Abstract
In the current study, we have identified Nε-thiocarbamoyl-lysine (TuAcK) as a general sirtuin inhibitory warhead which was shown to be able to confer potent sirtuin inhibition. This inhibition was also shown to be mechanism-based in that the TuAck residue was able to be processed by a sirtuin enzyme with the formation of a stalled S-alkylamidate intermediate.
Keywords: sirtuin, deacetylase, mechanism-based, inhibitor, Nε-thiocarbamoyl-lysine
Sirtuin enzymes are a family of cellular proteins able to catalyze the acetyl group removal from the specific Nε-acetyl-lysine (AcK, Fig. 1) side chains on both histone and non-histone proteins.1–7 These enzymes have been found in all the living organisms from bacteria to humans, with the yeast silent information regulator (Sir2) as the founding family member whose protein lysine deacetylase activity was discovered about one decade ago.3,8–12 The available research indicated that the lower prokaryotic species have less sirtuin isoforms than the higher eukaryotic species such as humans that have 7 identified sirtuin isoforms (SIRT1–7). As illustrated in Figure 1, this enzymatic acetyl group removal (or deacetylation) reaction employs β-nicotinamide adenine dinucleotide (β-NAD+ or NAD+) as its cosubstrate. Because of this use of NAD+, in addition to regulating cellular processes such as gene transcription via the functional modulation of histone proteins and various transcription factors and coregulators, the sirtuin-catalyzed lysine deacetylation reaction has also been shown to be important in regulating metabolism via the functional modulation of multiple metabolic enzymes in response to the cellular [NAD+]/[NADH] ratio fluctuations.11,13–20 The studies performed in the past a few years have also started revealing the therapeutic benefits of the chemical modulation of this enzymatic deacetylation reaction for such disease states as diabetes, cancer, and neurodegenerative diseases.11,13,21–34 In order to achieve a fuller exploration of the sirtuin pharmacology as well as biology, there is still a need for a continued search for novel chemical modulators for the sirtuin-catalyzed protein lysine deacetylation reaction that was discovered not long ago, yet has already revealed its (path)physiological prominence.
Figure 1.
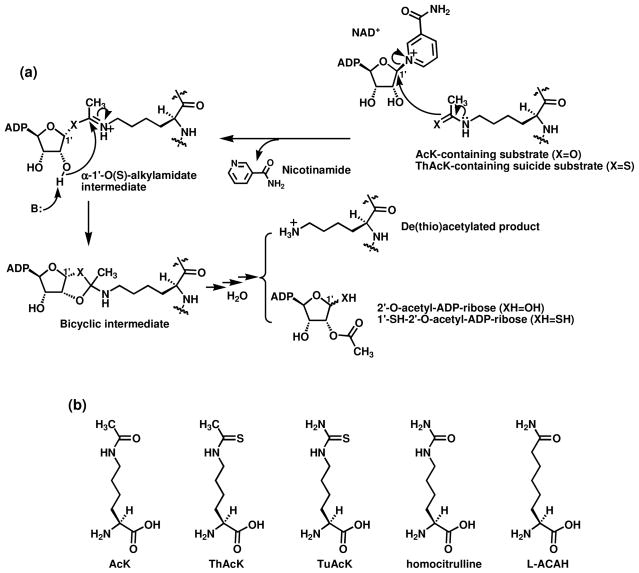
(a) The proposed chemical mechanism for the sirtuin-catalyzed lysine Nε de(thio)acetylation reaction. When a ThAcK-containing suicide substrate (or mechanism-based inhibitor) (X = S) is used, the corresponding α-1′-S-alkylamidate intermediate is stalled along the reaction coordinate. ADP, adenosine diphosphate; B: refers to a general base. (b) Structural comparison of Nε-acetyl-lysine (AcK) and its analogs.
Because the AcK residue could be regarded as the “minimal” substrate for the sirtuin-catalyzed lysine deacetylation reaction, our laboratory has been interested in identifying AcK analogs as the inhibitory warheads for this enzymatic deacetylation reaction.35–39 Previously, we found that very potent sirtuin inhibition could be obtained by a series of peptidic and peptidomimetic compounds containing Nε-thioacetyl-lysine (ThAcK, Fig. 1).39 The mechanistic studies by us and others demonstrated that ThAcK bestowed this potent sirtuin inhibition via a mechanism-based fashion in that the ThAcK-containing inhibitor could be recognized by a sirtuin enzyme as a substrate and processed to furnish a longer-lived (or stalled) α-1′-S-alkylamidate intermediate (Fig. 1a, X=S) that presumably behaves as a bisubstrate sirtuin inhibitory species.35,39–42 While the use of the thioamide-containing ThAcK has represented a simple yet very powerful technology to quickly identify very potent sirtuin inhibitors, the therapeutic use of a thioamide compound could be compromised by the potential cellular toxicity, especially the hepatotoxicity, after the metabolic S-oxidation and activation of such compound.43–47 It is possible to ameliorate this potential toxic side effect to certain extent via managing the drug administration route (e.g. i.v. versus p.o.),48 developing alternative potent sirtuin inhibitory AcK analogs would be another avenue to take.
In the current study, we have identified the novel AcK analog Nε-thiocarbamoyl-lysine (TuAcK, Fig. 1b) as a potent sirtuin inhibitory warhead.
Our pursuit of TuAcK as a potential novel sirtuin inhibitory warhead was based on the following considerations. First, since Nε-carbamoyl-lysine (or homocitrulline, Fig. 1b) was shown previously to be processed by the yeast sirtuin Hst2 to form the corresponding analog of α-1′-O-alkylamidate intermediate shown in Figure 1a following the Hst2-catalyzed nicotinamide cleavage from NAD+,49,50 we reasoned that TuAcK could also be processed similarly. However, unlike the intermediate formed from Nε-carbamoyl-lysine which was suggested to be intercepted by water to regenerate Nε-carbamoyl-lysine,49,50 the corresponding intermediate formed from TuAcK could be longer-lived just like that formed from ThAcK, thus exerting a potent sirtuin inhibition also presumably as a bisubstrate analog inhibitor. Secondly, some thiourea compounds have been known to be strong scavengers of the hydroxyl radical or the superoxide radical anion.51–53 This potential anti-oxidant effect of a thiourea compound constitutes another appeal for developing the thiourea-containing TuAcK as a basic sirtuin inhibitory motif since a compromised cellular anti-oxidation defense system could occur following the inhibition of the sirtuin-catalyzed deacetylation of the FoxO family of transcription factors. Specifically, the SIRT1- and SIRT2-catalyzed deacetylation of FoxO3/4 and FoxO3a, respectively, was shown to lead to the activation of these transcription factors and the consequent enhanced expression of their target genes whose products are components of the cellular anti-oxidation defense system, e.g., manganese superoxide dismutase.54–56 This sirtuin inhibition-induced potential attenuation of the cellular anti-oxidation defense system could be of a particular concern when developing therapeutics for the neurodegenerative diseases, for example SIRT2 inhibitors for the Parkinson’s disease.57 Within this context, a diminished cellular ATP level has been recently observed in PC12 cells following the SIRT2 silencing by RNAi, and the pharmacological SIRT2 inhibition has also been shown in the same study to exacerbate the ATP level diminishing from the H2O2 treatment.58
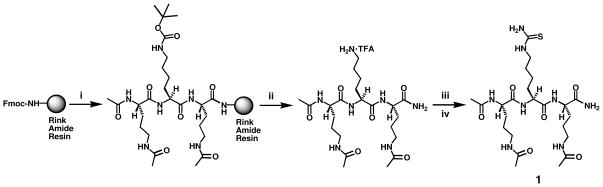
To assess the sirtuin inhibitory power of TuAcK, we prepared compound 1 as shown in Scheme 1.59 Following the assembly of a tripeptide on the Rink Amide resin via the 9-fluorenylmethoxycarbonyl (Fmoc) chemistry-based solid phase peptide synthesis (SPPS),60 the corresponding unprotected tripeptide amide was released from the resin with an 85% (v/v) solution of trifluoroacetic acid (TFA) in dichloromethane (DCM). This intermediate was then treated with thiophosgene followed by a 7 M solution of ammonia in MeOH, as literature procedures.61,62 The resulting product was isolated with the preparative high pressure liquid chromatography (HPLC).
Scheme 1.
Synthesis of the TuAcK-containing compound 1. Reagents: (i) Fmoc-based SPPS; (ii) 85% (v/v) TFA in DCM; (iii) Thiophosgene (CSCl2), Et3N; (iv) 7 N NH3 in MeOH.
It should be pointed out that the structural context of TuAcK in compound 1 was the peptidomimetic scaffold that we used previously, i.e. the tripartite structure with a central AcK analog and two immediately flanking Nδ-acetyl-ornithine (AcOrn).37,39 The use of this peptidomimetic scaffold in compound 1 also allowed us to examine whether this compound would also be able to inhibit a human sirtuin enzyme inside cells since this peptidomimetic scaffold was shown by us previously to be proteolytically stable and to be able to confer cell membrane permeability on the compound harboring ThAcK at the central position.37
When compound 1 was subjected to a HPLC-based SIRT1 inhibition assay,63 we obtained an IC50 of 89.5 ± 16.1 μM, a value which is ~15-fold greater than that of its ThAcK counterpart (Table 1). However, this IC50 value was ~3-fold lower than that for another peptidomimetic sirtuin inhibitor that we reported previously, i.e. CH3CONH-AcK-(L-ACAH)-AcK-CONH238 in which L-ACAH stands for L-2-amino-7-carboxamidoheptanoic acid (Fig. 1b) (Table 1). Of note, this AcK-based peptidomimetic scaffold is structurally very similar to the AcOrn-based scaffold in compound 1, since the AcOrn side chain is only one methylene (CH2) less than that of AcK. Considering that the AcOrn-based peptidomimetic scaffold was found by us previously to be ~3-fold weaker than the AcK-based one in bestowing SIRT1 inhibition,37 in our opinion, TuAcK is a more powerful SIRT1 inhibitory warhead than L-ACAH.
Table 1.
Sirtuin Inhibitiona
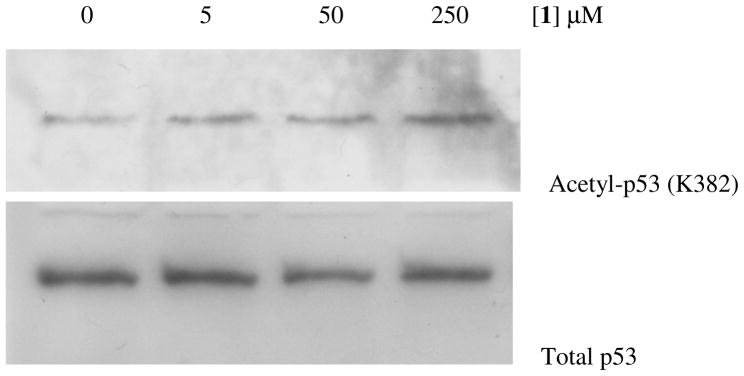
Encouraged by this in vitro SIRT1 inhibition result, compound 1 was further assessed for its capability of inhibiting the SIRT1 inside the HCT116 human colon cancer cells that express the wild-type p53 protein.65 The SIRT1 inhibitory potency in this cellular assay was assessed by the extent of the acetylation level increase for this SIRT1 endogenous substrate protein at the K382 position66 following the treatment with compound 1. As shown in Figure 2, compound 1 was observed to be able to enhance the p53 K382 acetylation level in a dose-dependent manner. Specifically, while a slight acetylation level increase was observed at 5 μM, compound 1 was found to be able to more dramatically enhance the p53 acetylation level at higher concentrations (50 and 250 μM).
Figure 2.
Western blot analysis of the p53 protein acetylation level change in HCT116 human colon cancer cells following the treatment with compound 1.
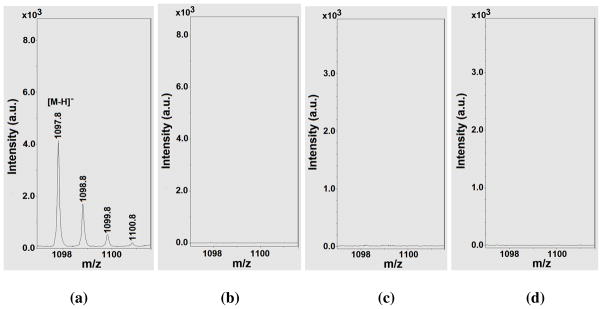
As mentioned above, it is possible that TuAcK could also be processed as ThAcK by a sirtuin enzyme to form a stalled alkylamidate intermediate following the nicotinamide cleavage from NAD+. To explore this possibility, we used mass spectrometry in an attempt to detect any possibly longer-lived catalytic species from an incubation of compound 1 with NAD+ in the presence of SIRT1.67 When the incubation mixture was subjected to a matrix-assisted laser desorption ionization (MALDI) mass spectral analysis in negative reflector mode, a species with m/z 1097.8 was observed, as shown in Figure 3a, from this incubation mixture under a non-quenching condition which was also used by Smith and Denu to detect the stalled α-1′-S-alkylamidate intermediate from an incubation of a ThAcK-containing peptide and NAD+ in the presence of the yeast sirtuin Hst2.40 We further found that the cluster of the peaks starting at m/z 1097.8 were not detectable under the same experimental conditions by omitting SIRT1 or compound 1 from the incubation mixture (Figs. 3b and 3c), and were not detectable from the MALDI matrix 3-Hydroxypicolinic acid (3-HPA) either (Fig. 3d). Therefore, the species with m/z 1097.8 appeared to be exclusively formed from the SIRT1-catalyzed reaction of NAD+ with compound 1. The species with m/z 1097.8 could well represent the [M-H]− anion derived from the corresponding alkylamidate intermediate (chemical formula (neutral form): C38H63N13O19P2S; exact mass (neutral form): 1099.4 Da) which is an analog of the α-1′-O-alkylamidate intermediate in Figure 1a, just as the α-1′-S-alkylamidate intermediate was detected with the MALDI-MS analysis in the study of Smith and Denu.40
Figure 3.
The MALDI-MS analysis of the following samples: (a) The assay mixture from compound 1 with SIRT1 and NAD+; (b) The assay mixture from compound 1 with NAD+ in the absence of SIRT1; (c) The assay mixture from NAD+ with SIRT1 in the absence of compound 1; (d) the MALDI matrix 3-hydroxypicolinic acid (3-HPA) only. All the MALDI-MS analyses were performed in a negative reflector mode. See ref. 67 for further experimental conditions.
In order to assess whether TuAcK could be a general sirtuin inhibitory warhead, we subsequently evaluated the inhibitory potency of compound 1 against human SIRT2 and SIRT3. As shown in Table 1, compound 1 was found to exhibit a more or less comparable inhibitory potency (within ~3-fold) against SIRT1, SIRT2, and SIRT3, thus suggesting that TuAcK could confer potent sirtuin inhibition as a general inhibitory warhead. Table 1 also shows the comparable inhibitory potency against SIRT1, SIRT2, and SIRT3 that we previously found for the peptidomimetic sirtuin inhibitors containing ThAcK or L-ACAH.37,38 However, a closer inspection of the inhibitory data for compound 1 and the ThAcK-containing inhibitor in Table 1 revealed their different rank order of potency. In specific, while both compounds were shown to exhibit a slightly stronger inhibition against SIRT3 than SIRT1, compound 1 exhibited the strongest inhibition against SIRT3, whereas the ThAcK inhibitor exhibited the strongest inhibition against SIRT2. Since these two inhibitors have the same peptidomimetic scaffold, their different rank order of sirtuin inhibitory potency suggested that different sirtuin active sites respond differently to the AcK side chain structural variations.
Taken together, in the current study, we have identified TuAcK as another general sirtuin inhibitory warhead which was shown to be able to confer potent sirtuin inhibition. Experiments with SIRT1 suggested that a TuAck sirtuin inhibitor also behaved as a mechanism-based inhibitor in that the TuAck residue was also able to be processed by a sirtuin enzyme with the formation of a stalled S-alkylamidate intermediate, like that for ThAcK. Given the potential for a thiourea compound to be a strong anti-oxidant, further development of peptidomimetic or non-peptidic human sirtuin inhibitors containing TuAcK or its homologs should be warranted in the continued search for novel sirutin inhibitors. For the TuAcK homologs, those bearing alkyl groups (methyl, ethyl, etc.) on the terminal NH2 group of TuAcK side chain could be considered, given the capability of the sirtuin active site to accommodate bulkier acyl groups than the acetyl group of AcK side chain.39 The study on TuAcK homologs is under way in our laboratory and the corresponding findings will be reported in due course.
Acknowledgments
The financial support from the U.S. National Institutes of Health (R15CA152972 and R01CA127590) is highly appreciated. We thank Prof. Tony Kouzarides (University of Cambridge, UK) for the GST-SIRT1 plasmid. We would like to thank the high resolution mass spectrometry facility of the University of California, Riverside for the HRMS data. We also thank Prof. Chrys Wesdemiotis and his research group at the University of Akron for the assistance with mass spectrometric analysis (ESI-MS) of the peptidomimetic compound 1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Sauve AA, Schramm VL. Curr Med Chem. 2004;11:807. doi: 10.2174/0929867043455675. [DOI] [PubMed] [Google Scholar]
- 2.Sauve AA, Wolberger C, Schramm VL, Boeke JD. Annu Rev Biochem. 2006;75:435. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 3.Smith BC, Hallows WC, Denu JM. Chem Biol. 2008;15:1002. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders BD, Jackson B, Marmorstein R. Biochim Biophys Acta. 2010;1804:1604. doi: 10.1016/j.bbapap.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauve AA. Biochim Biophys Acta. 2010;1804:1591. doi: 10.1016/j.bbapap.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirsch BM, Zheng W. Mol Biosyst. 2011;7:16. doi: 10.1039/c0mb00033g. [DOI] [PubMed] [Google Scholar]
- 7.Cen Y, Youn DY, Sauve AA. Curr Med Chem. 2011;18:1919. doi: 10.2174/092986711795590084. [DOI] [PubMed] [Google Scholar]
- 8.Frye RA. Biochem Biophys Res Commun. 2000;273:793. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 9.Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Ann N Y Acad Sci. 2003;983:84. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 10.Gregoretti IV, Lee YM, Goodson HV. J Mol Biol. 2004;338:17. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Saunders LR, Verdin E. Oncogene. 2007;26:5489. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 12.Greiss S, Gartner A. Mol Cells. 2009;28:407. doi: 10.1007/s10059-009-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu T, Liu PY, Marshall GM. Cancer Res. 2009;69:1702. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 14.Finkel T, Deng CX, Mostoslavsky R. Nature. 2009;460:587. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, Auwerx J. Ann N Y Acad Sci. 2009;1173:E10. doi: 10.1111/j.1749-6632.2009.04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai S, Guarente L. Trends Pharmacol Sci. 2010;31:212. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haigis MC, Sinclair DA. Annu Rev Pathol: Mech Dis. 2010;5:253. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verdin E, Hirschey MD, Finley LW, Haigis MC. Trends Biochem Sci. 2010;35:669. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan KL, Xiong Y. Trends Biochem Sci. 2011;36:108. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakshminarasimhan M, Steegborn C. Exp Gerontol. 2011;46:174. doi: 10.1016/j.exger.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 21.Porcu M, Chiarugi A. Trends Pharmacol Sci. 2005;26:94. doi: 10.1016/j.tips.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Gan L. Drug News Perspect. 2007;20:233. doi: 10.1358/dnp.2007.20.4.1101162. [DOI] [PubMed] [Google Scholar]
- 23.Elliott PJ, Jirousek M. Curr Opin Investig Drugs. 2008;9:371. [PubMed] [Google Scholar]
- 24.Outeiro TF, Marques O, Kazantsev A. Biochim Biophys Acta. 2008;1782:363. doi: 10.1016/j.bbadis.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Deppert W. Cell Cycle. 2008;7:2947. doi: 10.4161/cc.7.19.7010. [DOI] [PubMed] [Google Scholar]
- 26.Milne JC, Denu JM. Curr Opin Chem Biol. 2008;12:11. doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 27.Sauve AA. Curr Pharm Des. 2009;15:45. doi: 10.2174/138161209787185797. [DOI] [PubMed] [Google Scholar]
- 28.Imai S, Kiess W. Front Biosci. 2009;14:2983. doi: 10.2741/3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baur JA. Biochim Biophys Acta. 2010;1804:1626. doi: 10.1016/j.bbapap.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camins A, Sureda FX, Junyent F, Verdaguer E, Folch J, Pelegri C, Vilaplana J, Beas-Zarate C, Pallàs M. Biochim Biophys Acta. 2010;1799:740. doi: 10.1016/j.bbagrm.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Balcerczyk A, Pirola L. Biofactors. 2010;36:383. doi: 10.1002/biof.112. [DOI] [PubMed] [Google Scholar]
- 32.Blum CA, Ellis JL, Loh C, Ng PY, Perni RB, Stein RL. J Med Chem. 2011;54:417. doi: 10.1021/jm100861p. [DOI] [PubMed] [Google Scholar]
- 33.Chen L. Curr Med Chem. 2011;18:1936. doi: 10.2174/092986711795590057. [DOI] [PubMed] [Google Scholar]
- 34.Bonda DJ, Lee HG, Camins A, Pallàs M, Casadesus G, Smith MA, Zhu X. Lancet Neurol. 2011;10:275. doi: 10.1016/S1474-4422(11)70013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fatkins DG, Monnot AD, Zheng W. Bioorg Med Chem Lett. 2006;16:3651. doi: 10.1016/j.bmcl.2006.04.075. [DOI] [PubMed] [Google Scholar]
- 36.Fatkins DG, Zheng W. Int J Mol Sci. 2008;9:1. doi: 10.3390/ijms9010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirsch BM, Gallo CA, Du Z, Wang Z, Zheng W. MedChemComm. 2010;1:233. [Google Scholar]
- 38.Hirsch BM, Du Z, Li X, Sylvester JA, Wesdemiotis C, Wang Z, Zheng W. MedChemComm. 2011;2:291. [Google Scholar]
- 39.Hirsch BM, Zheng W. Mol Biosyst. 2011;7:16. doi: 10.1039/c0mb00033g. [DOI] [PubMed] [Google Scholar]
- 40.Smith BC, Denu JM. Biochemistry. 2007;46:14478. doi: 10.1021/bi7013294. [DOI] [PubMed] [Google Scholar]
- 41.Hawse WF, Hoff KG, Fatkins DG, Daines A, Zubkova OV, Schramm VL, Zheng W, Wolberger C. Structure. 2008;16:1368. doi: 10.1016/j.str.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin L, Wei W, Jiang Y, Peng H, Cai J, Mao C, Dai H, Choy W, Bemis JE, Jirousek MR, Milne JC, Westphal CH, Perni RB. J Biol Chem. 2009;284:24394. doi: 10.1074/jbc.M109.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neal RA, Halpert J. Annu Rev Pharmacol Toxicol. 1982;22:321. doi: 10.1146/annurev.pa.22.040182.001541. [DOI] [PubMed] [Google Scholar]
- 44.Ruse MJ, Waring RH. Toxicol Lett. 1991;58:37. doi: 10.1016/0378-4274(91)90188-c. [DOI] [PubMed] [Google Scholar]
- 45.Cox DN, Davidson VP, Judd CE, Stodgell C, Traiger GJ. Toxicol Appl Pharmacol. 1992;113:246. doi: 10.1016/0041-008x(92)90121-8. [DOI] [PubMed] [Google Scholar]
- 46.Coppola GM, Anjaria H, Damon RE. Bioorg Med Chem Lett. 1996;6:139. [Google Scholar]
- 47.Kang JS, Wanibuchi H, Morimura K, Wongpoomchai R, Chusiri Y, Gonzalez FJ, Fukushima S. Toxicol Appl Pharmacol. 2008;228:295. doi: 10.1016/j.taap.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 48.Holford NHG. In: Basic and Clinical Pharmacology. 11 Katzung BG, Masters SB, Trevor AJ, editors. McGraw-Hill Medical; USA: 2009. pp. 37–51. [Google Scholar]
- 49.Khan AN, Lewis PN. J Biol Chem. 2006;281:11702. doi: 10.1074/jbc.M511482200. [DOI] [PubMed] [Google Scholar]
- 50.Smith BC, Denu JM. J Biol Chem. 2007;282:37256. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 51.Wasil M, Halliwell B, Grootveld M, Moorhouse CP, Hutchison DC, Baum H. Biochem J. 1987;243:867. doi: 10.1042/bj2430867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong Y, Venkatachalam TK, Narla RK, Trieu VN, Sudbeck EA, Uckun FM. Bioorg Med Chem Lett. 2000;10:87. doi: 10.1016/s0960-894x(99)00581-8. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi H, Nishina A, Fukumoto RH, Kimura H, Koketsu M, Ishihara H. Life Sci. 2005;76:2185. doi: 10.1016/j.lfs.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 54.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Science. 2004;303:2011. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 55.Van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. J Biol Chem. 2004;279:28873. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 56.Wang F, Nguyen M, Qin FXF, Tong Q. Aging Cell. 2007;6:505. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 57.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, Young AB, Abagyan R, Feany MB, Hyman BT, Kazantsev AG. Science. 2007;317:516. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 58.Nie H, Chen H, Han J, Hong Y, Ma Y, Xia W, Ying W. Int J Physiol Pathophysiol Pharmacol. 2011;3:65. [PMC free article] [PubMed] [Google Scholar]
- 59.Synthesis of compound 1 (Scheme 1). (a) The solid phase synthesis part of the synthesis of this compound was performed with the Fmoc chemistry-based solid phase peptide synthesis (SPPS)60 at a 0.05 mmol scale on a PS3 peptide synthesizer (Protein Technologies Inc., Tucson, AZ, USA) from Rink Amide AM resin. For the amino acid coupling reactions, 4 equivalents of Nα-Fmoc-Nε-Boc-L-lysine or 2 equivalents of Nα-Fmoc-Nδ-acetyl-ornithine (synthesized as described previously37) were used together with 3.8 or 1.9 equivalents, respectively, of the coupling reagent HBTU and the additive HOBt in the presence of 0.4 M NMM/DMF, and the corresponding coupling reaction was allowed to proceed at room temperature for 1h or 2h, respectively. A 20% (v/v) piperidine/DMF solution was used for Fmoc removal. The resin-bound intermediate was treated with acetic anhydride (to acetylate the N-terminal α-amino group) before it was cleaved from the resin at room temperature for 4 h by a solution of TFA in DCM (85% v/v). This solution was then evaporated at room temperature on a Rotavapor to dryness. The resulting residue was further dried overnight under high vacuum (oil pump) and used for the next reaction without further purification. (b) To a cooled (ice-water bath) and stirred solution of the above-obtained residue (~ 0.05 mmol) in ddH2O (373 μL) was added dropwise triethylamine to neutralize the solution, which was followed by the further addition of 28 μL (0.2 mmol) of triethylamine dropwise at 0 °C. A solution of thiophosgene (CSCl2) (5.7 μL, 0.0744 mmol) in acetone (373 μL) was then added dropwise at 0 °C. After the addition was complete, the reaction mixture was stirred at room temperature for 3 h, and was concentrated to dryness with a Rotavapor. The resulting residue was used for the next reaction immediately. (c) The residue from the previous step was cooled down with an ice-water bath, and to which was added dropwise while stirring 1.5 mL of a 7 N solution of ammonia in methanol. The whole reaction mixture was then stirred at 0 °C for 3 h, and then concentrated to dryness with a Rotavapor. Compound 1 was isolated from the resulting residue by the reversed-phase HPLC on a preparative C18 column (100 Å, 2.14 × 25 cm). The column was eluted with a gradient of ddH2O containing 0.05% (v/v) TFA and acetonitrile containing 0.05% (v/v) TFA at 10 mL/min and monitored at 214 nm. The pooled HPLC fractions were stripped of acetonitrile and lyophilized to give compound 1 as a puffy white solid. The purified compound 1 was >95% pure as verified by the reversed-phase HPLC on an analytical C18 column (100 Å, 0.46 × 25 cm). The column was eluted with a gradient of ddH2O containing 0.05% (v/v) TFA and acetonitrile containing 0.05% (v/v) TFA at 1 mL/min and monitored at 214 nm. The exact mass of the purified compound 1 was confirmed by the electrospray ionization (ESI)-MS analysis with a Bruker Esquire-LC ion trap mass spectrometer at the University of Akron and the high resolution mass spectrometry (HRMS) analysis with an Agilent 6210 LCMS instrument at the high resolution mass spectrometry facility of the University of California, Riverside: MS (ESI) calcd. for C23H43N8O6S ([M + H]+) 559.3; found: 559.6. HRMS (Multimode) calcd. for C23H43N8O6S ([M + H]+) 559.3026; found: 559.3010. Calcd. for C23H42N8O6SNa ([M + Na]+) 581.2846; found: 581.2846. Calcd. for C23H42N8O6SK ([M + K]+) 597.2585; found: 597.2561.
- 60.Wellings DA, Atherton E. Methods Enzymol. 1997;289:44. doi: 10.1016/s0076-6879(97)89043-x. [DOI] [PubMed] [Google Scholar]
- 61.Narayanan K, Griffith OW. J Med Chem. 1994;37:885. doi: 10.1021/jm00033a004. [DOI] [PubMed] [Google Scholar]
- 62.Zheng W, Jing L, Patil PN, Lei L, Feller DR, Miller DD. Med Chem Res. 2001;10:318. [Google Scholar]
- 63.Sirtuin inhibition assay. The HPLC-based sirtuin inhibition assay was performed as described previously.35–38,64 Briefly, a sirtuin inhibition assay solution (50 μL) contained the following components: 25 mM (or 50 mM for the SIRT2 assay) Tris•HCl (pH 8.0), 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1 mg/mL BSA (Sigma Cat. No. A3803 with reduced fatty acid content, for the SIRT2 assay only), β-NAD+ (0.5 mM for the SIRT1 and the SIRT2 assay, 3.5 mM for the SIRT3 assay), a peptide substrate (0.3 mM of the SIRT1 substrate: H2NHK-AcK-LM-COOH; 0.5 mM of the SIRT2 substrate: H2N-MPSD-AcK-TIGG-COOH; or 0.03 mM of the SIRT3 substrate: H2N-KRLPKTRSG-AcK-VMRRLLRKII-COOH), a sirtuin enzyme (GST-SIRT1, 350 nM; SIRT2, 150 nM; or SIRT3, 500 nM), and compound 1 with varied concentrations including 0. Of note, the above peptides were also used in our previous studies as the sirtuin substrates.64,68 While GST-SIRT1 was expressed and purified from Escherichia coli as described previously using pGEX2TK-P•SIRT1 (human full length) (a kind gift from Prof. Tony Kouzarides),64 SIRT2 (human full length) and SIRT3 (human amino acid 102–399) were purchased from Enzo Life Sciences (Product Nos. BML-SE251-0500 and BML-SE270-0500, respectively). It should be pointed out that the same [S]/Km ratios for both substrates (~3.2 for the peptide substrates and ~5.6 for NAD+) were used for the inhibition assays with SIRT1, SIRT2, and SIRT3 for assessing the inhibitory selectivity of compound 1 among these three human sirtuins. An enzymatic reaction was initiated by the addition of a sirtuin enzyme at 37 °C and was allowed to be incubated at 37 °C for 5 min (for the SIRT1 and the SIRT3 assay) or 60 min (for the SIRT2 assay) until quenched with the following stop solution: 100 mM HCl and 0.16 M acetic acid. The quenched assay solutions were directly injected into a reversed-phase C18 column (100 Å, 5 μm, 0.46 × 25 cm), eluting with a gradient of ddH2O containing 0.05% (v/v) TFA and acetonitrile containing 0.05% (v/v) TFA at 1 mL/min, and UV monitoring at 214 nm. The enzymatic deacetylation product was quantified with HPLC peak integration. Turnover of the limiting substrate was maintained at ≤ 15%. The stock solution of compound 1 was prepared in ddH2O. IC50 values were estimated from the Dixon plots (1/v0 vs. [inhibitor])69 as an indication of the inhibition potency.
- 64.Jamonnak N, Hirsch BM, Pang Y, Zheng W. Bioorg Chem. 2010;38:17. doi: 10.1016/j.bioorg.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 65.Western blot analysis. This assay was also performed as described previously.37,38 HCT116 human colon cancer cells were cultured in McCoy5A culture medium containing 10% FBS with penicillin and streptomycin. HCT116 cells (2 × 105) were treated for 10 h with compound 1 at different concentrations (0, 5, 50, and 250 μM) and then collected and extracted with RIPA buffer (50 mM Tris•HCl (pH 8.0), 0.5% triton X-100, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 50 mM NaF, 1 mM PMSF, 1x Roche complete protease inhibitor cocktail). Equivalent amounts of proteins from each lysate were resolved in 10% SDS-polyacrylamide gel and then transferred onto PVDF membranes (Bio-Rad Laboratories). After having been blocked with Tris-buffered saline (TBS) containing 5% milk for 1 h, the transblotted membrane was incubated overnight at 4 °C with acetylated p53 antibody (Cell Signaling) (1:1000 dilution). After washed twice with water, the membranes were incubated with the rabbit anti-mouse IgG-horseradish peroxidase conjugate (diluted 1:5000) for 2 h at room temperature, and again washed twice with water. The immunoblots were visualized by enhanced chemiluminescence. Stripping the membrane and then blot it with p53 antibody (Cell Signaling) (1:1000 dilution) overnight at 4 °C.
- 66.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. Cell. 2001;107:149. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 67.Mass spectrometry detection of the catalytic species with compound 1 and SIRT1. This assay was also performed as described previously.37,38 (a) An assay solution (20 μL) contained the following components: 20 mM pyridinium formate (pH 7.0), 1 mM DTT, 0.5 mM β-NAD+, 0.6 mM compound 1, and 4 μM GST-SIRT1. An enzymatic reaction was initiated by the addition of GST-SIRT1 at 25 °C and was allowed to be incubated at 25 °C for 1 min before the reaction mixture was snap frozen in liquid nitrogen and stored at −70 °C until the mass spectral analysis by MALDI-MS (negative reflector mode with 3-HPA as the matrix). (b) The MALDI-MS experiments were carried out on a Bruker Ultraflex III tandem time-of-flight (ToF/ToF) mass spectrometer (Bruker Daltonics, Billerica, MA), equipped with a Nd:YAG laser emitting at a wavelength of 355 nm. All spectra were recorded in a negative reflector mode. The solution of the matrix 3-HPA was prepared in acetonitrile/water (1/1 (v/v)) at 5 mg/mL. The sample preparation followed the sandwich method. Briefly, 0.5 μL of the matrix solution was initially deposited on the wells of a 384-well ground-steel plate, allowing the spots to dry in the air. A SIRT1 assay sample was thawed and 0.5 μL of it was deposited on the top of a dried matrix spot. Another 0.5 μL of the matrix solution was then deposited on the top of the air-dried sample spot. Data analysis was conducted with the flexAnalysis software.
- 68.Robinson SJ, Hirsch BM, Jamonnak N, Zheng W. Undergraduate Student Research Poster Session, 41st ACS Central Regional Meeting; May 2009; Cleveland, Ohio, USA. [Google Scholar]
- 69.Dixon M. Biochem J. 1953;55:170. doi: 10.1042/bj0550170. [DOI] [PMC free article] [PubMed] [Google Scholar]