Abstract
The success of genome-wide association studies has paralleled the development of efficient genotyping technologies. We describe the development of a next-generation microarray based on the new highly-efficient Affymetrix Axiom genotyping technology that we are using to genotype individuals of European ancestry from the Kaiser Permanente Research Program on Genes, Environment and Health (RPGEH). The array contains 674,517 SNPs, and provides excellent genome-wide as well as gene-based and candidate-SNP coverage. Coverage was calculated using an approach based on imputation and cross validation. Preliminary results for the first 80,301 saliva-derived DNA samples from the RPGEH demonstrate very high quality genotypes, with sample success rates above 94% and over 98% of successful samples having SNP call rates exceeding 98%. At steady state, we have produced 462 million genotypes per week for each Axiom system. The new array provides a valuable addition to the repertoire of tools for large scale genome-wide association studies.
Keywords: Microarray, Genome-wide association study, Coverage, Throughput, Single nucleotide polymorphism
1. Introduction
The past decade has witnessed a revolution in genomics technology, enabling the correlation of common genetic variation with a variety of human traits and diseases through genome-wide association studies (GWAS) [1–4]. The large majority of these associations are not detectable by conventional linkage analysis, which is well powered primarily for Mendelian diseases and/or genes with high relative risks. While there has been recent debate regarding the success and significance of GWA studies [5,6], it is unequivocal that these studies have produced a large number of clearly replicated novel genetic associations with many diseases for which there had previously been no specific genes identified. It is also clear that the tools used to date have been based primarily on common genetic variation (minor allele frequency (MAF) of 0.10 or greater), leaving the door open for the potential detection of numerous additional disease-associated variants that are less common, provided that the necessary tools and sample sizes can be developed. This has also recently been made evident through consortia whose combined large sample sizes have given rise to a significantly increased number of validated associations [7,8].
The success of GWAS has been driven by the emergence and evolution of reliable and high-throughput genotyping platforms capable of simultaneously assaying hundreds of thousands of SNPs. Of the various genotyping technologies currently available, Affymetrix Axiom™ array plates and GeneChips™ and Illumina's Infinium BeadChips™ offer the highest SNP density and are mostly commonly used for GWAS [9].
The content of these high-density SNP arrays generally comes from the work of large SNP discovery consortia, including The SNP Consortium (TSC), followed by the International HapMap Project, and most recently the 1000 Genomes Project (KGP). As the SNP catalogs produced by the consortia grew, an early question was the extent of linkage disequilibrium (LD) among common variants in the human population and the degree to which untyped variants associated with disease could be detected through LD with genotyped SNPs [10,11]. As dense genetic maps emerged it became evident that the human genome is comprised of regions with extended LD and limited haplotype diversity, depending on the particular population studied. Within such regions the genotypes of common SNPs could be inferred from the genotypes of a smaller number of “tag” SNPs [12]. This meant that commercial chip designs assaying approximately 500,000 SNPs could potentially capture the majority of common genetic variation in most human populations. It was also recognized that African-derived populations have greater genetic diversity and lower levels of LD, requiring a greater density of SNPs to provide genome-wide coverage of common variation. With advances in technology for massively parallel genotyping of SNPs, the capacity of commercial arrays evolved to deliver very high density SNP capabilities, thus enabling highly powered GWAS [13].
Until recently the International HapMap project (http://www.hapmap.org) has been the primary data source for commercial SNP panels. The objective of the HapMap project when initiated in 2002 was to genotype common (MAF>0.05) SNPs across the genomes of 270 individuals from European, African, and Asian populations. Genotypes for more than 3 million SNPs were released by the Consortium by 2007, as well as the characterization of local LD patterns across the human genome. These Phase I and Phase II HapMaps guided SNP selection strategies for commercial SNP panels to “capture” the common variation in these particular populations.
The progression of Affymetrix arrays for massively parallel genotyping of SNPs (Table 1) illustrates the general progress of the field, beginning with the Mapping 10 K array [14]. The technology for the first array overcame two bottlenecks: the requirement for locus-specific SNP amplification and locus-specific allele discrimination. The source of variants was the TSC project and SNPs discovered by Perlegen Science's resequencing project, and criteria for selecting SNPs for the array resulted in a bias towards those with high (>0.1) MAF. The Mapping 100 K array set [15] was the first in a family of products suitable for association studies, and in fact produced the first GWAS finding, for adult onset macular degeneration and complement factor H [16]. The SNP selection process required at least two minor alleles in the 108 chromosomes genotyped, and consequently 90% of the SNPs are common.
Table 1.
Historical progression of Affymetrix SNP genotyping arrays. The number of SNPs/array has increased ~100× in five years. Significant increases in throughput have also occurred during this time period. These advances were the result of innovations in array feature size, assay, hardware and genotyping algorithms resulting in increased SNP density per unit area and overall productivity.
| Year | 2003 | 2004 | 2005 | 2006 | 2007 | 2009 |
|---|---|---|---|---|---|---|
| Product | Mapping 10 K | Mapping 100 K | Mapping 500 K | SNP 5.0 | SNP 6.0 | Axiom Genotyping Solution |
| # SNPs | 10,000 | 100,000 | 500,000 | 500,000 | 900,000 | up to 690,000 |
| # SNPs/mm2 | 60 | 300 | 1,500 | 3,000 | 6,000 | 19000 |
| # Samples/week | Not defined | Not defined | >96 | >96 | >96 | >750 |
The Mapping 500 K array (http://media.affymetrix.com/support/technical/datasheets/500k_datasheet.pdf) was the first array set with sufficient density to enable highly powered GWAS. SNPs were selected to be polymorphic in the HapMap populations, and average heterozygosity in those populations was 0.29. The Affymetrix SNP 5.0 array has essentially the same SNP content as the Mapping 500 K, but the design involves multiple replicates of the most informative probes rather than a single copy of many different probe sequences [17], leading to reduced cost and increased throughput. Affymetrix SNP 6.0 was the next generation array. The design was based on screening more than two million additional SNPs (chosen from HapMap and dbSNP) and selecting 906 K SNPs to optimize coverage of common HapMap Phase II variants in Europeans, East Asians and West Africans. As we show later, the SNP 6.0 array provides excellent whole genome coverage for genetic variants with MAF greater than 0.1.
The Axiom™ Genotyping Solution (http://media.affymetrix.com/support/technical/datasheets/axiom_genotyping_solution_datasheet.pdf) is the newest genotyping product developed to increase the power of a study by substantially increasing sample throughput with arrays that maximize genetic coverage for at least one population. Here we describe a newly developed Axiom array intended to provide coverage of both common and lower frequency variation in the European population; similar arrays for other ethnic groups are also under development.
The Axiom genotyping platform is a two color, ligation-based assay utilizing 30-mer oligonucleotide probes synthesized in situ on a microarray substrate, with automated, parallel processing of 96 samples per plate. Features are 3 µm squares, at a pitch of 5 µm center-to-center, with a total of ~1.38 million features available for experimental content. Each SNP feature contains a unique oligonucleotide sequence complementary to the genomic sequence flanking the SNP site on either the forward or reverse strand. Solution probes bearing attachment sites for one of two dyes, depending on the 3' (SNP-site) base (A or T, versus C or G) hybridize to the glass probe/target complex, and are then ligated for specificity. Features are typically replicated twice on the array, so that each SNP is interrogated by two features; A/T and C/G SNPs require four features, because the two alleles match the same dye and therefore distinct probe sequences in different physical locations on the array are required to distinguish them. A maximum of ~690,000 SNPs may be accommodated by this format; this number is reduced if A/T or C/G SNPs are included, or if additional features are used to improve the resolution of specific SNPs, but may also be increased if high resolution SNPs are tiled with a single feature. The platform can be leveraged for designing arrays at previously unattainable levels of SNP density with respect to customization of variant selection.
Here we describe the development and quality characteristics of a new microarray for the Axiom system tailored to the European population. This is the first in a series of customarrays being developed for a genome-wide genotyping analysis of 100,000 participants fromthe Kaiser Permanente Northern California Research Program on Genes, Environment and Health. The rationale for developing these new arrays is to maximize the number of high quality SNPs for genome-wide coverage; to provide coverage down to a minor allele frequency of 0.01 in gene-based regions; to saturate regions previously identified as disease associated from prior GWA studies for both replication and fine mapping; to improve coverage of both common and uncommon variants by making use of data from the low pass and high pass phases of the 1000 Genomes Project; and to incorporate redundant coverage of SNPs with known strong associations with disease or trait outcomes.
2. Results
2.1. Array statistics
Because the described Axiom array was developed as part of the Kaiser Permanente-UCSF collaborative RC2 project specific for European ancestry populations, it has been given the designation Axiom Genome-Wide EUR Array Plate. In total, there are 674,517 SNPs tiled onto the microarray. Among these, 116 are mitochondrial, 289 are on the Y chromosome, 388 are pseudoautosomal, 12,735 are on the X chromosome, and the remaining 660,989 SNPs are autosomal. Even though it is optimized for individuals of European ancestry, because of close kinship we expect it to also have high utility for individuals of West Asian, North African and South Asian ancestry.
2.2. Performance on HapMap samples
Performance of the array was assessed by assaying the Caucasian and Yoruban HapMap2 populations (90 individuals each with cell line DNA). Results are shown in Table 2. Call rates, sample concordance, reproducibility and Mendelian consistency are all extremely high. Furthermore, a large majority of SNPs (98.5%) have overall call rates of 97% or greater.
Table 2.
Performance of the array on 180 HapMap 2 individuals.
| Metric | Performance |
|---|---|
| Average Sample Call Rate | 99.69% |
| Average Sample Concordance to HapMap Reference Genotypes | 99.71% |
| Fraction of SNPs with Call Rate>97% | 98.5% |
| Average SNP Reproducibility | 99.89% |
| Mendelian Inheritance Accuracy | 99.94% |
2.3. Performance on the KP RPGEH GERA cohort
To date, we have completed genotyping of 80,301 saliva samples over a 9 month period, using 3 Affymetrix Gene Titan systems and 3 Beckman Biomek FXP Target Prep Express Systems, including 818 pairs of duplicate samples. That translates to approximately 685 samples per week per system, and 462 million genotypes per week per system. We used the Affymetrix Powertools Package version 1.12.0 [18] to make the genotype calls.
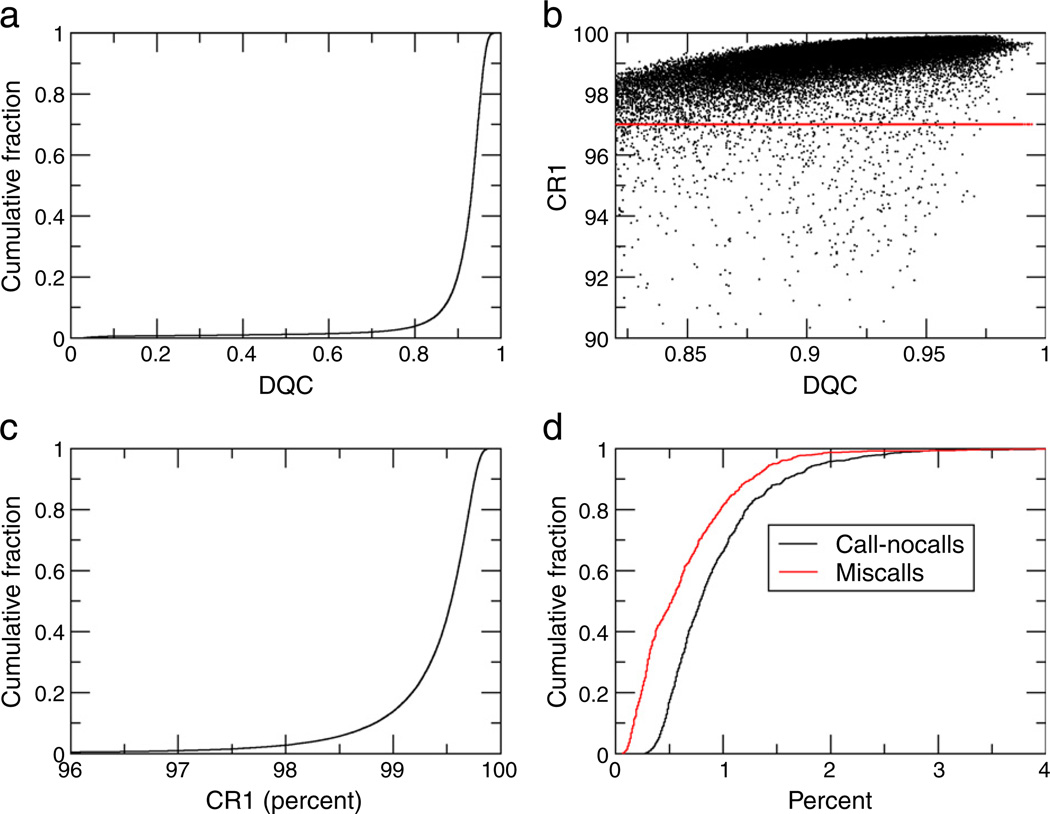
Fig. 1a is the cumulative distribution of DQC (quality control), scores for the 80,301 samples. Among these, 76,508 (95.3%) had passing DQC scores (>0.82). Fig. 1b is a scatter plot showing the distribution of sample call rate vs. DQC for samples with DQC>0.82. We see a strong correlation between the two measures, and that the great majority of the samples fall along a “main sequence” of behavior. Below the main sequence is a diffuse halo of samples whose call rate is below expected. Although few in number, these samples below the call rate threshold of 97% show the necessity for a second filtering and genotyping step in the standard workflow.
Fig. 1.
Results of QC analysis on the Kaiser Permanente RPGEH GERA cohort using the Affymetrix Axiom system. (a) Cumulative distribution of DQC scores for 80,301 genotyped saliva samples. (b) Sample call rate versus DQC derived from 76,412 genotyped saliva samples. Only samples passing a DQC threshold of 0.82 are included. Red line indicates threshold for passing call rate. (c) Cumulative distribution of sample call rates for 76,412 genotyped samples. Only samples passing a DQC threshold of 0.82 are included. (d) Cumulative distributions for call-nocalls and miscalls based on 818 duplicate samples.
Fig. 1c shows the cumulative distribution of sample call rates for sample assays with DQC>0.82. Among the 76,508 samples with DQC>0.82, 75,740 (99.0%) have a “passing” call rate of 97% or greater. Overall, 87% of the samples have a call rate ≥99% and over 98% of samples have a call rate ≥98%. From the original group of 80,301 samples, 94.3% pass both DQC and call rate criteria.
Genotype reproducibility was assessed from the 818 duplicate samples. There are two types of disagreement between duplicate samples. The first we refer to as a “call–no call” (CNC) — where one sample has a genotype call, while the other is considered a no call. The second we refer to as a “miscall.” This occurs when both samples receive genotype calls, but the calls differ. We calculated both the CNC rate (total number of disagreements out of total number of SNPs) and the miscall rate (total number of miscalls out of total number of SNPs) for each pair of duplicate samples. The majority of samples had CNC rates below 1%, with 0.7% of samples having a rate of 3% or higher and 4.8% with rates between 2% and 3% (Fig. 1d). Similarly, the majority of samples had miscall rates below 0.5%, with 1% having a miscall rate greater than 2% (Fig. 1d).
There was also a strong correlation between CNC and miscall rates for the duplicate samples (r=0.93), indicating that samples with lower DQC and overall call rates are also likely to have a higher rate of miscalls. These miscall rates are conservative overestimates because they are based on first pass genotyping, and presumably can be brought down further based on more refined genotyping procedures and higher stringency for genotype calls. We also noted that for miscalls, the large majority (97.6%) occurred between homozygotes and heterozygotes, and rarely between the two different homozygotes. This is consistent with occasional difficulties in clearly assigning a genotype to a given sample when adjacent clusters may not be very well separated.
2.4. Genome-wide coverage
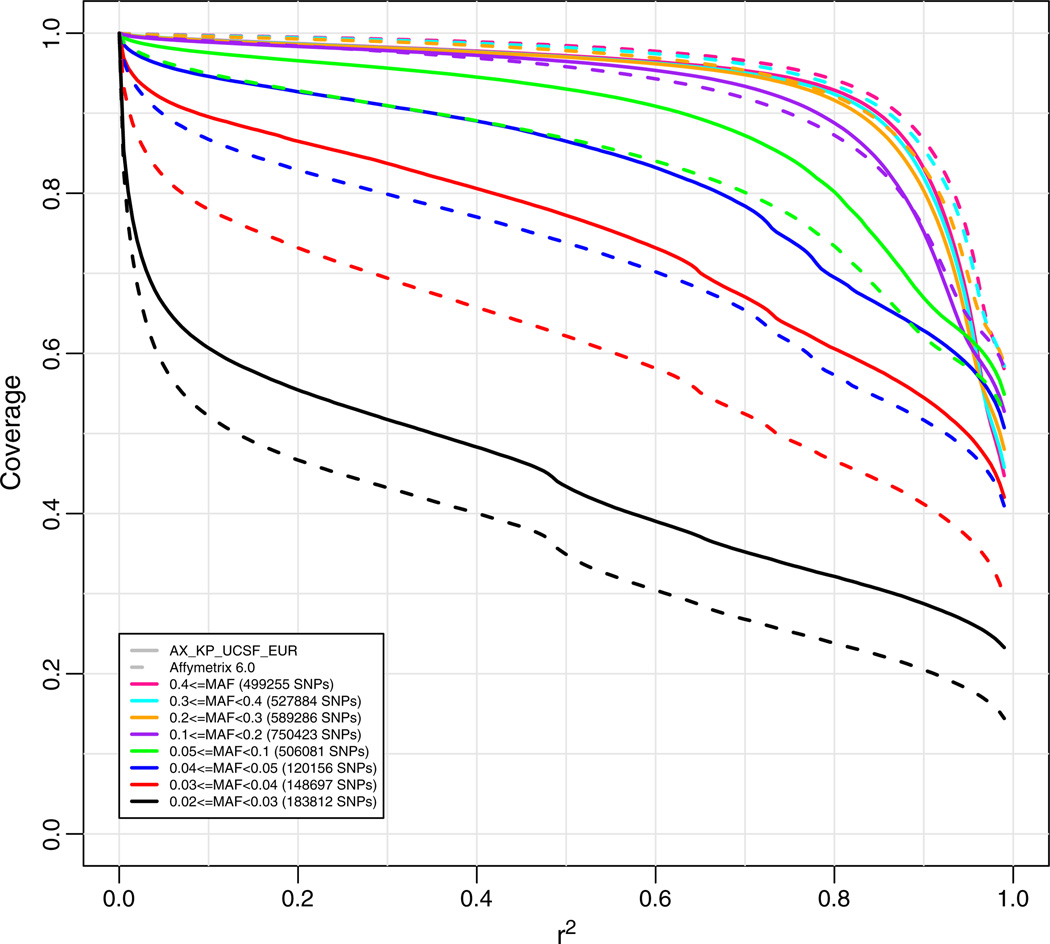
We first compared the coverage of our array to the Affymetrix 6.0 chip, which includes over 906,600 SNPs [17]. Because coverage is dependent on MAF, we stratified all coverage figures based on MAF. We first looked at the coverage of the “target set” that the array was designed for (3,431,598 SNPs with MAF≥0.02), stratified by MAF ranges. We see in Fig. 2 that that the new array provides coverage comparable to Affymetrix 6.0 for common SNPs (MAF≥0.1), but has higher coverage for lower allele frequencies, down to 0.03 frequency, despite having fewer SNPs on the microarray. This is because the Affymetrix 6.0 chip was designed from a smaller target set and was not an ethnic-specific chip. It is significant that coverage is still good down to 0.03 MAF, where greater than 60% of SNPs are covered with an r2 of 0.8.
Fig. 2.
Genome-wide coverage for the new Axiom Genome-Wide EUR Array (solid lines) versus the Affymetrix 6.0 array (dashed lines) for a target set of Affymetrix validated CEU SNPs using Affymetrix genotypes, stratified by minor allele frequency. Coverage based on imputation with “leave-one-out cross validation.” The numbers in parentheses in the legend are the numbers of markers in the target set in each particular minor allele frequency range.
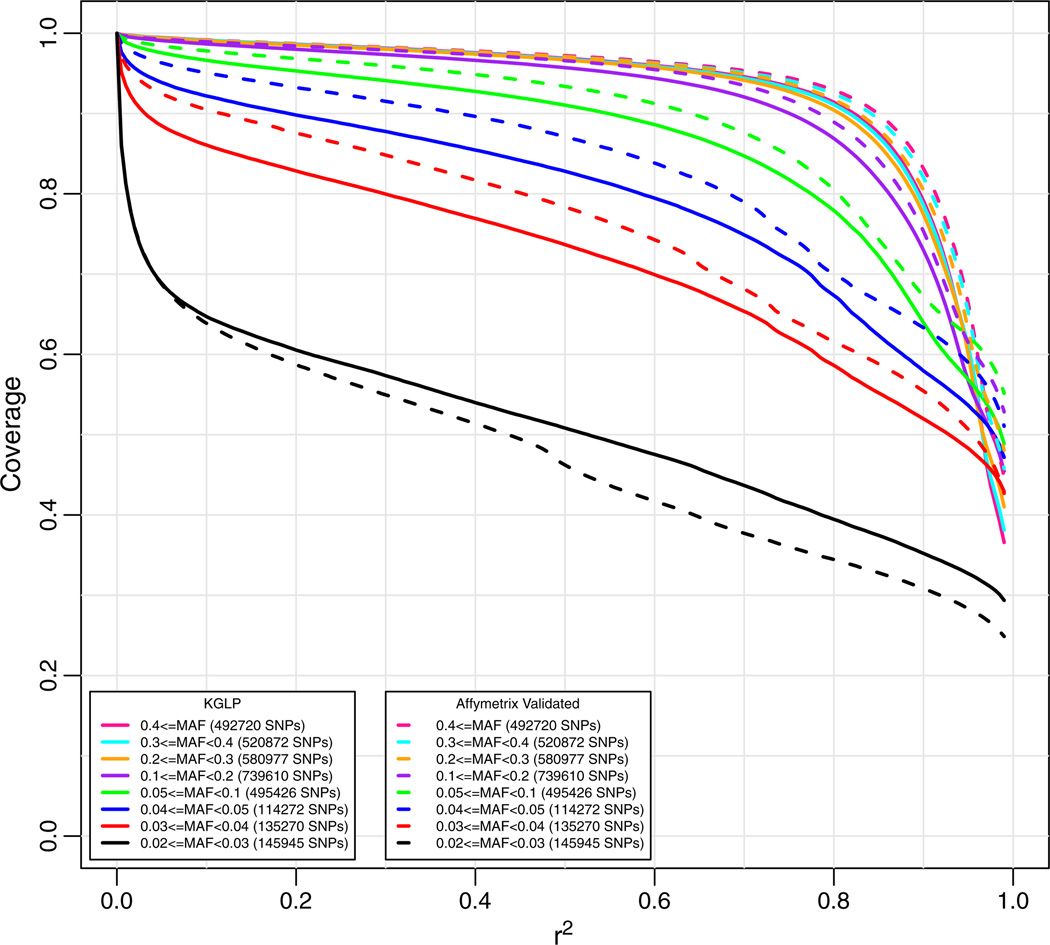
We know that this target set is incomplete (e.g., not all the KGP data were screened at design time). A larger set of 6,367,892 SNPs with MAF≥0.02 have been identified in the sequencing of 60 CEU individuals in the low pass sequencing phase (KGLP) of the KGP (http://www.1000genomes.org). This pilot dataset was sequenced at low (2–4X) coverage [19–22]. Some of the genotype calls for these 60 subjects available on the KGP website had been improved through imputation with HapMap 3 data [23]. Because of potential noise from the low pass sequencing, it was unclear whether the KGLP dataset could provide an accurate estimate of genome-wide coverage, so we first looked at coverage for the set of SNPs overlapping those in the target set for which the array was designed (Affymetrix data) and KGLP.
In Fig. 3 we see that the coverage calculated using these two sets of genotype data for the same target SNPs was very similar. Hence, we conclude that the KGLP genotype data serve as an adequate reference set for subsequent analyses.
Fig. 3.
Genome-wide coverage for the new array for a target set of Affymetrix validated CEU SNPs using either Affymetrix genotypes (dashed lines) or the 1000 Genomes Low Pass (KGLP) genotypes (solid lines). Coverage based on imputation with “leave-one-out cross validation.” The numbers in parentheses in the legend are the numbers of markers in the target set in each particular minor allele frequency range.
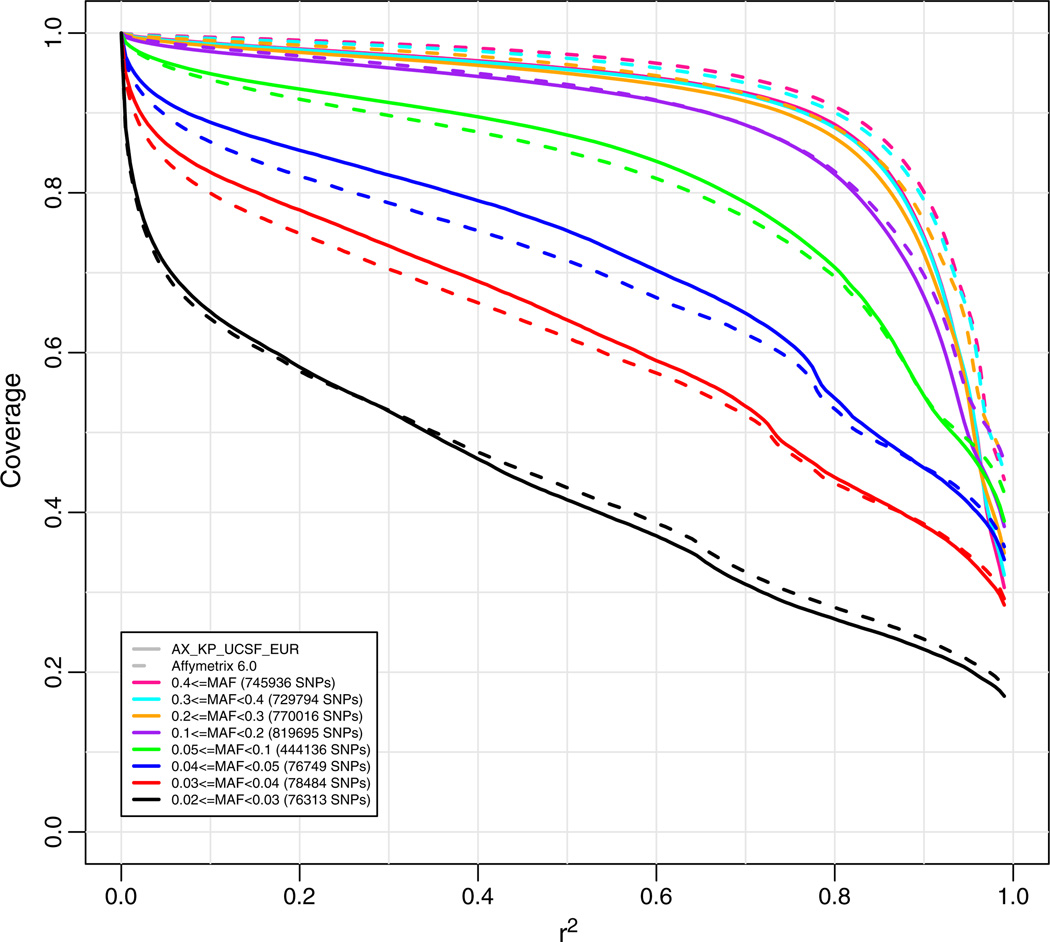
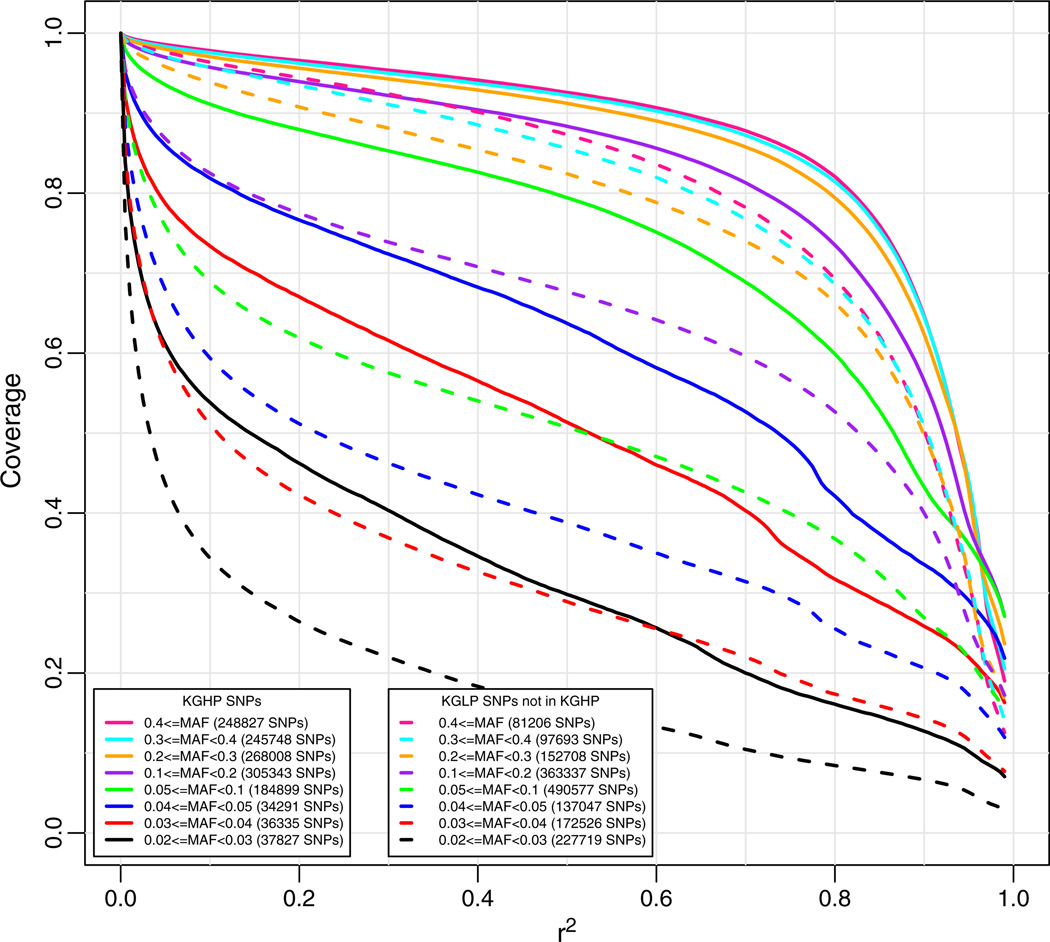
We were next concerned with estimating genome-wide coverage based on a random collection of valid SNPs, not limited to the SNPs used for the array design, to obtain an unbiased estimate of full genome coverage. To do so, in theory, we could focus simply on the totality of KGLP SNPs. However, because the KGLP SNP data were obtained from low pass sequencing, we were concerned about the possible role of false positives in these data. These will likely be particularly common for putative SNPs with low MAF. On the other hand, we know that the KGP Pilot high pass sequencing data (KGHP) were created with a much higher read depth (20–60×), although sequencing was only performed on 4 independent individuals (two CEU parents and two YRI parents). Sequence variations derived from these 4 founders can be considered a random sample of genotypes, albeit biased towards common variants due to the small number of individuals examined. Although low MAF SNPs are under-represented in this group overall, they still occur in this sample and represent a random sampling of low frequency variants. Therefore, to get another estimate of genome-wide coverage for a random set of valid SNPs, including common as well as rare alleles, we calculated coverage for KGHP-derived SNPs that also appeared in the KGLP data, and used the KGLP genotype data to determine coverage. We see that our array has good coverage when using all markers in KGHP as the target set (Fig. 4), although not as strong as for the Affymetrix target set for which it was designed (Fig. 2). For the original Affymetrix coverage set, at an r2 of 0.6, coverage ranged from about 74% at MAF of 0.03 to 96% at MAF of 0.1 or greater, while for the KPHP target set, coverage ranged from about 60% at MAF of 0.03 to 94% at MAF of 0.2. However, we also note that these coverage estimates are likely to be conservative, as they are based on a relatively small sample of 60 KGLP individuals. Therefore, the coverage of SNPs with MAF of 0.03 is likely to be higher than 60% for a larger reference panel.
Fig. 4.
Genome-wide coverage for the new array (solid lines) versus the Affymetrix 6.0 array (dashed lines) for a target set of 1000 Genome High Pass (KGHP) SNPs using 1000 Genomes Low Pass (KGLP) genotypes. Coverage based on imputation with “leave-one-out cross validation.” The numbers in parentheses in the legend are the numbers of markers in the target set in each particular minor allele frequency range.
We were also interested in comparing coverage of those SNPs that appeared in the KGLP data but did not appear in the KGHP or the Affymetrix database compared with those that did appear in the KGHP data (but not the Affymetrix database). For SNPs missing from KGHP, coverage was significantly worse (Fig. 5), in comparison to those that did appear in KGHP. This indicates that many of these KGLP SNPs are likely false positives, leading to absence of LD with neighboring SNPs and therefore a lack of coverage. Hence, caution needs to be exercised in terms of genome-wide SNP coverage estimates based solely on KGLP data.
Fig. 5.
Genome-wide coverage of the new array for two complementary target sets: KGHP SNPs with Affymetrix validated SNPs removed (solid lines), and KGLP SNPs with KGHP and Affymetrix validated SNPs removed (dashed lines). Coverage based on imputation with “leave-one-out cross validation” using KGLP genotypes. The numbers in parentheses are the numbers of markers in the two target sets in each particular minor allele frequency range.
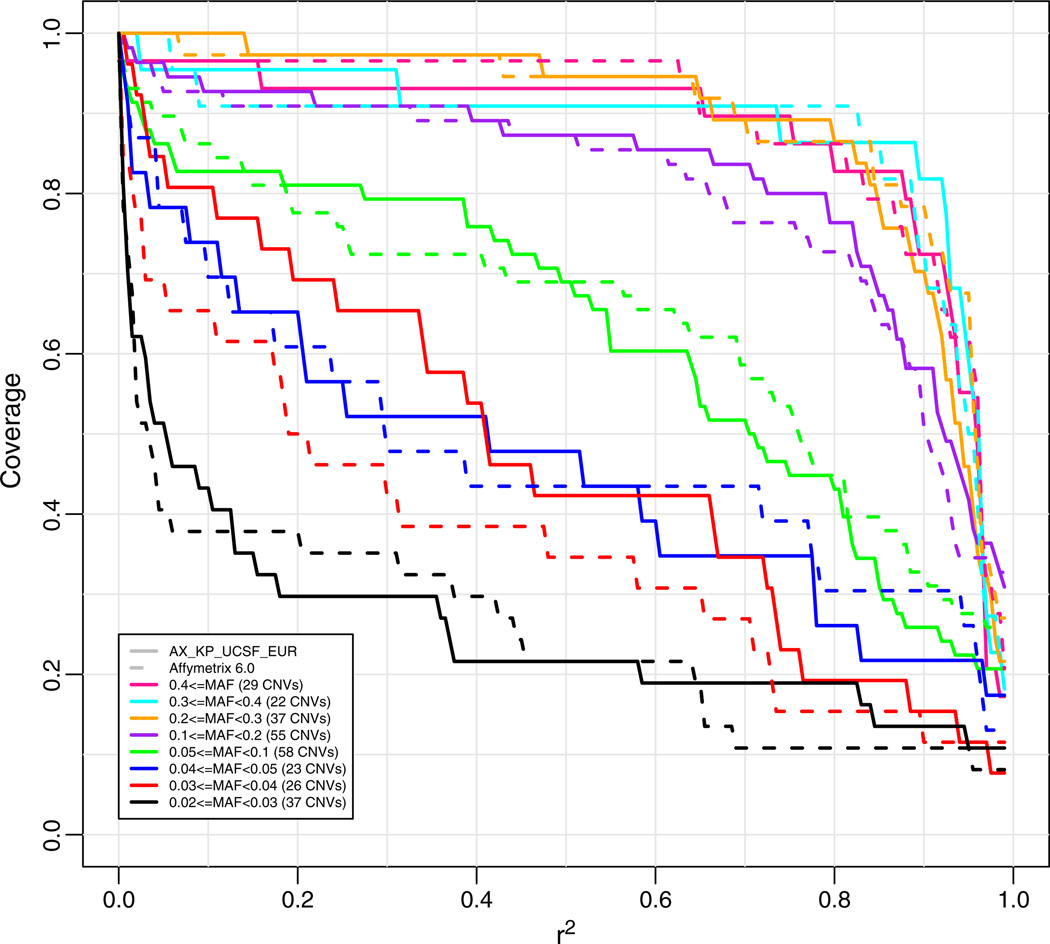
Lastly, we looked at the coverage of the CNV data detectable on the Affymetrix 6.0 chip [17,24,25] by our newly designed EUR array (Fig. 6). The dataset used for this analysis was based on 59 independent CEU HapMap individuals; because CNVs were not included in the EUR chip design and the SNP 6.0 chip uses a very different assay compared to Axiom (hybridization-based instead of ligation-based), this analysis also provides an independent estimate of how well the chip can cover variants not included in the target set used in the chip design. For this analysis we discarded 216 “multiallelic” CNVs (i.e., CNVs that could not be treated as SNPs, e.g., a CNV that could have 0, 1, 2, or 3 copies) and 632 “bi-allelic” CNVs (i.e., can be treated as a bi-alellic SNP) with MAF<0.02, and report coverage of 293 “bi-allelic” CNVs with MAF≥0.02. We would anticipate CNV coverage to be comparable to the random SNP coverage as depicted in Fig. 4. Such is the case down to a MAF of 0.1. However, below that point, CNV coverage is reduced compared to SNPs. As yet, we are uncertain as to the cause of this decrease; possibilities include reduced accuracy of the CNV calling for low frequency CNVs, and heterogeneous origins of lower frequency CNVs, both of which would reduce neighboring linkage disequilibrium and hence imputation ability. Again, these coverage estimates are likely to be conservative because of the small size of the reference data set of genotypes for this analysis.
Fig. 6.
Coverage of the new array (solid lines) versus the Affymetrix 6.0 array (dashed lines) for a target set of Canary CNVs using Canary CNV and Affymetrix genotype data. Coverage based on imputation with “leave-one-out cross validation.” The numbers in parentheses are the numbers of CNVs in the target set in each particular minor allele frequency range.
3. Discussion
While genotyping chips with up to 1 million SNPs have now been extensively applied in numerous GWA studies, questions about coverage of low frequency variation remain. Also, throughput and expense have previously been limitations for large scale application of high density genotyping chips.
The current Axiom system and the associated genotyping arrays, as described here for the European population, offer a new opportunity to rectify some of these previous limitations. We have shown the array described here, with approximately 675,000 SNPs, offers a new solution to high throughput, reduced cost genotyping that also provides coverage of low frequency variation. This development was required due to the large scale genotyping necessary for the KP RPGEH.
In considering the genotyping platform for the project to obtain genome-wide SNP data for 100,000 individuals of diverse ethnicity in 14 months, we chose the Axiom/GeneTitan platform over the Affymetrix HuSNP 6.0 and the Illumina Omni1-Quad Infinium HD platforms available in September 2009 because of 4 main reasons. First, the Axiom system allowed us to design custom SNP panels with great flexibility. Once the SNP panel is designed, fully manufactured Axiom arrays are shipped in less than 6 weeks. Second, the Axiom assay protocol is highly automated using microtiter plates with substantially less hands on time than that required on the other two platforms. The tasks performed by technicians involve simple manipulations of microtiter plates placed onto robotic equipment and reagents into reservoirs. In contrast, the Infinium and Affy 6.0 protocols require handling of glass slides and DNA chips that require more sophisticated skill and mental concentration. Technician burnout was a serious concern when considering the size of our project. Third, the GeneTitan is a compact system that has a small footprint. The amount of space required to house the iScans and robotic workstations needed for the Infinium platform is about 3 times that required for the Axiom platform. Fourth, sample tracking is robust in a 96-array format of the Axiom system. With the samples arrayed randomly according to gender, checking for male/female mismatches in the genotyping data is a simple way to confirm that there are no sampling errors.
To date, we have completed genotyping of approximately 80,000 RPGEH samples in a 9 month period, using three Axiom Gene Titan systems and three Beckman FXP Target Prep Express Systems. This translates to approximately 685 samples and 462 million genotypes per week per Gene-Titan system. The high throughput of this system will enable us to complete the successful genotyping of 100,000 RPGEH subjects in a period of 14 months or less.
As expected, the array demonstrates excellent coverage down to a MAF of 0.03 for the millions of SNPs it was designed to cover; but we have also shown excellent coverage for other random SNPs down to a MAF of 0.05 and reasonable coverage to even lower MAF of 0.03. Special attention was paid in the design process to coverage of SNPs with known or plausible disease associations, with the result that the chip provides very good coverage of SNPs with potential biological importance, such as SNPs in the regions of ADME genes, MHC genes, or genes associated with cardiovascular disease or cancer [26].
We believe that the next generation of genotyping chips, for example the Axiom based chip described here, will provide novel associations with both low frequency and common variants, especially on large scale, well phenotyped studies. The first 100,000 samples from the Kaiser Permanente Research Program on Genes, Environment and Health to be genotyped, as described here, is but one such example. From its design, the custom chip will also facilitate replication and extension studies of previously identified associations, and because of its focus on SNPs of pharmacogenetic interest, will also be a valuable tool for studying genetic effects on treatment response and side effects.
The high throughput and reduced cost with the Axiom system are attained at a tradeoff with ethnic-specific coverage. Lower frequency variants, for example those with frequency below 0.1, have an enhanced probability of being race/ethnic group specific. Thus, in a multi-ethnic study, different custom chips are required. We are currently designing other arrays specific for East Asians, African Americans and Latinos, which will be described in a subsequent publication. While we expect a large amount of overlap in SNP content on these different arrays for common variants (i.e., those with MAF>0.1), we also anticipate inclusion of a sizable number of SNPs that are unique to that ethnic group at a MAF of 0.02 or greater.
It is likely that genome-wide genotyping chips will continue to be more efficient and less expensive to apply than whole genome sequencing. As the field focuses more on studying non-European populations, the importance of diverse ethnic groups will lead to next generation arrays that provide significantly improved coverage of more relevant variants. The development of new platforms and arrays as described here will offer enhanced and complementary tools for genome-wide analysis to identify the genetic basis of complex diseases and traits, as has already been demonstrated for the earlier generation genotyping tools.
4. Materials and methods
4.1. SNP validation (conversion)
SNPs screened using the Axiom™ technology were validated (“converted”) according to several metrics encompassing the following general principals:
4.1.1. Resolution
Cluster resolution was assessed by cluster separation and call rate. The Axiom GT1 algorithm adapts pre-positioned clusters to the data using a probability-based method. Clustering is carried out in two dimensions, log ratio (log2(A) − log2(B)) and size (log2(A + B)/2). The algorithm is very similar to the modified version of BRLMM-P described in [24], except that no manual adjustment of priors was performed, only bi-allelic SNPs and indels are considered, and posterior estimation is dynamic using multiple samples in combination with predetermined priors. Cluster separation or resolution was measured by the minimum pairwise Fisher's Linear Discriminant (FLD) [27] between either homozygous cluster and the heterozygote cluster, in the log ratio dimension. A minimum threshold of 3.6 has been empirically determined to eliminate the great majority of poorly resolved SNPs. SNP call rate was also calculated, with a minimum threshold for conversion of 98% in the screen of KGP SNPs discussed below. Additional measures of relative cluster position are used to eliminate a variety of rare mis-clustering phenotypes that are not caught by the FLD and call rate thresholds.
4.1.2. Polymorphism
A minimum of three observed examples of the minor allele (e.g., three heterozygotes, or one heterozygote and one minor allele homozygote) was required for validation of a SNP. This provided evidence for both proper assay function and resolution and the existence of an actual polymorphism at the indicated site.
4.1.3. Accuracy
Accuracy was assessed by concordance, reproducibility and consistency with Mendelian inheritance. SNPs with HapMap reference calls in the individuals genotyped were assessed for concordance between array-derived and reference genotypes, with a minimum concordance threshold set at 96% and a minimum call rate threshold of 96%. For SNPs without HapMap reference genotype data, a minimum threshold for the call rate of 98% was imposed. Reproducibility was calculated for each SNP as the fraction of genotype calls concordant to the consensus call for all replicates of each sample. The threshold for reproducibility was 97%. Mendelian inheritance consistency was calculated as the fraction of offspring genotype calls in keeping with parental genotypes. The threshold for Mendelian inheritance consistency was 98.5%.
4.2. SNP selection for inclusion on the array
Two considerations guided SNP selection. First, we wished to maximize the number of SNPs on the array. More SNPs allow for greater genomic coverage. The array accommodates a fixed total of 1.38 million features. The majority of SNPs require two features. Limiting SNP selection to two-feature sets would, in theory, provide room for 690,000 SNPs. However, because certain SNPs were deemed as high priority (as described below), exceptions were made to allow for higher multiple-feature and multiple-probe SNPs. In addition, some high-value SNPs were tiled on the array in multiple replicates to improve the chances for high quality genotype calls.
The second consideration is that not all SNPs are equal in importance. SNPs based on strongly confirmed trait or disease associations are of highest value, while SNPs chosen for genomic coverage may have no particular significance beyond their ability to predict genotypes of other SNPs. We therefore divided SNPs into selection tiers based on a hierarchy of importance. Some SNPs were included because of known function or disease/trait association; some SNPs were selected to “cover” or “tag” SNPs of known importance which could not be converted in the Axiom assay, or for which we desired redundant coverage; and finally others were selected algorithmically to optimize the coverage of general genetic variation across the entire genome. SNP selection proceeded progressively down tiers of importance, as described below.
4.2.1. Stage 1: A preselected set of SNPs
We first created a list of “preselected” SNPs of varying levels of importance for inclusion on the array, defined as primary, secondary, tertiary, and gene enrichment, in decreasing level of significance. Because sources for these various tiers of SNPs often produced the same SNPs, we assigned each SNP uniquely to its highest tier. Some of these SNPs met QC standards of inclusion (and were directly tiled), while others did not. For those that did not, we algorithmically included neighboring SNPs to infer these high priority SNPs, as described below. For some of the highest priority SNPs, we also algorithmically included redundant coverage if possible.
There were 241 Primary SNPs. These were SNPs of highest importance, based on strongly confirmed trait or disease associations derived from the literature and other database sources, such as the NHGRI GWAS database [28] and the Human Genome Epidemiology (HuGE) Navigator [29].
The secondary group consisted of 7929 SNPs that were obtained from various literature and public database sources as suggestive of association with a disease or trait, but not yet as strongly replicated. Each SNP in the secondary set was included on the array if it produced high quality genotypes on the Axiom platform. The SNPs in this secondary group were identified from a variety of sources, including: Pharmacogenetics Knowledge Base (PharmGKB) [30] and Pharmacogenetics of Membrane Transporter (PMT) database [31] for SNPs of pharmacogenetic interest; Candidate SNPs compiled from the literature; NHGRI GWAS database for SNPs with association P-values <1e − 5 (appearing in the literature as of January, 2010); HuGE Navigator database for the most-cited candidate gene association SNPs.
The 72,324 tertiary SNPs were mined from various database sources and were based on potential functional significance. The list of sources for these SNPs included miRNA SNPs; splice site SNPs; MHC (chromosome 6p) SNPs; coding synonymous and non-synonymous SNPs; other SNPs preselected for high value from the Affymetrix commercial CEU array; and KGLP SNPs in functionally important regions.
At a similar level of importance to the tertiary set was the gene enrichment set defined as 198,771 SNPs chosen from exonic and flanking regions of genes. The flanking regions included 10Kb upstream of the first exon, 10Kb downstream of the last exon, and ± 50 bp flanking regions for all intermediate exons for 4246 genes that were confirmed or suggestive of potential disease or trait associations. The gene sources for these SNPs included the HuGE database (most often cited genes), genes of neuro-endocrine interest, HLA genes, telomere genes, genes of pharmacogenetic interest, genes of environmental interest identified from the NIEHS website, and genes identified in GWAS studies from the NHGRI database.
To screen out variants that are not polymorphic in Europeans and false positives in the tertiary and gene-enrichment sets, we required at least two heterozygotes and ≥0.01 MAF from 180 Caucasian individuals in a database generated by Affymetrix during their SNP screening for the Axiom platform. For the tertiary set, this resulted in a total of 59,063 SNPs for inclusion. For the gene-enrichment set, this resulted in a total of 107,706 SNPs.
The number of validated primary, secondary and tertiary SNPs that were directly tiled on the array was 153, 4,094 and 48,252, respectively. Our strict validation criteria were previously outlined. A few very high priority SNPs that were not validated were also tiled on the array, to allow for the possibility that they would still provide useable genotype data. We did not directly tile all the validated gene enrichment SNPs, but rather included them in the first target set for greedy algorithm SNP coverage as described below.
4.2.2. Stage 2: Additional coverage of preselected SNPs
To insure successful inference of the primary SNPs, layers of additional high QC SNPs were added. First, a single 'tagging' SNP in sufficiently high LD (r2>0.6) was included, when available. Subsequently, we greedily constructed a multiple marker “imputation tag” (r2 calculation details described in the genome-wide coverage section below) from a 100 kb window around each primary SNP, both those that could not be directly tiled and those that could, adding respective coverage and redundant coverage. SNPs increasing r2 by more than 3% were continuously added, prioritizing SNPs with better performance first, until no further SNPs could be added.
For secondary SNPs that could not be directly tiled, we assigned a single tagging SNP (with r2>0.6), if available. If no tagging SNP was available, imputations were performed and the most predictive SNPs were added as described above for primary SNPs.
No tagging or imputation SNPs were added for the tertiary or gene-enrichment SNPs. However, additional coverage was provided as the first step of the whole genome coverage stage as described below.
4.2.3. Stage 3: Redundant and genomewide coverage
SNPs selected for genome-wide coverage were obtained from several database resources, including HapMap, dbSNP, and the KGP. Genotypes from the HapMap database were included in the coverage analysis for SNP selection. Genotypes of SNPs identified by the KGP or in dbSNP, but not HapMap, were determined by screening on the Axiom™ platform in the HapMap2 CEU, CHB, JPT, and YRI populations (previous work by Affymetrix, results not shown).
An incremental greedy heuristic was used in the genome-wide coverage rounds, in which SNPs were chosen for their ability to cover one or more set of target SNPs according to multiple competing criteria. As with the preselected set, the most important coverage goal was pursued in the first selection round, followed by a descending sequence of less important goals in subsequent selection rounds.
The need to optimize across multiple criteria led to several rounds of coverage optimization. Within each round, SNPs were chosen from a set of candidate SNPs one at a time. Selection was based on multiple criteria, including: the marginal increase in coverage of the Caucasian population gained from inclusion of the candidate SNP; the observed robustness and accuracy of the assay for the candidate SNP; and the absence of nearby SNPs within the 30-mer probe sequence. These optimality criteria were set forth in a selection process, which reflected the hierarchical set of optimization priorities. At each level a configurable range of optimization measures were considered to be a tie, so that, e.g., a SNP would not necessarily be selected only for providing negligibly greater marginal coverage than a more robust alternative. If a single SNP was optimal for the criterion under consideration, with no others close enough to be tied, it was chosen. Otherwise, the subset of SNPs that were tied was tested in the same manner against the next criterion level, until a unique optimal SNP was identified. If no optimal SNP could be chosen, one was randomly selected from the remaining candidates. By controlling the size of the region of optimization measures that was considered a tie, we could investigate tradeoffs among multiple criteria.
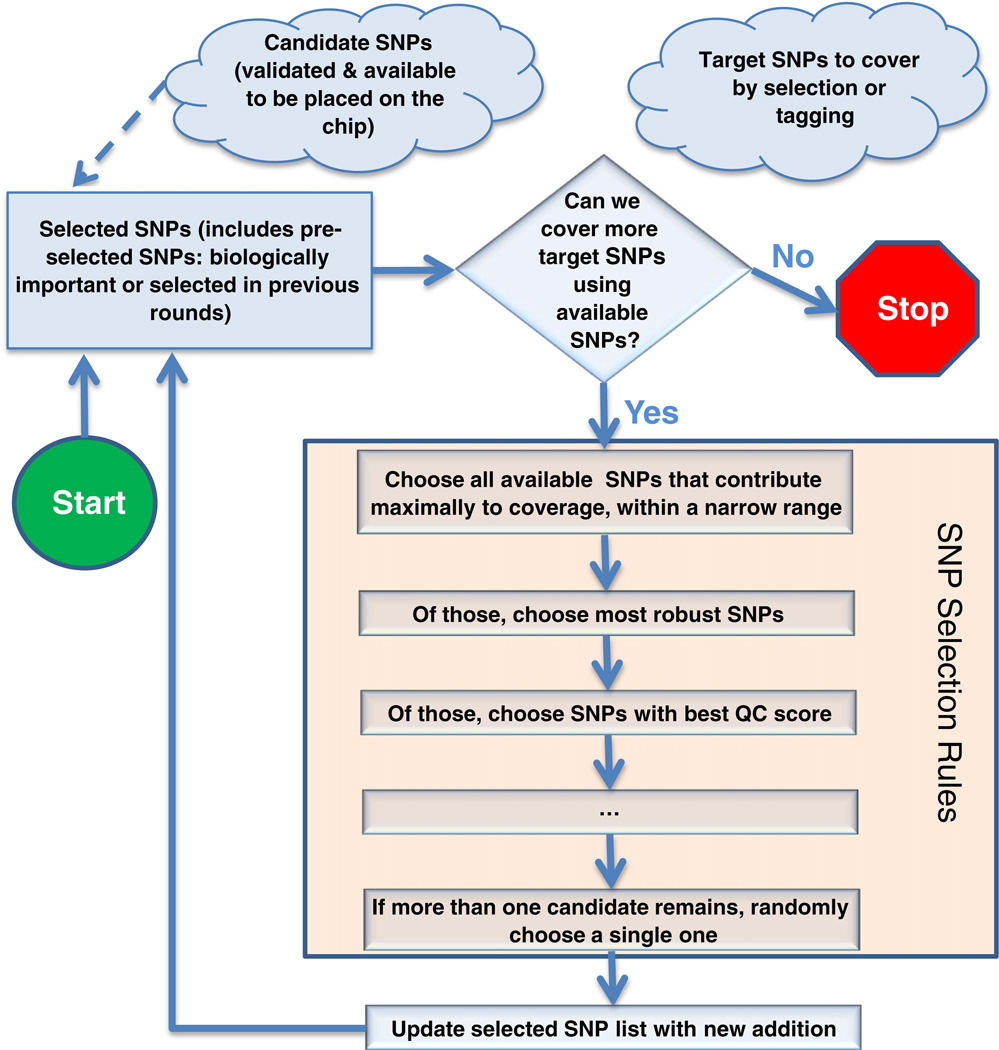
A guiding principal for SNP selection was the concept of a “target set” (SNPs to be included or covered by LD on the array), a “selected” set (SNPs chosen to be included on the array), and a “candidate” set (SNPs that passed our validation criteria for inclusion, as previously defined). The first step was to include the preselected set of SNPs described above under Stage 1 in the “selected” set. The second step was to include SNPs chosen to provide coverage or redundant coverage of the preselected set in the “selected” set. The next round of SNP selection was used to maximize the coverage across the whole genome. Here, a SNP was considered covered if it was 'tagged' by at least one selected SNP with r2 greater than 0.8. This round contributed the bulk of the array's SNPs. A final round again maximized genomic coverage, but allowed coverage with a lower r2 threshold of 0.6 to capture SNPs that could not be covered at high correlation. Fig. 7 shows a summary of the coverage algorithm in the form of a flow chart.
Fig. 7.
Greedy SNP selection algorithm. A set of SNPs are chosen for reasons of biological importance, significance in published GWAS, etc., or as the result of previous rounds of greedy SNP selection. The “target” set of SNPs to be covered by tagging is established to fit the purpose of the current round of SNP selection, e.g., maximize coverage of SNPs in coding regions, or maximize general coverage of the genome. Then, SNPs which are available to be placed on the microarray are assessed for their ability to increase coverage of the target set. If coverage can be increased, a set of decision rules is applied to select the best single SNP to add to the selected list, as described in the text. This process continues until maximum coverage of the target set is achieved, or no space for additional SNPs on the microarray remains.
4.2.4. Definition of the target set
The target set for the first round of greedy tag SNP selection (166,769 SNPs) consisted of SNPs with LD data and with MAF≥0.01 from the tertiary and the gene enrichment set of SNPs In this round, 32,066 additional SNPs were moved from the “candidate set” to the “selected set.” The second round of greedy tag SNP selection was carried out for redundant coverage for the primary, secondary, tertiary and gene enrichment SNPs. SNPs with LD data and with MAF≥0.01 from all four categories of SNPs were included in the target set (173,549 SNPs). A total of 37,776 SNPs were moved from the “candidate set” to the “selected set” in this round. For the next round of greedy tag SNP selection all SNPs in the genome with LD data and with MAF≥0.02 were included in the target set (3,505,510 SNPs), excluding SNPs already in the “selected” set. In this round, 544, 607 SNPs were added. The final round of greedy tag SNP selection used the same target set for genome-wide coverage with SNPs in or covered by the “selected” set removed from the target set; for this round, a lower threshold of r2 = 0.6 was used, and 9,294 SNPs were added. For all target sets, SNPs with only one homozygous genotype for the minor allele and no heterozygous genotypes in CEU samples were excluded.
4.3. Quality assessment for samples run on the final microarray product
The primary chip-level quality metric for Axiom™ microarrays is “DQC,” a measure of interference between the channel-to-channel signal contrast distributions of probes that complement genomic sequence with no expected polymorphism, half of which have foreground signal in one channel and half in the other. The DQC value is highly correlated with sample call rate. Loss of this correlation indicates sample or assay problems such as contamination or mixing of DNA, or technical issues with hybridization. Low DQC values likewise indicate problems such as low input DNA mass. DQC ranges from 0 to 1; we employed a minimum threshold of 0.82 for further analysis of a sample; samples with low DQC were not included in genotyping analysis. Subsequently, for a genotyped sample, the call rate was determined across all SNPs for an individual. A minimum threshold of 97% was employed to declare that a sample was successfully genotyped. After removal of failed samples, re-genotyping was performed to obtain more accurate genotypes for the remaining samples.
4.4. Quality assessment of SNPs on the final microarray product
SNP performance was assessed in the experimental work primarily on the basis of call rate and reproducibility of genotype calls in replicated samples. As described below, a single duplicate sample was included on each plate for QC purposes. Reproducibility of genotypes was then calculated based on these duplicates.
4.5. Genotyping the Kaiser Permanente Research Program on Genes, Environment and Health (RPGEH) Genetic Epidemiology Research in Adult Health and Aging (GERA) cohort
The Axiom array development we describe here derives from an NIH funded RC2 program entitled “A Resource for Genetic Epidemiology Research in Adult Health and Aging.” As part of this program, we will be performing GWAS genotyping on 100,000 members of the Kaiser Permanente Northern California (KPNC)membership who volunteered for the Research Program on Genes, Environment and Health (RPGEH). Participants provided a saliva sample in a whole-saliva collection kit (Oragene™ DNA collection kit, DNA Genotek, Inc., Ottawa, Ontario, Canada), from which DNA was extracted and normalized. Participants provided broad written informed consent for use of their DNA and resulting genetic data in health studies. The study received approval from the KPNC Institutional Review Board. This is a multi-ethnic cohort, including substantial numbers of Caucasians, African Americans, Asians and Latinos. To maximize genomic coverage for low frequency variation in each of these ethnic groups, we are creating four ethnic-specific arrays, one for each of the above mentioned ethnic groups. We are taking advantage of the high density customization enabled by the Axiom platform. In this first publication, we describe the array developed for European Americans. To characterize the performance of the newly developed array, we perform QC analysis on the first 838 plates, including ~80,000 participants of European ancestry from this cohort.
As part of the experimental design for this project, we have included duplicate samples on each plate. The plate contains space for 96 separate samples, one of which was a duplicate sample that was included on a previously processed plate. In each case, we randomly selected a sample that had been successfully genotyped on a prior plate as a duplicate for the current plate. The analysis of reproducibility is based on a total of 818 duplicates.
4.6. Calculating genome-wide coverage for the final microarray product
To assess the coverage performance of our array, we estimated the correlation between each marker in a “target” set to the markers on our array. In a GWAS analysis, typically SNPs not on the array will be imputed from SNPs that are on the array, when possible. Therefore, rather than using the maximum pairwise correlation within a fixed window around the SNP [12], we used imputation with regional markers. Normally, one would use the same set of individuals for the reference and predicted sets of genotypes in this analysis. However, leaving the same individual in both sets leads to overfitting, and hence overestimation of coverage. Therefore, we employed “leave-one-out cross validation” to compute an unbiased multiple-marker estimate of the correlation [32], as follows. For each individual in the predicted set, we removed him (her) from the reference set and used all other individuals to impute his (her) genotype via the software package Beagle v3.3.0 [33]. In this fashion, the imputed value was not influenced by an individual's actual genotype value. For each marker and individual, we used the imputed probabilities of each genotype to compute the expected value of the genotype under an additive coding (i.e., E(X)=pAa+2pAA). Then, using all individuals in the predicted set, the squared correlation r2 was calculated between the actual additive genotype value and the expected value from the imputed genotypes. For each SNP in the “target” set, the calculated r2 represents its coverage by SNPs present on the array. Finally, we computed the coverage of the full “target set” by the array for a given r2 threshold T as the proportion of the markers in the “target” set with r2>T. One thing to note is that imputation performance can be affected by the size of the reference panel (especially for lower minor allele frequencies); therefore, to facilitate comparability between coverage estimates on different sets of reference genotype data, we used 59 or 60 independent CEU HapMap individuals. For most analyses, these were the CEU founders, but in a few analyses it included a child instead of parents.
Acknowledgements
This work was supported by grant RC2 AG036607 from the National Institutes of Health, grants from the Robert Wood Johnson Foundation, the Ellison Medical Foundation, the Wayne and Gladys Valley Foundation, Kaiser Permanente, and NIH postdoctoral training grant R25 CA112355. We are grateful to the KPNC members who have generously agreed to participate in the Kaiser Permanente Research Program on Genes, Environment and Health.
Contributor Information
Thomas J. Hoffmann, Email: tjhoffm@gmail.com.
Andrea Finn, Email: andrea_finn@affymetrix.com.
Catherine Schaefer, Email: cathy.schaefer@nsmtp.kp.org.
Pui-Yan Kwok, Email: pui.kwok@ucsf.edu.
Neil Risch, Email: rischn@humgen.ucsf.edu.
References
- 1.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273(5281):1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 2.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manolio TA, Brooks LD, Collins FS. A hapmap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118(5):1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witte JS. Genome-wide association studies and beyond. Annu. Rev. Public Health. 2010;31:9–20. doi: 10.1146/annurev.publhealth.012809.103723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein DB. Common genetic variation and human traits. N. Engl. J. Med. 2009;360(17):1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 6.Kraft P, Hunter DJ. Genetic risk prediction — Are we there yet? N. Engl. J. Med. 2009;360(17):1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 7.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen HL, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ragoussis J. Genotyping technologies for genetic research. Annu. Rev. Genomics Hum. Genet. 2009;10:117–133. doi: 10.1146/annurev-genom-082908-150116. [DOI] [PubMed] [Google Scholar]
- 10.Müller-Myhsok B, Abel L. Genetic analysis of complex diseases. Science. 1997;275(5304):1328–1329. author reply 1329–30. [PubMed] [Google Scholar]
- 11.Risch N, Merikangas K. Genetic analysis of complex diseases: Reply to Muller-Myhsok and Abel. Science. 1997;275:1329–1330. [PubMed] [Google Scholar]
- 12.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74(1):106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al. A second generation human haplotype map of over 3.1 million snps. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy GC, Matsuzaki H, Dong S, Min Liu W, Huang J, Liu G, et al. Large-scale genotyping of complex dna. Nat. Biotechnol. 2003;21(10):1233–1237. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- 15.Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E, Law J, et al. Genotyping over 100,000 snps on a pair of oligonucleotide arrays. Nat. Methods. 2004;1(2):109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 16.Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, et al. Complement Factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, et al. Integrated detection and population-genetic analysis of snps and copy number variation. Nat. Genet. 2008;40(10):1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 18.Affymetrix. [Accessed March 6, 2010];Affymetrix power tools. http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx?hightlight=true&rootCategoryId=34002#1_3.
- 19.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentley D. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16(6):545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Mardis ER. Next-generation dna sequencing methods. Annu. Rev. Genomics Hum. Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 22.Smith AM, Heisler LE, Onge RPS, Farias-Hesson E, Wallace IM, Bodeau J, et al. Highly-multiplexed barcode sequencing: An efficient method for parallel analysis of pooled samples. Nucleic Acids Res. 2010;38(13):e142. doi: 10.1093/nar/gkq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.1000 Genomes Project Consortium. Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Affymetrix. Analysis methodology for the DMET Plus product, white paper. http://media.affymetrix.com/support/technical/whitepapers/dmet_plus_algorithm_whitepaperv1.pdf.
- 25.Affymetrix. [Accessed November 13, 2008];Affymetrix Canary algorithm version 1.0, white paper. http://media.affymetrix.com/support/technical/whitepapers/canary_algorithm_whitepaper.pdf.
- 26.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat. Rev. Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher R. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936;7:179–188. [Google Scholar]
- 28.Hindorff L, Junkins H, Hall P, Mehta J, Manolio T. [Accessed January 12 2010];A catalog of published genome-wide association studies. http://www.genome.gov/gwastudies.
- 29.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat. Genet. 2008;40(2):124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 30.Hewett M, Oliver DE, Rubin DL, Easton KL, Stuart JM, Altman RB, et al. Pharmgkb: The pharmacogenetics knowledge base. Nucleic Acids Res. 2002;30(1):163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroetz DL, Yee SW, Giacomini KM. The pharmacogenomics of membrane transporters project: Research at the interface of genomics and transporter pharmacology. Clin. Pharmacol. Ther. 2010;87(1):109–116. doi: 10.1038/clpt.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The International HapMap 3 Consortium. Altshuler DM, Gibbs RA, Peltonen L, Schaffner SF, Yu F, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]