Abstract
The HIV-1 long terminal repeat (LTR) is present on both ends of the integrated viral genome and contains regulatory elements needed for transcriptional initiation and elongation. Post integration, a highly ordered chromatin structure consisting of at least five nucleosomes is found at the 5′ LTR, the location and modification state of which controls the state of active viral replication as well as silencing of the latent HIV-1 provirus. In this context, the chromatin remodeling field is rapidly emerging as having a critical role in the control of viral gene expression. In the current study, we focused on unique Baf subunits that are common to the most highly recognized of chromatin remodeling proteins, the SWI/SNF complexes. We find that at least two Baf proteins, Baf53 and Baf170, are highly regulated in HIV-1 infected cells. Previously, studies have shown that the depletion of Baf53 in uninfected cells leads to the expansion of chromosomal territories and the decompaction of the chromatin. Baf53, in the presence of HIV-1 infection, co-elutes off of a chromatographic column as a different sized complex when compared to uninfected cells and appears to be predominantly phosphorylated. The innate function of Baf53-containing complexes appears to be transcriptionally suppressive, in that knocking down Baf53 increases viral gene expression from cells both transiently and chronically infected with HIV-1. Additionally, cdk9/Cyclin T in the presence of Tat is able to phosphorylate Baf53 in vitro, implying that this post-translationally modified form relieves the suppressive effect and allows for viral transcription to proceed.
Keywords: Chromatin remodeling, retrovirus, infection, phosphorylation, transcription
Introduction
The transcription of cellular genes is regulated by the highly organized chromatin structure within a cell’s nucleus; chromatin is folded into compact fibers (30 to 400 nm thick), which physically prevents the accessibility of cellular promoters by transcriptional machinery. A variety of enzymes and protein complexes are present in the cell to mediate the ‘opening’ and ‘closing’ of chromatin structure to allow for cellular processes to occur. This chromatin remodeling occurs through two major mechanisms, covalent epigenetic modifications of N-terminal histone tails and ATP-dependent chromatin remodeling complexes (CRCs). Ordered nucleosomal structures include DNA wrapped around core histones with a linker histone to hold the complex together, however, the N-terminal tails of the core histones are exposed and available for the addition of post-translational modifications. Extensive modifications are known to occur with multiple combinations of phosphorylation, acetylation, and methylation. A general rule of histone tail modifications argues that histone acetylation induces a euchromatin state and transcriptional activation whereas methylation induces a heterochromatin state and transcriptional repression. Notable cellular enzymes responsible for these events are the histone acetyltransferases (HATs) which covalently modify histone tails by acetylating lysine and arginine residues, inducing a conformational change at the nucleosome and, conversely, histone deacetylases (HDACs) which remove the acetyl groups from histone tails, reverting the nucleosomes back to a closed state. Similarly, histone methyltransferases (HMTs) and histone demethylases covalently methylate and demethylate histone tail residues, respectively. The manipulation of these post-translational modifications is critically linked to the availability of cellular gene promoters for transcription. CRCs are powerful molecular motors that use energy derived from ATP hydrolysis to disrupt histone–DNA contacts, thereby exposing nucleosomal DNA. Several classes of CRC have been isolated from eukaryotic cells, and all of these CRCs contain a related ATPase-motor subunit. The best-defined mammalian CRCs are the BAF (Brahma-related gene (BRG)/Brahma (BRM)-associated factor) complexes, which are related to the yeast SWI/SNF (switching-defective–sucrose non-fermenting) complex (Figure 1A). The mammalian BAF complexes are biochemically heterogeneous and functionally diversified, reflecting the elaborate mode of gene regulation that is unique to higher organisms. Finally, some of the BAF complexes associate into unique clusters (i.e. BRG can associate with Baf180-containing complex required for activated transcription; PBAF) and some are tissue specific and important for development (i.e. Baf60C expressed in the heart and somites of early mouse embryos1). A modern view of chromatin remodeling is that transcriptional activators must first recruit chromatin-remodeling enzymes to create a chromatin environment permissive for the preinitiation complex (PIC) assembly and ultimately transcriptional elongation2; 3; 4. Given that the HIV-1 provirus is integrated into the human genome, viral transcription and activation is likewise modulated by chromatin structure and will need to recruit the appropriate enzymes and chromatin remodeling proteins to preferentially direct cellular transcription to the viral LTR.
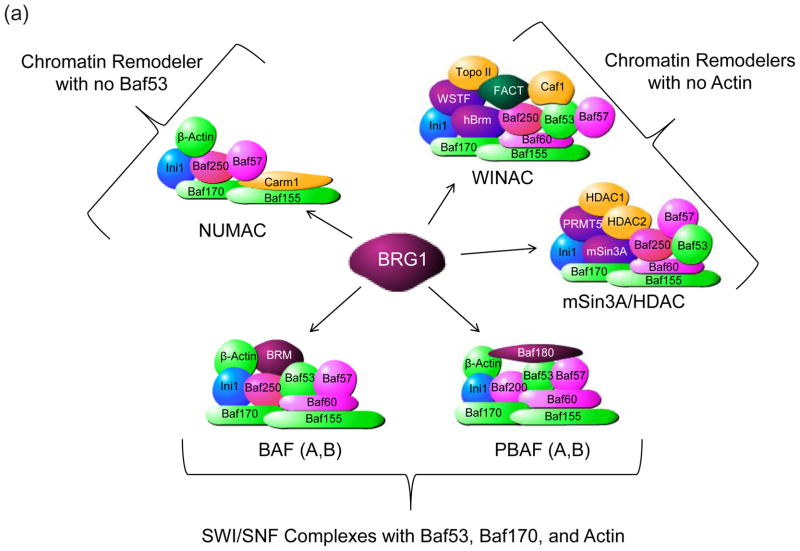
Figure 1. Differential organization of BRG1-associated chromatin-remodeling complex containing subunits.
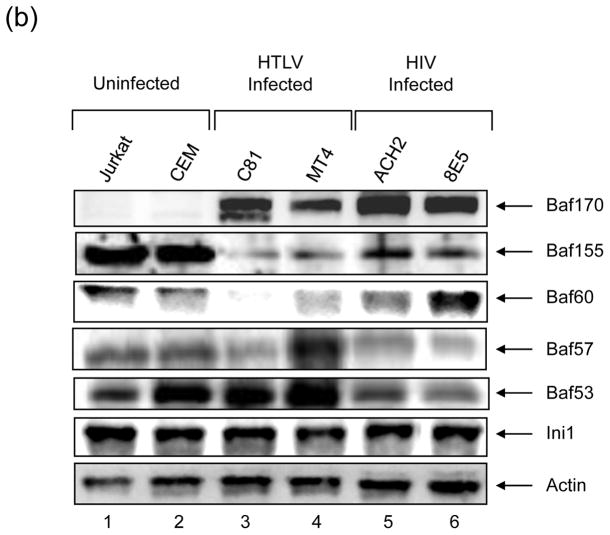
A) Chromatin remodeling complexes share subunits that have a variety of functions. All complexes shown contain Ini1, Baf170, and Baf155. The two BAF and PBAF complexes differ only by the incorporation of Brm and Baf180, respectively; all four complexes contain Baf53, Baf170, and Actin. The WINAC and mSIin3A/HDAC complexes do not contain Actin and the NUMAC complex does not contain Baf53. Subunits are organized as follows: Green – core proteins, Pink – nuclear receptor associated, Gold – DNA replication associated, Light Green – Actin related proteins. As chromatin remodelers, these complexes can act as either activators or repressors depending on the association of subunits. B) Identification of subunits between uninfected (Jurkat and CEM), HTLV-1 infected (C81 and MT4), and HIV-1 infected (ACH2 and 8E5) cells. One hundred microgram of each total extract was run on a 4–20% SDS-PAGE and western blotted with a specific Baf antibody. Ini1 and Actin are constitutively expressed in all cell lines.
Brahma-related gene 1 (BRG1) is the core catalytic subunit of a variety of SWI/SNF chromatin remodeling complexes, including BAF, PBAF, WINAC, NCoR, NUMAC, and mSin3A/HDAC 5; 6; 7; 8 (Figure 1A). While BRG1 does activate these complexes, it is important to note that other subunits of the complexes help determine whether a complex will act as a transcriptional activator or a repressor. For instance, core subunits that are essential to these complexes include Baf170, and Baf155; while subunits that are associated with nuclear receptors include Baf250, Baf60, and Baf53.
Baf53 is an Actin-related protein (Arp) that belongs to the Arp4 subfamily and can be found under the nomenclature Actin-like 6A, Act16, Arp4, Baf53a, and INO80K 9; 10. In Saccharomyces cerevisiae, Arp4 is a subunit of the Ino80 ATP-dependent chromatin-remodeling complex and the NuA4 histone acetyltransferase complex11; 12. In mammals, Baf53 is a subunit of the SWI/SNF ATP-dependent chromatin-remodeling complex13; 14; 15 the TIP60 histone acetyltransferase complexes, the TRRAP complex16 and the TIP48/49 helicase complex17. Arp4 is an essential gene in S. cerevisiae18 and disruption of Arp4 causes defects in NuA4 integrity and gene-specific transcription11,19. Baf53 is also indispensable for cell survival in mammals; knockdown of Baf53 with siRNA induces cell cycle arrest, the expansion of chromosome territories, and the decompaction of the chromatin20. These findings strongly support the idea that Baf53 plays a role in constructing higher-order chromatin structure such as chromosomal subdomains.
We and others have previously shown that both BRM and BRG are intricately involved in Tat-mediated activation of the HIV-1 LTR21; 22; 23; 24. Recently, a published interactome manuscript identified number of chromatin remodeling subunits that interact with the viral protein Tat, including those of the SWI/SNF, NURD, and FACT complexes25. Additionally, other cellular cofactors have been linked to Tat interaction, such as the histone chaperone protein hNAP-1 (Nucleosome Assembly Protein-1), which acts to promote viral transcription26. Very recently, we also have addressed which SWI/SNF family of chromatin remodeling complexes, BAF or PBAF, regulated Tat activated transcription in an acetylation dependent manner by modifying the nucleosome at the transcription start site (nuc-1)27; 28.
Here, in an attempt to define which components of the SWI/SNF complexes are important for activated transcription, we utilized a series of knock down experiments and found that some of the Baf members suppress HIV-1 transcription and some activate the promoter, stimulating viral replication and release. More importantly, we were surprised to find that some of the Baf components, such as Baf53, activated transcription in knockdown experiments and some were highly up-regulated after virus infection, such as Baf170. We found that Baf53 may play a role in genes regulating cell cycle progression (i.e. S-phase progression) and regulation of genes responsible for apoptosis. Baf53 was also differentially phosphorylated in HIV-1 infected cells and mostly observed in the supernatant (released complexes) after activated transcription with Tat. The implications of Baf53 effect will be discussed in the current manuscript.
Results
Varying levels of Bafs in virally infected cells
We previously have observed that retroviral infection of cells led to deregulation of some of the Baf subunits, therefore changing the stoichiometry of the SWI/SNF complexes in infected cells27; 29. Here we decided first to perform a series of western blots from uninfected and infected cells to see whether the Baf levels are differentially expressed in virally infected cells. We utilized uninfected, HIV-1, and HTLV-1 infected (control) cells and western blotted for key Baf protein subunits. Results are shown in Figure 1B, where there are clear changes in Baf subunit levels due to viral infection. For instance, Baf170 levels were virtually undetectable in uninfected cells, but dramatically increased in HIV-1 or HTLV-1 infected cells. Also, Baf60 levels were down regulated in HTLV-1 infected cells, but mostly unchanged in HIV-1 infected cells. Similarly, lower levels of Baf155 were observed in HTLV-1, but not HIV-1 infected cells. On the other hand, levels of Ini1 and Actin did not change in infected as compared to uninfected cells. Collectively, these data reinforce the notion that components of SWI/SNF complexes have varying levels of expression and dynamics in virally infected cells.
Effect of Baf53 knock down in retrovirally infected cells
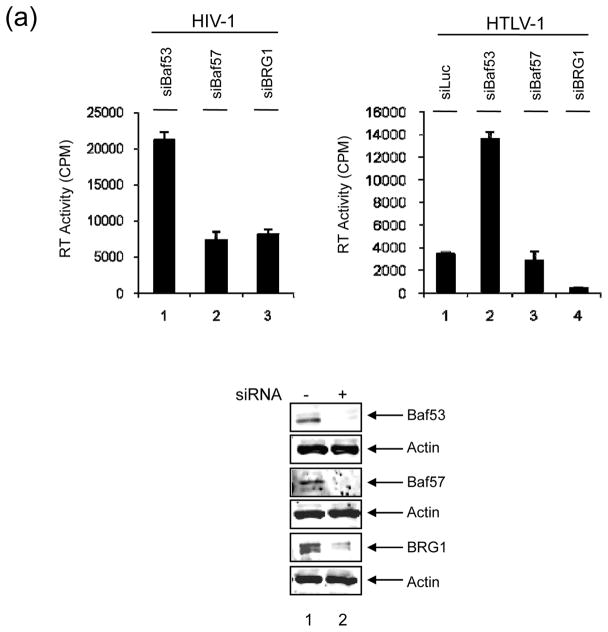
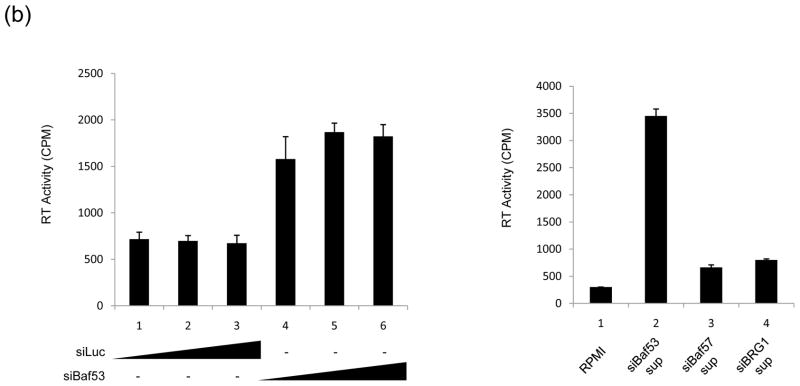
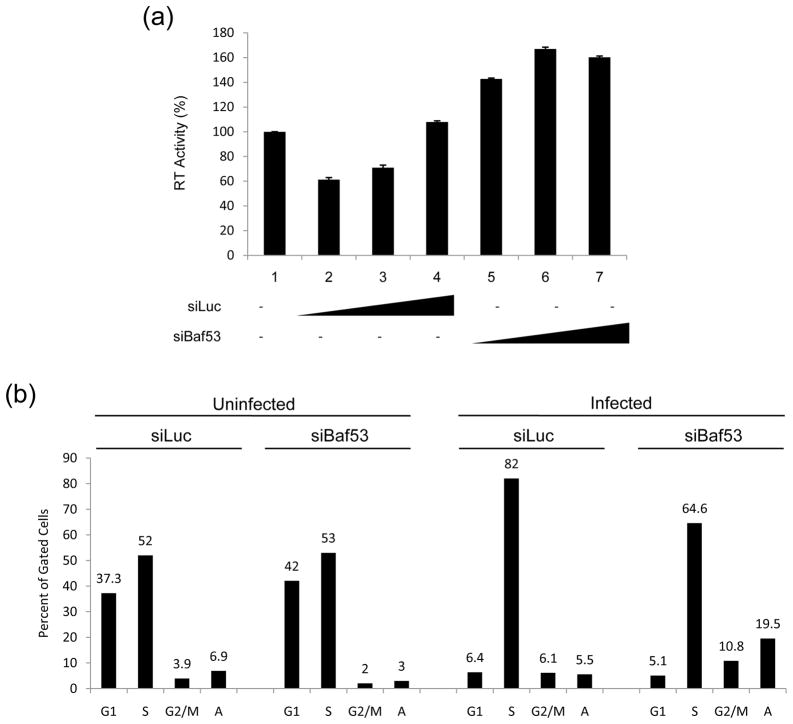
To determine which one of these complexes may be important for HIV-1 transcription, we utilized a number of siRNAs against specific Bafs in 293T transfection experiments. Cells were transfected with HIV-1 infectious clone, pNL4-3 and the indicated siRNAs. Results are shown in Figure 2A, where siRNA against Baf53 resulted in an up-regulation of HIV-1 in the supernatant, as compared to siBaf57 or siBRG1. This is consistent with our previous findings where knocking down of BRG1 resulted in a downregulation of HIV-1 gene expression27. We then asked whether this was unique to HIV-1 or if other viruses were also affected by Baf53 expression. We utilized a fully infectious clone of HTLV-1 (ACH, generous gift from Lee Ratner) in 293T transfection and found that, similar to HIV-1, knockdown of Baf53 increased the RT levels (infectious HTLV-1 particles) in the supernatant by more than fourfold (Figure 2A, upper right panel). This effect was specific since both siBaf57 and siBRG1 down-regulated HTLV-1 expression. Next, we subsequently performed a titration experiment in 293T cells with pNL4 construct and various concentrations of siLuc (control) or siBaf53 (experimental) RNA molecules. Results of such an experiment are shown in Figure 2B, where increasing concentrations of siBaf53, but not siLuc, showed a slight increase in RT levels in the supernatants. We then asked whether viral particles released from siBaf53 supernatants (Panel B, left side, Lane 6) or supernatants from siBaf57 or siBRG1 treated cells (Panel A, left side, lanes 2&3) were capable of reinfecting fresh 293T cells. Here we asked whether viral particles released from si-treated cells were functionally infectious. Results in Figure 2B, (Right side) indicated that at least some viruses released from siBaf53 treated cells were infectious as compared to siBaf57 or siBRG1 treated cell supernatants. Collectively, these results indicate that Baf53 may have an inhibitory effect on HIV-1 and HTLV-1 viral production.
Figure 2. SWI/SNF associated complexes involved in HIV-1 and HTLV-1.
A) 293T cells were transfected with pNL4-3 (HIV-1 clone, 10 μg, left panel) or ACH (HTLV-1 clone, 10 μg, right panel) along with siRNA against Baf53, Baf57 and BRG1 (100 nM each). Supernatants were collected after 48 hrs and used in an RT assay. Western blot analyses are shown below with α-Baf53, α-Baf57, α-BRG1 and α-Actin antibodies. B) Left panel: Three concentration of siRNA (50, 100 and 150 nM) were transfeced (electroporation) into 293T cells along with pNL4-3 (10 μg). Sups were collected for RT after 48hrs. Right panel: Supernatants (100 ul) from lane 6 of siBaf53 treated cells, and siBaf57 and siBRG1 (panel A) treated cells were used for infection of fresh 293T cells. Samples were collected 5 days after infection and used in RT assay.
Cell cycle effects of Baf53 knockdown in HIV-1 infected cells
Experiments above utilized a transfection of viral DNA into 293T cells and were scored for changes in RT levels in the supernatant. We next wanted to see if Baf knockdown could in fact activate silent virus already integrated into the genome. For this, we transfected siLuciferase (control, lanes 2–4) and siBaf53 (lanes 5–7) into HIV-1 latently infected J1.1 cells and collected supernatants after 48 hrs. Results in Figure 3A show that siBaf53 was able to increase viral expression in the supernatants of transfected cells (lanes 5–7) as detected by RT assay. However, the levels of activation in these cells were not as dramatic as when using drugs such as HDAC1 or HDAC3 inhibitors (data not shown).
Figure 3. Baf53 increases viral expression and regulates cell cycle in infected cells.
A) A titration of siRNAs against luciferase or Baf53 (100, 200, 300 nM) were performed by electroporated into J1.1 cells and supernatants were collected 48 hrs post-transfection for RT activity screening. B) CEM and ACH2 (infected cells) were transfected with 100 nM of either siBaf53 or siLuc as control. Cells were processed for cell cycle analysis using Propidium Iodine staining (PI) followed by FACS analysis. Each individual cell cycle phase population is graphed here. Samples were processed two days post siRNA treatment. “A” represents cells that had sub G-1 (apoptotic) peaks.
We then asked whether siRNAs against Baf53 had an effect in cell cycle progression in uninfected, as well as infected cells. Here we used uninfected T-cells as well as HIV-1 infected cells in transfections with siRNA followed by PI staining. Results of such an experiment are shown in Figure 3B. siBaf53 had minimal alterations in uninfected cells. There was a pronounced decrease in the G1 population, a slight increase in the S-phase population, and a pronounced increase in the apoptotic population (3 vs. 19.5) in infected cells. These results collectively indicate that Baf53 may play a role in genes responsible for apoptosis in infected cells.
A phosphorylated form of Baf53 co-elutes with Baf57 and Actin in HIV-1 infected cells
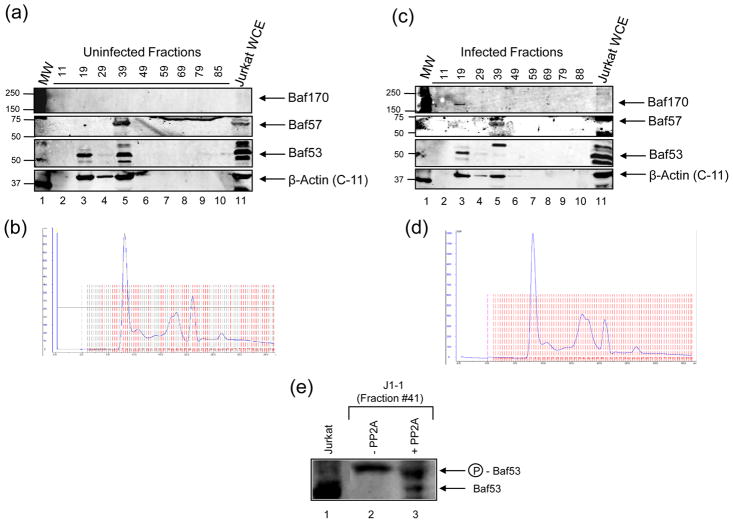
In order to determine the elution profile of the different chromatin remodeling complexes present in uninfected compared to HIV-1 infected cells, we performed size exclusion chromatography to separate native protein complexes by size and subsequently detect the proteins with western blots. Five mg of protein extract from Jurkat and J1.1 cell lines was subjected to column chromatography and fractions were collected as protein complexes eluted off of the column from high molecular weight to small complexes. The chromatograph associated with each elution is represented in Figure 4, B and D. A sampling of the proteins collected off of the column was performed by examining roughly every 10 fractions for the presence of Baf170, Baf57, Baf53, and Actin. Both Jurkat (Figure 4A) and J1.1 (Figure 4C) showed Baf53 and Actin co-eluting in the same high molecular weight fraction # 19 (lane 3A, lane 3C). Baf170 was also present in this high molecular weight fraction, but only in the infected cells. Both cell lines also showed Baf53, Actin, as well as Baf57 co-eluting in a slightly lower molecular weight fraction # 39 (lanes 5). Interestingly, Baf53 western blot indicated multiple forms in uninfected cells, mainly a possible unphosphorylated form. On the other hand, the majority of the Baf53 in infected cells appears to be a slightly higher, possibly phosphorylated form that could account for the size difference. We then asked whether Baf53 in HIV-1 infected J1-1 cells were indeed phosphorylated. For these experiments we used J1-1 fraction #41 which contained mostly the phosphorylated form of Baf53 and treated the fraction with phosphatase PP2A, followed by western blot against Baf53. Results of such an experiment are shown in Figure 4E, where PP2A treatment partially dephosphorylated the upper band to where the unphosphorylated Baf53 from Jurkat fraction #41 is detected. This implies that at least part of the modification for the upper Baf53 band in infected cells is the phosphorylated form of the protein.
Figure 4. Co-elution of Baf170, Baf57, and Baf53 in different complexes in uninfected vs. infected cells.
Size exclusion chromatography of native protein complexes of uninfected (A, B) compared to HIV-1 infected (C, D) T-cells. Fraction numbers correspond to elution of proteins from column as indicated by the chromatographs, from highest molecular weight complexes to lowest. Jurkat whole cell extracts are used as a positive control for WBs against Baf170, Baf57, Baf53, and Actin from a panel of fractions. Panel E represents PP2A treatment of J1-1 fraction #41 which contained mostly phosphorylated Baf53.
We also asked whether BRG1/Baf53 or its associated members in the BAF or PBAF complexes contained other post translational modifications including phosphatidylinositol that could control nuclear association14. Here both infected and uninfected cells were treated with a mixture of PIP2, a major mediator of signaling in lymphocytes, and western blotted for free or bound BRG1. It has previously been shown that PIP2 controls the nuclear association of the entire BAF complex and its association with chromatin14. Our results indicated that BRG1 complex which associates with BAF and PBAF components were mostly free in both infected and uninfected cells and upon treatment with PIP2, most of the BRG1 associated complexes were nuclear bound (Supplemental Figure 1) in both cell types. Therefore we concluded that the modification of BAF or PBAF components by PIP2 enhanced nuclear chromatin interaction irrespective of infection. These data collectively suggested that Baf53 may be differentially modified (i.e., phosphorylated) in HIV-1 infected T-cells as compared to uninfected cells, which may also contribute to the incorporation of different subunits into chromatin remodeling complexes.
Phosphorylation of Baf53 in vitro
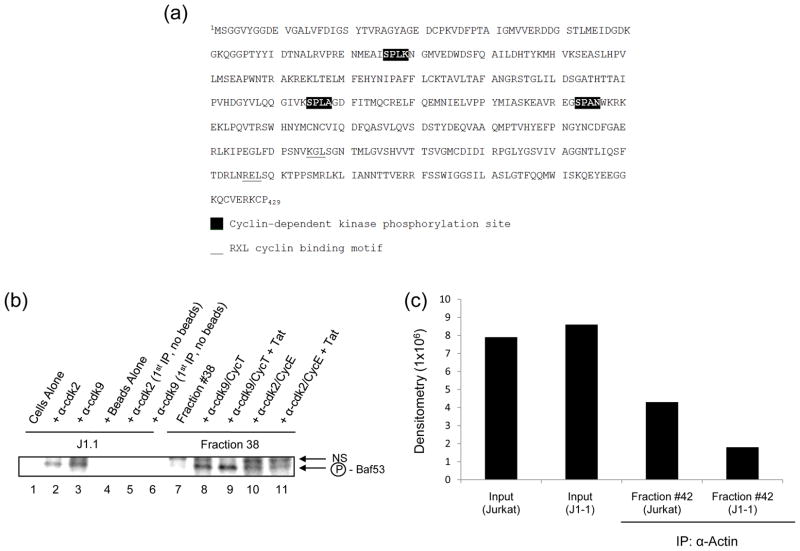
Baf155 and Ini1 are members of the SWI/SNF-like chromatin remodeling complexes and have been shown to be candidate substrates for phosphorylation by Akt30. We were interested to determine if Baf53 could serve as a kinase substrate. Full length Baf53 sequences were scanned (NCBI NP_004292.1) using the PROSITE web tool (http://au.expasy.org/prosite) for potential phosphorylation sites. A total of 12 sites were identified for PKC, CK2, and CAMP-dependent kinases. In addition, Baf53 harbored two RXL Cyclin binding motifs and three (S/T)PX(K/R) cdk phosphorylation sites31 (Figure 5A). These cdk/Cyclin binding and phosphorylation sites became of considerable interest, since HIV-1 basal and activated transcription requires multiple cdk/Cyclin complexes for optimal transcription.
Figure 5. Phosphorylation of Baf53 by cdk2/Cyclin E and cdk9/Cyclin T.
A) Baf53 primary structure contains multiple putative phosphorylation sites. Baf53 amino acid sequence (NCBI NP_004292.1) is shown to have three potential (S/T)PX(K/R) cdk phosphorylation sites (black box) and two RXL Cyclin binding motifs (underlined). B) Two milligrams of J1-1 total extract was IPed with α-cdk2 or α-cdk9 (10 μg) overnight. The next day, a 30% protein A/G bead slurry was added and samples were washed twice with TNE50 + 0.1% NP-40. Samples were then treated with Jurkat FPLC fraction # 38 and γ-32P ATP to initiate the kinase reaction (1 hr at 37°C). Following incubation, samples were diluted with RIPA buffer, centrifuged, and supernatants were IPed with α-Baf53 antibody overnight. The next day beads were added and samples were washed 3x with TNE50 + 0.1% NP-40. Separation of labeled proteins was carried out on 4–20% SDS-PAGE gels as follows: Lane 1: no ab; Lane 2: α-cdk2; Lane 3: α-cdk9; Lane 4: no beads; Lane 5: α-cdk2 and no beads; Lane 6: α-cdk9 and no beads; NS refers to non-specific bands. Samples 5 and 6 did not contain beads for the first IP (negative control). Samples 7–11 contained in vitro purified cdk2/Cyclin E or cdk2/Cyclin T as opposed to IPed material. One hundred nanograms of cdk2/Cyclin E with or without purified Tat (200 ng) were used in these assays. Cdk9/Cyclin T and cdk9/Cyclin T/Tat were used at 150 ng per reaction in these assays. Seventy microliters of fraction # 38 was mixed with cdk/Cyclin complexes for 1hr at 37°C along with γ-32P ATP. Next, samples were treated with 5x RIPA and IPed overnight with α-Baf antibody. The next day IPed material was recovered with a 30% protein A/G bead slurry, washed with TNE50 + 0.1%NP-40, heat denatured, and separated on a 4–20% SDS-PAGE gel as follows: Lane 7: Fraction #38 alone; Lane 8: #38 + α-cdk9/Cyclin T; Lane 9: #38 + α-cdk9/Cyclin T/Tat; Lane 10: #38 + α-cdk2/Cyclin E; Lane 11: #38 + α-cdk2/Cyclin E/Tat; NS refers to non-specific bands. Dried gels were exposed to PhosphoImager cassettes and subjected to dosimetry. C) FPLC fractions # 42 from J1-1 and Jurkat cells (~200 μg) were cross-linked with 0.5 mM dithiobis-succinimidylpropionate (DSP) for 30 min at 37°C. The extracts were then denatured by resuspension in 5.5 M urea. The denatured extracts were subsequently diluted 5-fold in RIPA buffer and immunoprecipitated with α-Actin (10 μg). Samples were IPed for 48 hrs at 4°C and then a 30% protein A/G bead slurry was added for 2 hrs at 4°C. Samples were washed three times with TNE50 + 0.1 % NP40. The cross-linking was cleaved by boiling the sample in Laemmli buffer and proteins were analyzed by 4–20% SDS-PAGE gels and western blotted with α-Baf53 antibody.
We next asked whether Baf53 could be phosphorylated in vitro. For these experiments, we utilized either cdk2 or cdk9 immunoprecipitated from J1-1 infected cells (source of kinase) and used the unphosphorylated form of Baf53 from the Jurkat uninfected fraction #38 as the substrate. The experimental design of the IP included first using whole cell extract from J1-1 cells and immunoprecipitating with α-cdk2 or α-cdk9. The IPed material (which contained kinase active cdk2 or cdk9 from infected cells) was then mixed with FPLC fraction # 38 (from Jurkat cells which contained mostly the unphosphorylated form of Baf53) and with γ-32P ATP for 1hr at 37°C. Samples were then treated with RIPA buffer and then centrifuged. Soluble radioactive-labeled proteins were then IPed overnight with α-Baf53 antibody. Subsequently, beads were added and radiolabeled proteins were separated on a 4–20% Tris-Glycine SDS-PAGE gel. Results of such an experiment is shown in Figure 5B (Lanes 1–6) where both cdk2 and cdk9 IPed material were able to show phosphorylation of Baf53 (“NS” refers to a non-specific band). Next, a somewhat similar set of experiments were performed with purified cdk2/Cyclin E and cdk9/Cyclin T in the presence or absence of Tat (Supplemental Figure 2). Here, Jurkat fraction # 38 was directly incubated with purified cdk/Cyclin complexes along with γ-32P ATP in a kinase reaction. Samples were then IPed with α-Baf53 antibody overnight and complexes were resolved on a 4–20% Tris-Glycine SDS-PAGE gel. Results of this experiment are shown in lanes 7–11, where cdk9/Cyclin T and cdk9/Cyclin T/Tat more effectively phosphorylated Baf53 as compared to cdk2/Cyclin E or cdk2/Cyclin E/Tat. Collectively, these results suggest that the cdk9/Cyclin T complex more efficiently phosphorylates Baf53 in vitro.
It has previously been shown that BRG1 was required for Actin and Baf53 to associate with the BAF complex. This study utilized cross linking of the nuclear extracts first, and then treated their Jurkat extracts with 5.5 M urea for 30 min at 25°C, diluted with RIPA, and finally immunoprecipitated with Actin antibody. They observed Actin and Baf53 to directly interact with BRG1 in the BAF complex in vitro. Along these lines we used a similar approach of cross linking our infected and uninfected fractions first, and then IPing with α-Actin antibody followed by western blots for Baf53. The hypothesis here was that the unphosphorylated form of Baf53 may be in complex with Actin, but the phosphorylated form of Baf53 (from infected cells) may no longer be associated with Actin. Results of such an experiment is shown in Panel C, where Actin and Baf53 were found to coimmunoprecipitate in the Jurkat fraction (# 42), however much less Baf53 were found to complex with Actin from the J1-1 infected fraction (# 42, counts from IP/WB). Collectively these results indicate that Baf53 may be phosphorylated in cells with kinases including cdk2/Cyclin E and/or cdk9/Cyclin T complexes. The phosphorylation event may be important for release of the Baf53 from its Actin-associated partners.
Phosphorylation of Baf53 can occur during Tat activated transcription
The lab of Emma Lees has previously identified a number of proteins co-precipitating with Cyclin E and not with others including Cyclin A, B1, or D32; 33. In these assays, it was shown that both BRG1 (which contains DNA-dependent ATPase activity) and Baf155 (both components of SWI/SNF chromatin remodeling complex) were heavily phosphorylated in the Cyclin immunoprecipitates (off DNA experiments). They also showed that cdk2/Cyclin E associated with SWI/SNF complexes and appeared to be relatively stable and resistant to stringent washing conditions (750 mM NaCl washes).
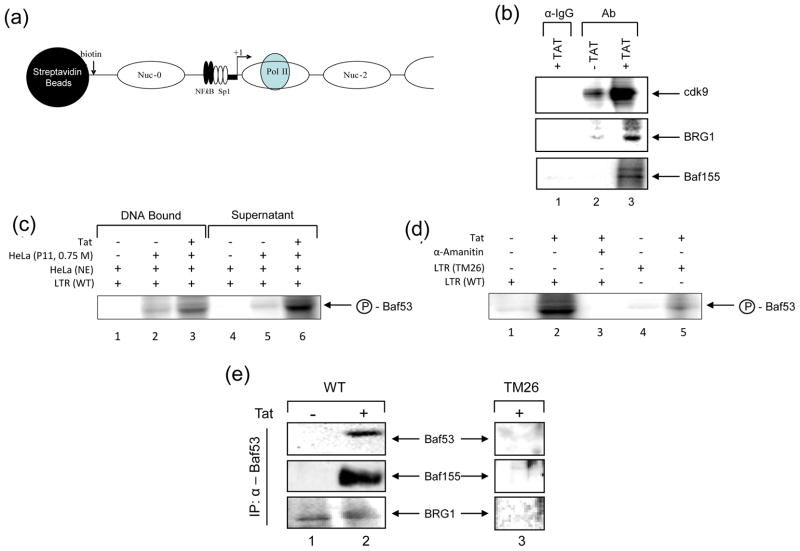
We and others have previously shown that the HIV-1 promoter utilizes SWI/SNF complexes for activated transcription both in vitro and in vivo. To validate the previously published phosphorylation data, we first asked if either BRG1 or other components of SWI/SNF (i.e. Baf155) could be phosphorylated by cdk2/Cyclin E or cdk9/Cyclin T kinases on the HIV-1 LTR. For these experiments we utilized a biotin pull-down assay where the HIV-1 LTR (U3/R/U5) was chromatinized with all 4 histones and subsequently used for in vitro transcription (IVT) using active extracts (Figure 6A). We reasoned that in the presence of cold nucleotides for RNA synthesis and 32p γ-ATP, cdk/Cyclin complexes may be able to phosphorylate BRG1, and Baf155 (or other proteins) and we could subsequently pull-down the DNA associated complexes with streptavidin beads, dissociate protein complexes using RIPA buffer, and immunoprecipitate (IP) with specific antibodies to cdk9, BRG1 and Baf155. Results of such an experiment are shown in Figure 6 panel B, where the IgG negative control IP did not bring down cdk9, BRG1 or Baf155 phosphorylated proteins (Lane 1). Interestingly in the absence of Tat, the IVT reaction pulled down very little cdk9 or BRG1 with the indicated antibodies (Lane 2). However, in presence of Tat we observed much better phosphorylation activity in the pull-down assay (Lane 3). All three proteins exhibited a varying degree of phosphorylation when Tat was present.
Figure 6. Substrates of cdk/Cyclin complexes on HIV-1 DNA.
A) In vitro transcription was performed with CEM whole-cell G1/S extracts (50 μg total) on immobilized HIV-1 LTR chromatin templates. The DNA fragments were biotinylated, gel purified, and reconstituted with core histones by step dilution. In vitro transcription reactions in the presence or absence of Tat (500 ng) were incubated for 1 hr at 30°C and contained the nucleoside triphosphates cold ATP (0.05 μM), GTP, UTP, and CTP at a final concentration of 50 μM and [32P] γ-ATP (20 μCi; 400 Ci/mmol; Amersham, Piscataway, NJ) in buffer D. B) Transcription reactions were washed twice after 1hr with TNE 50 + 0.1% NP-40 and subsequently addition of 200 μl of RIPA buffer to strip proteins off of the DNA. Radioactively labeled proteins were then used for immunoprecipitations with specific antibodies to cdk9 (top panel), BRG1 (middle panel), Baf155 (bottom panel) (10 μg each) and other Baf antibodies (data not shown). IPed material was ran on a 4–20% gel, dried and exposed to a PhosphoroImager cassette. C) In vitro transcription was performed with wild type chromatinized LTR, HeLa nuclear extracts (12 μg total), 25 μg of HeLa phosphocellulose nuclear fraction (0.75 M NaCl) that contained both BAF and PBAF complexes, and Tat protein (200 ng). D) Same as panel C, except LTR TAR mutant (TM26) was used as control. α-Amanitin at 0.1 μg/ml was used to inhibit transcription. E) Same experiment as panel C, however following immunoprecipitation of the hot Baf53 from the supernatants, samples were washed in TNE 50 + 0.1% NP-40, ran on a 4–20% SDS/PAGE and western blotted with antibodies against Baf 53 (positive control), Baf155, and BRG1. Lanes 1 and 2 are transcription from HIV-1 wild type LTR and lane 3 used TAR mutant TM26.
We next performed a similar set of experiments using either wild type or TAR mutant DNA. HeLa nuclear extracts were incubated with LTR DNA, HeLa phosphocellulose nuclear fractions (0.75 M NaCl) that contained both BAF and PBAF complexes, and Tat protein. Following the IVT reaction, both the DNA (panel C, lanes 1–3) and supernatant of the IVT (panel C, lanes 4–6) were mixed with RIPA buffer, and then immunoprecipitated with α-Baf53 antibody. Interestingly we observed more labeled Baf53 in the supernatant of the IVT reaction (compare lanes 3 and 6) which may indicate that phosphorylated Baf53 following transcription is mostly dissociated from DNA. Finally, we performed an experiment using an LTR TAR mutant DNA (TM26) and observed minimal levels of Baf53 release into the IVT supernatant reaction (Panel D, compare lanes 2 and 5). The phosphorylation of Baf53 was transcription dependent as the presence of α-Amanitin did not show any phosphorylated Baf53 (Lane 3).
We finally asked whether the phosphorylated form of Baf53 was disassociating away from the whole BAF complex or was it still associated with at least some members of the BAF complex after Tat activated transcription. We performed similar IVT experiments as above and used the radioactive supernatant (off DNA complexes) first by immunoprecipitating with α-Baf53 antibody followed by western blots for Baf53, Baf155 and BRG1. Results of such an experiment are shown in panel E, where there was a clear association of Baf53 with Baf155 but very little with BRG1 protein. This complex was specific to the wild type LTR and not the TAR mutant (TM26). Collectively, these results imply that phosphorylation of Baf53 by transcriptionally competent complex is disassociated from the LTR and it most likely is in complex with other subunit of BAF including Baf155. Collectively, these data indicates that Baf53 may be phosphorylated in the presence of Tat activated transcription and that the majority of the Baf53 may be released from DNA following active transcription.
Absence of Baf53 on activated LTR promoter
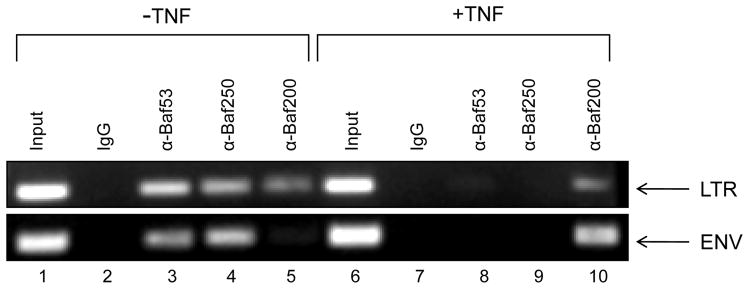
We then asked whether Baf53 could associate with the LTR before and after activation. Here we used J1-1 cells and treated these cells with TNFα to activate the virus. We then performed chromatin immunoprecipitation experiments using various Baf antibodies and PCR amplified two target regions including LTR (nuc-1 region) and ENV. We have previously shown that SWI/SNF BAF and PBAF complexes are both present on the LTR, but once activated, the BAF complex (or at least some of its components) are removed and the predominant remaining complex is PBAF27. These two complexes can be separated based on the two unique protein components where BAF contains Baf250 and PBAF contains Baf200 and Baf180. Results of such an experiment are show in Figure 7, where all three Baf53, Baf250 and Baf200 are present on the LTR in the absence of induction. However, upon addition of TNFα, the majority of Baf53 and Baf250 were removed from the promoter and only Baf200 remain. Similar results were also seen with the ENV primers indicating that both Baf53 and Baf250 may be insulating the entire viral genome and assist in general repression. Taken together these data imply that DNA associated with active transcription may have much less inhibitory proteins and a minimal selective set of activators present on the viral genome.
Figure 7. Absence of Baf53 protein on activated HIV-1 LTR promoter.
Chromatin Immunoprecipitation (ChIP) assays were performed in J1-1 cells (5% serum) either untreated or treated with TNF-α (10 ng/ml) for 2 hrs. Twenty four hours post treatment, cells were cross linked and DNA sonicated for ChIP assays. Antibodies (10 ug of each) against Baf53, Baf250 and Baf200 were used in the ChIP assays and samples were processed for presence of HIV-1 LTR (+ 10 to + 165) or Env (+ 8990 to + 9120) using specific primers. Following PCR reaction 1/5 of the samples were ran on a 2% agarose gel and stained.
Discussion
As shown in Figure 1, BRG1 uses energy from its ATP hydrolysis properties to power a variety of complexes toward chromatin remodeling. While BRG1 does power these complexes, it is important to note that other subunits of the complexes help determine whether a complex will act as a transcriptional activator or repressor. This issue becomes even more significant when looking at viral infections. For instance we have consistently observed varying levels of Bafs in HIV-1 or HTLV-1 infected cells. A more dramatic result has been with Baf170, but its role is still unclear to us. We have performed some preliminary experiments in HIV-1 infected cells and have found that siBaf170 in fact suppresses viral activation in promyelocytic cells (Supplemental Figure 3). However, we find much more consistent data with siBaf53 activating virus partially in cancer cells. In addition to the increase in viral replication in the absence of Baf53 possibly due to release of chromatin modulation mechanisms, it is probable that this effect is also due to indirect cellular responses to the loss of Baf53. Baf53 may play an additional non-chromatin remodeling related role in the suppression of HIV-1 viral gene and cellular gene expression. It remains to be seen if these results reproduce in primary T-cells and/or monocyte/macrophages, since their chromatin structure is much more condensed as compared to the cancer cells used here. In fact, the Baf53 knockdown data may be more significant and dramatic in primary cells when scoring for either a better integration (Ini1, a component of SWI/SNF was originally discovered for its involvement in integration) or transcriptional activation, since Baf53 presumably is involved in closing the chromatin structure in quiescent cells. It may also be very possible that there are a collective combination of effects including removal of histone H1, recruitment of activating factors, such as NFκB, or modulation of chromatin complexes on histone tails, in siBaf53 treated cells which results in the activation of the LTR.
Our other virus data with HTLV-1 also indicates that the Baf53 effect may not be so specific to HIV-1 but toward most retroviral infections in general. Although we don t have a complete set of data to predict such a general function, we expect that the opening up of host chromatin may be significant for viruses that require nuclear transcription and nuclear signal transduction.
Our chromatography data also suggests that the SWI/SNF components elute from two distinct places. It is interesting to note that the large complex (Fraction #19) in uninfected cells has Baf53 and Actin but no Baf170. In contrast, Baf170 is present in Fraction #19 in infected cells. The second complex that eluted is slightly smaller (Fraction #39) which contains no Baf170, but has Baf57, 53, and Actin. More importantly it seems that the majority of the Baf53 in infected cells is phosphorylated. Potentially, this phosphorylation event is occurring by kinases such as cdk9/Cyclin T and/or cdk2/Cyclin E (Supplemental Figure 4). An intriguing result from our studies is that a combination of cdk9/Cyclin T/Tat is able to phosphorylate Baf53 more efficiently which presumably is now released from DNA, allowing for elongation of transcription to take place.
In addition, Baf53 is a component of the TRRAP complex that is essential for c-Myc- and E1A-mediated oncogenic transformation34. Although not proven, the proposed role of Baf53/b-Actin complex is to regulate dynamic motile processes via Actin polymerization, in a manner analogous to the Arp2/3 complex. Therefore, an intriguing possibility is that Baf53/β-Actin binds to exporting complexes and contributes to their motility. In recent years the control of subcellular localization has emerged as one of the main mechanisms that regulate the activity of a variety of molecules including beta-catenin, NF-κB, and NFAT. It will be interesting to determine whether the subcellular distribution of Baf53 changes in particular physiological conditions after infection.
As BRG1 is necessary for HIV-1 transcription, future studies are needed to understand which BRG1 chromatin complexes are critical or whether different complexes are important for certain stages of the viral life cycle (i.e. active vs. latent populations). It is also important to note that the BRG1-containing BAF complex has been shown to play a role in transcriptional activation and gene regulation35; 36. It has been shown that a catalytically inactive mutant of BRG1 is still able to activate transcription in the complete absence of ATP-dependent chromatin remodeling activity. It is necessary to determine whether or not Baf53 regulates HIV-1 gene expression and transcription in a chromatin-independent manner in primary latent cells.
Our results from the ChIP assays are also interesting in that it appears that a number of large complexes including BAF and PBAF are normally present on the inactive LTR. However, once transcription is activated, most of the inhibitory complexes including BAF (i.e. Baf250) are removed from the genome and mostly replaced by active complexes such as PBAF (i.e. Baf200). It is unclear at this point whether Baf53 that is phosphorylated may be ejected from the LTR as a complex (i.e. as part of BAF complex) or it is simply removed by itself away from any BRG1 containing complexes that may be associated with LTR. However, one interesting observation is that both Baf53 and some Baf155 may be released as sub-complexes as seen in the in vitro transcription assays.
Future studies could also focus on determining the contribution of the less well-defined Baf subunits to viral replication as well as the possibility of viral infection creating various stoichiometrically novel complexes of SWI/SNF that benefits the virus over host gene expression. It would also be worth pursuing other epigenetic markers in these Baf53 knockdown uninfected and infected cells. For instance, H3-K9 and H3-K79 methylation are two key histone modifications closely linked to the formation of silent chromatic regions with the opposite behaviors: H3-K9 is hypermethylated and H3-K79 is hypomethylated in silent chromatic regions. The hypermethylated H3-K9 and the hypomethylated H3-K79 could promote the association of non-histone architectural proteins such as HP1 making the chromatin more compact. Indeed, we have previously shown that in latently HIV-1 infected OM10.1 cells, there is a dramatic increase in the methylation state of Histone H3 at residues K4 and K36 upon activation with TNF-α 37. We expect that this may be the case in cells that have nuclear and DNA bound Baf53.
Another set of experiments that may shed more light on various Baf expressions in HIV-1 infected cells (i.e. Baf170 and Baf53) is to perform a simple microarray. Genes that could be initially tested include cell cycle targets including Mdm2, p21/waf1, and Cyclin D1. These genes have previously been shown to be upregulated in Baf53 knockdown cells38. Finally, ChIP-chip or high-throughput sequencing ChIP-Seq could define all the targets that are modulated by some of the critical Bafs that regulate HIV-1 in infected cells.
Materials and Methods
Cell Culture and Antibodies
Jurkat cell line is an uninfected T-cell line and is grown in RPMI media containing 10% FBS, 1% L-glutamine, and 1% streptomycin/penicillin. ACH2 and J1.1 cell lines are latently HIV-1 infected T-cells, U1 is a latently HIV-1 infected monocytic cell line, and OM10.1 is a latently infected promyelocytic cell line. Cells are grown in RPMI-1640 media containing 10% FBS, 1% L-glutamine, and 1% streptomycin/penicillin (Quality Biological). For cell lines used in Flow Cytometry and RT assays, cells were grown in RPM1-1640 media containing 5% FBS, 1% L-glutamine, and 1% streptomycin/penicillin (Quality Biological). All cells were incubated at 37°C and 5% CO2. α-Baf53 (N-19), α-Baf57 (C-20), α-Baf170 (H-116) and α-Actin (C-11) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Size Exclusion Chromatography
Jurkat and J1.1 cell lines (5×108) were cultured to early-mid log phase of growth and pelleted at 4°C for 5 min at 1,800 rpm. The cell pellets were washed twice with phosphate buffered saline (PBS) without Ca2+ and Mg2+ (Quality Biological). Cell pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 120 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, 0.2 mM Na3VO4, 1 mM DTT, one complete protease cocktail tablet/50 mL) and incubated on ice for 20 min, with a gentle vortexing every 5 min. Cell lysates were centrifuged at 4°C at 10,000 rpm for 10 min. Supernatants were transferred to a fresh tube, where protein concentrations were determined using Bradford protein assay (BioRad, Hercules, CA). Five milligram of protein from each cell line were acetone precipitated using 4 volumes of ice cold 100% acetone, incubating for 15 min on ice. Lysates were centrifuged at 4°C for 10 min at 12,000 rpm, supernatants were removed, and the pellet allowed to dry. Pellets were resuspended, equilibrated, and degassed in chromatography running buffer (0.2 M Tris-HCl, pH 7.5, 0.5 M NaCl, 5% Glycerol). Five mg of lysates were run on a Superose 6 HR 10/30 size exclusion chromatography column using the AKTA purifier system (GE Healthcare, Piscataway, NJ, USA). Flow-through was collected at 4°C at a flow rate of 0.3 mL/min at 0.5 mL for approximately 90 fractions. Every 10th fraction was analyzed by immunoblotting for Baf53, Baf57, Baf170 and Actin in order to determine point of protein complex elution.
Western Blots
Chromatography fractions and input lysates were resolved by SDS PAGE on a 4–20% Tris-Glycine gel (Invitrogen, Carlsbad, CA, USA). Proteins were transferred to PVDF membranes overnight. Western blot visualization and analysis was performed with the Odyssey IR imaging system (LI-COR Biosciences) following the manufacturer’s instructions. Briefly, membranes were blocked with 5 mL of LI-COR blocking buffer (LI-COR Biosciences, Cambridge, UK) for 1 hour at room temperature and incubated overnight at 4°C with primary antibodies in LI-COR buffer containing 0.1% Tween-20. Membranes were washed three times with PBS+0.1% Tween-20 and incubated with donkey α-goat IRDye 800CW (LI-COR) or goat α-rabbit IRDye 680 (LI-COR) for 30 min. Membranes were washed four times with PBS+0.1% Tween-20 and once with PBS prior to scanning. Membranes were stripped and re-processed with appropriate antibodies when needed.
Electroporation/Transfections
Transfections were performed using the Neon transfection system using manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Briefly, ACH2, J1.1, U1, and OM10.1 cell lines were electroporated using 10 μL tips, 5×104 cells, 100 nM (or other concentrations) of siRNA, in RPMI-1640 media containing 5% FBS and 5% L-glutamine. The cell lines were transfected with the following parameters: Pulse voltage of 1,325 V, Pulse width of 10 ms, and a pulse number of 3.
Flow Cytometry
Cells were harvested and washed in PBS without Ca2+ and Mg2+ twice. Cells were fixed in 2 mL of cold 100% ethanol at 4°C for at least 30 min. Cells were spun at 4°C at 2,500 rpm and supernatants were discarded. Cells were rehydrated on ice in PBS without Ca2+ and Mg2+ for at least 15 min. Cells were spun at 4°C at 2,500 rpm for 5–10 min and supernatants were discarded. Cells were resuspended in 1 mL of propidium iodide (PI) solution (PBS without Ca2+ and Mg2+, 25 μg/mL PI (Sigma), 10 μg/mL RNase A (Sigma), and 0.1% NP-40). Cells were kept at 4°C in the dark until ready to analyze by flow cytometry. Cells were analyzed using the Accuri C6 Flow Cytometer (Accuri, Ann Arbor, MI, USA). Aggregates and debris were excluded by gating on the Width and FL2A parameters. The sub-G1 cells were considered as apoptotic population.
Reverse-Transcriptase Assay
Supernatants were collected at various time points post infection to test for the presence of virus post transfection. Viral supernatants (10 μl) were incubated in a 96-well plate with reverse transcriptase (RT) reaction mixture containing 1X RT buffer (50 mM Tris-HCl, 1 mM DTT, 5 mM MgCl2, 20 mM KCl), 0.1% Triton, poly(A) (1U/ml), pd(T) (1U/ml), and [3H]TTP). The mixture was incubated overnight at 37°C, and 5 μl of the reaction mix was spotted on a DEAE Filtermat paper, washed four times with 5% Na2HPO4, three times with water, and then dried completely. RT activity was measured in a Betaplate counter (Wallac, Gaithersburg, MD).
Kinase Assay
Immunoprecipitated samples were assessed for kinase activity. Kinase assays were performed with immunoprecipitates from both infected and uninfected cells using α-cdk2, and α-cdk9 antibodies in a 100 μl reaction volume containing 0.5 μM ATP, 3 μCi of [γ-32P]ATP in TTK kinase buffer containing 50 mM HEPES (pH 7.9), 10 mM MgCl2, 6 mM EGTA, and 2.5 mM dithiothreitol for 60 min at 37 °C. The samples were separated by reducing SDS-PAGE on a 4–20% Tris–Glycine gel. Gels were stained with Coomassie blue, destained, and then dried for 2 h. Following drying, the gels were exposed to a PhosphorImager cassette and analyzed utilizing Molecular Dynamic’s ImageQuant Software.
Immunoprecipitation and in vivo Cross-Linking
FPLC fractions # 40 and 41 from Jurkat and J1-1 cells (~200 μg) were cross-linked with 0.5 mM dithiobis-succinimidylpropionate (DSP) for 30 min at 37°C. The extracts (200 μl) were then denatured by resuspending in 5.5 M urea. The denatured extracts were subsequently diluted 5-fold in RIPA buffer (10 mM Tris/Cl [pH 7.4], 1 mM EDTA [pH 7.4], 300 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM DTT, 1 mM PMSF) and immunoprecipitated with antibodies against Actin (10 μg). Samples were IPed 48hrs at 4°C and then 50 μl of 30% A + G beads were added for 2 hrs at 4°C. Samples were washed three times with TNE 50 and 0.1 % NP40. The cross-linked material was dissociated by boiling the sample in 2x Laemmli sample buffer for 3 min, and proteins were analyzed by 4–20% SDS-PAGE and western blot.
PP2A phosphatase assay
PP2A (Upstate, Millipore) is a type 2 protein Ser/Thr phosphatase characterized by high activity against the α subunit of Phosphorylase Kinase and a low sensitivity to Inhibitor-2. PP2A is distinguishable from PP2B/Calcineurin and PP2C by its partial activity in the absence of divalent cations, and its high sensitivity to Okadaic Acid. Fractions from both infected and uninfected cells were diluted in PPase buffer (50 mm HEPES, pH 7.2, 60 mm NaCl, 60 mm KCl, and protease inhibitors) to a final volume of 50 μl. After incubation with PP2A (50 units) at 30°C for 30 min, samples were precipitated, ran on a 4–20% gel, and western blotted with α-Baf53 antibody.
PIP2 assay
Uninfected (Jurkat) and infected (J1-1) Lymphocytes (five million) were grown in complete media containing 10% serum. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. The released proteins were separated from nuclei by centrifugation, and both the supernatant and pellet were analyzed by SDS-PAGE and Western blotted. For the phosphotidylinositol phosphate experiments, the nuclei isolated from lymphocytes were incubated in the presence of three PIP2 mixtures (10 ug of each) including PI(3,5)P2 ((1,2-dihexanoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,5′-bisphosphate)), PI(3,4)P2 ((1,2-dioctanoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,4′-bisphosphate)), and PI(4,5)P2 ((1,2-dioctanoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate)). Samples were incubated for 4hrs prior to separation and western blot.
Cdk/Cyclin expression and purification
Cdk2 and Cyclin E were purified from lysates of Sf9 insect cells infected with baculoviruses producing tagged-cdk2 and Cyclin E viruses. Two separate cell cultures, one infected with cdk2-expressing baculovirus and the other one infected with Cyclin E-expressing baculovirus were used for purification. The Mono-Q fractions containing cdk2 or Cyclin E were mixed 1:1 and loaded onto Superdex column. Fractions containing cdk2/Cyclin E were concentrated using Microcon tubes. Expression of cdk9/Cyclin T and Tat (generous gift of Dr. David Price) have previously been described and used for crystal structure39. Briefly Sf9 insect cells were co-infected with three baculoviruses that individually directed the expression of C-terminally His-tagged human cdk9, human Cyclin T1 and HIV-1 Tat. The three coding sequences were individually cloned into the pENTR/SD/D-TOPO vector (Invitrogen) after PCR amplification. Large scale production of the Tat/P-TEFb complex was carried out using Sf-900 serum-free medium (GIBCO) with 1.5 × 106 Sf9 cells/ml (Supplement Figure 1).
Chromatin Immunoprecipitation (ChIP) Assay
J1-1 cells were treated with TNF-α (10 ng/ml) for 2 hrs, washed, and put back into 37°C with complete medium (RPMI). Cells were initially treated with 5% serum to completely silent the provirus and kept for 3 days prior to TNF-α treatment. Following TNF-α treatment, cells were put back in media with 5% serum for 24 hrs. Approximately 3–5 million cells were cross-linked (1% formaldehyde; 10 min at 37°C), and samples were sonicated to reduce DNA fragments to ~200 to 800 bp for ChIP assays, in which DNA bound to various proteins were immunoprecipitated with antibodies as indicated in the figure legends. Primer pairs for PCR analysis of chromatin immunoprecipitation amplified sequences flanking the HIV-1 LTR nuc-1 (+ 10 to + 165) and Env (+ 8990 to + 9120).
Supplementary Material
Acknowledgments
We would like to thank the members of the Kashanchi lab for experiments and assistance with the manuscript. Also, we are grateful to Dr. Lee Ratner for the HTLV-1 infectious clone and Dr. David Price for generous gift of cdk9/Cyclin T and cdk9/Cyclin T and Tat. Most of the data on the current manuscript was generated using funds from NIH grants AI078859, AI074410 and AI043894. Rachel Van Duyne is a predoctoral student in the Microbiology and Immunology Program of the Institute for Biomedical Sciences at the George Washington University
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Rachel Van Duyne, Email: rachel.vanduyne@gmail.com.
Irene Guendel, Email: iguendel@gmu.edu.
Aarthi Narayanan, Email: anaraya1@gmu.edu.
Edward Gregg, Email: edwardwgregg@gmail.com.
Nazly Shafagati, Email: nshafagati@gmail.com.
Mudit Tyagi, Email: mxt108@case.edu.
Rebecca Easley, Email: rleasl@gmail.com.
Zachary Klase, Email: klaseza@niaid.nih.gov.
Sergei Nekhai, Email: snekhai@howard.edu.
Kylene Kehn-Hall, Email: kkehnhal@gmu.edu.
Fatah Kashanchi, Email: fkashanc@gmu.edu.
References
- 1.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–12. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 2.Klochendler-Yeivin A, Muchardt C, Yaniv M. SWI/SNF chromatin remodeling and cancer. Curr Opin Genet Dev. 2002;12:73–9. doi: 10.1016/s0959-437x(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 3.Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 2002;16:2509–17. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–7. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–87. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 6.Sims RJ, 3rd, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr Opin Cell Biol. 2004;16:263–71. doi: 10.1016/j.ceb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Fan HY, Narlikar GJ, Kingston RE. Noncovalent modification of chromatin: different remodeled products with different ATPase domains. Cold Spring Harb Symp Quant Biol. 2004;69:183–92. doi: 10.1101/sqb.2004.69.183. [DOI] [PubMed] [Google Scholar]
- 8.Liu N, Balliano A, Hayes JJ. Mechanism(s) of SWI/SNF-induced nucleosome mobilization. Chembiochem. 2011;12:196–204. doi: 10.1002/cbic.201000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodson HV, Hawse WF. Molecular evolution of the actin family. J Cell Sci. 2002;115:2619–22. doi: 10.1242/jcs.115.13.2619. [DOI] [PubMed] [Google Scholar]
- 10.www.genecards.com
- 11.Galarneau L, Nourani A, Boudreault AA, Zhang Y, Heliot L, Allard S, Savard J, Lane WS, Stillman DJ, Cote J. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell. 2000;5:927–37. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 12.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–4. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 13.Sung YH, Choi EY, Kwon H. Identification of a nuclear protein ArpN as a component of human SWI/SNF complex and its selective association with a subset of active genes. Mol Cells. 2001;11:75–81. [PubMed] [Google Scholar]
- 14.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–36. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 15.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–73. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 17.Wood MA, McMahon SB, Cole MD. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell. 2000;5:321–30. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- 18.Harata M, Oma Y, Mizuno S, Jiang YW, Stillman DJ, Wintersberger U. The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol Biol Cell. 1999;10:2595–605. doi: 10.1091/mbc.10.8.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sunada R, Gorzer I, Oma Y, Yoshida T, Suka N, Wintersberger U, Harata M. The nuclear actin-related protein Act3p/Arp4p is involved in the dynamics of chromatin-modulating complexes. Yeast. 2005;22:753–68. doi: 10.1002/yea.1239. [DOI] [PubMed] [Google Scholar]
- 20.Lee K, Kang MJ, Kwon SJ, Kwon YK, Kim KW, Lim JH, Kwon H. Expansion of chromosome territories with chromatin decompaction in BAF53-depleted interphase cells. Mol Biol Cell. 2007;18:4013–23. doi: 10.1091/mbc.E07-05-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treand C, du Chene I, Bres V, Kiernan R, Benarous R, Benkirane M, Emiliani S. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 2006;25:1690–9. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agbottah E, Deng L, Dannenberg LO, Pumfery A, Kashanchi F. Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology. 2006;3:48. doi: 10.1186/1742-4690-3-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bukrinsky M. SNFing HIV transcription. Retrovirology. 2006;3:49. doi: 10.1186/1742-4690-3-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahmoudi T, Parra M, Vries RG, Kauder SE, Verrijzer CP, Ott M, Verdin E. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J Biol Chem. 2006;281:19960–8. doi: 10.1074/jbc.M603336200. [DOI] [PubMed] [Google Scholar]
- 25.Gautier VW, Gu L, O’Donoghue N, Pennington S, Sheehy N, Hall WW. In vitro nuclear interactome of the HIV-1 Tat protein. Retrovirology. 2009;6:47. doi: 10.1186/1742-4690-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vardabasso C, Manganaro L, Lusic M, Marcello A, Giacca M. The histone chaperone protein Nucleosome Assembly Protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology. 2008;5:8. doi: 10.1186/1742-4690-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Easley R, Carpio L, Dannenberg L, Choi S, Alani D, Van Duyne R, Guendel I, Klase Z, Agbottah E, Kehn-Hall K, Kashanchi F. Transcription through the HIV-1 nucleosomes: effects of the PBAF complex in Tat activated transcription. Virology. 2010;405:322–33. doi: 10.1016/j.virol.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Easley R, Van Duyne R, Coley W, Guendel I, Dadgar S, Kehn-Hall K, Kashanchi F. Chromatin dynamics associated with HIV-1 Tat-activated transcription. Biochim Biophys Acta. 2010;1799:275–85. doi: 10.1016/j.bbagrm.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Easley R, Carpio L, Guendel I, Klase Z, Choi S, Kehn-Hall K, Brady JN, Kashanchi F. Human T-lymphotropic virus type 1 transcription and chromatin-remodeling complexes. J Virol. 2010;84:4755–68. doi: 10.1128/JVI.00851-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foster KS, McCrary WJ, Ross JS, Wright CF. Members of the hSWI/SNF chromatin remodeling complex associate with and are phosphorylated by protein kinase B/Akt. Oncogene. 2006;25:4605–12. doi: 10.1038/sj.onc.1209496. [DOI] [PubMed] [Google Scholar]
- 31.Ammosova T, Berro R, Jerebtsova M, Jackson A, Charles S, Klase Z, Southerland W, Gordeuk VR, Kashanchi F, Nekhai S. Phosphorylation of HIV-1 Tat by CDK2 in HIV-1 transcription. Retrovirology. 2006;3:78. doi: 10.1186/1742-4690-3-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol Cell Biol. 1999;19:1460–9. doi: 10.1128/mcb.19.2.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendricks KB, Shanahan F, Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol Cell Biol. 2004;24:362–76. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–74. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 35.Jani A, Wan M, Zhang J, Cui K, Wu J, Preston-Hurlburt P, Khatri R, Zhao K, Chi T. A novel genetic strategy reveals unexpected roles of the Swi-Snf-like chromatin-remodeling BAF complex in thymocyte development. J Exp Med. 2008;205:2813–25. doi: 10.1084/jem.20080938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou M, Deng L, Lacoste V, Park HU, Pumfery A, Kashanchi F, Brady JN, Kumar A. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J Virol. 2004;78:13522–33. doi: 10.1128/JVI.78.24.13522-13533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee YS, Sohn DH, Han D, Lee HW, Seong RH, Kim JB. Chromatin remodeling complex interacts with ADD1/SREBP1c to mediate insulin-dependent regulation of gene expression. Mol Cell Biol. 2007;27:438–52. doi: 10.1128/MCB.00490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465:747–51. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.