Abstract
An optimal measurement of glomerular filtration rate (GFR) should minimize the number of blood draws, and reduce procedural invasiveness and the burden to study personnel and cost, without sacrificing accuracy. Equations have been proposed to calculate GFR from the slow compartment separately for adults and children. To develop a universal equation, we used 1347 GFR measurements from two diverse groups consisting of 527 men in the Multi center AIDS Cohort Study and 514 children in the Chronic Kidney Disease in Children cohort. Both studies used nearly identical two-compartment (fast and slow) protocols to measure GFR. To estimate the fast component from markers of body size and of the slow component, we used standard linear regression methods with the log-transformed fast area as the dependent variable. The fast area could be accurately estimated from body surface area by a simple parameter (6.4/body surface area) with no residual dependence on the slow area or other markers of body size. Our equation measures only the slow iohexol plasma disappearance curve with as few as two time points and was normalized to 1.73m2 body surface area. It is of the form: GFR=slowGFR/[1+0.12 (slowGFR/100)]. In a random sample utilizing a third of the patients for validation, there was excellent agreement between the calculated and measured GFR with low root mean square errors being 4.6 and 1.5ml/min per 1.73m2 for adults and children, respectively. Thus, our proposed simple equation, developed in a combined patient group with a broad range of GFRs, may be applied universally and is independent of the injected amount of iohexol.
Keywords: iohexol, glomerular filtration rate, plasma disappearance curves, renal function, kidney disease, nephrology
Introduction
Glomerular filtration rate (GFR) can be determined accurately by measuring the plasma clearance of a single intravenous injection of a contrast medium such as iohexol, calculated from plasma sampling at multiple time points over several hours [1;2]using an open 2-compartment (slope-intercept) mathematical model for the plasma disappearance curve. The protocols to measure GFR in the Multicenter AIDS Cohort Study (MACS) and Chronic Kidney Disease in Children (CKiD) study involved 4 venous blood samples after a single bolus injection of iohexol. To properly estimate the two compartments, referred to hereafter as “fast” for the first compartment, and “slow” for the second compartment, two of the four blood samples were collected within approximately 30 minutes of the injection (for the fast compartment) and the other two were obtained two or more hours after the injection (for the slow compartment). The ratio of the injected amount of iohexol to the area under the disappearance curve is a direct measure of GFR. This “gold-standard” protocol is time-intensive and requires multiple blood draws, thereby increasing complexity of large epidemiologic studies [2;3]. An optimal GFR measurement should minimize the number of blood draws, procedural invasiveness, burden to study personnel, and cost.
Restricting sampling to only the slow compartment of the model has been proposed in adults [4;5] and children[2;6]. GFR equations to quantify a universal relationship between the 1-compartment (slow) and the 2-compartment (slow + fast) plasma disappearance models have been published, based on diverse, but small, study samples (combining adults and children) representing different clinical populations (i.e., varying levels of renal function) [7;8;9]. Thus, the purpose of the current study was to develop a formula to determine the 2-compartment GFR for studies where samples in the fast compartment are not collected. To accomplish this, we used iohexol-based studies that measured both the slow disappearance curve and the fast disappearance curve in two large-scale, clinically disparate populations. Our analysis includes large populations with a broad range of GFRs and thus should provide a strong basis for a universally applicable equation.
The protocol for measuring GFR in the MACS and CKiD study used a 2-compartment, four blood sample model of plasma iohexol disappearance to calculate GFR (see Materials and Methods). Two GFR values were calculated: one GFR based on the slow compartment only (GFR0,2) and the other GFR based on both the fast and slow compartments (GFR2,2;see Variables subsection). Since there is a close relationship between GFR0,2 and GFR2,2, two approaches have been proposed to estimate GFR2,2 from GFR0,2; this estimate is denoted hereafter as . These approaches are: 1) the classical Brochner-Mortensen equation [4] that has been used typically in clinical guidelines [5] and epidemiological research [2]; and 2) a new equation based on theoretical underpinnings by Fleming [7] and Brochner-Mortensen & Jodal [8;9].
In this study, we aimed to develop a formula to estimate the fast component in GFR2,2 not measured in GFR0,2 (i.e., missing fast area). Once such a formula was developed, we derived the corresponding formula to determine GFR2,2 based on GFR0,2, and we position our proposed equation in the context of the recently improved principles [7;8;9]. The MACS and CKiD studies provided an excellent opportunity to develop a universal equation because the GFR measuring protocols were nearly identical (except for the time of the last blood sample: 240 minutes in the MACS and 300 minutes in CKiD) and included the same central biochemistry laboratory (P.I. GJS, University of Rochester Medical Center, Rochester, NY) and the same data coordinating center (co-P.I.s AM and LPJ, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD), while the two populations studied were extremely different. Agreement between and GFR2,2 within each study population, therefore, would assess the appropriateness of a truly universal equation. Such a robust equation would highlight any common dynamics between these two disparate groups and permit future iohexol GFR studies in any population to accurately determine GFR using only two plasma samples.
Results
Table 1 presents descriptive statistics of the 527 adult men from the MACS and the 514 children from the CKiD cohort who were studied. In the CKiD dataset, 63% of subjects underwent one GFR2,2 study, 36% had two studies, and 1% had three studies (with repeated studies at approximately one-year intervals), yielding a total of 820 GFR2,2 studies. The two cohorts differed substantially in most characteristics shown in Table 1. The MACS subjects were adult men with a median age of 51 years, and the CKiD subjects were children (62% male) with a median age of 11 years. HIV infection was an exclusion criterion for CKiD, but 70% of the MACS subjects were HIV-infected. In CKiD, a cohort of children with chronic kidney disease (CKD), 21% had glomerular kidney disease.
Table 1.
Descriptive statistics (percentor median [interquartile range]) of clinical and demographic characteristics of the MACS and CKiD populations at baseline.
| Characteristic | MACS (n= 527) | CKiD (n= 514) |
|---|---|---|
| Age (years) | 50.9 [46.0, 57.1] | 11.1 [7.7, 14.7] |
| Male | 100% | 62% |
| Race | ||
| White | 57% | 66% |
| Black | 35% | 23% |
| Other | 8% | 12% |
| Height (m) | 1.76 [1.71, 1.80] | 1.40 [1.20, 1.58] |
| Weight (kg) | 80.7 [72.6, 90.4] | 36.3 [23.7, 54.8] |
| BMI (kg/m2) | 26.2 [23.9, 28.8] | 18.3 [16.2, 22.0] |
| BSA (m2) | 2.00 [1.87, 2.14] | 1.19 [0.89, 1.57] |
| HIV infected | 70% | 0% |
| Serum creatinine (mg/dl) | 0.92 [0.79, 1.08] | 1.30 [1.00, 1.80] |
| GFR2,2 (ml/min|1.73m2) | 109.2[94.9, 125.2] | 44.0 [32.8, 56.0] |
BMI, body mass index; BSA, body surface area;
MACS, Multicenter AIDS Cohort Study; CKiD, Chronic Kidney Disease in Children Cohort Study
Table 2 presents the plasma disappearance parameters by study cohort and by training dataset (for model development using a 2/3 random sample) and validation dataset (for evaluation of agreement between observed and estimated GFR using the remaining 1/3 random sample). All parameters were significantly different between the cohorts, except for the injected iohexol which was approximately 3200 mg by the common protocol. As expected, randomization yielded similar parameters for the training and validation datasets for each cohort. For CKiD, these parameters indicated renal insufficiency. In contrast, most of the MACS had normal renal function: 81% of these subjects had a GFR2,2 at or above 90 ml/min|1.73m2.
Table 2.
Median [interquartile range]of plasma disappearance parameters of iohexol studies for the training and validation datasets from the MACS and CKiD populations.
| Training Dataset (2/3 random sample)
|
Validation Dataset (1/3 random sample)
|
||||
|---|---|---|---|---|---|
| MACS (n= 350) | CKID (n= 546) | p-valuea | MACS (n= 177) | CKiD (n= 274) | |
| Iohexol injection (mg) | 3180 [3151, 3243] | 3190 [3156, 3223] | 0.426 | 3185 [3127, 3223) | 3192 [3160, 3228] |
| Fast area b (mg min/ml) | 3.2 [2.6, 3.9] | 5.7 [4.0, 8.1] | <0.001 | 3.4 [2.8, 4.1] | 5.3 [3.7, 8.2] |
| Slow areac (mg min/ml) | 21.9 [18.6, 25.5] | 107.6 [73.9, 156.2] | <0.001 | 21.3 [18.7, 25.0] | 97.5 [63.8, 142.8] |
| GFR0,2d (ml/min | 1.73 m2) | 125.3 [105.5, 149.0] | 45.3 [33.4, 59.2] | <0.001 | 128.3 [110.0, 150.1] | 47.7 [36.2, 63.6] |
| GFR2,2e (ml/min | 1.73 m2) | 108.6 [93.8, 126.2] | 42.9 [32.0, 54.8] | <0.001 | 112.1 [96.3, 124.9] | 45.4 [34.6, 58.6] |
MACS, Multicenter Aids Cohort Study; CKiD, Chronic Kidney Disease in Children Cohort Study
Wilcoxon rank-sum test comparing MACS and CKiD in training dataset, bold indicates statistically significant (p< 0.05).
Based on plasma samples at 10 and 30 minutes after iohexol injection
Based on plasma samples at 120 and 240 for MACS and 120 and 300 for CKiD minutes after iohexol injection
(iohexol injection/ slow area) × (1.73 / BSA)
(iohexol injection/ total area) × (1.73 / BSA)
Predictors of the fast area
The left side of Table 3 shows the regression coefficients and R2 for the univariate regressions with the dependent variable being the fast area in the log scale. All variables showed substantial associations with the fast area. In particular, the fast area was inversely proportional to body surface area (BSA; i.e., coefficient of BSA was close to, and not statistically different from, −1). Furthermore, there was a strong positive relationship between the slow and fast areas, with the slow area explaining close to 34% of the variability in fast area. Given the strong relationship between the fast area and BSA, we explored the predictive power of variables in Table 3 on the variability of the fast area unexplained by BSA. The right side of Table 3 shows that none of the predictors explain the variability of residuals of the regression of fast area on BSA. In particular, the strong univariate association between slow and fast area completely disappeared when the dependent variable was BSA-adjusted fast area (regression coefficient= −0.018 (p= 0.194), R2= 0.2%). Further evidence of the lack of association between the fast and slow area conditional on BSA is the fact that the R2= 34% observed in the overall univariate relationship between the fast and slow area reduced to 0.6%, 0.1%, 0.7% and 1.6% in four strata defined by quartiles of BSA.
Table 3.
Univariate linear regression models used to determine the percent of explained variance of BSA-unadjusted and BSA-adjusted fast area for the combined MACS (n=350) and CKiD (n=546) training dataset.
| Dependent Variable
|
||||
|---|---|---|---|---|
| BSA-unadjusted fast areab |
BSA-adjusted fast areac |
|||
| Independent Variablea | Regression coefficient | R2 | Regression coefficient | R2 |
| BSA (m2) | −1.023 | 55.9% | -- | -- |
| Slow Area (mg min/ml) | 0.359 | 33.6% | −0.018 | 0.2% |
| GFR0,2 (ml/min/1.73m2) | −0.315 | 12.2% | 0.041 | 0.5% |
| Height (m) | −2.075 | 56.4% | −0.082 | 0.2% |
| Weight (kg) | −0.670 | 54.6% | −0.006 | <0.1% |
| Body Mass Index (kg/m2) | −1.187 | 32.2% | 0.128 | 0.8% |
| Age (years) | −0.430 | 49.9% | −0.022 | 0.3% |
| Male Gender | 0.335 | 6.6% | 0.032 | 0.1% |
All variables except gender in natural logarithmic scale.
Dependent variable is the natural log of the observed fast area.
Dependent variable is the residuals of the regression of fast area on BSA (log scale).
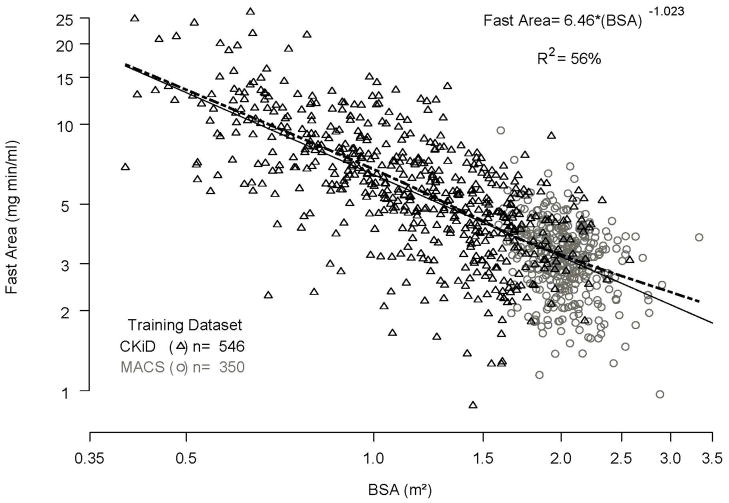
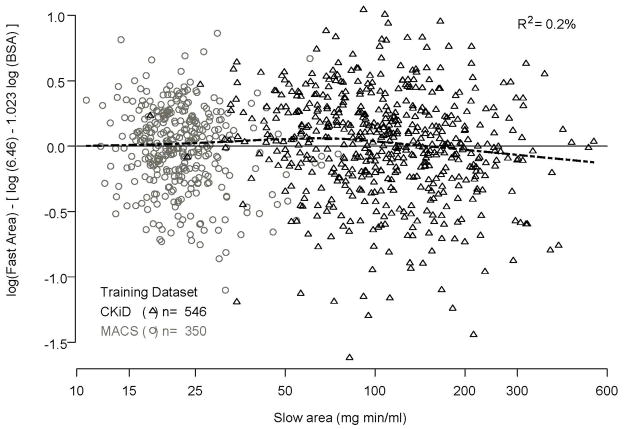
Figure 1a shows that the relationship between fast area and BSA (R2= 56%) is given by fast area = 6.46 × (BSA)−1.023. More importantly, Figure 1b shows that once the variability of fast area due to BSA has been accounted for, there is no additional information from the slow area (R2= 0.2%). Hence, the fast area depends on BSA, but not on the slow area once BSA has been taken into account.
Figure 1.
Figure 1a. Relationships between fast area, BSA, and slow area. Regression of fast area (y axis) on body surface area (x axis) in the log scale for the combined MACS and CKiD training dataset. The dashed line represents the nonparametric spline.
Figure 1b. Non-relationship of residuals from the regression of fast area on body surface area presented in Figure 1A (y axis) and slow area (x axis) from the training dataset.
Formula to determine GFR2,2 from GFR0,2
Not only does the fast area not depend on the slow area once BSA has been taken into account, but the relationship between the fast area and BSA can be simplified to
and we can derive an equation to determine GFR2,2 in terms of GFR0,2. Specifically, by simply dividing both sides of the above equation by the slow area and multiplying and dividing the right hand side of the equation by 1.73 × I (where I is the injected amount of iohexol), the equation becomes
Since by protocol, I is close to 3200 and has very low variability (see Table 2), 6.4/(1.73x I) is equal to 0.00116 (which hereafter we round to 0.0012). In addition, the ratio of fast to slow area is simply (GFR0,2/GFR2,2) −1.
Hence,
which, solving for GFR2,2 yields our proposed equation for from GFR0,2 as follows:
Or, in short,
This equation is of the same form as the one proposed by Fleming [7] and simpler than the one proposed by Brochner-Mortensen and Jodal [9]. More importantly, the form of the equation is consonant with the theoretical and physiological considerations put forward by these authors.
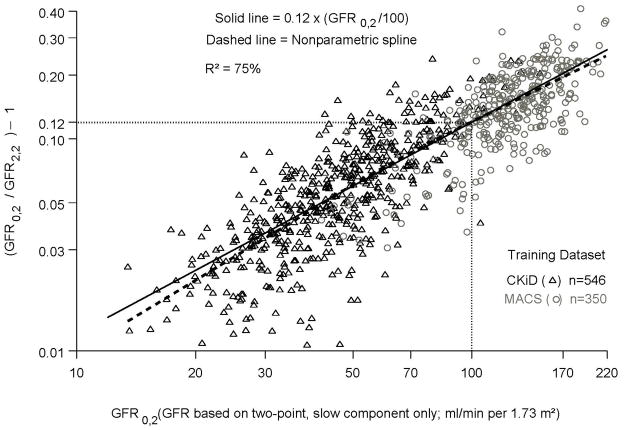
Figure 2 plots the data for (GFR0,2/GFR2,2) −1 (y axis) on GFR0,2 (x axis) and it indicates that the simple proportionality factor of 0.12 in the equation not only fits the data well for both cohorts but also explains 75% of the variability for the combined training dataset.
Figure 2.
Relationship between GFR0,2 (x axis) and (GFR0,2/GFR2,2) −1 (y axis) in the log scale, for the combined MACS and CKiD training dataset. The equation, (GFR0,2/GFR2,2) − 1= 0.12 x (GFR0,2/100), is depicted as the solid line.
Evaluation of equations to calculate and agreement between and GFR2,2
For the validation datasets of each study population (1/3 random sample), Tables 4a and 4b describe the agreement of GFR2,2 (measured) and the (estimated) from four different GFR0,2- based equations. The equations evaluated included the original Brochner-Mortensen equations for adult [4] and pediatric [6] populations; those published by Brochner-Mortensen & Jodal [8] and Fleming [7]; and our proposed equation.
Table 4.
Tables 4a and 4b. Agreement of the estimated GFR ( ) based on two-point GFR from the slow curve only (GFR0,2) and selected models with the observed four-point GFR (GFR2,2) in the MACS (n= 177) and CKiD (n= 274) validation datasets.
| Table 4a
| ||||||
|---|---|---|---|---|---|---|
| MACS Validation Dataset (n= 177) | ||||||
| Model of as a function of GFR0,2 and BSA | Source | Bias Ratio (95% CI) | Ratio of SDsa (95% CI) | Correlation (95% CI) | RMSEb | % of within 5% of GFR2,2 |
| Original Brochner-Mortensen C1 (GFR0,2/100) + C2 (GFR0,2/100)2 C1 = 98.31; C2 = −0.1218 |
Brochner- Mortensen (1972)[4] | 0.967 (0.961, 0.973) | 0.937 (0.909, 0.965) | 0.978 (0.971, 0.984) | 6.49 | 62.7% |
| GFR0,2 / [1+ B0 × (GFR0,2/100) B1 × BSAB2] B0 = 0.185; B1 = 1, B2 = −0.3 |
Brochner- Mortensen & Jodal (2009) [8,9] | 0.972 (0.967, 0.978) | 0.960 (0.934, 0.986) | 0.979 (0.972, 0.984) | 5.80 | 66.1% |
| GFR0,2 / [1+B0 × (GFR0,2/100) B1] B0 = 0.17; B1 = 1 |
Fleming (2007) [7] | 0.953 (0.947, 0.959) | 0.941 (0.916, 0.967) | 0.979 (0.972, 0.984) | 7.49 | 49.7% |
| MACS & CKiD Proposed Equation GFR0,2 / [1+B0 × (GFR0,2/100)B1] B0 = 0.12; B1 = 1 |
Training Set CKiD + MACS 2/3 random sample (n=896) | 1.005 (1.000, 1.011) | 0.987 (0.962, 1.013) | 0.980 (0.972, 0.985) | 4.59 | 79.1% |
| Table 4b
| ||||||
|---|---|---|---|---|---|---|
| CKiD Validation Dataset (n= 274) | ||||||
| Model of as a function of GFR0,2 and BSA | Source | Bias Ratio (95% CI) | Ratio of SDsa (95% CI) | Correlation (95% CI) | RMSEb | % of within 5% of GFR2,2 |
| Original Brochner-Mortensen C1 (GFR0,2/100) + C2 (GFR0,2/100)2 C1 = 101.0; C2 = −0.17 |
Brochner- Mortensen (1974)[6] | 0.983 (0.980, 0.986) | 0.968 (0.959, 0.976) | 0.996 (0.995, 0.997) | 2.04 | 91.2% |
| GFR0,2 / [1+ B0 × (GFR0,2/100)B1 × BSAB2] B0 = 0.185; B1 = 1, B2 = −0.3 |
Brochner- Mortensen & Jodal (2009) [8,9] | 0.978 (0.975, 0.981) | 0.984 (0.976, 0.992) | 0.996 (0.995, 0.997) | 1.92 | 86.5% |
| GFR0,2 / [1+B0 × (GFR0,2/100)B1] B0 = 0.17; B1 = 1 |
Fleming (2007) [7] | 0.980 (0.978, 0.983) | 0.983 (0.974, 0.991) | 0.996 (0.995, 0.997) | 1.87 | 90.9% |
| MACS & CKiD Proposed Equation GFR0,2 / [1+B0 × (GFR0,2/100) B1] B0 = 0.12; B1 = 1 |
Training Set CKiD + MACS 2/3 random sample (n=896) | 1.004 (1.001, 1.006) | 1.005 (0.996, 1.013) | 0.996 (0.995, 0.997) | 1.46 | 96.0% |
Bold indicates statistically significantly different from 1 (i.e., p < 0.05);
SDs, standard deviations
RMSE, root mean square error
The original Brochner-Mortensen equations showed significant underestimation of GFR2,2 in the MACS (Bias: −3.3%, 95% Confidence Interval: −3.9%, −2.7%) and in the CKiD (−1.7%, 95%CI:−2.0%, −1.4%). The equations also produced significant under-dispersion in both populations and higher RMSEs and lower percentages of within 5% of GFR2,2 than those of our proposed equation. Although these estimates were statistically significant, the effect size was relatively small. The equations proposed by Brochner-Mortensen & Jodal [9] and Fleming [7] each yielded a slight systematic underestimation and under-dispersion in both populations. Our proposed equation, based upon the MACS and CKiD, showed good agreement in each validation dataset. In the MACS dataset, there was no significant bias or difference in dispersion associated with compared to GFR2,2. In the CKiD dataset, there was a minor, but significant, over estimation (Bias: +0.4%, 95%CI: 0.1%, 0.6%) and no significant over-dispersion (Ratio of SDs: +0.5%, 95%CI: −0.2%, 1.3%), but these effects were small (≤ 1%). The proposed equation also provided the lowest root mean square error (RMSE) and the highest % of being within 5% of GFR2,2 of the equations tested. The correlations between all four equations were high and essentially the same in both the MACS (r= 0.980) and CKiD (r= 0.996) validation datasets.
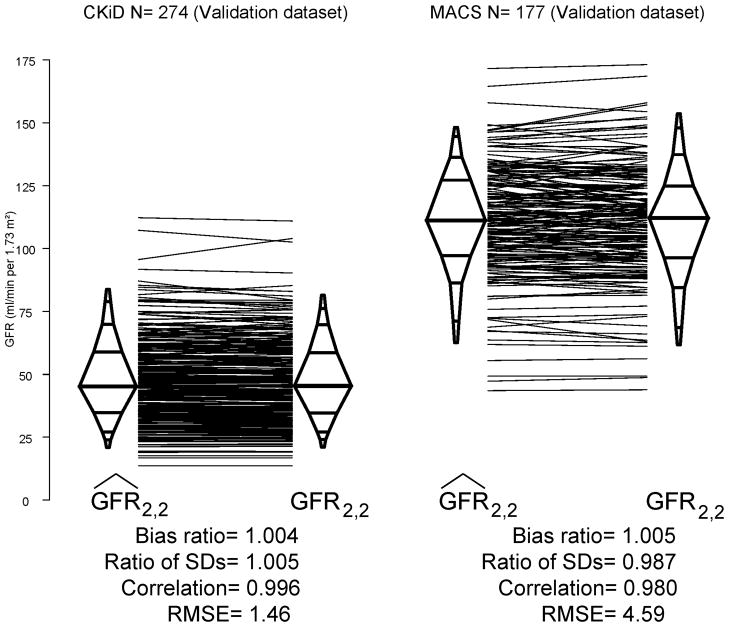
Figure 3 depicts agreement of calculated using the MACS & CKiD proposed equation with GFR2,2. The values were essentially identical to GFR2,2 with minimal bias, a preservation of dispersion, low RMSE and very high correlation in these two distinct populations.
Figure 3.
Comparison of based on GFR0,2 and proposed equation with four-point GFR2,2 in the validation datasets for MACS (n=177) and CKiD (n=274). Percentile (2.5th, 5th, 10th, 25th, 50th, 75th, 90th, 95th, and 97.5th) box plots showing a high agreement within each study validation dataset.
Discussion
Current guidelines provide separate equations to calculate for adults and children, but the present analysis shows that a universal equation is applicable indiverse populations. The literature often cautions that equations to calculate are appropriate only in populations similar to those in which they were developed. In the present study, two large studies of different populations, demographically diverse and with a broad range of renal function, underwent essentially the same iohexol GFR2,2 measurement protocols; this allowed us to develop an equation suitable for both populations. Crucial findings for the proposed universal equation were that the fast area was inversely proportional to BSA and that the BSA-adjusted fast area was independent of the slow area. Thus, BSA was a key consideration when evaluating the common dynamic between these two populations, and must routinely be accounted for when measuring GFR.
The simple relationship between the fast area and BSA allowed the derivation of a simple equation to calculate from GFR0,2, as measured by an iohexol-based GFR protocol. To confirm this derivation, a direct regression analysis was performed on the data in Figure 2. Indeed, the slope (= 1.025) was not significantly different from 1 (95%CI: 0.987, 1.063), and the intercept (i.e., at GFR0,2= 100 ml/min|1.73m2) was equal to 0.117. Our proposed equation provides an accurate and reliable GFR measurement from as few as two blood samples collected between 120 minutes and 300 minutes after injection of iohexol. This modification of the iohexol-based protocol represents an important reduction in study burden for subjects and personnel, and should facilitate GFR measurements in large-scale clinical and epidemiological studies.
Our analysis assessed the agreement between GFR2,2 and of our proposed equation as well as of previously published equations [4;8;9;7]. While the previously published equations showed good agreement, our proposed equation was even better: RMSEswere4.59 and 1.46 ml/min|1.73m2 for the MACS and CKiD validation datasets, respectively. Furthermore, our proposed equation had the highest proportion of within 5% of the GFR2,2 (79% for MACS and 96% of CKiD).
Importantly, the equation is consistent in form with the equations proposed by Brochner-Mortensen & Jodal [8,9] and Fleming [7] and coheres with the theoretical principles discussed by these authors. Specifically, using the general expression:
the Brochner-Mortensen & Jodal [8;9]equation corresponds to B0= 0.185, B1= 1 and B2= −0.3; the Fleming [7] equation corresponds to B0 = 0.17, B1 = 1 and B2 = 0; and our proposed equation corresponds to B0= 0.12, B1= 1 and B2= 0.
Another feature of our proposed equation is that it is invariant with respect to the injected amount of iohexol. Indeed, the relationship describing GFR0,2 and the ratio of GFR0,2 to GFR2,2 in Figure 2 is not dependent on the dose of iohexol and the equation with0.12 provided an excellent fit to the data. In contrast, the constant of 6.4 relating fast area to BSA is directly proportional to the dose of iohexol and in general, if fast area = c / BSA, c will be equal to 0.00116 × 1.73 × I, where c= 6.4 for I= 3200. Thus, our proposed equation may be applicable to other iohexol protocols (with variations in amounts of iohexol injection), while the estimation of fast area by 6.4/BSA is specific to this protocol (i.e., when I=3200 mg).
The difference between the Fleming equation and our equation was meaningful: the Fleming equation had relatively high RMSE in our study populations and it systematically underestimated GFR2,2, although this underestimation was modest (about 2% in the CKiD and 5% in the MACS). This slight discrepancy may be due to differences in the exogenous clearance markers used (i.e., iohexol versus 51Cr-EDTA); however, previous studies have shown comparable GFR measurement performance with either marker [10;11;12]. It is also possible that this discrepancy may be due to other factors altogether (i.e., study design or model development).
The relationship between fast area and BSA determined in Table 3 and depicted in Figure 1a (R2= 56%) may be re-expressed to form the basis of our proposed equation whose fit to the data is depicted in Figure 2 (R2= 75%). Given that the fast area depends solely on BSA, we compared our proposed equation with the agreement in an equation that directly imputes the fast area as inversely proportional to BSA:
This equation yielded RMSEs of 5.81 and 1.48 ml/min|1.73m2 in the MACS and CKiD, respectively; which, for the MACS, was 27% higher than the RMSE of 4.59 of our proposed equation ( as a function of GFR0,2). Since the main assumption from the analysis is that the fast area depends only on BSA, the proposed equation does not take into account the between-subject variability in the fast area among those with the same BSA. Despite this limitation, the assumption works well because the fast area does not contribute much to the overall GFR2,2.
As a secondary analysis, we also investigated how a previously published Brochner-Mortensen-like equation that was developed exclusively in the CKiD population [2] performed when applied separately to the validation datasets in the two studies. In the CKiD validation dataset, there was no significant bias or difference in dispersion, and a very high correlation (r= 0.998, 95%CI: 0.995, 0.997) between and GFR2,2. This was expected since the equation was derived in the same clinical population. However, when the equation was applied to the MACS validation dataset, the measures of agreement were much poorer: there was significant underestimation (bias= −2.6%, p< 0.01), shrinking of dispersion (ratio of SDs= 0.932, p< 0.01) and lower correlation (r= 0.978, 95%CI: 0.971, 0.984) than had been observed in the CKiD validation dataset. This finding highlights a limitation of developing an equation within a specific population and then applying it to a different one. As such, a limitation of our study is the lack of adult female subjects studied, with whose data we could have assessed the validity of the proposed equation. Nevertheless, ours is the only study that has developed an accurate iohexol equation using two disparate, large-scale cohorts (total n= 1347) that underwent essentially identical GFR protocols.
Although the fast area can be estimated from the body surface area, it should be noted that the intercept and slope of the fast curve cannot be determined with our method. Determination of these parameters (needed for measures like the extracellular volume [13]) would require collecting samples within 60 minutes of iohexol injection. Another limitation to our method is that our data did not take into account obese or edematous subjects, populations for which it is unclear whether our approach will work. However, in spite of the substantial differences in weight and BMI between the two study populations (i.e., MACS men with a median BMI of 26 kg/m2 versus CKiD children being overweight relative to their height), the analysis showed excellent agreement in each population.
In conclusion, due to the fast area being dependent on BSA but not on the slow area, determination of GFR using only the slow component showed a remarkable consistency between the MACS and CKiD populations. A simple equation was derived from a training dataset that included both cohorts and showed excellent agreement when applied to validation sets from both studies. Using this validated equation, GFR can be accurately measured across populations with diverse demographics and renal function using only the slow iohexol plasma disappearance curve with as few as two time points.
MATERIALS AND METHODS
Variables
GFRx,y denotes the glomerular filtration rate calculated from disappearance curves of iohexol-based studies. The subscript x,y refers to the number of time points sampled in the first (fast) and second (slow) compartments, respectively. Specifically, GFR2,2 denotes the GFR derived from sampling times of 10and30minutes (i.e., two time points in the fast compartment), and 120 and 240 (or 300) minutes (i.e., two time points in the slow compartment)after iohexol injection; and GFR0,2 denotes the GFR based only on the slow compartment. The area under the curve of the second compartment is derived from the plasma iohexol concentrations at 120 and 240 minutes for the MACS and 120 and 300 minutes for the CKiD, and is referred to as the slow area. Likewise, based on concentrations at 10 and 30 minutes after injection, we derived the disappearance curve of the first compartment, whose area under the curve is hereafter referred as the fast area. GFR2,2 was calculated as the ratio of the amount of iohexol injected (I) to the total area under the iohexol disappearance curve and calibrated to a BSA of 1.73m2. That is, GFR2,2= (I/(fast area + slow area)) × (1.73/BSA). If the fast area in the above expression is omitted, it yields GFR0,2= (I /slow area) (1.73/BSA) which always overestimates GFR2,2.
(with circumflex) denotes the estimate of GFR2,2 from an equation based on GFR0,2 in studies where the two time points in the fast compartment are not taken into account. A central aim of this study was to determine the agreement between and GFR2,2.
Study Participants
The MACS is a prospective observational cohort study of the natural and treated histories of HIV infection among homosexual/bisexual men over the age of18 in foursites in the US. To investigate the natural history of renal function as it relates to HIV infection and use of highly active antiretroviral therapy, a sub-group of the cohort (n=565) participated in an iohexol-based GFR measurement between August 2008and May 2010. Of these, 527 (94%) had a successful GFR2,2 study; we excluded individual patient studies if there were problems with any plasma concentration measurements (n= 32), if an incorrect amount of iohexol was administered (n= 3) or if the study indicated an extremely high GFR2,2 (i.e., GFR2,2 > 180 ml/min|1.73m2; n= 3).
The CKiD study enrolled 586 children between 1 and 16 years of age with mild to moderate CKD in 47sites in the US and Canada. Enrollment criteria included an estimated GFR (eGFR) range of 30 to 90 ml/min|1.73 m2, based on the original Schwartz formula [14]. From these subjects, there were 915 subject-visits at which an iohexol injection for a GFR2,2 protocol occurred. For our analysis, 514 subjects (88%of 586) contributed a total of 820 GFR2,2 studies (90%); we excluded studies if there were problems with plasma concentration measurements or blood sampling time values for any of the 4 samples (n= 38), the wrong amount of iohexol was administered (<2000 mg or >4500; n= 5), if the GFR2,2 was significantly different (>50%) from the corresponding eGFR [15] (n= 45), or if the BSA calculation was based on only one measurement of height and weight instead of the average of 3 measurements (n= 7). BSA for both populations was calculated from the Haycock et al. [16] formula.
The two studies used nearly identical iohexol GFR protocols: the only difference was that the last blood sample after injection of iohexol was collected at 240 minutes in the MACS and at 300 minutes in the CKiD. This difference in the protocol was justified by the expectation of relatively normal kidney function among the MACS men.
Both the MACS and CKiD studies were approved by Research Review Boards at all participating sites in the US and Canada.
Statistical analysis
To overcome the limitation of studies that do not measure concentrations of iohexol at time points in the fast compartment, formulas to predict the fast area are needed. We used standard linear regression methods with the log of the fast area as the dependent variable and we examined the predictive power of the following independent variables: BSA, slow area, GFR0,2, height, weight, body mass index, age and male gender. The prediction formulas can be used to directly impute the missing fast area and also to derive formulas to estimate GFR2,2 as a function of GFR0,2, as this is the form that numerous investigators have previously used[7;8;9]. In order to develop a formula to predict the fast area, we randomly selected a 2/3 sample of each cohort and combined these two subgroups into a training dataset in which the models were developed. Once the predicted formula for the fast area was developed, and in order to independently test the agreement between GFR2,2 and we applied the equation for to the remaining 1/3 random sample from each study (validation dataset).
In all regression analyses, a generalized estimating equation (GEE) was used to account for correlation between repeated measurements and to obtain correct standard errors[17]. Results were unchanged when we performed the same analysis using only the first GFR2,2 study contributed by each CKiD subject (n=514; training dataset n= 343, validation dataset n= 171). To maximize the use of all available GFR studies, the results from the GEE analyses using all the studies are reported.
Agreement between and GFR2,2
Agreement was assessed in the validation dataset using methods described by Bland and Altman [18]and extended by regressing the difference on the average of the two measurements centered around the overall mean[1]. Absence of bias corresponds to the intercept of the regression being equal to zero and equal dispersion corresponds to the slope being equal to zero[1]. Robust methods were used to account for repeated measurements. 95% confidence intervals for correlations were determined by Fisher’s method. Agreement plots were used to visually depict each data point and the box-percentile plots for and GFR2,2 in each cohort[19]. To compare with other published equations, in each MACS and CKiD dataset, was calculated from the published population-specific Brochner-Mortensen-equations [4,6], the equations by Brochner-Mortensen & Jodal [9] and Fleming [7], and our proposed equation derived from the training dataset. The calculated values based on these four equations were then compared with the observed GFR2,2 based on both fast and slow compartments in the validation dataset.
Acknowledgments
Data in this article were collected by the Chronic Kidney Disease in Children prospective cohort study (CKiD) with clinical coordinating centers (principal investigators) at Children’s Mercy Hospital and the University of Missouri—Kansas City (Bradley Warady), The Children’s Hospital of Philadelphia (Susan Furth), data coordinating center at the Johns Hopkins Bloomberg School of Public Health (Alvaro Muñoz), and the Central Biochemistry Laboratory at the University of Rochester (George J. Schwartz). The CKiD is funded by the National Institute of Diabetes and Digestive and Kidney Diseases, with additional funding from the National Institute of Neurological Disorders and Stroke, the National Institute of Child Health and Human Development, and the National Heart, Lung, and Blood Institute (U01 DK82194, U01-DK-66143, U01-DK-66174, and U01-DK-66116). The CKID website is located at http://www.statepi.jhsph.edu/ckid.
Data in this manuscript were also collected by the Multicenter AIDS Cohort Study (MACS) with centers (Principal Investigators) at The Johns Hopkins Bloomberg School of Public Health (Joseph B. Margolick, Lisa P. Jacobson), Howard Brown Health Center, Feinberg School of Medicine, Northwestern University, and Cook County Bureau of Health Services (John P. Phair, Steven M. Wolinsky), University of California, Los Angeles (Roger Detels), and University of Pittsburgh (Charles R. Rinaldo) and the Central Biochemistry Laboratory at the University of Rochester (George J. Schwartz). The MACS is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute. UO1-AI-35042, UL1-RR025005 (GCRC), UO1-AI-35043, UO1-AI-35039, UO1-AI-35040, UO1-AI-35041. Website located at http://www.statepi.jhsph.edu/macs/macs.html. We are grateful to GE Healthcare, Amersham Division, for providing the CKiD and MACS studies with iohexol (Omnipaque) for the GFR measurements.
Footnotes
Disclosure: No interests to disclose.
References
- 1.Schwartz GJ, Furth SL, Cole SR, et al. Glomerular filtration rate via plasma iohexol disappearance: Pilot study for chronic kidney disease in children. Kidney Int. 2006;69:2070–2077. doi: 10.1038/sj.ki.5000385. [DOI] [PubMed] [Google Scholar]
- 2.Schwartz GJ, Abraham AG, Furth SL, et al. Optimizing iohexol plasma disappearance curves to measure the glomerular filtration rate in children with chronic kidney disease. Kidney Int. 2010;77 :65–71. doi: 10.1038/ki.2009.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Work DF, Schwartz GJ. Estimating and measuring glomerular filtration rate in children. Current Opinion in Nephrology and Hypertension. 2008;17:320–325. doi: 10.1097/MNH.0b013e3282fb77f2. [DOI] [PubMed] [Google Scholar]
- 4.Brochner-Mortensen JA. A simple method for the determination of glomerular filtration rate. Scand J Clin Lab Invest. 1972;30:271–274. doi: 10.3109/00365517209084290. [DOI] [PubMed] [Google Scholar]
- 5.Fleming JS, Zivanovic MA, Blake GM, et al. Guidelines for the measurement of glomerular filtration rate using plasma sampling. Nuclear Medicine Communications. 2004;25:759–769. doi: 10.1097/01.mnm.0000136715.71820.4a. [DOI] [PubMed] [Google Scholar]
- 6.Brochner-Mortensen JA, Haahr J, Christoffersen J. A simple method for accurate assessment of the glomerular filtration rate in children. Scand J Clin Lab Invest. 1974;33:139–143. [PubMed] [Google Scholar]
- 7.Fleming JS. An improved equation for correcting slope-intercept measurements of glomerular filtration rate for the single exponential approximation. Nuclear Medicine Communications. 2007;28:315–320. doi: 10.1097/MNM.0b013e328014a14a. [DOI] [PubMed] [Google Scholar]
- 8.Jodal L, Brochner-Mortensen J. Reassessment of a classical single injection n51Cr-EDTA clearance method for determination of renal function in children and adults. Part I: Analytically correct relationship between total and one-pool clearance. Scand J Clin Lab Invest. 2009;69:305–313. doi: 10.1080/00365510802566882. [DOI] [PubMed] [Google Scholar]
- 9.Brochner-Mortensen J, Jodal L. Reassessment of a classical single injection n51Cr-EDTA clearance method for determination of renal function in children and adults. Part II: Empirically determined relationships between total and one-pool clearance. Scand J Clin Lab Invest. 2009;69 :314–322. doi: 10.1080/00365510802653680. [DOI] [PubMed] [Google Scholar]
- 10.Rydstrom M, Tengstrom B, Cederquist I, et al. Measurement of glomerular filtration rate by single-injection, single-sample techniques, using 51Cr-EDTA or iohexol. Scand J Urol Nephrol. 1995;29:135–139. doi: 10.3109/00365599509180553. [DOI] [PubMed] [Google Scholar]
- 11.Brandstrom E, Grzegorczyk A, Jacobsson L, et al. GFR measurement with iohexol and 51Cr-EDTA. A comparison of the two favoured GFR markers in Europe. Nephrol Dial Transplant. 1998;13:1176–1182. doi: 10.1093/ndt/13.5.1176. [DOI] [PubMed] [Google Scholar]
- 12.Bird NJ, Peters C, Michell AR, et al. Comparison of GFR measurements assessed from single versus multiple samples. Am J Kid Dis. 2009;54:278–288. doi: 10.1053/j.ajkd.2009.03.026. [DOI] [PubMed] [Google Scholar]
- 13.Abraham AG, Muñoz A, Furth SL, et al. Extracellular volume and disease progression in children with chronic kidney disease. Clin J Am Soc Nephrol. 2011;6:741–747. doi: 10.2215/CJN.08020910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz GJ, Haycock GB, Edelmann CM, Jr, et al. A simple estimate of glomerular filtration rate in children derived from body length and plasma creatinine. Pediatrics. 1976;58:259–263. [PubMed] [Google Scholar]
- 15.Schwartz GJ, Muñoz A, Schneider MF, et al. New equations to estimate GFR in children with CKD. J Am Soc Nephrol. 2009;20:629–637. doi: 10.1681/ASN.2008030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haycock GB, Schwartz GJ, Wisotsky DH. Geometric method for measuring body surface area: A height-weight formula validated in infants, children, and adults. J Pediatr. 1978;93:62–66. doi: 10.1016/s0022-3476(78)80601-5. [DOI] [PubMed] [Google Scholar]
- 17.Liang KY, Zeger SL. Longitudinal Data Analysis Using Generalized Linear Models. Biometrika. 1986;73:13–22. [Google Scholar]
- 18.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 19.Esty W, Banfield J. The box-percentile plot. J Stat Software. 2008;8:1–14. [Google Scholar]