Abstract
Impaired regulation of the transforming growth factor-β (TGFβ) signaling pathway has been linked to thoracic aortic aneurysm (TAA). Previous work has indicated that differential splicing is a common phenomenon, potentially influencing the function of proteins. In the present study we investigated the occurrence of differential splicing in the TGFβ pathway associated with TAA in patients with bicuspid aortic valve (BAV) and tricuspid aortic valve (TAV). Affymetrix human exon arrays were applied to 81 intima/media tissue samples from dilated (n = 51) and nondilated (n = 30) aortas of TAV and BAV patients. To analyze the occurrence of alternative splicing in the TGFβ pathway, multivariate techniques, including principal component analysis and OPLS-DA (orthogonal partial least squares to latent structures discriminant analysis), were applied on all exons (n = 614) of the TGFβ pathway. The scores plot, based on the splice index of individual exons, showed separate clusters of patients with both dilated and nondilated aorta, thereby illustrating the potential importance of alternative splicing in TAA. In total, differential splicing was detected in 187 exons. Furthermore, the pattern of alternative splicing is clearly differs between TAV and BAV patients. Differential splicing was specific for BAV and TAV patients in 40 and 86 exons, respectively, and splicings of 61 exons were shared between the two phenotypes. The occurrence of differential splicing was demonstrated in selected genes by reverse transcription–polymerase chain reaction. In summary, alternative splicing is a common feature of TAA formation. Our results suggest that dilatation in TAV and BAV patients has different alternative splicing fingerprints in the TGFβ pathway.
INTRODUCTION
Thoracic aortic aneurysm (TAA) is a pathological widening of the aorta resulting from degeneration of the extracellular matrix and loss of smooth muscle cells in the tunica media. TAA is an asymptomatic disease before the actual rupture of the aorta, a condition that is lethal if not treated in time. There are several different etiologies of TAA involving monogenic syndromes (such as Marfan and Loeys-Diezt syndromes) that predispose individuals to TAA, aneurysm associated with bicuspid aortic valve (BAV) disease, and idiopathic causes of TAA. TAA is a widespread complication in individuals who have BAV disease, which is a common congenital disorder present in 1–2% of the population (1). The prevalence of aortic dilatation in patients with BAV without significant valve dysfunction has been estimated to be 50–70% (2).
The pathogenesis of aneurysm formation in the monogenic syndromes has been extensively studied (3), whereas the molecular mechanisms of the other forms, which constitute the vast majority of TAAs, remain largely unknown. A common key feature of the monogenic syndromes appears to be dysregulation of transforming growth factor-β (TGFβ) signaling. TGFβ, a crucial player in vascular remodeling, alters both structure and composition of the extracellular matrix. Marfan syndrome is caused by mutations in the fibrillin-1 gene, which has been suggested to influence the bioavailability of active TGFβ. Moreover, mutations in the TGFβ receptors also impair the signaling cascade in other Marfan syndrome–related disorders, including Loeys-Dietz syndrome as well as familial TAA and aortic dissection. Furthermore, mutations in NOTCH1, a gene that cross-talks with TGFβ via the Notch1 pathway (4), have been found in TAA patients with BAVs (5).
Recently, our group has shown a link between the TGFβ activity and aneurysm formation in BAV patients (6). In particular, we observed that BAV patients, in contrast to TAV patients, have an impaired mechanism for TGFβ-mediated splicing of the extra domain A (exon 33, EDA) of fibronectin (FN1). The present study was based on further investigation of the importance of alternative splicing in the TGFβ signaling pathway with respect to dilated and nondilated thoracic aorta tissues in patients with TAV and BAV. The selection of genes in the present study is composed of 26 genes from the TGFβ pathway, including FN1. All the selected genes and their functions are summarized in Supplemental Table 1.
Our approach to alternative splicing analysis is based on a microarray technology (GeneChip Human Exon 1.0 ST Affymetrix arrays) that measures the expression of single exons to identify alternatively spliced events. With this kind of array it is possible to investigate both alternative splicing patterns and differential gene expression. Multivariate data analysis tools were adopted for the statistical analysis of alternative splicing events. The findings in the present study demonstrate that the two different types of aorta tissues, dilated and nondilated, show different alternative splicing patterns in the TGFβ pathway with respect to TAV and BAV. Diverging alternative splicing patterns observed between the two valve types indicate that the underlying molecular mechanisms of dilatation in TAV patients differ from the mechanisms of dilatation in BAV patients.
EXPERIMENTAL PROCEDURES
Clinical Samples
The Advanced Study of Aortic Pathology (6) biobank materials were generated after written informed consent from all participants had been obtained according to the declaration of Helsinki and with approval by the ethics committee of the Karolinska Institute (application number 2006/784-31/1). The study included BAV and TAV patients undergoing aortic valve surgery with a reconstitution of a dilated aorta and patients who underwent only aortic valve surgery (nondilated controls) at the Karolinska University Hospital, Stockholm, Sweden. Aortic biopsies were taken from the anterior (convex) part of the aorta, that is, the site of aortotomy a few centimeters above the aortic valve. According to coronary angiography results, none of the patients had significant coronary artery disease. Marfan syndrome patients and monocuspid valve patients were excluded from the study. Definition of dilation was based on an aorta dilatation of >45 mm (dilated) or <40 mm (nondilated). Thoracic aorta dilatation measurements were taken at the point of the aorta showing maximal dilatation. The measurements were performed during anesthesia using transesophageal echocardiography.
RNA Extraction and Array Hybridization
The intima-medial layer of the vascular specimens were isolated by adventicectomy, incubated with RNAlater (Ambion, Austin, TX, USA) and homogenized by using the FastPrep method (Qbiogene, Irvine, CA, USA) with Lysing Matrix D tubes (MP Biomedicals, Hessen, Germany). The RNA samples were hybridized and scanned at the Karolinska Institute microarray core facility. Affymetrix GeneChip® Human Exon 1.0 ST arrays and protocols were used.
Data Analysis
A battery of different statistical tools has been used on both meta probe set (gene expression) and core probe set (exon expression) levels. These tools include principal component analysis (PCA) (7), orthogonal projections to latent structures discriminant analysis (OPLS-DA) (8,9) and false discovery rate (FDR)-corrected t tests. The exon and gene level data were preprocessed with robust multichip average (RMA) (10), followed by splice index (SI) (SI = log2[probe set intensity/transcript expression level]) calculation only on the exon level, unit variance (UV) scaling and mean centering. Thus, the exon level data were normalized with respect to whole-transcript (gene) expression. The PCA and OPLS-DA were performed by using Simca P + 12.0.1 × 64 (Umetrics, Umeå, Sweden) software. All multivariate models were mean centered and UV scaled, and model complexity was estimated according to cross-validation (11) in Simca p + 12.0.1 × 64 (Umetrics) software, unless otherwise stated. Import of Affymetrix GeneChip Exon ST 1.0 CEL files, RMA normalization and SI calculations were made by using the OneChannelGUI package for R (12). FDR and t test calculations were performed in Partek Genomics Suite software. The complete statistical analysis procedure and different statistical methods for all the analyses included in the manuscript were performed as described in the following text.
Multidimensional data obtained by screening several patient samples with regard to gene/exon level expression can be considered as a set of patient vectors, for which the expression of all genes/ exons in a particular row defines the position of the vector in the multivariable space, thereby describing the particular patient sample with regard to its gene/ exon expression. Each patient sample will thus have a unique expression profile that is a summary of all the genes/ exons present in the data set, an expression fingerprint for that particular patient. PCA explains the variance-covariance structure of a set of variables by using linear combinations (7). The linear combination will represent a new coordinate system that is obtained by rotating the original space. The new axes obtained in this way represent the directions with maximum variability, where PC1 indicates the direction of the highest variability and PC2 is the direction of the next-to-highest variability in a direction orthogonal to the first PC. All the subsequent PCs will describe diminishing variability in a sequential manner as well as being orthogonal to all previous directions.
OPLS-DA is a multivariate analysis method used when a quantitative relationship between a data matrix X (in our case expression levels of different genes/exons) and a vector (or matrix), Y containing qualitative values (class belonging), are analyzed together (8). The main idea of OPLS-DA is to separate the systematic variation in X into two parts, one that is linearly related to Y and one that is unrelated (orthogonal) to Y. In this way it is possible to remove systematic variation from X not correlated to the response set Y, thereby giving the opportunity to study only the variation that is correlated to the classification of interest. Two new units from both analyses (PCA and OPLS-DA), scores and loadings, can be obtained. The scores give information about the new observations projected onto a lower-dimensional plane that summarizes the behavior of the original ones. The loadings, on the other hand, reveal how the PC model plane is inserted into the variable space and thereby can be used to interpret the meaning of the scores. Quality parameters for all multivariate analysis models performed in this study are shown in Supplemental Table 3.
In the present study, PCA (7) was applied to meta probe set (gene) expression data for which all human genes were included that had been filtered with respect to signal levels of Y-chromosome genes on array analyses of female samples. Genes that expressed below a threshold of 5.7 in intensity were removed from subsequent analysis. This filtering resulted in inclusion of 11,417 genes. PCA was applied to a dataset with 95 patients included with dilated aortas (n = 58) and nondilated aortas (n = 37). Prior to PCA, the gene expression data (meta probe set) were RMA (10) normalized, scaled to UV and mean centered. Dilated and nondilated samples were color coded in the scores plot, and the first three PCs (which contributed the most to the variation in the multidimensional data space) from the nonsupervised PCA were examined visually. In the present study, we chose to base the inclusion criteria for further alternative splicing analysis on samples having similar gene expression profiles, based on all expressed genes, in a PCA score plot (three dimensional) and correct assignment based on clinical data (aorta diameter and BAV or TAV). If both criteria were fulfilled, the sample was included, and if not, the sample was excluded from further exon level analysis. Of 95 consecutively collected clinical samples, 14 did not fulfill both inclusion criteria and were therefore excluded from further exon level analysis. This exclusion resulted in a dataset composed of 81 patients, 51 with dilated (15 TAV, 36 BAV) and 30 with nondilated aortas (14 TAV, 16 BAV). In addition, PCA on that gene expression level was made on TAV and BAV patients separately for the initial data set of 95 patients (analysis not shown). The same inclusion criteria have been used as described above for the new analysis (similarity in gene expression profiles and clinical assignment). The new analysis showed that the results were largely in concordance with the results in which TAV and BAV patients were analyzed together. The TGFβ exon level was examined by applying PCA and OPLS-DA on the 81 samples chosen according to the analysis of gene-expression data described above. The analyses were performed on the 81 samples, including TAV and BAV, as well as on TAV and BAV sample sets separately, with dilated and nondilated samples as two discriminant Y-variable groups. The number of exons (throughout this study probe sets are called exons) in the TGFβ pathway was 614. The data were RMA normalized, and then subjected to SI calculation (SI = log2[probe set intensity/transcript expression level]) on the core level, UV scaling and mean centering. The genes included in these analyses were filtered with respect to signal levels of Y-chromosome genes on array analyses of female samples. The FDR (q-value) significance level 0.05 was calculated for TAV and BAV data sets separately, based on a two-sided Student t test.
The significance of differentially expressed exons was also analyzed by means of loadings in OPLS-DA models for TAV and BAV, including jack-knife confidence levels (9,11) of the loadings. Furthermore, the TAV model loadings were plotted against BAV model loadings in a combined model scatter plot to identify TAV- and BAV-specific alternative-splicing events. A handful of exons that were statistically significant according to both FDR and jack-knife confidence levels were chosen for validation. A mean value of SI data was calculated for dilated and nondilated samples for TAV and BAV groups separately and plotted for each gene of interest in a gene view plot. OPLS-DA followed by combined model scatter plot and analysis of jack-knife confidence levels was performed on the meta probe set level of genes included in the TGFβ pathway (26 genes) on TAV and BAV models separately, including dilated and nondilated samples as two discriminant Y-variable groups to identify differentially expressed genes. OPLS-DA was also performed at the level of all exons in the human genome. Prior to analysis, the core level exons were normalized with RMA and filtered according to detection above background (DABG) P value < 0.05 in 100% of the samples (81 patients), followed by inclusion of only genes present in the Ensembl Genome Browser (http://useast.ensembl.org/index.html). This filtering resulted in 4631 genes and 76,677 exons. Data for the same group of 81 patients from the TGFβ pathway alternative splicing analysis were included. OPLS-DA was performed on dilated versus nondilated samples as a Y vector in both TAV and BAV separately. Prior to the analysis the splice index was calculated, whereupon the gene level–normalized expression data were subjected to UV scaling and mean centering.
Reverse Transcription–Polymerase Chain Reaction Validation of Alternative Splicing Events
Reverse transcription–polymerase chain reaction (RT-PCR) using exons spanning FN1_EDA, FN1_EDB (exon 25), LTBP3_E19 and FN1_E09 was performed on cDNA isolated from thoracic aorta tissues (medial layer in BAV and TAV patients, with and without dilatation). The patients chosen for RT-PCR experiments were included in the array data analysis. The primer sequences are shown in Supplemental Table 2. The thermal conditions were as follows: 95ºC for 2 min, followed by a total of 35 cycles (95ºC for 30 sec, 52°C for 1 min, 72°C for 1 min) and 72°C for 2 min.
All supplementary materials are available online at www.molmed.org
RESULTS
Alternative Splicing Pattern in the TGFβ Pathway Is Different between Dilated and Nondilated Aortas
A global gene expression level PCA was performed on gene expression data (meta probe set level) (Supplemental Figure 1) of 95 patients to investigate discrimination between the two groups, patients with dilated aortas and those with nondilated aortas. Given the fact that each patient has a unique gene expression profile in the 11,417 (the number of genes included in the analysis) variable space, data for those patients who showed similar profiles to each other were grouped together in the PC scores plot (Supplemental Figure 1A). The scores plot (Supplemental Figure 1A) shows that the data for most dilated patients form a cluster, whereas the data for nondilated samples spread more in the PC1–PC3 variable space. This result indicates that the dilated samples are more homogeneous in their expression profiles than the nondilated counterparts. To obtain two well-separated groups, a subset of both dilated and nondilated samples that separates well in the scores plot in Supplemental Figure 1A was chosen based on visual inspection of the scores plot. This subset includes 81 patients, 51 dilated and 30 nondilated (Supplemental Figure 1B). The scores plot of PCA of the 81 chosen patients (Supplemental Figure 1C) shows that the two groups now are better separated. The subset of patients chosen for subsequent alternative splicing analysis on the probe set level (exon expression) thus was selected not only on the basis of medical phenotypes (dilated, nondilated thoracic aorta) but also on the basis of gene expression signatures (meta probe set level) for each individual in the study; that is, the inclusion criteria of patients for the subsequent exon level analysis were based on both similarity in gene expression profiles and clinical assignment of a certain patient.
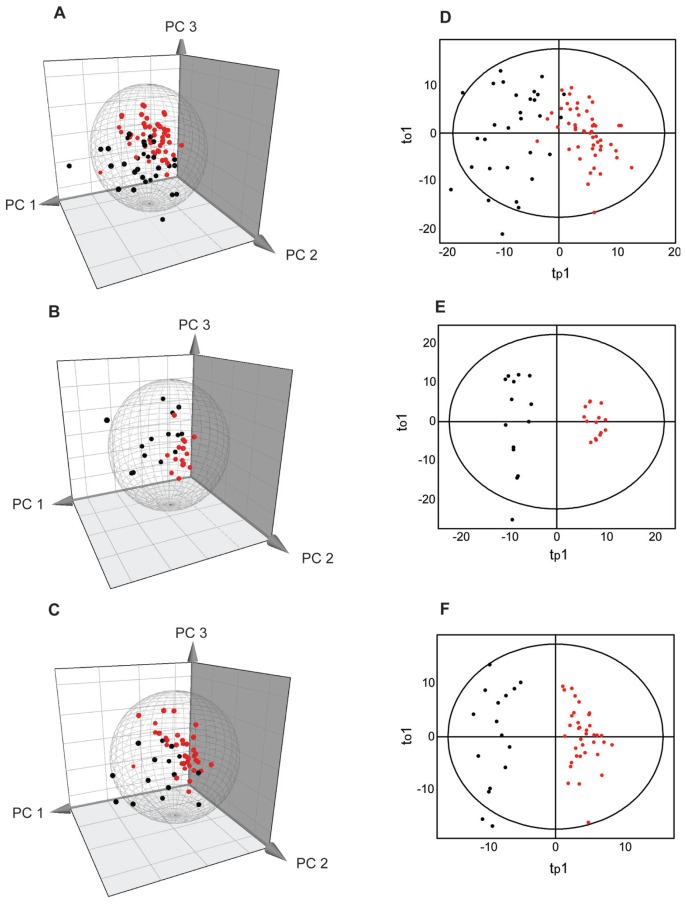
Data for the 81 patients who met both selection criteria were subjected to alternative splicing analysis of probe sets covering all known exons of the TGFβ pathway genes. To start with, PCA was performed on the gene level normalized exon expression (splice indices) of TGFβ exons including all the 81 patients (Figure 1A). The scores plot based on the splice index of individual exons shows separate clusters of patients with dilated and patients with nondilated aorta (Figure 1A). Furthermore, to examine differences in the splicing profiles of BAV and TAV patients, the PCA was applied to the two subgroups of patients separately (Figure 1B, C). According to the PC1–PC3 plane scores plot, it is clear that the two groups, dilated and nondilated aorta tissues, show tendencies to cluster separately from each other in both TAV (Figure 1B) and BAV (Figure 1C). In addition, the dilated and nondilated samples, including both TAV and BAV as well as separated TAV and BAV groups, were subjected to a supervised OPLS-DA (Figure 1D–F). In contrast to PCA that is unsupervised, in OPLS-DA the group differences are investigated by using a discriminant Y variable and thereby identifying differences between dilated and nondilated aortas. The analysis results show that the dilated and nondilated patients separated well along the first predictive component (tp1) in the first model, in which both TAV and BAV patients were included. The dilated and nondilated samples were well separated along the first predictive component (tp1) in the two models, in which TAV and BAV samples are investigated separately. These results indicate that the gene level–normalized TGFβ exons discriminate between the dilated and nondilated samples in both TAV and BAV. A clear separation in the OPLS-DA model between dilated and nondilated patients is possible only if the transcript-normalized exon expression is significant (according to jack-knife confidence intervals) for a number of exons, thereby being able to build up the model (Figure 2). Alternative splicing is thus an important mechanism in describing dilated and nondilated patients, thereby separating them from each other in both TAV and BAV. At this point, it is interesting to find out what exons are responsible for the group separation seen in TAV and BAV.
Figure 1.
PCA (A–C) and OPLS-DA (D–F) of gene level normalized exon expression (splice index) in the TGFβ pathway. The analyses were performed on 81 patients chosen according to medical data and selection based on meta probe set level PCA (Supplemental Figure 1). The Hotelling’s T2 (based on 95% confidence level) tolerance ellipsoid and ellipse are shown in the scores plots of PCA and OPLS-DA, respectively. Three-dimensional scores plot showing the PC1–PC3 plane of nondilated (black) and dilated (red) thoracic aorta samples with BAV and TAV together (A), as well as samples from TAV (B) and BAV (C) patients separately. Two-dimensional scores plot of an OPLS-DA showing the first predictive component (tp1) and orthogonal component (to1) plane of nondilated (black) and dilated (red) thoracic aorta samples with BAV and TAV patients together (D), as well as TAV (E) and BAV (F) patients separately.
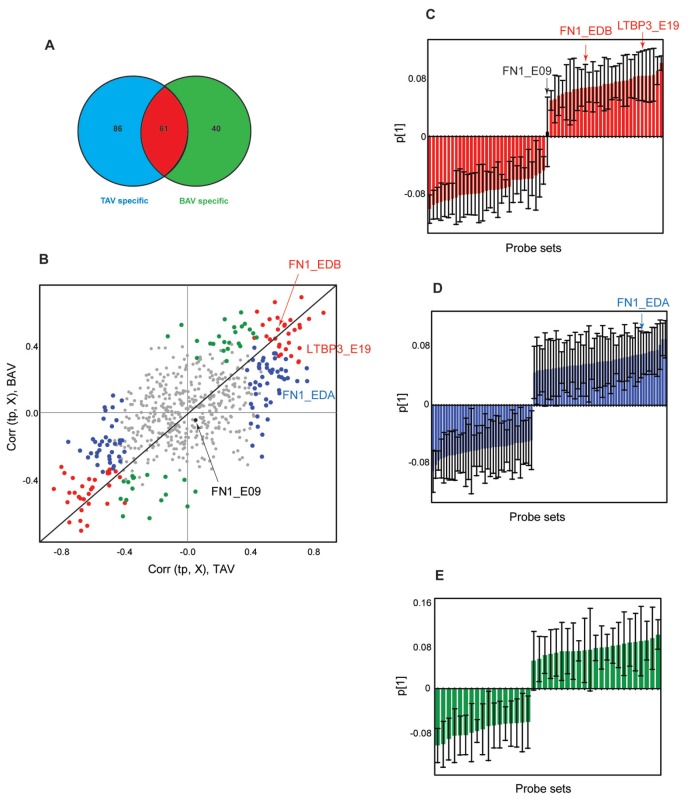
Figure 2.
Venn diagram and OPLS-DA of gene level–normalized exon expression (splice-index) data included in the TGFβ pathway. (A) Venn diagram based on an FDR-corrected t test of dilated versus nondilated samples in TAV and BAV patients separately. TAV-specific (blue), BAV-specific (green), and common (red) exons are marked in the diagram. (B) Combined model scatter plot based on TAV dilated versus nondilated and BAV dilated versus nondilated OPLS-DA models. The exons found to be significant in (A) are color coded accordingly in the combined model scatter plot. The black diagonal is aimed for interpretation purposes. Bar plots showing the loadings of each of the significant exons and indicating their contribution to the first PC are shown in (C–E) and color coded according to (A). The confidence levels for each data point were estimated by a jack-knife algorithm. Exons chosen for RT-PCR validation are marked in (B–E).
Diverging Alternative Splicing Fingerprints Are Associated with Dilatation in TAV and BAV Patients
When only TAV patients were subjected to an FDR-corrected t test between dilated and nondilated samples, 147 exons showed significant differences in splicing, with a cutoff P value of 0.023. The corresponding analysis for BAV patients resulted in 101 significant exons with a cutoff P value of 0.013. The results were applied to a Venn diagram, and it can be seen in Figure 2A that 40 and 86 exons, respectively, are BAV and TAV specific, and 61 exons are shared between the two phenotypes. The contribution of each exon to the separation seen among the patients (Figure 1E–F) can be visualized as loadings of the two separate OPLS-DA models, patients with TAV and patients with BAV in a combined model scatter plot (Figure 2B). Loadings for the TAV model versus the BAV model were thus plotted together (Figure 2B) to enhance interpretation of how different exons contribute to the model. In Figure 2B the significant exons that were TAV specific according to the FDR-corrected t test described above are marked with blue dots and those found to be BAV specific are marked with green dots, whereas shared exons between TAV and BAV are marked with red dots. All the other exons that are not significant according to the FDR-corrected t test are marked with gray dots. The level of contribution of each exon to the two models can be interpreted from the combined model scatter plot; the further away an exon is situated from the origin, the higher impact it has on the model. Furthermore, shared and discriminative exons between the two models can be identified. Exons that have high loading values and are positioned along each side of the diagonal far away from the center of the figure (red dots in Figure 2B) are indicative of a shared correlation pattern for both models. Exons that show high gene level normalized expression (splice index) in dilated samples (for example, FN1_EDB and LTBP3_E19) are situated in the upper right hand corner of the combined model scatter plot, whereas the corresponding exons with low gene level normalized expression (splice index) in dilated samples are situated on the opposite side along the diagonal. The variables that show high gene level normalized expression and are located close to one or the other axis and have a zero or low coordinate along the other axis (blue dots, for example, FN1_EDA) or BAV (green dots) are indicative of TAV-specific probe sets (Figure 2B). The combined model scatter plot facilitates identification of both common and specific exons for patients with TAV and BAV as well as which exons show higher gene level normalized expression in dilatation compared with their nondilated counterparts. The pattern of alternative splicing is, according to the combined model scatter plot, clearly different between TAV and BAV, which indicates that dilatation in the two different valve types adopts different alternative splicing mechanisms.
Alternatively Spliced Exons Are Present in Both TAV and BAV Groups of Patients
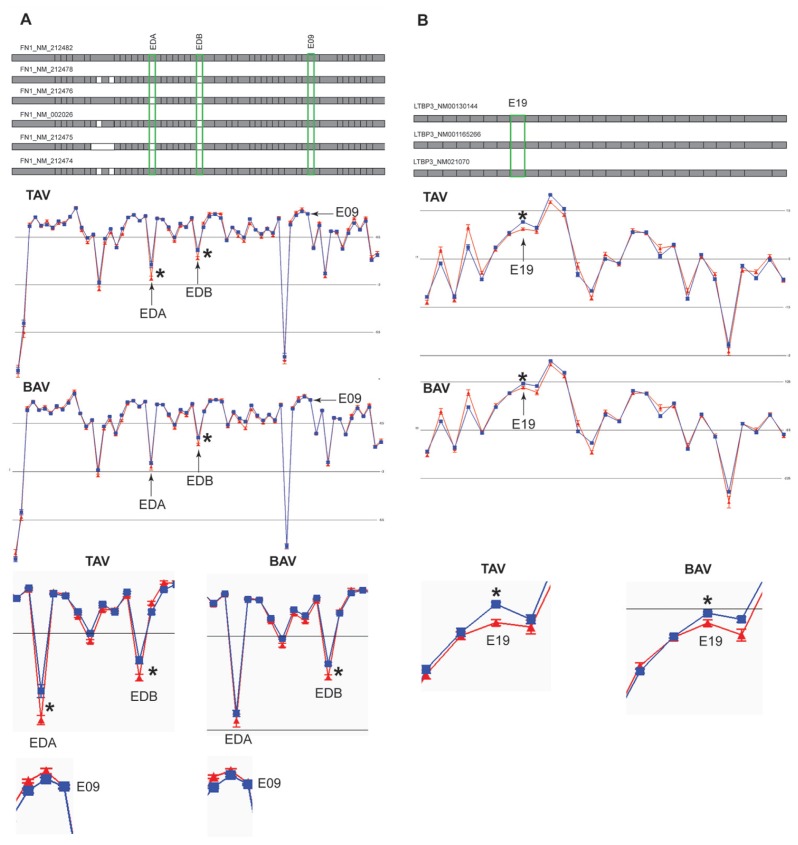
To find out how much each exon contributes to the model it is useful to plot the loadings with jack-knife confidence intervals (9) that will aid in the search for the significant variables that contribute the most to the two models. The numbers of significantly differentially expressed exons (normalized with respect to the whole transcript level) according to jack-knife confidence levels and loading values from OPLS-DA were 230 and 179 in TAV and BAV, respectively (Supplemental Table 4). Of exons found to be differentially expressed (when normalized with respect to whole transcript level) according to the FDR-corrected t test, 99.2% and 97.8% can also be found among the jack-knife confidence level significant exons in the TAV and BAV models, respectively (Supplemental Table 4). The loadings in the first principal component were plotted for the FDR-corrected t test significant exons for shared (Figure 2C), TAV specific (Figure 2D) and BAV specific (Figure 2E) exons. It is clear from the analysis of the loadings with the corresponding jack-knife confidence interval that not all the FDR-corrected t test significant variables were also significant in the multivariate analysis models. The selection of the exons that showed differential expression was thus based on both the analyses, for which those that are significant according to jack-knife confidence levels and FDR-corrected P values can be chosen for further validation (Table 1). In this case, two exons shared between TAV and BAV (FN1_EDB and LTBP3_E19), one exon specific for TAV (FN1_EDA) and one control exon (FN1_E09) that did not show any significant differential expression, were chosen for RT-PCR validation of alternative splicing events. The chosen exons for validation are marked in Figure 2B–E, and shown as part of the gene they belong to in Figure 3. The mean values of every gene expression normalized exon in dilated and nondilated samples of TAV and BAV separately in FN1 and latent TGFβ binding protein 3 (LTBP3) are shown. It is clear that FN1_EDA shows higher expression in dilated samples in only TAV patients but not in BAV patients, whereas FN1_EDB shows higher expression in both TAV and BAV patients. E19 in LTBP3 also shows higher expression in dilated than nondilated samples in both TAV and BAV. The control exon, E09 in FN1, does not show any changes in the transcript normalized exon expression between dilated and nondilated patients (Figure 3).These findings are in concordance with the OPLS-DA performed.
Table 1.
Splice variants chosen for alternative splicing validation in TAV and BAV tissue samples.a
| Gene symbol | FN1 | FN1 | FN1 | LTBP3 |
|---|---|---|---|---|
| Probe set | 2598304 | 2598321 | 2598356 | 3377684 |
| Probe set location | FN1_E33 (EDA) | FN1_E25 (EDB) | FN1_E09 | LTBP3_E19 |
| TAV | ||||
| Pb | 0.0014 | 0.0012 | 0.9644 | 2.60E-05 |
| Loading valuec | 0.0694 | 0.0682 | 0.0062 | 0.0847 |
| Jack-knife confidence intervald | 0.0320 | 0.0328 | 0.0487 | 0.0348 |
| ABS(loading) – ABS(jack-knife confidence interval)e | 0.0373 | 0.0354 | −0.0424 | 0.0499 |
| ΔSI (D – C)f | 0.7224 | 0.4288 | 0.0020 | 0.2242 |
| BAV | ||||
| Pb | 0.2533 | 5.63E-05 | 0.5047 | 0.0026 |
| Loading valuec | 0.0192 | 0.0809 | −0.0069 | 0.0630 |
| Jack-knife confidence intervald | 0.0512 | 0.0286 | 0.0639 | 0.0288 |
| ABS(loading) – ABS(jack-knife confidence interval)e | −0.0320 | 0.0523 | −0.0570 | 0.0341 |
| ΔSI (D – C)f | 0.1924 | 0.3565 | −0.0208 | 0.1447 |
The statistical analyses are based on controls (C) versus dilated (D) tissue samples. Values in bold indicate significant exons.
FDR-corrected t test.
Loading values from OPLS-DA multivariate model.
Jack-knife confidence interval derived from cross-validation in OPLS-DA models.
Subtraction of the jack-knife confidence intervals from the corresponding loading values, if positive indicates significance.
Difference in average mean splice index between the groups indicated.
Figure 3.
The expression plot of alternative splicing events of genes chosen for RT-PCR validation. The expression is shown as mean log2 intensity of splice index data for each probe set (exon) for dilated (blue) and nondilated (red) samples in TAV and BAV patients separately. The isoforms known from the RefSeq database are shown on top with exons present in the particular isoform (gray box) and if absent (white box). The exons that were validated with RT-PCR are marked with green boxes in the RefSeq plot, by arrows in the expression plots and zoomed in to the particular exon of interest in the lower part of the figure. The RefSeq exons are position-matched with the expression plots probe sets (exons) below. FN1 is shown in (A) and LTBP3 in (B). The three isoforms in (B) do not show any difference at core level. The significance level (*) was calculated according to two methods, one null-hypothesis–driven t test for one variable at a time, by which the resulting P values have been corrected according to the FDR q-method, as well as a multivariate method by which jack-knife confidence levels and loading values have been calculated from OPLS-DA models (Figure 2, Table 1).
Alternative Splicing Analysis of All Exons in the Human Genome Reveals the Importance of TGFβ Pathway Exons
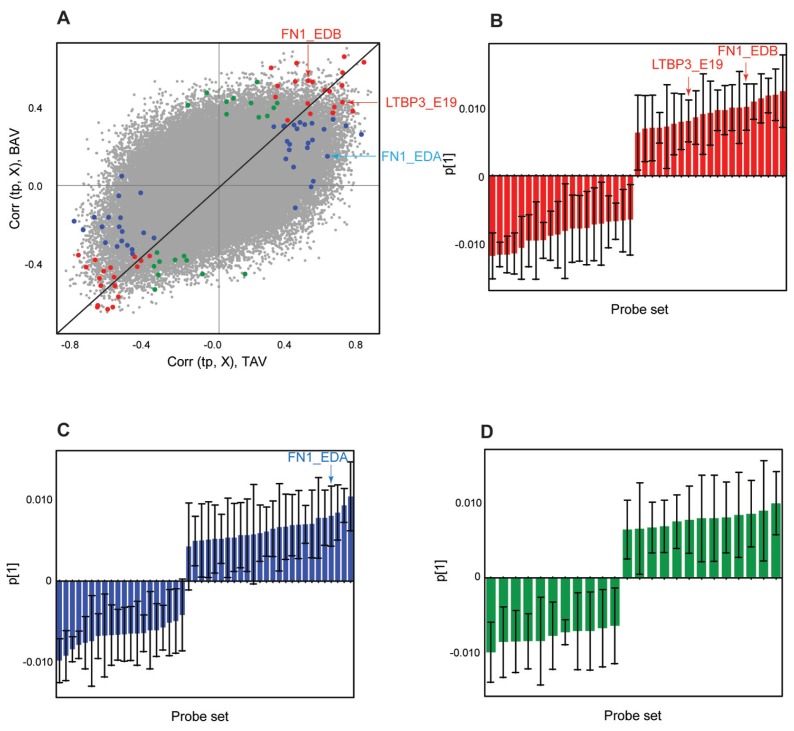
Thus far we have described the analysis performed on exons belonging to one specific pathway, that is, the TGFβ pathway. OPLS-DA was thereafter performed with the inclusion of all exons in the human genome. Prior to analysis, the probe set level data were normalized with RMA and filtered according to the DABG P value < 0.05 in 100% of the samples (81 patients), followed by inclusion of only genes present in Ensembl. This filtering resulted in 4631 genes and 76,677 exons. The same group of 81 patients from the TGFβ pathway alternative splicing analysis was included. OPLS-DA on dilated versus nondilated samples in both TAV and BAV separately was performed. Loadings for the two models, TAV and BAV, were plotted together in a combined model scatter plot (Figure 4A). Exons found to be significant in the TGFβ pathway alternative splicing analysis (according to FDR-corrected t test) were marked in the plot according to Figure 2B. This analysis revealed that the exons that were shown to have high impact in the TGFβ pathway models, that is, situated far away from the origo in the combined model scatter plot, also have high impact in Figure 4A. The exons found to be important in the TGFβ pathway are thus among the most significant ones if all the genes are included (Figure 4B, C). This finding indicates that alternative splicing in TGFβ pathway is one of the important processes that, together with other alternatively spliced genes, is able to discriminate between dilated and nondilated samples in both TAV and BAV patients (Figure 4).
Figure 4.
OPLS-DA of gene level normalized exon expression (splice index) of the filtered data set for all human exons. The exons are color coded according to significant exons included in TGFβ analysis in Figure 2: TAV-specific (blue), BAV-specific (green), and common (red) probe sets. (A) Combined model scatter plot based on TAV dilated versus nondilated and BAV dilated versus nondilated OPLS-DA models. The black diagonal is aimed for interpretation purposes. Bar plots showing the loadings of each of the significant exons indicating their contribution to the first PC are shown in (B–D) and color coded according to (A). The confidence levels for each data point were estimated by a jack-knife algorithm. Exons chosen for RT-PCR validation are marked in (A–D).
RT-PCR Validation of Differentially Spliced Transcripts
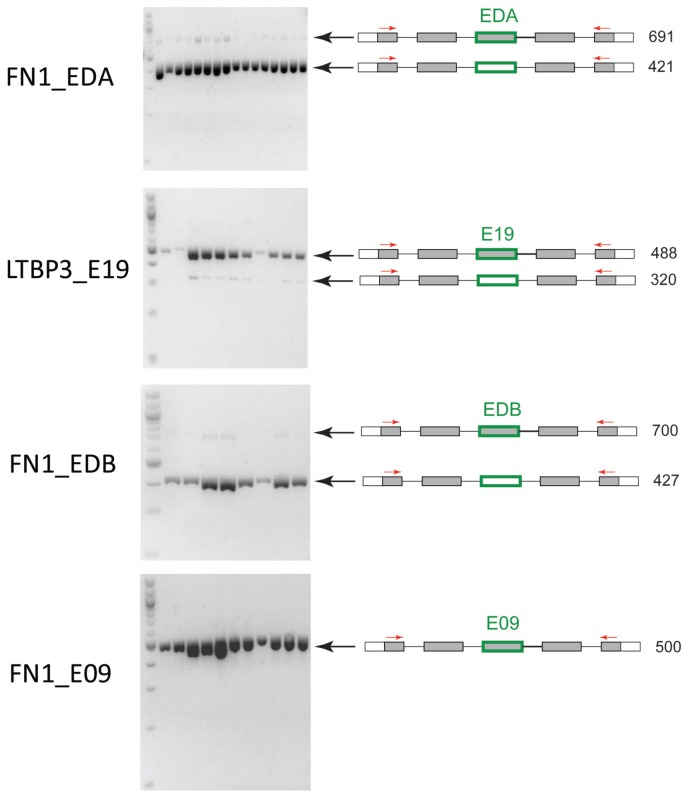
A handful of exons found to be differentially spliced according to the statistical analysis (Figure 2, Table 1) were chosen for RT-PCR validation. Those exons include FN1_EDB and LTBP3_E19, which display common differential expression in patients having TAV and BAV; FN1_EDA, which is differentially expressed between dilated and nondilated patients only in TAV; and FN1_09, which does not show any differential expression and serves as a control (Figure 5). The three exons FN1_EDA, LTBP3_E19 and FN1_EDB clearly show two different iso-forms present according to RT-PCR gels, one including and one excluding the exon of interest. The control (FN1_E09), however, shows only one isoform in which the exon is included (Figure 5). FN1_EDA and FN1_EDB represent already known alternative splicing events (13), whereas alternative splicing of exon 19 in LTBP3 (LTBP3_E19) has not previously been shown.
Figure 5.
RT-PCR validation of alternative splicing events. A set of exons found to be differentially expressed by statistical analyses (Figure 2) were chosen for RT-PCR validation of alternative splicing. (A) FN1_EDA, (B) LTBP3_E19 and (C) FN1_EDB show alternative splicing, and FN1_E09 (D) is not alternatively spliced and serves as a control. The gray boxes represent flanking exons in closest proximity to the exon that was investigated (marked in green around the box). Red arrows represent the position of the primers for RT-PCR. The length of the PCR product is shown in numbers. Black arrows indicate the position of the two isoforms on the gel.
Gene Expression Patterns of Differentially Spliced Genes
OPLS-DA was performed on the meta probe set level to find out how many genes in the TGFβ pathway are differentially expressed. Two separate OPLS-DA models were made on the RMA normalized data set of TAV patients with dilated and nondilated aortas and BAV patients with dilated and nondilated aortas; 29 and 52 patients were included in the TAV and BAV data sets, respectively (the same set of samples used in the exon level analysis). The two model loadings were plotted with jack-knife confidence levels (Supplemental Figure 2A, B) and in a combined model scatter plot (Supplemental Figure 2C). The total number of differentially expressed genes according to jack-knife confidence intervals was found to be 20, of which 2 genes were TAV specific, 6 genes were BAV specific, and 11 were common between TAV and BAV (Supplemental Figure 2). In summary, 76.9% of the genes included in this analysis were found to be differentially expressed between dilated and nondilated thoracic aortas.
DISCUSSION
In the present study, we used microarray technology to measure the expression of single exons to identify alternatively spliced events. We sought to analyze the importance of alternative splicing in dilated and nondilated thoracic aorta tissues in patients with TAV and BAV. Given that the TGFβ signaling pathway is believed to be important in aortic aneurysm (6,14), the focus of this study was an exploration of alternative splicing in all genes of this pathway. Our hypothesis leans on the concept that splicing might have an impact on the effectiveness of the TGFβ pathway itself. If the pathway is altered, the cells might have an improper response to injuries, for example, dilatation of the aortic wall.
To identify differentially spliced exons and their potential contribution to dilated aorta in patients with TAV and BAV, multivariate data exploration techniques such as PCA and OPLS-DA were adopted together with an FDR-corrected two-sided Student t test. The multivariate statistical techniques were able to identify alternative splicing associated with aneurysm of the ascending aorta. Furthermore, the jack-knife confidence levels calculated in a multivariate data analysis provided additional information about the significance of a certain exon in alternative splicing. Moreover, to find out how the two groups, dilated and nondilated, separated, we used gene expression data set of all genes in the human genome in a nonsupervised PCA. This analysis served, along with medical phenotype descriptions, as inclusion criteria for subsequent exon level analysis (Supplemental Figure 1). Alternative splicing analysis of genes included in the TGFβ pathway revealed different splicing patterns between patients with TAV and BAV. In particular, the difference between dilated and nondilated alternative splicing of aorta tissues illustrated divergence in the splice pattern of different exons. Although the number of significant differentially spliced exons that were shared between dilatation in TAV and BAV was 61, the numbers of TAV-specific and BAV-specific counterparts were 40 and 86, respectively. Comparison of dilated with nondilated tissues showed that TAV and BAV patients share many differentially spliced exons. However, four genes showed higher prevalence for alternative splicing in TAV compared with BAV (ENG, FN1, COL4A1 and FURIN), whereas two showed clear prevalence for BAV compared with TAV (EMILIN and LTPB3) (Supplemental Table 5). These findings suggest that alternative splicing in TGFβ pathway genes is important in the dilatation in both TAV and BAV patients. However, the pattern of alternative splicing, i.e., which exons are differentially expressed between TAV and BAV, is diverging between dilated and nondilated patients in the two groups TAV and BAV. Furthermore, the exons chosen for validation with RT-PCR represent different kinds of alternative splicing events, exon inclusion (FN1_EDA and FN1_EDB) as well as exon skipping (LTBP3_E19). This indicates that both events, a retained exon as well as a skipped exon, may have an impact on the function of the gene in question and thereby differentiate between two phenotypes.
In the present study we identified different splice forms of FN1 that may have an impact on TAA formation. We have previously shown that exons EDA and EDB show different alternative patterns in BAV and TAV patients (6). FN1 is a glycoprotein influencing cell migration, differentiation and growth and is a common extracellular matrix component in the vessel wall. The soluble form of FN1 present in plasma is dimeric and lacks the EDA and EDB exons. In tissue, multimeric forms containing the EDA and/or EDB domains are present in addition to the soluble form. In humans, EDA-containing FN1 (EDA-FN1) is associated with a stable plaque phenotype in atherosclerosis (15), whereas in mice it has been shown that EDA-FN1 has an essential role in pulmonary fibrosis (16) and in skin wound healing (17). It has also been shown that EDA-FN1 and EDB-FN1 expression levels are associated with restenosis in a rat model (18). Furthermore, both EDA and EDB appear to have critical roles in vascular morphogenesis during embryogenesis (19). Together, these studies show that FN1 has different function depending on which one of the two exons EDA and EDB that is included in the protein isoform.
The importance of splicing of the TGFβ pathway genes and the ability to separate dilated from nondilated aorta samples in a PCA is further illustrated by the fact that many of the exons found to be alternatively spliced in TGFβ exon analysis also showed high impact in discrimination between the two groups when all human exons (filtered with several different criteria prior to the analysis) were examined (Figure 4). TGFβ pathway is thus clearly important in alternative splicing of patients with a dilated compared with patients with a nondilated aorta, and shows typical TAV and BAV alternative splicing patterns in an overall exon level analysis. However, the analysis of alternative splicing of all exons (filtered with several criteria) in the human genome also showed common and distinct spliced exons in addition to the TGFβ pathway in TAV and BAV patients. It is thus clear that many exons are able to separate dilated and nondilated aortas from each other and it is important to stress that TGFβ alone is not responsible for the diverging alternative splicing fingerprints identified between dilated and nondilated aortic tissues of TAV and BAV patients. It can thereby be concluded that alternative splicing, in general, is an important process in this disease, with many important pathways, one being the TGFβ-signaling pathway.
An interesting aspect to investigate is whether the genes that show alternative splicing also are differentially expressed. The analysis of TGFβ gene expression revealed that the majority of genes (76.9%) in this signal pathway indeed are differentially expressed. The 11 genes that show the highest level of alternative splicing (Supplemental Table 5), with 1 exception (FKBP1A), are all differentially expressed, with higher expression in dilatation of both TAV and BAV patients. This finding further highlights the importance of alternative splicing in thoracic aortic aneurysm.
In our study, we demonstrated that alternative splicing in the TGFβ pathway can be used to characterize patients with dilated and nondilated thoracic aortic aneurysms. The characterization of the two different types of aortas revealed diverging alternative splicing fingerprints in TAV and BAV patients, which indicates that the underlying mechanisms of dilatation differ in TAV compared with BAV. Interestingly, epigenetic modification of the chromatin structure has been demonstrated to participate in the regulation of alternative splicing of fibronectin mRNA in respect to exon 33 (EDA) (20). Therefore, it can be speculated that different epigenetic fingerprints in BAV and TAV patients might be the cause of the observed different splicing patterns. The importance of alternative splicing in this respect is further explained by the fact that almost all the genes that are alternatively spliced also are differentially expressed. The diverging alternative splicing fingerprints in patients with thoracic aortic aneurysm are important targets for understanding the underlying mechanisms of aneurysm formation.
Supplemental Data
ACKNOWLEDGMENTS
This work was supported by the Swedish Research Council (12660), the Stockholm County Council (20090077), the Swedish Heart-Lung Foundation (20090541), the European Commission (Fighting Aneurysmal Disease [FAD], Health F2 2008 200647), a donation by Fredrik Lundberg, and a Swedish Research Council postdoctoral fellowship (21629 to S Kurtovic).
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Friedman T, Mani A, Elefteriades JA. Bicuspid aortic valve: Clinical approach and scientific review of a common clinical entity. Expert Rev Cardiovasc Ther. 2008;6:235–48. doi: 10.1586/14779072.6.2.235. [DOI] [PubMed] [Google Scholar]
- 2.Cecconi M, et al. Aortic dilatation in patients with bicuspid aortic valve. J Cardiovasc Med (Hagerstown) 2006;7:11–20. doi: 10.2459/01.JCM.0000199777.85343.ec. [DOI] [PubMed] [Google Scholar]
- 3.Mizuguchi T, Matsumoto N. Recent progress in genetics of Marfan syndrome and Marfan-associated disorders. J Hum Genet. 2007;52:1–12. doi: 10.1007/s10038-006-0078-1. [DOI] [PubMed] [Google Scholar]
- 4.Kluppel M, Wrana JL. Turning it up a Notch: Cross-talk between TGF beta and Notch signaling. Bioessays. 2005;27:115–8. doi: 10.1002/bies.20187. [DOI] [PubMed] [Google Scholar]
- 5.McKellar SH, et al. Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J Thorac Cardiovasc Surg. 2007;134:290–6. doi: 10.1016/j.jtcvs.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 6.Paloschi V, et al. Impaired splicing of fibronectin is associated with thoracic aortic aneurysm formation in patients with bicuspid aortic valve. Arterioscler Thromb Vasc Biol. 2011;31:691–7. doi: 10.1161/ATVBAHA.110.218461. [DOI] [PubMed] [Google Scholar]
- 7.Jackson JE. A User’s Guide to Principal Components. New York: Wiley; 1991. p. 569. [Google Scholar]
- 8.Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) J Chemometrics. 2002;16:119–28. [Google Scholar]
- 9.Martens H, Hoy M, Westad F, Folkenberg D, Martens M. Analysis of designed experiments by stabilised PLS Regression and jack-knifing. Chemometr Intell Lab Syst. 2001;58:151–70. [Google Scholar]
- 10.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 11.Wold S. Cross-validatory estimation of number of components in factor and principal components models. Technometrics. 1978;20:397–405. [Google Scholar]
- 12.Wettenhall JM, Simpson KM, Satterley K, Smyth GK. affylm GUI: A graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22:897–9. doi: 10.1093/bioinformatics/btl025. [DOI] [PubMed] [Google Scholar]
- 13.Schwarzbauer JE, Tamkun JW, Lemischka IR, Hynes RO. Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell. 1983;35:421–31. doi: 10.1016/0092-8674(83)90175-7. [DOI] [PubMed] [Google Scholar]
- 14.Jones JA, Spinale FG, Ikonomidis JS. Transforming growth factor-beta signaling in thoracic aortic aneurysm development: A paradox in pathogenesis. J Vasc Res. 2009;46:119–37. doi: 10.1159/000151766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Keulen JK, et al. Levels of extra domain A containing fibronectin in human atherosclerotic plaques are associated with a stable plaque phenotype. Atherosclerosis. 2007;195:e83–91. doi: 10.1016/j.atherosclerosis.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Muro AF, et al. An essential role for fibronectin extra type III domain A in pulmonary fibrosis. Am J Respir Crit Care Med. 2008;177:638–45. doi: 10.1164/rccm.200708-1291OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muro AF, et al. Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan. J Cell Biol. 2003;162:149–60. doi: 10.1083/jcb.200212079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubin D, et al. Balloon catheterization induced arterial expression of embryonic fibronectins. Arterioscler Thromb Vasc Biol. 1995;15:1958–67. doi: 10.1161/01.atv.15.11.1958. [DOI] [PubMed] [Google Scholar]
- 19.Astrof S, Hynes RO. Fibronectins in vascular morphogenesis. Angiogenesis. 2009;12:165–75. doi: 10.1007/s10456-009-9136-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allo M, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–24. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.