. [DOI] [Google Scholar]
- Waller AS, Krajmalnik-Brown R, Löffler FE, Edwards EA. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl Environ Microbiol. 2005;71(12):8257–64. doi: 10.1128/AEM.71.12.8257-8264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung JM, Morris RM, Adrian L, Zinder SH. Expression of reductive dehalogenase genes in Dehalococcoides ethenogenes strain 195 growing on tetrachloroethene, trichloroethene, or 2,3-dichlorophenol. Appl Environ Microbiol. 2007;73(14):4439–45. doi: 10.1128/AEM.00215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahm BG, Richardson RE. Correlation of respiratory gene expression levels and pseudo-steady-state PCE respiration rates in Dehalococcoides ethenogenes. Environ Sci Technol. 2008;42(2):416–21. doi: 10.1021/es071455s. [DOI] [PubMed] [Google Scholar]
- Johnson DR, Brodie EL, Hubbard AE, Andersen GL, Zinder SH, Alvarez-Cohen L. Temporal Transcriptomic Microarray Analysis of Dehalococcoides ethenogenes Strain 195 During the Transition into the Stationary Phase. Appl Environ Microbiol. 2008;74(9):2864–2872. doi: 10.1128/AEM.02208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Adrian L, Kleinsteuber S, Andreesen J, Lechner U. Transcription analysis of genes encoding homologues of reductive dehalogenases in Dehalococcoides sp. strain CBDB1 using terminal restriction fragment length polymorphism and quantitative PCR. Appl Environ Microbiol. 2009;75(7):1876–1884. doi: 10.1128/AEM.01042-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y, Ritalahti KM, Apkarian RP, Löffler FE. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl Environ Microbiol. 2006;72(3):1980–7. doi: 10.1128/AEM.72.3.1980-1987.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes VF, He J, Lee PKH, Alvarez-Cohen L. Discrimination of multiple Dehalococcoides strains in a trichloroethene enrichment by quantification of their reductive dehalogenase genes. Appl Environ Microbiol. 2006;72(9):5877–83. doi: 10.1128/AEM.00516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajmalnik-Brown R, Hölscher T, Thomson IN, Saunders FM, Ritalahti KM, Löffler FE. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl Environ Microbiol. 2004;70(10):6347–6351. doi: 10.1128/AEM.70.10.6347-6351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Behrens SF, Holmes S, Spormann AM. Unusual codon bias in vinyl chloride reductase genes of Dehalococcoides species. Appl Environ Microbiol. 2007;73(8):2744–2747. doi: 10.1128/AEM.02768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Hsiao WWL, Brinkman FSL. Detecting genomic islands using bioinformatics approaches. Nat Rev Microbiol. 2010;8(5):373–82. doi: 10.1038/nrmicro2350. [DOI] [PubMed] [Google Scholar]
- Keiler KC. Biology of trans-translation. Annu Rev Microbiol. 2008;62:133–51. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271(5251):990–3. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Williams KP. Traffic at the tmRNA gene. J Bacteriol. 2003;185(3):1059–70. doi: 10.1128/JB.185.3.1059-1070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major DW, McMaster ML, Cox EE, Edwards EA, Dworatzek SM, Hendrickson ER, Starr MG, Payne JA, Buonamici LW. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ Sci Technol. 2002;36(23):5106–16. doi: 10.1021/es0255711. [DOI] [PubMed] [Google Scholar]
- Duhamel M, Mo K, Edwards EA. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl Environ Microbiol. 2004;70(9):5538–5545. doi: 10.1128/AEM.70.9.5538-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RE, Bhupathiraju VK, Song DL, Goulet TA, Alvarez-Cohen L. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ Sci Technol. 2002;36(12):2652–62. doi: 10.1021/es0157797. [DOI] [PubMed] [Google Scholar]
- Yu S, Semprini L. Comparison of trichloroethylene reductive dehalogenation by microbial communities stimulated on silicon-based organic compounds as slow-release anaerobic substrates. Water Res. 2002;36(20):4985–96. doi: 10.1016/S0043-1354(02)00222-1. [DOI] [PubMed] [Google Scholar]
- Lorah MM, Majcher EH, Jones EJ, Voytek MA. Microbial consortia development and microcosm and column experiments for enhanced bioremediation of chlorinated volatile organic compounds, West Branch Canal Creek wetland area, Aberdeen Proving Ground, Maryland. U.S. Geological Survey Scientific Investigations Report. 2007;2007-5165:1–79. [Google Scholar]
- Grostern A, Edwards EA. Growth of Dehalobacter and Dehalococcoides spp. during degradation of chlorinated ethanes. Appl Environ Microbiol. 2006;72:428–36. doi: 10.1128/AEM.72.1.428-436.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regeard C, Maillard J, Dufraigne C, Deschavanne P, Holliger C. Indications for acquisition of reductive dehalogenase genes through horizontal gene transfer by Dehalococcoides ethenogenes strain 195. Appl Environ Microbiol. 2005;71(6):2955–61. doi: 10.1128/AEM.71.6.2955-2961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajmalnik-Brown R, Sung Y, Ritalahti KM, Saunders FM, Löffler FE. Environmental distribution of the trichloroethene reductive dehalogenase gene (tceA) suggests lateral gene transfer among Dehalococcoides. FEMS Microbiol Ecol. 2007;59:206–14. doi: 10.1111/j.1574-6941.2006.00243.x. [DOI] [PubMed] [Google Scholar]
- Wozniak RAF, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8(8):552–63. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999;43(6):1449–58. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, van Embden JDA, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–75. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol. 2002;3(11):859–70. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- Hayes F, Barillà D. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat Rev Microbiol. 2006;4(2):133–43. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Funnell BE. Structures of ParB bound to DNA reveal mechanism of partition complex formation. Nature. 2005;438(7067):516–9. doi: 10.1038/nature04149. [DOI] [PubMed] [Google Scholar]
- Toussaint A. The DNA modification function of temperate phage Mu-1. Virology. 1976;70:17–27. doi: 10.1016/0042-6822(76)90232-4. [DOI] [PubMed] [Google Scholar]
- Allet B, Bukhari AI. Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol. 1975;92(4):529–40. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- Swinton D, Hattman S, Crain PF, Cheng CS, Smith DL, McCloskey JA. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2'-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc Natl Acad Sci USA. 1983;80(24):7400–4. doi: 10.1073/pnas.80.24.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Keiler KC. Beyond ribosome rescue: tmRNA and co-translational processes. FEBS Lett. 2010;584(2):413–9. doi: 10.1016/j.febslet.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Bhardwaj K, Sharkady S, Fish R, Driscoll T, Wower J, Zwieb C, Sobral B, Williams K. Variations on the tmRNA gene. RNA Biol. 2009;6(4) doi: 10.4161/rna.6.4.9172. [DOI] [PubMed] [Google Scholar]
- Charif D, Lobry J. In: Structural approaches to sequence evolution: Molecules, networks, populations, Biological and Medical Physics, Biomedical Engineering. U Bastolla HR M Porto, Vendruscolo M, editor. New York: Springer Verlag; 2007. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis; pp. 207–232. [ISBN: 978-3-540-35305-8] [Google Scholar]
- 2011. http://services.cbu.uib.no/tools/kaks
- Moe WM, Yan J, Nobre MF, da Costa MS, Rainey FA. Dehalogenimonas lykanthroporepellens gen. nov., sp. nov., a reductively dehalogenating bacterium isolated from chlorinated solvent-contaminated groundwater. Int J Syst Evol Microbiol. 2009;59(11):2692–7. doi: 10.1099/ijs.0.011502-0. [DOI] [PubMed] [Google Scholar]
- Strous M, Jetten MSM. Anaerobic oxidation of methane and ammonium. Annu Rev Microbiol. 2004;58:99–117. doi: 10.1146/annurev.micro.58.030603.123605. [DOI] [PubMed] [Google Scholar]
- Pollard PC, Moriarty DJ. Validity of the tritiated thymidine method for estimating bacterial growth rates: measurement of isotope dilution during DNA synthesis. Applied and environmental microbiology. 1984;48(6):1076–1083. doi: 10.1128/aem.48.6.1076-1083.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock T. Microbial growth rates in nature. Bacteriological Reviews. 1971;35:39–58. doi: 10.1128/br.35.1.39-58.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler F, Borchers R, Pracht J, Rheinberger S, Scholer HF. Natural formation of vinyl chloride in the terrestrial environment. Environmental Science & Technology. 2002;36(11):2479–2483. doi: 10.1021/es015611l. [DOI] [PubMed] [Google Scholar]
- Hagège J, Pernodet JL, Sezonov G, Gerbaud C, Friedmann A, Guérineau M. Transfer functions of the conjugative integrating element pSAM2 from Streptomyces ambofaciens: characterization of a kil-kor system associated with transfer. J Bacteriol. 1993;175(17):5529–38. doi: 10.1128/jb.175.17.5529-5538.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagège J, Pernodet JL, Friedmann A, Guérineau M. Mode and origin of replication of pSAM2, a conjugative integrating element of Streptomyces ambofaciens. Molecular Microbiology. 1993;10(4):799–812. doi: 10.1111/j.1365-2958.1993.tb00950.x. [DOI] [PubMed] [Google Scholar]
- Maphosa F, Smidt H, de Vos WM, Röling WFM. Microbial community- and metabolite dynamics of an anoxic dechlorinating bioreactor. Environ Sci Technol. 2010;44(13):4884–90. doi: 10.1021/es903721s. [DOI] [PubMed] [Google Scholar]
- Pavlovic G, Burrus V, Gintz B, Decaris B, Guédon G. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology (Reading, Engl) 2004;150(Pt 4):759–74. doi: 10.1099/mic.0.26883-0. [DOI] [PubMed] [Google Scholar]
- Futagami T, Morono Y, Terada T, Kaksonen AH, Inagaki F. Dehalogenation activities and distribution of reductive dehalogenase homologous genes in marine subsurface sediments. Appl Environ Microbiol. 2009;75(21):6905–9. doi: 10.1128/AEM.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble GW. The diversity of naturally produced organohalogens. Chemosphere. 2003;52(2):289–297. doi: 10.1016/S0045-6535(03)00207-8. [DOI] [PubMed] [Google Scholar]
- Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, Staphylococcus Cassette Chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44(6):1549–55. doi: 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen AM, Sollid JUE. SCCmec in Staphylococci : genes on the move. FEMS Immunol Med Microbiol. 2006;46:8–20. doi: 10.1111/j.1574-695X.2005.00009.x. [DOI] [PubMed] [Google Scholar]
- Noto MJ, Kreiswirth BN, Monk AB, Archer GL. Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J Bacteriol. 2008;190(4):1276–83. doi: 10.1128/JB.01128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre H, Oliveira D, Tomasz A. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr Opin Microbiol. 2007;10(5):428–35. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nübel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, Coombs G, Ip M, Westh H, Skov R, Struelens MJ, Goering RV, Strommenger B, Weller A, Witte W, Achtman M. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2008;105(37):14130–5. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7(9):629–41. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327(5964):469–74. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Archer GL. Roles of CcrA and CcrB in excision and integration of staphylococcal cassette chromosome mec, a Staphylococcus aureus genomic island. J Bacteriol. 2010;192(12):3204–12. doi: 10.1128/JB.01520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow M. What antimicrobial resistance has taught us about horizontal gene transfer. Methods Mol Biol. 2009;532:397–411. doi: 10.1007/978-1-60327-853-9_23. [DOI] [PubMed] [Google Scholar]
- Montero CI, Lewis DL, Johnson MR, Conners SB, Nance EA, Nichols JD, Kelly RM. Colocation of genes encoding a tRNA-mRNA hybrid and a putative signaling peptide on complementary strands in the genome of the hyperthermophilic bacterium Thermotoga maritima. J Bacteriol. 2006;188(19):6802–7. doi: 10.1128/JB.00470-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West K, Johnson D, Hu P, Desantis T, Brodie E, Lee P, Feil H, Andersen G, Zinder S, Alvarez-Cohen L. Comparative Genomics of Dehalococcoides ethenogenes 195 and an Enrichment Culture Containing Unsequenced Dehalococcoides Strains. Appl Environ Microbiol. 2008;74(11):3533–3540. doi: 10.1128/AEM.01835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23(2):254–67. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–63. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Ochman H, Elwyn S, Moran NA. Calibrating bacterial evolution. PNAS. 1999;96(22):12638–43. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148(4):1667–86. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum C, Ohsumi TK, Gomez J, Aquadro J, Victor TC, Warren RM, Hung DT, Birren BW, Lander ES, Jaffe DB. Sensitive, specific polymorphism discovery in bacteria using massively parallel sequencing. Nat Methods. 2009;6:67–9. doi: 10.1038/nmeth.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria; 2009. [ISBN 3-900051-07-0] [Google Scholar]
- Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–6. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319(5):1059–66. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31(13):3423–8. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann SE, Gorodkin J, Backofen R. Unifying evolutionary and thermodynamic information for RNA folding of multiple alignments. Nucleic Acids Res. 2008;36(20):6355–62. doi: 10.1093/nar/gkn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CH, Ochman H. Inferring clocks when lacking rocks: the variable rates of molecular evolution in bacteria. Biol Direct. 2009;4:35. doi: 10.1186/1745-6150-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009. pp. D205–10. [DOI] [PMC free article] [PubMed]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubakishita S, Kuwahara-Arai K, Baba T, Hiramatsu K. Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob Agents Chemother. 2010;54(4):1469–75. doi: 10.1128/AAC.00575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K, Delong E. Genomic patterns of recombination, clonal divergence and environment in marine microbial populations. ISME J. 2008. [DOI] [PubMed]
- He J, Sung Y, Krajmalnik-Brown R, Ritalahti KM, Löffler FE. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ Microbiol. 2005;7(9):1442–50. doi: 10.1111/j.1462-2920.2005.00830.x. [DOI] [PubMed] [Google Scholar]
- Ellis D, Lutz E, Odom J, Buchanan R, Bartlett C, Lee M, Harkness M, Deweerd K. Bioaugmentation for accelerated in situ anaerobic bioremediation. Environ Sci Technol. 2000;34(11):2254–2260. doi: 10.1021/es990638e. [DOI] [Google Scholar]
- Freedman DL, Gossett JM. Biological reductive dechlorination of tetrachloroethylene and trichloroethylene to ethylene under methanogenic conditions. Appl Environ Microbiol. 1989;55(9):2144–51. doi: 10.1128/aem.55.9.2144-2151.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak J, Kirsch N, Hegemann W, Stan HJ. Total reductive dechlorination of chlorobenzenes to benzene by a methanogenic mixed culture enriched from Saale river sediment. Appl Microbiol Biotechnol. 1996;45(5):700–709. doi: 10.1007/s002530050751. [DOI] [Google Scholar]
- Adrian L, Manz W, Szewzyk U, Görisch H. Physiological characterization of a bacterial consortium reductively dechlorinating 1,2,3- and 1,2,4-trichlorobenzene. Appl Environ Microbiol. 1998;64(2):496–503. doi: 10.1128/aem.64.2.496-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian L, Szewzyk U, Wecke J, Görisch H. Bacterial dehalorespiration with chlorinated benzenes. Nature. 2000;408(6812):580–583. doi: 10.1038/35046063. [DOI] [PubMed] [Google Scholar]
- He J, Sung Y, Dollhopf ME, Fathepure BZ, Tiedje J, Löffler FE. Acetate versus hydrogen as direct electron donors to stimulate the microbial reductive dechlorination process at chloroethene-contaminated sites. Environ Sci Technol. 2002;36(18):3945–3952. doi: 10.1021/es025528d. [DOI] [PubMed] [Google Scholar]
- Rosner B, McCarty P, Spormann A. In Vitro Studies on Reductive Vinyl Chloride Dehalogenation by an Anaerobic Mixed Culture. Appl Environ Microbiol. 1997;63(11):4139–4144. doi: 10.1128/aem.63.11.4139-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupples, Spormann, McCarty. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl Environ Microbiol. 2003;69(2):953–959. doi: 10.1128/AEM.69.2.953-959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Alignment and Predicted Secondary Structure of Putative tRNA-gly. These tRNA-gly are strongly conserved in 16 Dehalococcoides ssrA-GI integration modules. Bases are shaded according to the Vienna RNA conservation coloring schema in both the alignment (A) and secondary structure cartoon indicating the majority consensus with degeneracy (B). Secondary structure prediction was unanimous from three independent secondary structure prediction servers [80-82]. Free energy of the thermodynamic ensemble is -54.26 kcal/mol [80]. Substructure labels correspond to classical tRNA, including the apparent anti-codon 'DCC'.
Figure S2: Primers Mapped onto an Alignment of 16 ssrA Integration Modules. (A) Annotated alignment of the 16 integration modules discussed in this study. Individual sequences are shown as a thick black line, with gaps indicated by a thin horizontal line. Plot of average nucleotide identity (14 bp window) for all 16 sequences is shown along the top of the alignment. Three main target locations for primer design are indicated with downward-pointing black triangles, numbered beginning at ssrA (left). (B) Zoomed-in view of the alignment at the three target locations for primer binding. The 75% Consensus sequence is depressed slightly at the region targeted by primers, which are annotated along the top. Exact position of putative tRNA-gly is also shown.
Figure S3: Phylogenetic Tree of ssrA Versus 16S rRNA gene. The most likely of 100 bootstrap Maximum Likelihood trees with bootstrap support shown at nodes. Support not shown at nodes with poor or ambiguous support. (A) Phylogenetic tree of ssrA, the ~350 bp gene encoding tmRNA. (B) Similarly calculated tree based on the 16S rRNA gene (~1500 bp), reflected relative to typical tree orientation to emphasize topological similarity with (A). Other Chlorofiexi are included, with Staphylococcus aureus as an outgroup. Full name and accession number correspond to the following abbreviations: Dehalococcoides - Dhc; CBDB1 - Dhc CBDB1 NC_007356; GT - Dhc GT NC_013890; BAV1 - Dhc BAV1 NC_009455; 195 - Dhc ethenogenes 195 NC_002936; VS - Dhc VS NC_013552; Deha lyk - Dehalogenimonas lykanthroporepellens BL-DC-9 NC_014314; Staph aur - Staphylococcus aureus NC_002952; Rose cast - Ro-seiflexus castenholzii DSM 13941 NC_009767; Rose RS-1 - Roseiflexus sp. RS-1 NC_009523; Chlo aur - Chloroflexus aurantiacus J-10-fl NC_010175; Chlo agg - Chloroflexus aggregans DSM 9485 NC_011831.
Table S1: Parameters and example calculations utilized in divergence age estimates. (Top table) Summary of age estimates for Dehalococcoides-related genetic divergence utilizing four different models for rate of evolution: (1) estimated universal bacterial rate of evolution in nature [74], (2) in vitro E. coli empirically derived rate of evolution [75], (3) empirical Dehalococcoides rate based on observed mutations in the whole genomes of strain 195 and its resequenced variant in the DONNA2 sister culture (see Methods), and (4) the 16S rRNA gene clock model. For ages based on the first two rates of evolution, we further considered six different values for doubling time that span a range relevant to Dehalococcoides, including four published values for Dehalococcoides growth in laboratory culture [4,6,88,96], other anaerobic bacterial growth rates [47], and values derived from environmental anaerobic systems [48,49], as well as one arbitrarily large value (130 days) intended to represent general substrate-limited conditions. The left two columns indicate the divergence being considered and the tree calculation method, respectively. Ages are presented in units of 1 million years. (Middle Two Tables) Referenced summary of growth rates utilized for the age estimate calculations. (Bottom Table) Sample calculation for length of time to a single mutation, given rates of evolution taken from literature and the averaged Dehalococcoides growth rate.
Figure S4: KB-1 variant at vcr-GI module transition. (A) Cartoon representation of the vcr-GI observed in all 8 versions, as shown in Figure 3. (B) Alignment of the region at the transition between integration and vcrABC cargo modules, including reads in the KB-1 metagenome dataset that disagree with the main consensus at this location. All 3 of these variant reads are perfectly identical to the VS, GT, WL, and WBC-2 vcr-GIs at this position.
Figure S5: Genetic Map of Putative Fixed rdhA Region Downstream of Direct Repeats. (Top) Genetic map output from a Mauve alignment of the portion of High Plasticity Region 2 (HPR2) downstream of any ssrA direct repeats in the Dehalococcoides genomes. Each sequence was first aligned at tRNA-Ala-3 previously defining the boundary of HPR2 closest to the Ori [9], with local collinear blocks (LCBs) indicating large collinear homologous region that are free from rearrangements, but not necessarily indels. Large gaps were manually inserted such that vertical positions also containing the identity graph indicate aligned positions within the LCB. The darker grey LCB is the putative 'fixed' region of HPR2 downstream of any ssrA direct repeats. The lighter grey LCB is a portion of the Dehalococcoides core genome that surrounds the Ori. Annotated genes are shown beneath each LCB, with genes on the forward and reverse strands drawn as rectangles above or below the midline, respectively. rdhA are shaded red for emphasis. Scale bar shown in top left corner. Note that two different contigs from the ANAS genome are included. (Bottom) Phylogenetic trees of three semi-core (missing strain BAV1) rdhA that share a syntenic neighborhood within the putative fixed region. Each orthologous rdhA group recapitulates the topology and approximate genetic distances of the whole-genome tree (Figure 4). HPR2 was deleted in strain BAV1 [9], save for a ~600 bp rdhA fragment (DehaBAV1_1302) that is the basis for the tree on the right-hand side.
Figure S6: Genetic Map of a dsiB-Containing Deep-Sea Environmental Fosmid. The fosmid, EU016565, contains the most similar non-Dehalococcoides integration module(s) detected in the public database. EU016565 is part of an environmental shotgun sequencing dataset of genomic DNA obtained from a 4000 m sub-seafloor sediment [87]. Two partial Dehalococcoides ssrA integration modules are detectable, one of which contains an ssrA direct repeat at the expected location within a dsiB homolog. It also contains 4 of the 6 protein encoding genes typically found in integration modules as well as the putative tRNA embedded within mom homolog. The reverse-complement of EU016565 is displayed for consistent orientation with other figures. Light grey, dark grey, and black indicate protein encoding genes for which the annotation is hypothetical, identifiable, or part of the integration module, respectively.
Figure S7: Geographic locations of Dehalococcoides strains and cultures mentioned in this article. The underlying map was created using Google Earth. Labels have a dark red border if they are cultures/strains for which high throughput sequencing data is available and vinyl chloride respiration is reported. Blue borders indicate the vinyl chloride respiring cultures for which genomic island data was obtained during this study. White stars indicate cultures/strains for which no high throughput sequencing data was available at the time of this publication. The origin of the Dehalococcoides isolate FL2 [88] and the Dehalococcoides enrichment culture 'Pinellas' [89] are also shown. The following isolated bacterial strains were discussed in the manuscript: Dehalococcoides ethenogenes 195 - Ithaca Wastewater Treatment Plant, Ithaca, NY, USA [6,90]; CBDB1 - Saale River, Jena, Germany [91-93]; BAV1 - Bachman Road Site, Oscada, MI, USA [94]; VS - Contaminated Site, Victoria, Texas, USA [95]; GT - Hydrite Chemical Co., Cottage Grove, WI, USA [17]; Dehalogenimonas lykanthroporepellens BL-DC-9 [46]. The following Dehalococcoides enrichments were discussed. An asterisk indicates that no high-throughput sequence data is currently available: KB-1 - Southern Ontario, Canada [25]; ANAS - Alameda Naval Air Station, CA, USA [27] *PM - Point Mugu Naval Weapon Facility, CA, USA [28]; *EV - Evanite contaminated site, Corvallis, Oregon, USA [28]; *WBC-2 - West Branch Canal Creek, Aberdeen Proving Ground, MD [29] *WL - contaminated site, Western Louisiana, USA [30].