Abstract
The explosive growth of the human neuroimaging literature has led to major advances in understanding of human brain function, but has also made aggregation and synthesis of neuroimaging findings increasingly difficult. Here we describe and validate an automated brain mapping framework that uses text mining, meta-analysis and machine learning techniques to generate a large database of mappings between neural and cognitive states. We demonstrate the capacity of our approach to automatically conduct large-scale, high-quality neuroimaging meta-analyses, address long-standing inferential problems in the neuroimaging literature, and support accurate ‘decoding’ of broad cognitive states from brain activity in both entire studies and individual human subjects. Collectively, our results validate a powerful and generative framework for synthesizing human neuroimaging data on an unprecedented scale.
INTRODUCTION
The development of non-invasive neuroimaging techniques such as functional magnetic resonance imaging (fMRI) has spurred explosive growth of the human brain imaging literature in recent years. In 2010 alone, over 1,000 fMRI articles were published1. This proliferation has led to substantial advances in understanding of human brain and cognitive function; however, it has also introduced important new challenges. In place of too little data, researchers are now besieged with too much. Because individual neuroimaging studies are often underpowered and exhibit relatively high false positive rate2-4, multiple studies are required to achieve consensus regarding even broad relationships between brain and cognitive function. Distilling the extant literature thus necessitates development of new techniques for large-scale aggregation and synthesis of human neuroimaging data4-6.
Here we describe and validate a novel framework for brain mapping, NeuroSynth, that takes an instrumental step towards automated large-scale synthesis of the neuroimaging literature. NeuroSynth combines text mining, meta-analysis, and machine learning techniques to generate probabilistic mappings between cognitive and neural states that can be used for a broad range of neuroimaging applications. Whereas previous approaches have relied heavily on researchers’ manual efforts (e.g., 7,8)—a constraint that limits the scope and efficiency of resulting analyses1—the present framework is fully automated, enabling rapid and scalable synthesis of the neuroimaging literature. We demonstrate the capacity of this framework to generate large-scale meta-analyses for hundreds of broad psychological concepts; support quantitative inferences about the consistency and specificity with which different cognitive processes elicit regional changes in brain activity; and decode and classify broad cognitive states in new data based solely on observed brain activity.
RESULTS
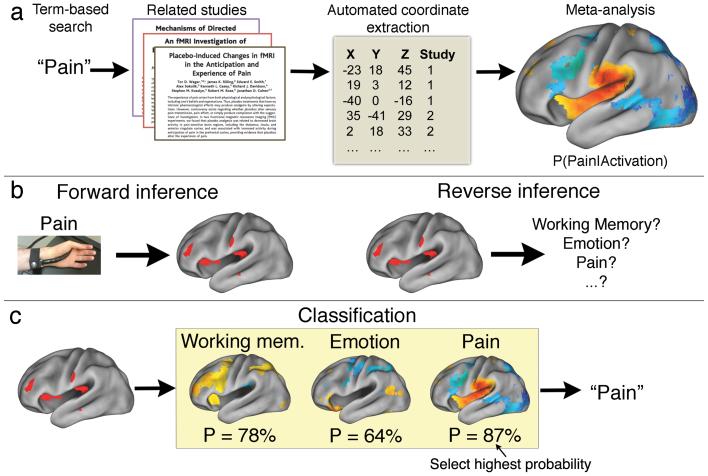
Our methodological approach includes several steps (Fig. 1a). First, we used text-mining techniques to identify neuroimaging studies that used specific terms of interest (e.g., ‘pain’, ‘emotion’, ‘working memory’, etc.) at a high frequency (>1 in 1,000 words) within the article text. Second, we automatically extracted activation coordinates from all tables reported in these studies. This approach produced a large database of term-to-coordinate mappings; we report results based on 100,953 activation foci drawn from 3,489 neuroimaging studies published in more than 15 journals (Online methods). Third, we conducted automated meta-analyses of hundreds of psychological concepts, producing an extensive set of whole-brain images quantifying brain-cognition relationships (Fig. 1b). Finally, we used a machine learning technique (naïve Bayes classification) to estimate the likelihood that new activation maps were associated with specific psychological terms, enabling relatively open-ended decoding of psychological constructs from patterns of brain activity (Fig. 1c).
Figure 1.
Schematic overview of NeuroSynth framework and applications. (a) Schematic of NeuroSynth approach. The full text of a large corpus of articles is retrieved and terms of scientific interest are stored in a database. Articles are retrieved from the database based on a user-entered search string (e.g., the word ‘pain’), and peak coordinates from the associated articles are extracted from tables. A meta-analysis of the peak coordinates is automatically performed, producing a whole-brain map of the posterior probability of the term given activation at each voxel (i.e P(Pain|Activation)). (b) Two types of inference in brain imaging. Given a known psychological manipulation, one can quantify the corresponding changes in brain activity and generate a forward inference; however, given an observed pattern of activity, drawing a reverse inference about associated cognitive states is more difficult, because multiple cognitive states could have similar neural signatures. (c) Given meta-analytic posterior probability maps for multiple terms (e.g., working memory, emotion, pain), one can classify a new activation map by identifying the class with the highest probability P given the new data (in this example, pain).
Automated coordinate extraction
Our approach differs from previous work in its heavy reliance on automatically extracted information, raising several potential data quality concerns. For example, the software might incorrectly classify non-coordinate information in a table as an activation focus (i.e., a false positive); different articles report foci in different stereotactic spaces, resulting in potential discrepancies between anatomical locations represented by the same set of coordinates and the software did not discriminate activations from deactivations.
To assess the impact of these issues on data quality, we conducted an extensive series of supporting analyses (Supplementary Note). First, we compared automatically extracted coordinates with a reference set of manually-entered foci in the SumsDB database7,9, revealing high rates of sensitivity (84%) and specificity (97%). Second, we quantified the proportion of activation increases versus decreases reported in the neuroimaging literature. We found that decreases constitute a small proportion of results and have minimal effect on the results we report. Third, we developed a preliminary algorithm for automatically detecting and correcting (based on ref. 10) for between-study differences in stereotactic space (Supplementary Fig. 1). Collectively, these results indicate that while automated extraction misses a minority of valid coordinates, and work remains to be done to increase the specificity of the extracted information, the majority of coordinates are extracted accurately, and a number of factors of a priori concern have relatively small influences on the results.
The database of studies and coordinates, software, and meta-analysis maps for several hundred terms used in the present study are made available via a web interface (http://neurosynth.org) that provides instant access to, and visualization of, thousands of whole-brain images.
Large-scale automated meta-analysis
We used the database of automatically extracted activation coordinates to conduct a comprehensive set of automated meta-analyses for several hundred terms of interest. For each term, we identified all studies that used the term at high frequency anywhere within the article text11 and submitted all associated activation foci to a meta-analysis. This approach generated whole-brain maps displaying the strength of association between each term and every location in the brain, enabling multiple kinds of quantitative inference (e.g., if the term ‘language’ was used in a study, how likely was the study to report activation in Broca’s area? If activation was observed in the amygdala, what was the probability that the study frequently used the term ‘fear’?).
To validate this automated approach—which rests on the assumption that simple word counts are a reasonable proxy for the substantive content of articles—we conducted a series of supporting analyses (Supplementary Note). First, we demonstrated that NeuroSynth accurately recaptured conventional boundaries between distinct anatomical regions by comparing lexically defined regions-of-interest (ROIs) to anatomically defined ROIs (Supplementary Fig. 2). Second, we used NeuroSynth to replicate previous findings of visual category-specific activation in regions such as the fusiform face area (FFA12) and visual word form area (VWFA13; Supplementary Fig. 3). Third, we demonstrated that more conservative meta-analyses restricting the lexical search space to only article titles yielded similar, though less sensitive, meta-analysis results (Supplementary Fig. 4).
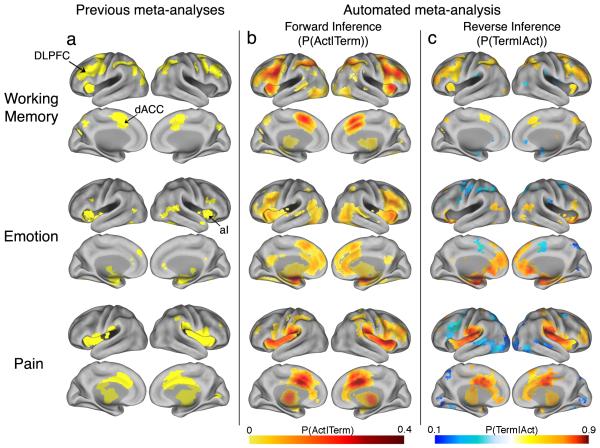
Finally, we compared our results with those produced by prior manual approaches. Comparison of automated meta-analyses of three broad psychological terms (‘working memory’, ‘emotion’, and ‘pain’) with previously published meta- or mega-analytic maps14-16 revealed a marked convergence between approaches both qualitatively (Fig. 2) and quantitatively (Supplementary Fig. 5). To directly test the convergence of automated and manual approaches when applied to similar data, we manually validated a set of 265 automatically extracted pain studies and performed a standard multilevel kernel density analysis (MKDA15) contrasting experimental pain stimulation with baseline (n = 66 valid studies). Direct comparison between automated and manual results revealed a striking overlap (correlation across voxels = .84; Supplementary Fig. 6). Thus, these results demonstrated that, at least for broad domains, an automated meta-analysis approach generates results comparable in sensitivity and scope to those produced more effortfully in previous studies.
Figure 2.
Comparison of previous meta-analysis results with forward and reverse inference maps produced automatically using the NeuroSynth framework. Meta-analyses were carried out for working memory (top row), emotion (middle row), and physical pain (bottom row), and mapped to the PALS-B12 atlas45. (a) Meta-analytic maps produced manually in previous studies14-16. (b) Automatically generated forward inference maps displaying the probability of observing activation given the presence of the term (i.e., P(Activation|Term)). (c) Automatically generated reverse inference maps display the probability of the term given observed activation (i.e., P(Term|Activation)). Thus, regions in (b) are consistently associated with the term, and regions in (c) are selectively associated with the term. To account for base differences in term frequencies, reverse inference maps assume uniform priors (i.e., equal 50% probabilities of Term and No Term). Activation in orange/red regions implies a high probability that a term is present, and activation in blue regions implies a high probability that a term is not present. Values for all images are displayed only for regions that are significant for a test of association between Term and Activation, with a whole-brain correction for multiple comparisons (FDR = .05). DLPFC = dorsolateral prefrontal cortex; dACC = dorsal anterior cingulate cortex; aI = anterior insula.
Quantitative reverse inference
The relatively comprehensive nature of the NeuroSynth database enabled us to address a long-standing inferential problem in the neuroimaging literature—namely, how to quantitatively identify cognitive states based on patterns of observed brain activity. This problem of ‘reverse inference’17 arises because most neuroimaging studies are designed to identify neural changes that result from known psychological manipulations, and not to determine what cognitive state(s) a given pattern of activity implies (Fig. 1b, ref.17). For instance, fear consistently activates the human amygdala, but this does not imply that people who show amygdala activation must be experiencing fear, because other affective and non-affective states have also been reported to produce amygdala activation4,18. True reverse inference requires knowledge of which brain regions and networks are selectively, and not just consistently, associated with particular cognitive states15,17.
Because the NeuroSynth database contains a broad set of term-to-activation mappings, our framework is well suited for drawing quantitative inferences about mind-brain relationships in both the forward and reverse directions. We were able to quantify both the probability of observing activation in specific brain regions given the presence a particular term (P(Activation|Term), or ‘forward inference’), and the probability of a term occurring in an article given the presence of activation in a particular brain region (i.e., P(Term|Activation), or reverse inference). Comparison of these two analyses provided a way to assess the validity of many common inferences about the relationship between neural and cognitive states.
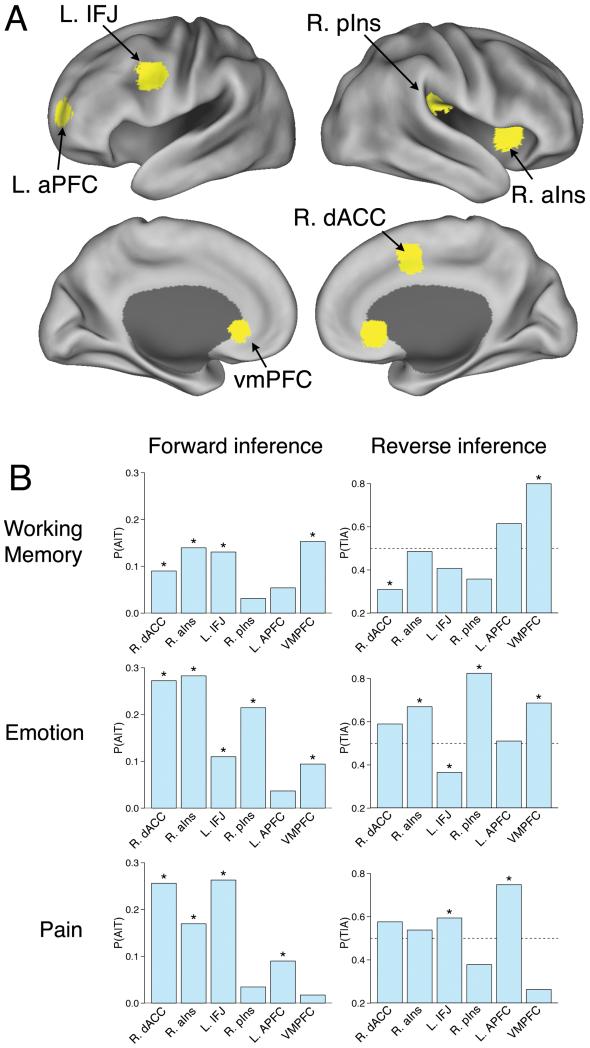
For illustration purposes, we focused on the sample domains of working memory, emotion, and pain, which are of substantial basic and clinical interest and have been extensively studied using fMRI (for additional examples, see Supplementary Fig. 7). These domains are excellent candidates for quantitative reverse inference, as they are thought to have somewhat confusable neural correlates, with common activation of regions such as dorsal anterior cingulate cortex (ACC)19 and anterior insula.
Results revealed important differences between the forward and reverse inference maps in all three domains (Fig. 2). For working memory, the forward inference map revealed the most consistent associations in dorsolateral prefrontal cortex (DLPFC), anterior insula and dorsal medial frontal cortex (MFC), replicating previous findings15,20. However, the reverse inference map instead implicated anterior PFC and posterior parietal cortex as the regions most selectively activated by working memory tasks.
We observed a similar pattern for pain and emotion. In both domains, frontal regions broadly implicated in goal-directed cognition21-23 showed consistent activation in the forward analysis, but were relatively non-selective in the reverse analysis (Fig. 2). For emotion, the reverse inference map revealed much more selective activations in the amygdala and ventromedial PFC (Fig. 3). For pain, the regions of maximal pain-related activation in insula and ACC shifted from anterior foci in the forward analysis to posterior ones in the reverse analysis (Fig. 3). This is consistent with nonhuman primate studies implicating dorsal posterior insula as a primary integration center for nociceptive afferents24 as well as human studies demonstrating that anterior aspects of the so-called ‘pain matrix’ respond non-selectively to multiple modalities25.
Figure 3.
Comparison of forward and reverse inference in selected regions of interest. (a) Labeled regions of interest displayed on lateral and medial brain surfaces. (b) Comparison of forward inference (i.e., probability of activation given term P(T|A)) and reverse inference (probability of term given activation P(A|T)) for the domains of working memory (top), emotion (middle), and pain (bottom). Bars with asterisks denote statistically significant effects (whole-brain FDR, q = .05). dACC = dorsal anterior cingulate cortex (coordinates: +2, +8, +50); aIns = anterior insula (+36, +16, +2); IFJ = inferior frontal junction (−50, +8, +36); pIns = posterior insula (+42, −24, +24); aPFC = anterior prefrontal cortex (−28, +56, +8); vmPFC = ventromedial prefrontal cortex (0, +32, −4). L and R refer to the left and right hemispheres, respectively.
Perhaps most strikingly, several frontal regions that showed consistent activation for emotion and pain in the forward analysis were actually associated with a decreased likelihood that a study involved emotion or pain in the reverse inference analysis (Fig. 3). This seeming paradox reflected the fact that even though lateral and medial frontal regions were consistently activated in studies of emotion and pain, they were activated even more frequently in studies that did not involve emotion or pain (Supplementary Fig. 8). Thus, the fact that these regions showed involvement in pain and emotion likely reflected their much more general role in cognition (e.g., sustained attention or goal-directed processing22,23) rather than pain- or emotion-specific process.
These results demonstrate that without the ability to distinguish consistency from selectivity, neuroimaging data can produce misleading inferences. For instance, neglecting the high base rate of ACC activity might lead researchers in the areas of cognitive control, pain, and emotion each to conclude that the ACC plays a critical role in their particular domain. Instead, because the ACC is activated consistently in all of these states, its activation may not be diagnostic of any one of them—and conversely, might even predict their absence. The NeuroSynth framework can potentially address this problem by enabling researchers to conduct quantitative reverse inference on a large scale.
Open-ended classification of cognitive states
An emerging frontier in human neuroimaging is brain ‘decoding’: inferring a person’s cognitive state based solely on their observed brain activity. The problem of decoding is essentially a generalization of the univariate reverse inference problem addressed above: instead of predicting the likelihood of a particular cognitive state given activation at a single voxel, one can generate a corresponding prediction based on an entire pattern of brain activity. The NeuroSynth framework is well positioned for such an approach: whereas previous decoding approaches have focused on discriminating between narrow sets of cognitive states and have required extensive training on raw fMRI datasets (e.g., refs.26-28), the breadth of cognitive concepts represented in the NeuroSynth database affords relatively open-ended decoding, with little or no training on new datasets.
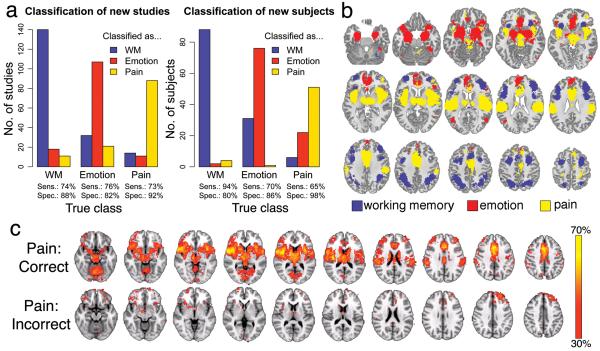
To assess the ability of our approach to decode and classify cognitive states, we trained a naïve Bayes classifier29 capable of discriminating between flexible sets of cognitive states given new images as input (Fig. 1c). First, we tested the classifier’s ability to correctly classify studies in the NeuroSynth database that were associated with different terms. In a 10-fold cross-validated analysis, the classifier discriminated between studies of working memory, emotion, and pain, with high sensitivity and specificity (Fig. 4a), demonstrating that each of these domains has a relatively distinct neural signature (Fig. 4b).
Figure 4.
Three-way classification of working memory (WM), emotion, and pain. (a) Naive Bayes classifier performance when cross-validated on studies in the database (left) or applied to entirely new subjects (right). Sens. = sensitivity; Spec. = specificity. (b) Whole-brain maximum posterior probability map; each voxel is colored by the term with the highest associated probability. (c) Whole-brain maps displaying the proportion of individual subjects in the three pain studies (total n = 79) who showed activation at each voxel (P < .05, uncorrected), averaged separately for subjects who were correctly (n = 51; top row) or incorrectly (n = 28; bottom row) classified. Regions are color-coded according to the proportion of subjects in the sample who showed activation at each voxel.
To assess the classifier’s ability to decode cognitive states in individual human subjects, we applied the classifier to 281 single-subject activation maps derived from contrasts between: N-back working memory performance vs. rest (n = 94), negative vs. neutral emotional photographs (n = 108); and intense vs. mild thermal pain (n = 79). The classifier performed substantially above chance, identifying the originating study type with sensitivities of 94%, 70%, and 65%, respectively (chance = 33%), and specificities of 80%, 86%, and 98% (Fig. 4a). Moreover, there were systematic differences in activation patterns for correctly vs. incorrectly classified subjects. For example, incorrectly classified subjects in physical pain tasks (e.g., Fig. 4c) systematically activated lateral orbitofrontal cortex and dorsomedial prefrontal cortex, but not SII/posterior insula, suggesting that the discomfort due to noxious heat in these subjects may have been qualitatively different (e.g., emotionally-generated vs. physically-generated pain). Thus, these findings demonstrate the viability of decoding cognitive states in new subjects without training while suggesting novel hypotheses amenable to further exploration.
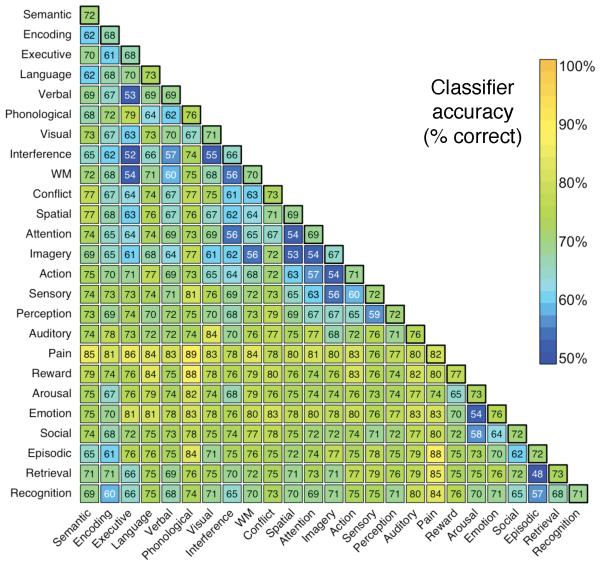
Next, to generalize beyond working memory, emotion, and pain, we selected 25 broad psychological terms used at high frequency in the database (Fig. 5). Classification accuracy was estimated in ten-fold cross-validated two-alternative and multi-class analyses. The classifier performed substantially above chance in both two-alternative classification (mean pairwise accuracy of 72%; Fig. 4) and relatively open-ended multi-class classification on up to ten simultaneous terms (Supplementary Fig. 9). Moreover, the results provide insights into the similarity structure of neural representation for different processes. For instance, pain was highly discriminable from other psychological concepts (all pairwise accuracies > 74%), suggesting that pain perception may be a distinctive state that is neither grouped with other sensory modalities or with other affective concepts like arousal and emotion. Conversely, conceptually related terms like ‘executive’ and ‘working memory’ could not be distinguished at a rate different from chance, reflecting their closely overlapping usage in the literature.
Figure 5.
Accuracy of the naive Bayes classifier when discriminating between all possible pairwise combinations of 25 key terms. Each cell represents a cross-validated binary classification between the intersecting row and column terms. Off-diagonal values reflect accuracy (in %) averaged across the two terms. Diagonal values reflect the mean classification accuracy for each term. Terms were ordered using the first two factors of a principal components analysis (PCA). All accuracy rates above 58% and 64% are statistically significant at P < .05 and P < .001, respectively.
DISCUSSION
The advent of modern neuroimaging techniques such as fMRI has spurred dramatic growth in the primary cognitive neuroscience literature, but has also made comprehensive synthesis of the literature increasingly difficult. The NeuroSynth framework introduced here addresses this problem in several ways. First, we validated a novel approach for conducting large-scale automated neuroimaging meta-analyses of broad psychological concepts that are lexically well represented in the literature. A key benefit is the ability to quantitatively distinguish forward inference from reverse inference, enabling researchers to assess the specificity of mappings between neural and cognitive function—a long-standing goal of cognitive neuroscience research. Although considerable work remains to be done to improve the specificity and accuracy of the tools developed here, we expect quantitative reverse inference to play an increasingly important role in future meta-analytic studies.
Second, we demonstrated the viability of decoding broad psychological states in a relatively open-ended way in individual subjects—to our knowledge, the first application of a domain-general classifier that can distinguish a broad range of cognitive states based solely on prior literature. Particularly promising is the ability to decode brain activity without prior training data or knowledge of the “ground truth” for an individual. Our results raise the prospect that legitimate ‘mind reading’ of more nuanced cognitive and affective states might eventually become feasible given additional technical advances. However, the present NeuroSynth implementation provides no basis for such inferences, as it distinguishes only between relatively broad psychological categories.
Third, the platform we introduce is designed to support immediate use in a broad range of neuroimaging applications. To name just a few potential applications, researchers could use these tools and results to define region-of-interest masks or Bayesian priors in hypothesis-driven analyses; conduct quantitative comparisons between meta-analysis maps of different terms of interest; use the automatically-extracted coordinate database as a starting point for more refined manual meta-analyses; draw more rigorous reverse inferences when interpreting results by referring to empirically established mappings between specific regions and cognitive functions; and extract the terms most frequently associated with an active region or distributed pattern of activity, thus contextualizing new research findings based on the literature.
Of course, the NeuroSynth framework is not a panacea for the many challenges facing cognitive neuroscientists, and a number of limitations remain to be addressed. We focus on two in particular here. First, the present reliance on a purely lexical coding approach, while effective, is suboptimal in that it relies on traditional psychological terms that may fail to carve the underlying neural substrates at their natural joints, fails to capitalize on redundancy across terms (e.g., ‘pain’, ‘nociception’, and ‘noxious’ overlap closely but are modeled separately), and does not allow closely related constructs to be easily distinguished (e.g., physical vs. emotional pain). Future efforts could overcome these limitations by using controlled vocabularies or ontologies for query expansion, developing extensions for conducting multi-term analyses, and extracting topic-based representations of article text (Supplementary Note).
Second, while our automated tools accurately extract coordinates from articles, they are unable to extract information about fine-grained cognitive states (e.g., different negative emotions). Thus, the Neurosynth framework is currently useful primarily for large-scale analyses involving broad domains, and should be viewed as a complement to, and not a substitute for, manual meta-analysis approaches. We are currently working to develop improved algorithms for automatic coding of experimental contrasts, which should substantially improve the specificity of the resulting analyses. In parallel, we envision a ‘crowdsourced’ collaborative model in which multiple groups participate in validation of automatically extracted data, thereby combining the best elements of both automated and manual approaches. Such efforts should further increase the specificity and predictive accuracy of the decoding model, and will hopefully lead to the development of many other applications that we have not anticipated here.
To encourage further application and development of a synthesis-oriented approach, we have publicly released most of the tools and data used in the present study via a web interface (http://neurosynth.org). We hope that cognitive neuroscientists will use, and contribute to, this new resource, with the goal of developing next-generation techniques for interpreting and synthesizing the wealth of data generated by modern neuroimaging techniques.
Supplementary Material
Acknowledgments
The authors wish to thank T Braver (Washington University), J Gray (Yale University), and K Ochsner (Columbia University) for providing data; E Reid for help with validation analyses; members of the Wager lab for manually coding the pain database; members of the Neuroimaging and Data Access Group (http://nidag.org), and particularly R Mar, for helpful suggestions; and R Bilder, R Raizada, and J Andrews-Hanna for helpful comments on a draft of this paper. This work was supported by awards NINR F32NR012081 to T.Y., NIMH R01MH082795 to R.P., NIH R01MH60974 to D.V.E., and NIMH R01MH076136, NIDA R01DA027794, and NIDA 1RC1DA028608 to T.D.W.
Appendix
ONLINE METHODS
Automated coordinate extraction
To automatically extract stereotactic coordinate information from published neuroimaging articles, we developed a software library written in the Ruby programming language. We have released the NeuroSynth Automated Coordinate Extraction (ACE) tools under an open source license, and encourage other researchers to contribute to the codebase (http://github.com/neurosynth). Because the code is freely available for inspection and use, we provide only a functional, non-technical overview of the tools here.
In brief, ACE consists of a parsing engine that extracts coordinate information from published articles by making educated guesses about the contents of the columns reported in tables in neuroimaging articles (at present, ACE does not attempt to extract coordinates that are reported in the main text of an article, or in supplementary materials). For each full-text HTML article provided as input, ACE scans all tables for rows that contain coordinate-like data. Rows or tables that do not contain values that correspond to a flexible template used to detect coordinates are ignored. Moreover, all extracted coordinates are subjected to basic validation to ensure that they reflect plausible locations in stereotactic space (e.g., all coordinates with absolute values > 100 in any plane are discarded).
Although neuroimaging coordinates are reported in a variety of stereotactic spaces in the neuroimaging literature30,31, for technical reasons, the results reported in the main text ignore such differences and collapse across different spaces. Moreover, the parser does not distinguish activations from deactivations, and aggregates across all reported contrasts within each article—i.e., it makes no attempt to code different tables within an article, or different contrasts within a table, separately. As extensive validation analyses show (Supplementary Note), these factors appear to exert only a modest influence on results, and in some cases can be automatically accounted for; however, for present purposes we simply note that the net effect of these limitations should be to reduce fidelity rather than to introduce systematic bias.
In addition to extracting coordinates, ACE parses the body of each article and generates a list of all words that appear at least once anywhere in the text, along with a corresponding frequency count for each word. All data are then stored in a relational (MySQL) database that maintains associations between words, articles, and activation foci, allowing flexible and powerful structured retrieval of information.
Database
The foci used to generate the results of the present study were extracted from 17 source journals, including Biological Psychiatry, Brain, Brain and Cognition, Brain and Language, Brain Research, Cerebral Cortex, Cognitive Brain Research, Cortex, European Journal of Neuroscience, Human Brain Mapping, Journal of Neurophysiology, Journal of Neuroscience, NeuroImage, NeuroLetters, Neuron, Neuropsychologia, and Pain. We deliberately focused on journals containing a high incidence of functional neuroimaging studies; thus, some important general neuroscience or science journals (e.g., Science, Nature, Nature Neuroscience, etc.) were not included. The range of years represented varied by journal, with the earliest studies dating to 2000, and the latest to early 2010. In total, the database contains 3,489 articles and 100,953 foci, representing the single largest extant database of neuroimaging foci—though still capturing only a minority of the published literature1.
Importantly, the database has considerable potential for additional growth. Because neuroimaging studies appear in dozens of different journals (besides those analyzed here), which typically require the use of publisher-specific or journal-specific filter to correctly obtain coordinates from tables, the database will continue to grow as new filters are added. An additional limitation is that many journals have not yet made full-text HTML versions of older articles available online; as such efforts proceed, the database will grow correspondingly. Finally, because authors report coordinate information in a variety of different formats, false negatives may occur (i.e., ACE may fail to extract real coordinate information). However, these limitations do not bias the present analyses in any particular way, and suggest that if anything, one can expect the sensitivity and specificity of the reported results to improve as the database grows and additional quality assurance procedures are implemented.
Statistical inference and effect size maps
In keeping with previous meta-analyses that employed multilevel kernel density analysis (MKDA15), we represented reported activations from each study by constructed a binary image mask, with a value of 1 (reported) assigned to each voxel in the brain if it was within 10 mm of a focus reported in that article, and 0 (not reported) if it was not within 10 mm of a reported focus (see ref. 15). These maps used 2×2×2 mm3 voxels, with nV=231,202 voxels in the brain mask. We denote the activation map for study i as Ai = (Aij), a length-nV binary vector.
A frequency cut-off of 0.001 was used in order to eliminate studies that only used a term incidentally (i.e., in order to be considered about pain, a study had to use the term ‘pain’ at a rate of at least one in every 1,000 words). Subsequent testing revealed that the results were largely insensitive to the exact cut-off used (including no cut-off at all), except that very high cut-offs (e.g., 0.01 or higher) tended to leave too few studies for reliable estimation of most terms. (As the database grows, even very conservative thresholds that leave no ambiguity at all about the topic of a study should become viable.) The total number of terms harvested was nT=10,000 (though the vast majority of these were non-psychological in meaning—e.g., ‘activation’, ‘normal’, etc.). We write term indicator for study i as Ti = (Tik), a length-nT binary vector marking each term “present” (i.e. has frequency above the cut-off) or “absent” (frequency below the cut off).
For each term of interest in the database (e.g., ‘pain’, ‘amygdala’, etc.) we generated whole-brain meta-analysis maps displaying the strength of statistical association between the term and reported activation at each voxel. For each voxel j and term k, every study can be cross-classified by activation (present or absent) and term (present or absent), producing a 2 × 2 contingency table of counts.
Statistical inference maps were generated using a Chi-Square test of independence, with a significant result implying the presence of a dependency between term and activation (i.e., a change in activation status would make the occurrence of the term more or less likely). This approach departs from the MKDA approach used in previous meta-analyses 3,15,16 in its reliance on a parametric statistical test in place of permutation-based family-wise error rate (FWE) correction; however, permutation-based testing was not computationally feasible given the scale of the present meta-analyses (multiple maps for each of several thousand terms).
To stabilize results and ensure all cells had sufficient observations for the parametric Chi-Square test, we excluded all voxels that were active in fewer than 3% of studies (Supplementary Fig. 10). The resulting p-value map was FDR-corrected for multiple comparisons using a whole-brain FDR threshold of 0.05, identifying voxels where there was significant evidence that term frequency varied with activation frequency. Intuitively, one can think of regions that survive correction as those that display differential activation for studies that include a term versus studies that do not include that term.
We also compute maps of the posterior probability that term k is used in a study given activation at voxel j:
(generically referred to as P(Term|Activation) in the text). We use the “smoothed” estimates for the likelihood of activation given term:
where p(·) reflects an estimated versus true probability, m is a virtual equivalent sample size, and p is a prior probability. The parameters m and p are set equal to 2 and 0.5 respectively; this smoothing is equivalent to adding two virtual studies that have term k present—one having an activation one, one without. This regularization prevents rare activations or terms from degrading accuracy32. For P(Tk=1) we impose a uniform prior for all terms, P(Tk=1)=P(Tk=0)=0.5. We use this uniform prior because terms differed widely in frequency of usage, leading to very different posterior probabilities for different terms. This is equivalent to making an assumption that the usage and non-usage of the term would be equally likely in the absence of any knowledge about brain activation. Note that this is a conservative approach; using uniform priors will tend to reduce classifier accuracy relative to using empirically estimated priors (i.e., allowing base rate differences to play a role in classification), because it increases the accuracy of rare terms at the expense of common ones. Nonetheless, we opted to use uniform priors because they place all terms on a level footing and provide more interpretable results.
The estimate of P(Aj=1) reflects the regularization and the prior on term frequency:
where
To ensure that only statistically robust associations were considered, all posterior probability maps were masked with the FDR-corrected p-value maps. For visualization purposes, thresholded maps were mapped to the PALS-B12 surface atlas33 in SPM5 stereotaxic space. Average fiducial mapping (AFM) values are presented. Datasets associated with Figure 2 and Supplementary Figure 3 are available in the SumsDB database (http://sumsdb.wustl.edu/sums/directory.do?id=8285126).
Naive Bayes classifier
We used a naive Bayes classifier29 to predict the occurrence of specific terms using whole-brain patterns of activation. In classifier terminology, we have nS instances of feature-label pairs (Ai,Ti). The use of a Naive Bayes classifier allows us to neglect the spatial dependence in the activation maps Ai. Since simultaneous classification for the presence/absence of all nT terms is impractical due to the larger number (2nT) of possible labels, for this work we only consider mutually exclusive term labels—ranging from binary classification of two terms (e.g., pain vs. working memory) to multiclass classification of ten terms (e.g., pain, working memory, language, conflict, etc.).
For this setting we revise notation slightly from the previous section describing calculation of the posterior probability maps, letting scalar Ti take values 1, …, nT* for subset of nT* terms under consideration. For a new study with activation map A, the probability of term t is
P(A) is computed as above for the studies under consideration, and by independence,
and we use a regularized estimate for voxel j,
where I(·) is the indicator function (1 if the operand is true, 0 otherwise).
Note that although the assumption of conditional independence is usually violated in practice, leading to biased posterior probabilities, this generally does not affect classification performance, because classification depends on the rank-ordered posterior probabilities of all classes rather than their absolute values. In complex real-world classification settings, naive Bayes classifiers often substantially outperform more sophisticated and computationally expensive techniques34.
In the context of the present large-scale analyses, the naive Bayes classifier has several advantages over other widely used classification techniques (e.g., support vector machines35,36). First, it requires substantially less training data than many other techniques, because only the cross-classified cell counts are needed. Second, it is computationally efficient, and can scale up to extremely large sets of features (e.g., hundreds of thousands of individual voxels) or possible outcomes without difficulty. Third, it produces easily interpretable results: the naive Bayes classifier’s assumption of conditional independence ensures that the strength of each feature’s contribution to the overall classification simply reflects the posterior probability of class membership conditioned on that feature (i.e., P(T=t|Ai).
Cross-validated classification of study-level maps
For cross-validated classification of studies included in the NeuroSynth database (Figs. 3 and 4), we used the NBC to identify the most probable term from among a specified set of alternatives (e.g., ‘pain’, ‘emotion’, and ‘working memory’) for each map. We used four-fold cross-validation to ensure unbiased accuracy estimates. Because the database was known to contain errors, we took several steps to obtain more accurate classification estimates. First, to improve the signal-to-noise ratio, we trained and tested the classifier only on the subset of studies with at least 5,000 “active” voxels, i.e. studies satisfying ΣjAij ≥ 5,000, occurring when there were more than about four reported foci; nS = 2,107 studies satisfied this criterion. This step ensured that studies with few reported activations (a potential marker of problems extracting coordinates) did not influence the classifier. Second, we only considered voxels that were activated in at least 3% of studies, i.e. ΣiAij/nS ≥ 0.03, ensuring that noisy features would not exert undue influence on classification. Third, any studies in which more than one target term was used were excluded in order to ensure that there was always a correct answer (e.g., if a study used both ‘pain’ and ‘emotion’ at a frequency of 0.001 or greater, it was excluded from classification). No further feature selection was used (i.e., all remaining voxels were included as features).
Classifier accuracy was calculated by averaging across classes (i.e., terms) rather than studies (e.g., if the classifier correctly classified 100% of 300 working memory studies, but 0% of 100 pain studies, we would report a value of 50%, reflecting the mean of 0% and 100%, rather than the study-wise mean of 75%, which allows for inflation due to differing numbers of studies). Using this accuracy metric, called balanced loss in the machine learning literature, eliminated the possibility of the classifier capitalizing on differences in term base rates, and ensured that chance accuracy was always 50%. Note that this is the appropriate comparison for a naive Bayes classifier when uniform prior probabilities are stipulated, because the classifier should not be able to capitalize on base rate differences even if they exist (as it has no knowledge of base rates beyond the specified prior).
For binary classification (nT*=2), we selected 25 terms that occurred at high frequency in our database (> 1 in 1,000 words in at least 100 different studies; see Figs. 5 and Supp. Fig. 3) and ran the classifier on all possible pairs. For each pair, the set of studies used included all those with exactly one term present (ns ranging from 23 to 794; mean = 178.2, median = 141). Statistical significance for each pairwise classification was assessed using Monte Carlo simulation. Across all comparisons, the widest 95% confidence interval was 0.4 – 0.6, and the vast majority of observed classification accuracies (283 / 300) were statistically significant (P < 0.05; 257/300 were significant at P < .001).
For multi-class analyses involving more than nT* > 2 terms (Supp. Fig. 5), an exhaustive analysis of all possible combinations was not viable due to combinatorial explosion and increased processing time required. We therefore selected 100 random subsets of nT* terms from the larger set of 25, repeating the process for values of nT* between 3 and 10. All procedures were otherwise identical to those used for binary classification.
Classification of single-subject data
For classification of single-subject data, we used data drawn from several previous studies, including a large study of N-back working memory37,38, five studies of emotional experience and reappraisal39-43, and three studies of pain14,44. Methodological details for these studies can be found in the corresponding references. For working memory, we used single-subject contrasts comparing N-back working memory blocks to a fixation baseline. For emotion studies, we used single-subject contrast maps comparing negative emotional pictures (from the IAPS set) to neutral pictures. For pain studies, we compared high and low thermal pain conditions.
Because the NBC was trained on binary maps (i.e., active vs. inactive), and single-subject p-maps varied continuously, all single-subject maps were binarized prior to classification. We used an arbitrary threshold of P < 0.05 to identify ‘active’ voxels. Because the maps the classifier was trained on did not distinguish activations from deactivations, only positively activated voxels (i.e., N-back > fixation, negative emotion > neutral emotion, or high pain > low pain) were considered active. Negatively activated voxels and non-significantly activated voxels were all considered inactive. To ensure that all single-subject maps had sufficient features for classification, we imposed a minimum cut-off of 1% of voxels—that is, for maps with fewer than 1% of voxels activated at P < 0.05, we used the top 1% of voxels, irrespective of threshold. Once the maps were binarized, all classification procedures were identical to those used for cross-validated classification of study-level maps.
It is important to note that the studies and contrasts included in the single-subject classification analysis were selected using objective criteria rather than on the (circular) basis of optimizing performance. The results we report include all studies that were subjected to the classifier (i.e., we did not selectively include only studies that produced better results), despite the fact that there was marked heterogeneity within studies. For instance, one of the pain studies produced substantially better results (n = 41; sensitivity = 80%) than the other two (total n = 34; sensitivity = 47%), likely reflecting the fact that the former study contained many more trials per subject, resulting in more reliable single-subject estimates. Thus, the accuracy levels we report are arguably conservative, as they do not account for the potentially lower quality of some of our single-subject data.
Similarly, the contrasts we used for the three sets of studies were selected a priori based on their perceived construct validity, and not on the basis of observed classification success. In fact, post-hoc analyses demonstrated that alternative contrasts would have produced better results in some cases. Notably, for the emotion studies, using single-subject maps contrasting passive observation of negative IAPS pictures with active reappraisal of negative pictures would have improved classifier sensitivity somewhat (from 70% to 75%). Nonetheless, we opted to report the less favorable results in order to provide a reasonable estimate of single-subject classifier accuracy under realistic conditions, uncontaminated by selection bias. Future efforts to optimize the classifier for single-subject prediction—e.g., by developing ways to avoid binarizing continuous maps, improving the quality of the automatically-extracted data through manual verification, etc.—would presumably lead to substantially better performance.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Derrfuss J, Mar RA. Lost in localization: the need for a universal coordinate database. Neuroimage. 2009;48:1–7. doi: 10.1016/j.neuroimage.2009.01.053. [DOI] [PubMed] [Google Scholar]
- 2.Yarkoni T. Big Correlations in Little Studies: Inflated fMRI Correlations Reflect Low Statistical Power-Commentary on Vul et al. Perspect Psycholog Sci. 2009;4:294–298. doi: 10.1111/j.1745-6924.2009.01127.x. 2009. [DOI] [PubMed] [Google Scholar]
- 3.Wager TD, Lindquist M, Kaplan L. Meta-analysis of functional neuroimaging data: current and future directions. Soc Cogn Affect Neurosci. 2007;2:150–158. doi: 10.1093/scan/nsm015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarkoni T, Poldrack RA, Van Essen DC, Wager TD. Cognitive neuroscience 2.0: building a cumulative science of human brain function. Trends Cogn Sci. 2010;14:496–489. doi: 10.1016/j.tics.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Horn JD, Grafton ST, Rockmore D, Gazzaniga MS. Sharing neuroimaging studies of human cognition. Nat Neurosci. 2004;7:473–481. doi: 10.1038/nn1231. [DOI] [PubMed] [Google Scholar]
- 6.Fox PT, Parsons LM, Lancaster JL. Beyond the single study: function/location metanalysis in cognitive neuroimaging. Curr Opin Neurobiol. 1998;8:178–187. doi: 10.1016/s0959-4388(98)80138-4. [DOI] [PubMed] [Google Scholar]
- 7.Van Essen DC. Lost in localization--But found with foci?! Neuroimage. 2009;48:14–17. doi: 10.1016/j.neuroimage.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laird AR, et al. ALE meta-analysis workflows via the BrainMap database: progress towards a probabilistic functional brain atlas. Front Neuroinformatics. 2009;3:23. doi: 10.3389/neuro.11.023.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson J, Drury HA, Van Essen DC. “The surface management system” (SuMS) database: a surface-based database to aid cortical surface reconstruction, visualization and analysis. Philos Trans R Soc Lond B Biol Sci. 2001;356:1277–1292. doi: 10.1098/rstb.2001.0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lancaster JL, et al. Bias between MNI and Talairach coordinates analyzed using the ICBM-152 brain template. Hum brain mapp. 2007;28:1194–205. doi: 10.1002/hbm.20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen FA, Hansen LK, Balslev D. Mining for associations between text and brain activation in a functional neuroimaging database. Neuroinformatics. 2004;2:369–80. doi: 10.1385/NI:2:4:369. [DOI] [PubMed] [Google Scholar]
- 12.Kanwisher N, McDermott J, Chun MM. The fusiform face area: a module in human extrastriate cortex specialized for face perception. J Neurosci. 1997;17:4302–4311. doi: 10.1523/JNEUROSCI.17-11-04302.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCandliss BD, Cohen L, Dehaene S. The visual word form area: expertise for reading in the fusiform gyrus. Trends Cogn Sci. 2003;7:293–299. doi: 10.1016/s1364-6613(03)00134-7. [DOI] [PubMed] [Google Scholar]
- 14.Atlas LY, Bolger N, Lindquist MA, Wager TD. Brain Mediators of Predictive Cue Effects on Perceived Pain. J Neurosci. 2010;30:12964. doi: 10.1523/JNEUROSCI.0057-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wager TD, Lindquist MA, Nichols TE, Kober H, Van Snellenberg JX. Evaluating the consistency and specificity of neuroimaging data using meta-analysis. Neuroimage. 2009;45:210–221. doi: 10.1016/j.neuroimage.2008.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kober H, et al. Functional grouping and cortical-subcortical interactions in emotion: A meta-analysis of neuroimaging studies. Neuroimage. 2008;42:998–1031. doi: 10.1016/j.neuroimage.2008.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poldrack RA. Can cognitive processes be inferred from neuroimaging data. Trends Cogn Sci. 2006;10:59–63. doi: 10.1016/j.tics.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Zald DH. The human amygdala and the emotional evaluation of sensory stimuli. Brain Res Brain Res Rev. 2003;41:88–123. doi: 10.1016/s0165-0173(02)00248-5. [DOI] [PubMed] [Google Scholar]
- 19.Shackman AJ, et al. The integration of negative affect, pain and cognitive control in the cingulate cortex. Nat Rev Neurosci. 2011;12:154–167. doi: 10.1038/nrn2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Owen AM, McMillan KM, Laird AR, Bullmore E. N-back working memory paradigm: a meta-analysis of normative functional neuroimaging studies. Hum Brain Mapp. 2005;25:46–59. doi: 10.1002/hbm.20131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dosenbach NU, et al. A core system for the implementation of task sets. Neuron. 2006;50:799–812. doi: 10.1016/j.neuron.2006.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duncan J. The multiple-demand (MD) system of the primate brain: mental programs for intelligent behaviour. Trends Cogn Sci. 2010;14:172–179. doi: 10.1016/j.tics.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Yarkoni T, Barch DM, Gray JR, Conturo TE, Braver TS. BOLD correlates of trial-by-trial reaction time variability in gray and white matter: a multi-study fMRI analysis. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0004257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craig AD. How do you feel? Interoception: the sense of the physiological condition of the body. Nat Rev Neurosci. 2002;3:655–666. doi: 10.1038/nrn894. [DOI] [PubMed] [Google Scholar]
- 25.Legrain V, Iannetti GD, Plaghki L, Mouraux A. The pain matrix reloaded: a salience detection system for the body. Prog Neurobiol. 2011;93:111–24. doi: 10.1016/j.pneurobio.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Norman KA, Polyn SM, Detre GJ, Haxby JV. Beyond mind-reading: multi-voxel pattern analysis of fMRI data. Trends Cogn Sci. 2006;10:424–430. doi: 10.1016/j.tics.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell TM, et al. Predicting human brain activity associated with the meanings of nouns. Science. 2008;320:1191. doi: 10.1126/science.1152876. [DOI] [PubMed] [Google Scholar]
- 28.Poldrack RA, Halchenko YO, Hanson SJ. Decoding the large-scale structure of brain function by classifying mental states across individuals. Psychol Sci. 2009;20:1364–1372. doi: 10.1111/j.1467-9280.2009.02460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis D. Naive (Bayes) at forty: The independence assumption in information retrieval. Machine Learning: ECML-98. 1998:4–15. [Google Scholar]
- 30.Van Essen DC, Dierker DL. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56:209–225. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 31.Laird AR, et al. Comparison of the disparity between Talairach and MNI coordinates in functional neuroimaging data: Validation of the Lancaster transform. Neuroimage. 2010;51:677–683. doi: 10.1016/j.neuroimage.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nigam K, McCallum AK, Thrun S, Mitchell T. Text classification from labeled and unlabeled documents using EM. Machine Learning. 2000;39:103–134. [Google Scholar]
- 33.Van Essen DC. A Population-Average, Landmark-and Surface-based (PALS) atlas of human cerebral cortex. Neuroimage. 2005;28:635–662. doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- 34.Langley P, Iba W, Thompson K. An analysis of Bayesian classifiers; Proceedings of the tenth national conference on Artificial intelligence; 1992.p. 223. [Google Scholar]
- 35.Mitchell TM, et al. Learning to decode cognitive states from brain images. Machine Learning. 2004;57:145–175. [Google Scholar]
- 36.Cox DD, Savoy RL. Functional magnetic resonance imaging (fMRI) “brain reading”: detecting and classifying distributed patterns of fMRI activity in human visual cortex. Neuroimage. 2003;19:261–270. doi: 10.1016/s1053-8119(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 37.DeYoung CG, Shamosh NA, Green AE, Braver TS, Gray JR. Intellect as distinct from openness: differences revealed by fMRI of working memory. J Pers Soc Psychol. 2009;97:883. doi: 10.1037/a0016615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shamosh NA, et al. Individual differences in delay discounting. Psychol Sci. 2008;19:904. doi: 10.1111/j.1467-9280.2008.02175.x. [DOI] [PubMed] [Google Scholar]
- 39.McRae K, et al. The neural bases of distraction and reappraisal. J Cogn Neurosci. 2010;22:248–262. doi: 10.1162/jocn.2009.21243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochsner KN, et al. For better or for worse: neural systems supporting the cognitive down- and up-regulation of negative emotion. Neuroimage. 2004;23:483–499. doi: 10.1016/j.neuroimage.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 41.Ochsner KN, Bunge SA, Gross JJ, Gabrieli JD. Rethinking feelings: an FMRI study of the cognitive regulation of emotion. J Cogn Neurosci. 2002;14:1215–1229. doi: 10.1162/089892902760807212. [DOI] [PubMed] [Google Scholar]
- 42.Wager TD, Davidson ML, Hughes BL, Lindquist MA, Ochsner KN. Prefrontalsubcortical pathways mediating successful emotion regulation. Neuron. 2008;59:1037–1050. doi: 10.1016/j.neuron.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McRae K, Ochsner KN, Mauss IB, Gabrieli JJD, Gross JJ. Gender differences in emotion regulation: An fMRI study of cognitive reappraisal. Group Processes & Intergroup Relations. 2008;11:143. doi: 10.1177/1368430207088035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kross E, Berman MG, Mischel W, Smith EE, Wager TD. Social rejection shares somatosensory representations with physical pain. Proc Natl Acad Sci U S A. 2011:1102693108. doi: 10.1073/pnas.1102693108. doi:10.1073/pnas.1102693108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lang PJ, Bradley MM, Cuthbert BN. International affective picture system (IAPS): Instruction manual and affective ratings. The Center for Research in Psychophysiology, University of Florida; 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.