Abstract
Acute cellular rejection (ACR) and hepatitis C virus (HCV) recurrence (HCVrec) are common complications after liver transplantation (LT) in HCV patients, who share common clinical and histological features, making a differential diagnosis difficult. Fifty-three liver allograft samples from unique HCV LT recipients were studied using microarrays, including a training set (n = 32) and a validation set (n = 19). Two no-HCV-ACR samples from LT recipients were also included. Probe set intensity values were obtained using the robust multiarray average method (RMA) method. Analysis of variance identified statistically differentially expressed genes (P ≤ 0.005). The limma package was used to fit the mixed-effects models using a restricted maximum likelihood procedure. The last absolute shrinkage and selection operator (LASSO) model was fit with HCVrec versus ACR as the dependent variable predicted. N-fold cross-validation was performed to provide an unbiased estimate of generalization error. A total of 179 probe sets were differentially expressed among groups, with 71 exclusive genes between HCVrec and HCV-ACR. No differences were found within ACR group (HCV-ACR vs. no-HCV-ACR). Supervised clustering analysis displayed two clearly independent groups, and no-HCV-ACR clustered within HCV-ACR. HCVrec-related genes were associated with a cytotoxic T-cell profile, and HCV-ACR–related genes were associated with the inflammatory response. The best-fitting LASSO model classifier accuracy, including 15 genes, has an accuracy of 100% in the training set. N-fold cross-validation accuracy was 78.1%, and sensitivity, specificity and positive and negative predictive values were 50.0%, 90.9%, 71.4% and 80.0%, respectively. Arginase type II (ARG2), ethylmalonic encephalopathy 1 (ETHE1), transmembrane protein 176A (TMEM176A) and TMEM176B genes were significantly confirmed in the validation set. A molecular signature capable of distinguishing HCVrec and ACR in HCV LT recipients was identified and validated.
INTRODUCTION
Liver transplantation (LT) is the chosen treatment for patients with advanced liver disease and cirrhosis (http://optn.transplant.hrsa.gov). End-stage liver disease associated with hepatitis C virus (HCV) infection constitutes the leading indication for orthotopic LT (OLT) in the U.S., Europe and Japan. Unfortunately, reinfection of the graft represents a universal phenomenon after LT. The majority of patients with virological recurrence will develop histological graft injury, with about one-third of patients developing cirrhosis at only 5 years of follow-up (1–5). In addition, time interval from HCV cirrhosis to decompensation is significantly shorter in OLT recipients than in nontransplant patients (1–5). Not surprisingly, HCV is associated with decreased patient and graft survival when compared with other indications of OLT (1,5).
Factors associated with accelerated HCV recurrence (HCVrec) after LT include high viremia, donor age and quality (older than 55 years old), prolonged cold ischemia time, cytomegalovirus and immunosuppressant protocols.
Acute cellular rejection (ACR) after LT is common and remains an important cause of morbidity and late graft failure in the liver transplant recipient (LTR). Recent data suggest a variable rate of ACR between 18% and 30% (6). This tendency may be explained by the introduction of new and more potent immunosuppressant drugs in the last decade, capable of reducing the risk of ACR (6,7).
ACR rate after LT is especially controversial in HCV-positive recipients because of the overlapping of clinical and histological features with HCVrec. Reports suggest that the incidence of ACR may be in the range of 40% to 49% in HCV-positive recipients 6 months after LT (7). The high discrepancy of ACR incidence in HCV patients is explained by the difficulty on the differential diagnoses, which represents one of the most critical challenges in the liver transplant community. Furthermore, accurate diagnosis remains a critical issue, since associated therapy may lead to worse graft and patient survival (6–10). It is well documented that pulse intravenous corticosteroid bolus treatment for ACR exacerbates HCV infection, inducing transient 1- to 2-log increased HCV RNA levels and thus increasing mortality and graft loss (5,7,11,12).
There is a critical need for biomarkers capable of accurately differentiating between these two entities after LT. Despite some promising results, discovery of reliable diagnostic biomarkers is still lacking. Whole genome oligonucleotide microarrays represent a powerful technology to study thousands of genes simultaneously and concomitantly to discover new biomarkers for diagnosis, prognosis and treatment of complex and multifactorial diseases. In the present report, we evaluate differential gene expression, using microarrays in liver samples from HCV transplant recipients with HCVrec disease, and ACR in the setting of HCV infection.
MATERIALS AND METHODS
Patients and Tissue Samples
The Institutional Review Board approved the study at Virginia Commonwealth University. Written informed consent was obtained from all patients. A total of 53 liver samples from unique HCV LT recipients were used for this study. As the training set, 32 fresh-frozen liver tissue samples were obtained for clinical indication: 22 tissues from patients with HCVrec and 10 with ACR in the setting of HCV infection (all diagnosed by histopathological examination). In addition, two fresh-frozen tissue samples from HCV seronegative post-LT recipients and with histological diagnosis of ACR were included. Furthermore, an independent set of 19 biopsy samples histologically diagnosed with HCVrec (n = 9) and HCV-ACR (n = 10) were evaluated as the validation set. Demographic and clinical characteristics of both training and validation sets of patients were collected.
RNA Isolation, cDNA Synthesis and In Vitro Transcription for the Labeled cRNA Probe
Tissue samples were snap-frozen in liquid nitrogen or collected in RNA Later solution (Ambion, Austin, TX, USA) and stored at −80°C until use. Total RNA was extracted using TRIzol (Life Technologies, Carlsbad, CA, USA) by strictly following the Affymetrix GeneChip® Expression Analysis Manual (Affymetrix, Santa Clara, CA, USA). Total isolated RNA sample preparation follows purity and integrity quality control parameter criteria previously established in our laboratory (13). cDNA synthesis, in vitro transcription for labeled cRNA probe, Affymetrix HGU133A version 2.0 GeneChip® hybridization, as well as array image generation and probe sets reading and expression analysis were performed as described previously (13).
Data Analysis
To identify probe sets significantly differentially expressed from hybridized microarrays and among the three samples groups, probe set level analysis of variance models were fit considering probe set expression as the response and tissue type (HCVrec, HCV-ACR and no-HCV-ACR) as the fixed effect of interest. A group’s mean parameterization of tissue type was included as the fixed effect term to facilitate extraction of linear contrasts of interest. To adjust data for multiple comparisons, probe sets with P values from the overall F test <0.005 were considered significant. Thereafter, among probe sets identified as differentially expressed via the F test, all pair-wise comparisons were performed. The limma package was used to fit the mixed-effects models using restricted maximum likelihood procedure. Also, an empirical Bayes method was applied to moderate probe set standard errors by borrowing information across the entire group.
Statistical Predictive Class Analysis
The last absolute shrinkage and selection operator (LASSO) L1-penalized method was performed as a classifier for HCVrec versus HCV-ACR as a predicted dependent variable. The relevance and effectiveness of penalized methods, more specifically for LASSO, when modeling microarray data to identify important genes, was previously described (14). The LASSO model was fit using the glmpath package (15) in the R programming environment (16) using an appropriate Bioconductor package (17). All probe sets identified as significantly differentially expressed when comparing HCVrec (n = 22) to HCV-ACR (n = 10) were included into the analysis. N-fold cross-validation was performed to provide an unbiased estimate of generalization error when classifying groups and thus to validate the accuracy and analytical performance of the best-fitted model.
Interaction Network and Functional Analysis
Gene ontology and interaction were analyzed with the Ingenuity Pathways Analysis (IPA) tool 8.7 (http://www.ingenuity.com). Lists of specific genes were generated from each corresponding supervised analysis obtained for each pair-wise comparison. Also, the IPA tool was used to analyze the biological role of those genes identified by the best-fitting LASSO model. All gene lists generated from pair-wise comparisons and containing probe set IDs, Entrez gene IDs as clone identifiers and fold-change values were mapped in the Ingenuity Knowledge Base (genes + endogenous chemicals) as the reference set. A similar analysis was performed for those differentially expressed probe sets representing genes identified as specifics for HCVrec versus HCV-ACR (Table 1). Biological role and disease relevance were established for each specific gene list through the associated network functions and canonical pathways analysis.
Table 1.
Exclusive genes differentially expressed between HCVrec and HCV-ACR.
| Affymetrix ID | Gene symbol | Entrez gene name | P |
|---|---|---|---|
| 206506_s_at | SUPT3H (includes EG:8464) | Suppressor of Ty 3 homolog (S. cerevisiae) | 0.0019 |
| 214473_x_at | PMS2L3 | Postmeiotic segregation increased 2-like 3 | 0.0008 |
| 211558_s_at | DHPS | Deoxyhypusine synthase | 0.0009 |
| 202120_x_at | AP2S1a | Adaptor-related protein complex 2, sigma 1 subunit | 0.0002 |
| 201765_s_at | HEXA | hexosaminidase A (α polypeptide) | 0.0003 |
| 216032_s_at | ERGIC3 | ERGIC and golgi 3 | 0.0005 |
| 216903_s_at | CBARA1 | Calcium binding atopy-related autoantigen 1 | 0.0027 |
| 204985_s_at | TRAPPC6A | Trafficking protein particle complex 6A | 0.0002 |
| 204601_at | N4BP1 | NEDD4 binding protein 1 | 0.0003 |
| 201600_at | PHB2 | Prohibitin 2 | 0.0002 |
| 212782_x_at | POLR2J | Polymerase (RNA) II (DNA directed) polypeptide J, 13.3 kDa | 0.0016 |
| 202387_at | BAG1 | BCL2-associated athanogene | 0.0003 |
| 214277_at | COX11 | COX11 cytochrome c oxidase assembly homolog (yeast) | 0.0004 |
| 218679_s_at | VPS28 | Vacuolar protein sorting 28 homolog (S. cerevisiae) | 0.0004 |
| 220089_at | L2HGDH | L-2-hydroxyglutarate dehydrogenase | 0.0001 |
| 218345_at | TMEM176A | Transmembrane protein 176A | 0.0001 |
| 210927_x_at | JTB | Jumping translocation breakpoint | 0.0003 |
| 213550_s_at | NDUFA2 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 2, 8 kDa | 0.0002 |
| 220532_s_at | TMEM176B | Transmembrane protein 176B | <0.0001 |
| 203927_at | NFKBIE | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | 0.0002 |
| 201119_s_at | COX8A | Cytochrome c oxidase subunit VIIIA (ubiquitous) | 0.0003 |
| 221488_s_at | CUTA | cutA divalent cation tolerance homolog (E. coli) | 0.0002 |
| 213540_at | HSD17B8 | Hydroxysteroid (17-beta) dehydrogenase 8 | 0.0003 |
| 202283_at | SERPINF1 | Serpin peptidase inhibitor, clade F (α-2 antiplasmin, pigment epithelium derived factor), member 1 | 0.0002 |
| 215084_s_at | LRRC42 | Leucine rich repeat containing 42 | 0.0003 |
| 203024_s_at | C5ORF15 | Chromosome 5 open reading frame 15 | 0.0002 |
| 219345_at | BOLA1 | bolA homolog 1 (E. coli) | 0.0005 |
| 209208_at | MPDU1 | Mannose-P-dolichol utilization defect 1 | 0.0003 |
| 207157_s_at | GNG5 | Guanine nucleotide binding protein (G protein), γ 5 | 0.0001 |
| 1861_at | BAD | BCL2-associated agonist of cell death | 0.0003 |
| 48825_at | ING4 | Inhibitor of growth family, member 4 | 0.0004 |
| 203478_at | NDUFC1 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6 kDa | 0.0001 |
| 218309_at | CAMK2N1 | Calcium/calmodulin-dependent protein kinase II inhibitor 1 | 0.0001 |
| 208074_s_at | AP2S1a | Adaptor-related protein complex 2, sigma 1 subunit | 0.0001 |
| 203857_s_at | PDIA5 | Protein disulfide isomerase family A, member 5 | 0.0003 |
| 201023_at | TAF7 | TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55 kDa | 0.0005 |
| 201812_s_at | TOMM7 | Translocase of outer mitochondrial membrane 7 homolog (yeast) | 0.0006 |
| 203781_at | MRPL33 | Mitochondrial ribosomal protein L33 | 0.0002 |
| 201560_at | CLIC4 | Chloride intracellular channel 4 | 0.0014 |
| 211732_x_at | HNMT | Histamine N-methyltransferase | 0.0022 |
| 204559_s_at | LSM7 | LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) | 0.0005 |
| 204880_at | MGMT | O-6-methylguanine-DNA methyltransferase | <0.0001 |
| 204034_at | ETHE1 | Ethylmalonic encephalopathy 1 | <0.0001 |
| 220016_at | AHNAK | AHNAK nucleoprotein | 0.0001 |
| 205750_at | BPHL | Biphenyl hydrolase-like (serine hydrolase) | 0.0004 |
| 220942_x_at | FAM162A | Family with sequence similarity 162, member A | 0.0001 |
| 219479_at | KDELC1 | KDEL (Lys-Asp-Glu-Leu) containing 1 | 0.0003 |
| 201028_s_at | CD99 (includes, for example, 4267) | CD99 molecule | 0.0001 |
| 217963_s_at | NGFRAP1 | Nerve growth factor receptor (TNFRSF16) associated protein 1 | 0.0003 |
| 217289_s_at | SLC37A4 | Solute carrier family 37 (glucose-6-phosphate transporter), member 4 | 0.0003 |
| 206262_at | ADH1C (includes, for example, 126) | Alcohol dehydrogenase 1C (class I), γ polypeptide | 0.0025 |
| 212282_at | TMEM97 | Transmembrane protein 97 | 0.0002 |
| 218459_at | TOR3A | Torsin family 3, member A | 0.0001 |
| 202427_s_at | BRP44 | Brain protein 44 | 0.0003 |
| 207621_s_at | PEMT | Phosphatidylethanolamine N-methyltransferase | 0.0001 |
| 201316_at | PSMA2 | Proteasome (prosome, macropain) subunit, α type, 2 | 0.0005 |
| 204284_at | PPP1R3C | Protein phosphatase 1, regulatory (inhibitor) subunit 3C | 0.0001 |
| 220445_s_at | CSAG2 | CSAG family, member 2 | 0.0003 |
| 207097_s_at | SLC17A2 | Solute carrier family 17 (sodium phosphate), member 2 | 0.0001 |
| 210797_s_at | OASL | 2′-5′-Oligoadenylate synthetase-like | 0.0004 |
| 203946_s_at | ARG2a | Arginase type II | <0.0001 |
| 219230_at | TMEM100 | Transmembrane protein 100 | 0.0001 |
| 205968_at | KCNS3 | Potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 | 0.0006 |
| 208241_at | NRG1 | Neuregulin 1 | 0.0007 |
| 211743_s_at | PRG2 | Proteoglycan 2, bone marrow (natural killer cell activator, eosinophil granule major basic protein) | 0.0001 |
| 204076_at | ENTPD4 | Ectonucleoside triphosphate diphosphohydrolase 4 | 0.0002 |
| 203168_at | ATF6B | Activating transcription factor 6 beta | <0.0001 |
| 202106_at | GOLGA3 | Golgin A3 | 0.0005 |
| 219944_at | CLIP4 | CAP-GLY domain containing linker protein family, member 4 | 0.0003 |
| 37986_at | EPOR | Erythropoietin receptor | 0.0004 |
| 218792_s_at | BSPRY | B-box and SPRY domain containing | 0.0002 |
| 203945_at | ARG2a | Arginase type II | 0.0001 |
| 221854_at | PKP1 | Plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) | 0.0007 |
Pair-wise comparison was performed as HCVrec versus HCV-ACR. Significant differential gene expression is indicated by P values for each molecule.
ARG2 gene is represented by two different probe sets.
Reverse Transcription and Real-Time PCR (RT-qPCR) for an Independent Validation Set of Samples
Reverse transcription reactions were performed using TaqMan® Reverse Transcription Reagents (Applied Biosystems, Foster City, CA, USA) and according to the manufacturer protocol. Real-time PCRs (qPCRs) were carried out in an ABI Prism 7700 Sequence Detection System, using TaqMan® Gene Expression Assays (Applied Biosystems) (ARG2: Hs00982833_m1, ETHE1: Hs00204752_m1, TMEM176A: Hs00218506_m1 and TMEM176B: Hs00962650_m1). Gene expression fold-changes were obtained using the ΔCt calculation model between the target gene mean cycle threshold (Ct) and β2-microglobulin (B2M) Ct as endogenous control, or ΔCt (gene) – ΔCt (B2M). A two-tailed t test was performed to compare the mean expression between HCVrec and HCV-ACR samples for all validated genes. A P value <0.05 was considered significant.
All supplementary materials are available online at www.molmed.org.
RESULTS
Patients
Clinicopathological characteristics of the 51 LT patients are fully described in Supplementary Table S1 (A,B). All patients (except the two no-HCV patients with ACR) included into this study were diagnosed with HCV infection, genotype 1b before LT, and were the primary transplant, and all patients received an HCV-negative graft from a standard deceased donor. The immunosuppression protocol was similar for all patients and consisted of calcineurin inhibitor (tacrolimus, cyclosporine)-based therapy and steroid induction. The steroid-based therapy was routinely discontinued 3 months after LT in all cases.
Genomic Clustering of Liver Biopsy Samples
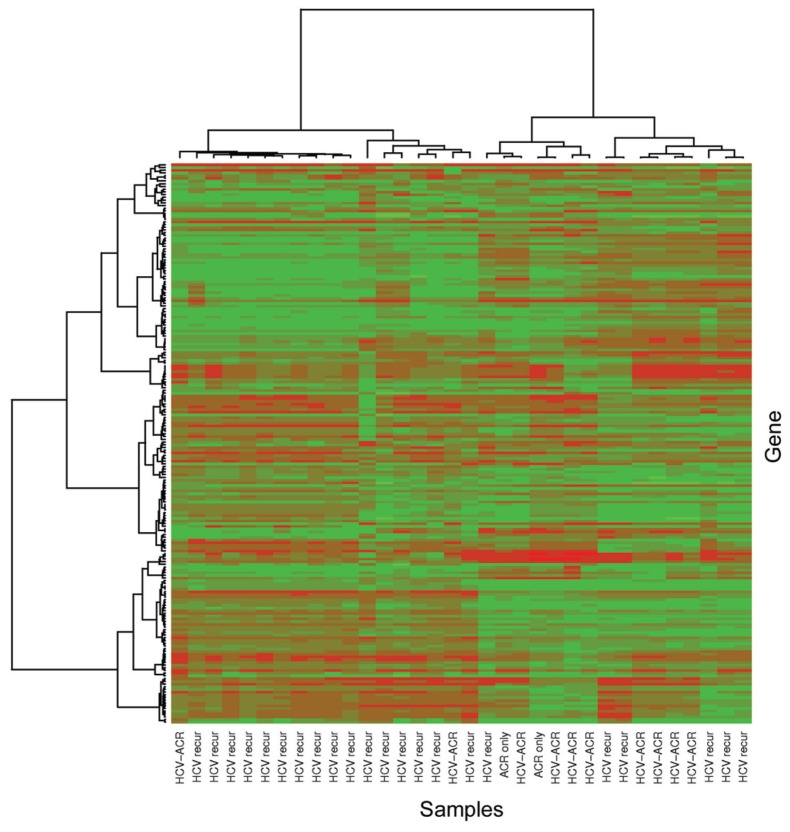
A total of 22,098 probe sets (>91%) were identified as present in each hybridized microarray generated for each tissue sample diagnosed with HCVrec (n = 32) and HCV-ACR and no-HCV-ACR (n = 19). Afterward, control probe sets were removed and the gene expression data set was further restricted to include only those probe sets declared to be either present or marginally present in at least 50% of the samples with an expression standard deviation among the 99th percentile or higher. Unsupervised hierarchical clustering analysis was performed to determine the natural sample clustering (Figure 1). The generated heatmap and dendrogram grouped most of HCVrec samples (68%; 15/22) in a defined cluster. A similar trend was observed for the majority of HCV-ACR samples. In addition, one HCV-ACR sample was found to be clustered independently from both main groups. The misclassified profile may be a result of intrinsic clinicopathological features particular for each patient. Nevertheless, this analysis further suggested the possibility to differentiate both pathological conditions through the study of the differential genomic profile.
Figure 1.
Unsupervised hierarchical clustering that includes all samples from the training group. Heatmap and dendrogram were generated using Ward’s method with 1 – |Pearson correlation| as the distance measure, including probe sets declared to be either present or marginally present in at least 50% of samples and having an expression standard deviation among ≥99th percentile. HCV recur, HCV recurrence; ACR only, no-HCV-ACR.
Identification of Differentially Expressed Probe Sets Among Sample Clusters
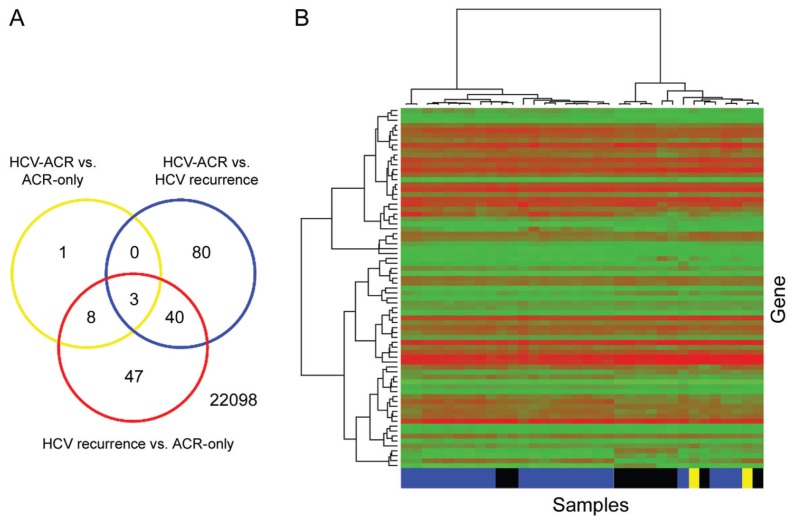
To better define a differential genomic profile within conditions, only probe sets exhibiting P values ≤0.001 were considered significant. A total of 179 probe sets were identified as significantly differentially expressed among three diagnostic groups (HCVrec, HCV-ACR and no-HCV-ACR). All multiple pair-wise comparisons were performed to better define the relevance of those significant probe sets (Figure 2A). From this analysis, 123 (116 genes), 98 (94 genes) and 12 (12 genes) probe sets were found to be significantly differentially expressed among HCVrec versus HCV-ACR, and HCVrec versus no-HCV-ACR, and HCV-ACR versus no-HCV-ACR, respectively. From the analysis of each pair-wise comparison, 80 probe sets (71 genes) were identified as exclusively differentially expressed between HCVrec and HCV-ACR. When the analysis was performed for HCVrec versus no-HCV-ACR, 47 probe sets (36 genes) were identified as significant (Figure 2A). Furthermore, only one probe set was exclusively identified between ACR tissue samples regarding the HCV infection state of the patient. Those ACR tissue samples were incorporated to identify genes specifically associated with this post-LT complication independent of HCV setting. Thus, this finding suggests independent characteristics for ACR, regardless of the recipient’s clinical feature.
Figure 2.
(A) Venn diagram illustrating the number of significant probe sets for the three pair-wise comparisons. 22098: total analyzed probe sets. (B) Supervised hierarchical clustering restricted to the 80 exclusive probe sets (71 genes) significantly differentially expressed between HCVrec and HCV-ACR samples (see Table 1). Heatmap and dendrogram were generated using Ward’s method with 1 – |Pearson correlation| as the distance measure. Blue boxes, HCV recurrence; black boxes, HCV-ACR; yellow boxes, no-HCV-ACR (ACR-only).
Supervised Genomic Clustering
Those specific probe sets identified by pair-wise comparison between HCVrec and HCV-ACR were analyzed to identify a characteristic molecular signature. From the analysis of those 71 genes specifically differentially expressed between groups, 59 genes (83%) were preferentially expressed in HCVrec samples, whereas the remaining 17% (12 genes) were identified in ACR-HCV (Table 1). The probe set intensity values representing those exclusive genes were used to conduct a supervised hierarchical clustering analysis (Figure 2B). The generated heatmap and dendrogram identified most HCVrec and HCV-ACR samples tightly clustered in two independent groups. Importantly, those no-HCV-ACR samples clustered within the HCV-ACR group in favor of the negligible differential gene expression between both ACR origins.
Biological Function Analysis of HCVrec and HCV-ACR Exclusive Genes
Core analysis was performed to interpret the data set in the context of biological processes, pathways and molecular networks those specific genes differentially expressed when comparing HCVrec with HCV-ACR. HCVrec-related genes were found to be preferentially associated with cell death regulation (P = 9.55E-04–4.55E-02), cell morphology (P = 1.59E-03–4.55E-02), carbohydrate (P = 3.32E-03–4.23E-02), amino acid metabolism (P = 3.32E-03–2.30E-02) and cellular development (P = 3.32E-03–2.30E-02). From the analysis of HCV-ACR–related genes, the identified canonical pathways were significantly implicated in cell growth and proliferation (P = 7.11E-04–4.93E-02), cell cycle (P = 7.76E-04–1.47E-02), cell-to-cell signaling and interaction (P = 7.76E-04–4.88E-02) and cellular assembly and organization (P = 7.76E-04–3.06E-02) (Supplementary Table S2). The overview of this analysis clearly suggests different cellular mechanisms involved in both post-LT pathological conditions.
Association of the Differentially Expressed Genes with Pathological Conditions
To better differentiate both conditions, the role and impact of these differentially expressed genes in disease development were analyzed using the IPA tool (www.ingenuity.com).
From the analysis, 32% (19/59) of HCVrec-deregulated genes were significantly associated with disease molecular pathways involved in cancer development (17 molecules; P = 1.42E-03 to 4.60E-02) and gastrointestinal disease (6 molecules; P = 6.42E-03 to 3.27E-02). A group of cancer-associated genes was found to be involved in T cytotoxic lymphocyte development (adaptor-related protein complex 2, sigma 1 subunit [AP2S1]) and adhesion to endothelium (CD99), T-cell lymphoma (serpin peptidase inhibitor, clade F [α-2 antiplasmin, pigment epithelium derived factor], member 1 [SERPINF1], and O-6-methylguanine-DNA methyltransferase [MGMT]), pro-apoptotic genes related to B and T lymphocytes regulation (BCL2-associated agonist of cell death [BAD]) and apoptosis of activated T cells (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon [NFKBIE]). The analysis result also identified other pro-apoptotic genes (inhibitor of growth family, member 4 [ING4]), transforming growth factor (TGF)-β–regulated genes involved in Histamine metabolism (histamine N-methyltransferase [HNMT]), nitric oxide and reactive oxygen species production in macrophages (protein phosphatase 1, regulatory [inhibitor] subunit 3C [PPP1R3C]) and immune modulation–related genes through proteasome degradation of the major histocompatibility complex (MHC) class I molecule (proteasome [prosome, macropain] subunit, α type [PSMA2]). HNMT and hexosaminidase A (HEXA) genes were found to be associated with rheumatoid arthritis, an inflammatory disease with high predominance of activated T cells. Moreover, the 2′-5′-oligoadenylate synthetase-like (OASL) encoding gene, positively related to high HCV load (18), was also differentially expressed in HCVrec disease patients.
Analysis of the HCV-ACR genomic pattern revealed most of the deregulated genes are further associated with inflammatory response and disease, such as airway hypersensitive reaction through induction of eosinophil, basophil and neutrophil degranulation (proteoglycan 2 [PRG2]) and nitric oxide degradation (ARG2). The neuregulin-1 encoding gene (NRG1) was also found to be associated with the development of hypersensitive reactions. Additionally, other genes were found to be involved in the development of atopic dermatitis (ectonucleoside triphosphate diphosphohydrolase 4 [ENTPD4]) and rheumatoid arthritis (potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 [KCNS3]; erythropoietin receptor [EPOR]; and activating transcription factor 6 beta [ATF6B]).
Overall, these results propose that a cytotoxic component may be exerted by a tuned T-cell-mediated cytolysis in response to virus-infected cells in HCVrec samples. The role of HCV-ACR–related genes suggests allograft damage may be caused by an adaptive immune response as an immediate hypersensitivity reaction and also may be through a T-cell immunoreaction trend.
Predictive Model that Uses the L1 Penalized Method
The LASSO (L1)-penalized method was performed as a classifier for HCVrec versus HCV-ACR to identify most important genes. The best-fitting LASSO model was subsequently confirmed using the N-fold cross-validation method as described in previous reports from our laboratory (14). The genomic pattern displayed by the best-fitting LASSO model included 15 probe sets differentially expressed for HCV-ACR (eight genes) and HCVrec disease (seven genes), listed in Table 2. Except nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (NFKBIA), all genes were also present in the differentially expressed probe sets identified by pairwise comparisons. More importantly, the N-fold cross- validation estimated error rate for the best-fitting model was 21.9%, which indicates a classifier accuracy rate of 78.1%. The classifier rates for sensitivity, specificity and positive and negative predictive values were 50.0%, 90.9%, 71.4% and 80.0%, respectively. Interestingly, 4 of those 15 genes (1 associated with HCV-ACR [ARG2] and 3 associated with HCVrec [ETHE1, TMEM176A and TMEM176B]) were also included into the characteristic molecular signature identified between HCVrec and HCV-ACR (Tables 1 and 2).
Table 2.
Genes identified by the best-fitting LASSO model.
| Affymetrix ID | Entrez ID | Gene symbol | Coefficient estimatea |
|---|---|---|---|
| 212619_at | 23306 | TMEM194A | −5.33 |
| 207367_at | 479 | ATP12A | −4.56 |
| 212056_at | 23199 | KIAA0182 | −3.89 |
| 213515_x_at | 3048 | HBG2 | −1.16 |
| 203946_s_at | 384 | ARG2a | −1.12 |
| 212856_at | 23151 | GRAMD4 | −0.82 |
| 212993_at | 138151 | BTBD14A | −0.55 |
| 206214_at | 7941 | PLA2G7 | −0.44 |
| 201502_s_at | 4792 | NFKBIA | 0.19 |
| 206293_at | 6822 | SULT2A1 | 0.26 |
| 204034_at | 23474 | ETHE1a | 0.39 |
| 205251_at | 8864 | PER2 | 0.52 |
| 202732_at | 11142 | PKIG | 0.61 |
| 220532_s_at | 28959 | TMEM176Ba | 1.36 |
| 218345_at | 55365 | TMEM176Aa | 2.00 |
Coefficients estimated by the LASSO model reflect the log odds of having HCV-ACR.
Genes identified as exclusive by the LASSO model and HCVrec versus HCV-ACR pair-wise comparison (see also Table 1).
Biological function analysis associated those genes with cell death, gene expression and cell cycle progression as top scored network functions (score >26), along with molecular transport, lipid metabolism, cellular movement and protein synthesis pathways. HCVrec differentially expressed genes were found to be associated with IFN-γ positive gene expression regulation (period homolog 2 (Drosophila) [PER2] [19]); NFKB signaling pathway involved in regulation of T-cell proliferation, activation and apoptosis (NFKBIA and ETHE1 [20,21]); dehydroepiandrosterone sulfation and detoxification of bile acids (SULT2A1), which was downregulated in the lipopolysaccharide (LPS)-induced acute-phase response (22); cAMP-dependent protein kinase (PKA) signaling inhibitor (PKIG [23]); and genes involved in the maintenance of the immature state of dendritic cells (TMEM176A and TMEM176B [24]) (Table 2). In a different trend, HCV-ACR–associated genes were found to be preferentially involved in the proinflammatory process related to hypersensitive reaction and apoptosis regulation (phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) [PLA2G7]; ARG2; GRAM domain containing 4 [GRAMD4]), oxygen transport activity and severe acute respiratory syndrome (hemoglobin, gamma G [HBG2]). Of note, the arginase 2 encoding gene (ARG2) exerts an anti-apoptotic role through the negative modulation of nitric oxide (25), and its expression was found to be increased during eosinophil inflammatory reaction in asthma via the Th2 lymphocyte profile induced by interleukin (IL)-4 and IL-13 (26,27). The platelet-activating factor–acetylhydrolase enzyme encoding gene (PLA2G7) is a proinflammatory factor associated with atherogenic plaque formation (28,29) and apoptosis inhibition through oxidized phospholipid hydrolization (30). The cellular function of GRAMD4 was postulated as similar to the p53 apoptosis regulatory factor (31). ATP12A is related with ATPase H(+) and K(+) ions exchange, and no specific cellular role has yet been established for TMEM194A, KIAA0182 and BTBD14A (Table 2).
To sum up, the HCVrec-associated genes identified by the LASSO predictive model are preferentially involved in an innate immune response, whereas HCV-ACR–related genes suggest a strong proinflammatory and antiapoptotic activity more related to an adaptive immune response that may be associated to a Th2 context
Gene Expression Validation in an Independent Set of Samples
A set of genes was selected for further evaluation in an independent set of samples using the following criteria: (a) differentially expressed genes between HCVrec and HCV-ACR and (b) identification in the best LASSO model. The expression levels of ARG2, ETHE1, TMEM176A and TMEM176B genes, all identified by supervised analysis and the LASSO model, were validated in an independent sample set by qPCR. From the analysis, all genes were found to be significantly and differentially expressed between both post-LT conditions. More importantly, there was a perfect correlation between both expression profiles identified by microarrays (training set) and RT-qPCR (validation set), as shown in Table 3.
Table 3.
Gene expression validation in an independent set of samples.
| Gene ID | qPCR fold-change | P |
|---|---|---|
| ARG2 | 2.1 ± 1.3 | 0.032 |
| ETHE1 | 4.2 ± 0.8 | 0.010 |
| TMEM176A | 2.0 ± 0.9 | 0.034 |
| TMEM176B | 2.1 ± 0.8 | 0.017 |
DISCUSSION
Liver biopsy is currently considered the gold standard for the histopathological evaluation of complications occurring on the graft after LT. However, differentiation of HCVrec from ACR in HCV LT recipients is especially challenging and provides conflicting results, even among experienced liver pathologists (32–37).
Importantly, therapy selection and efficacy further relies on the accuracy of the diagnosis. Actually, an ambiguous or uncertain diagnosis may induce a detrimental manner on the graft and patient survival rates (7,11).
In the recent years, a substantial number of markers were postulated as potential biomarkers of disease differentiation. Recent studies on immunohistochemistry assays proposed the end product of activated complement cascade component C4 (C4d) as a promising biomarker to differentially diagnose HCVrec from ACR (36,37). Unfortunately, these findings were not able to be validated by a prospective study using an enzyme-linked immunosorbent assay–based test (38). In addition, MxA and IFI16 interferon-inducible proteins (39,40) and the lymphocyte-expressed Mcm-2 protein (35) were also postulated as potential biomarkers, but prospective studies for further validation are lacking. The potential utility of the ImmunKnow assay for the post-LT immune function status of HCV recipients using biopsy samples was suggested by Cabrera et al. (41). Moreover, the findings from this report were further supported by Rubinas et al. (42) and others (43). However, the reliable availability and accuracy of biomarkers to support or replace histopathology is scarce.
Microarray technology represents an excellent tool to elucidate differential gene expression patterns and establish accurate molecular markers for differential disease diagnosis (44). There are two previous reports that aimed to differentiate ACR and HCVrec by genomic studies (45,46). These reports showed gene expression profiles characterizing the two subgroups of HCV patients. However, in the study performed by Sreekumar et al. (46), the signatures were not validated in an independent set of patients. Also, both reports lack inclusion of ACR samples from recipients with no HCV infection.
In the present study, we successfully performed whole genome analysis using GeneChip microarrays to differentiate biopsy-proven HCVrec disease from ACR in the setting of HCV infection. From this study, we identified a gene signature composed of 71 molecules differentially expressed between HCVrec (59 genes) and ACR (12 genes). The biological analysis of the cellular function of those genes associated with HCVrec revealed a strong hepatocyte cytotoxicity component represented by genes involved in regulation of proliferation, growth, adhesion and apoptosis of cytotoxic T lymphocytes and regulated by canonical pathways such as IFN-γ and nuclear factor κB. In comparison, the biological role of genes identified as exclusive for ACR in the setting of HCV infection suggested an association of the allograft damage through an immediate hypersensitivity reaction. This result may be triggered by eosinophil and neutrophil cell degranulation (major basic protein-1 encoding gene [PRG2]), along with the possibility of atherosclerosis development, mainly due to the presence of macrophage-related metabolic genes. It is interesting to note that some genes exclusively identified in ACR were associated with atherosclerosis development (such as PLA2G7 [28,29]) and nitric oxide degradation (such as ARG2 [25]), which may be involved in the hepatic vein endothelium injury mechanism characteristic of this post-LT complication.
Sreekumar et al. (46) previously reported a differential gene expressed between well-characterized HCVrec and ACR, but this report suggested that the HCVrec pathological component is more related to HCV infection cytopathic effects rather than the immune-mediated process. Also, the authors related the differential expression of IL-2, IFN-γ and tumor necrosis factor (TNF)-α encoding genes to ACR, whereas other authors found increased IL-4 and IL-10 gene expression (47). Furthermore, other reports suggest that the pathophysiological mechanisms of both entities are mainly characterized by a T-lymphocyte infiltrate, and although the distinction is difficult when overlapping occurs, the fine immune cell functions tuning and localization indicate that those conditions are completely different (4,7). In a timeline, HCVrec mostly begins 3–8 weeks after LT, whereas ACR begins mostly within 60–90 days. However, it is well known that the time of occurrence of these two complications overlaps (4).
Histopathologically, HCVrec is characterized by lobular hepatitis featured by mottled apoptotic hepatocytes and inflammation generated by a Th-1 type lymphocytic immune reaction (INF-γ, TNF-α, IL-2 and IL-12) with predominant cytotoxic T cells (mostly CD8+) aimed at directly eliminating infected hepatocytes. In combination, high HCV viral replication induces oxidative stress and ineffective antiviral immune response, contributing to the inflammatory scenario through activation of the Fas ligand apoptotic pathway and inducing hepatocyte lysis through the perforin/granzyme system (4,48,49). ACR is histologically characterized by inflammatory infiltrates of portal tracts, bile ducts and hepatic vein endothelium by CD4+ and CD8+ T lymphocytes and macrophages, as dominant cell types and together with eosinophil cells (4,6). Furthermore, in addition to T cells, B lymphocytes and plasma cells were also suggested to be present in ACR infiltrates (7,50). The identified gene expression profiles in the present study correlate with the predicted signatures for the cells described as involved in the characteristic histological features for both HCVrec and HCV-ACR.
Little is known about the ACR condition in the presence and absence of a viral immune reaction scenario. We performed genomic comparative studies to further characterize differential gene expression between ACR alone and in the setting of HCV graft infection, but interestingly, no further significant differences were identified (Figure 2A). These results are of high interest because they strongly indicate that ACR immune reaction is independent of viral infection backgrounds, at least through gene expression modulation. Nevertheless, it was postulated that the generated inflammatory microenvironment by acute HCV reinfection in combination with partial donor-recipient MHC class I compatibility may trigger an ACR response by enabling MHC- restricted HCV- specific cytotoxic T cells to target alloantigens (4,7). In this sense, immune-modulatory–related genes by the MHC class I molecule proteasome degradation (PSMA2) and maintenance of the immature state of dendritic cells (TMEM176A, TMEM176B) were identified as specific for HCVrec. The immature state of dendritic cells was related to the silencing of antigen- specific T cells, thus avoiding an autoimmune reaction (51). Thus, an early imbalance of the expression of this kind of gene might contribute to the ACR triggering in the acute graft HCV infection.
To gain insight into the diagnostic impact of the generated genomic signature, supervised hierarchical clustering was performed, and all patients were classified into two major groups as HCVrec and HCV-ACR (Figure 2B). It is interesting to note that all cases of ACR from patients with no HCV infection were clustered within HCV-ACR samples. This clustering pattern is in agreement with the observed null differential genomic expression between both ACR origins and further corroborates the unique nature of this particular post-LT condition. Also, from the analysis, it is possible to distinguish six samples (two from HCV-ACR and four from HCVrec) as misclassified. Interestingly, reinspection of the pathology report of those HCV-ACR samples revealed mixed histological features with more prominent pan-lobular inflammation and hepatocytes apoptosis and resolved ACR. In contrast, those misclassified HCVrec samples showed histological characteristics of extensive fibrosis and inflammation together with macrovesicular steatosis. In addition, the HCV serum load was found one log lower with respect to other patients, and two of them were directly negative. In conclusion, these cases represent why diagnosis is difficult and show the risk of misdiagnosing post-LT patients because of unclear histopathological reports. These findings further support the potential utility of molecular markers in the differential diagnosis.
Hereby, we proposed a set of genes capable of differentiating both conditions with high accuracy (LASSO), which was further validated in an independent group of well-diagnosed patients. Furthermore, it was possible to determine through the absence of major genomic differences that ACR is a HCV-independent immunological reaction post-LT. More importantly, we proposed a set of four validated genes as potential biomarkers for differential diagnosis of both complications: HCVrec and ACR. Altogether, our findings are of high importance for the accurate diagnosis of both entities, with a concomitant high impact on the selection of suitable therapy. Nevertheless, subsequent cohort studies must be performed and aimed to further validate the identified molecular markers.
Supplemental Data
ACKNOWLEDGMENTS
This work was presented in part at the 15th Annual International Congress, New York, New York, 8–11 July 2009.
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Garcia-Retortillo M, et al. Hepatitis C virus kinetics during and immediately after liver transplantation. Hepatology. 2002;35:680–7. doi: 10.1053/jhep.2002.31773. [DOI] [PubMed] [Google Scholar]
- 2.Gane EJ, et al. Long-term outcome of hepatitis C infection after liver transplantation. N Engl J Med. 1996;334:815–20. doi: 10.1056/NEJM199603283341302. [DOI] [PubMed] [Google Scholar]
- 3.Roche B, Samuel D. Risk factors for hepatitis C recurrence after liver transplantation. J. Viral Hepat. 2007;14(Suppl 1):89–96. doi: 10.1111/j.1365-2893.2007.00920.x. [DOI] [PubMed] [Google Scholar]
- 4.Demetris AJ. Evolution of hepatitis C virus in liver allografts. Liver Transpl. 2009;15(Suppl 2):S35–41. doi: 10.1002/lt.21890. [DOI] [PubMed] [Google Scholar]
- 5.Watt K, Veldt B, Charlton M. A practical guide to the management of HCV infection following liver transplantation. Am J Transplant. 2009;9:1707–13. doi: 10.1111/j.1600-6143.2009.02702.x. [DOI] [PubMed] [Google Scholar]
- 6.Knechtle SJ, Kwun J. Unique aspects of rejection and tolerance in liver transplantation. Semin Liver Dis. 2009;29:91–101. doi: 10.1055/s-0029-1192058. [DOI] [PubMed] [Google Scholar]
- 7.Burton JR, Jr, Rosen HR. Acute rejection in HCV-infected liver transplant recipients: the great conundrum. Liver Transpl. 2006;12:S38–47. doi: 10.1002/lt.20944. [DOI] [PubMed] [Google Scholar]
- 8.Kurian S, et al. Applying genomics to organ transplantation medicine in both discovery and validation of biomarkers. Int Immunopharmacol. 2007;7:1948–60. doi: 10.1016/j.intimp.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelb B, Feng S. Management of the liver transplant patient. Expert Rev Gastroenterol Hepatol. 2009;3:631–47. doi: 10.1586/egh.09.58. [DOI] [PubMed] [Google Scholar]
- 10.Manousou P, et al. Outcome of recurrent hepatitis C virus after liver transplantation in a randomized trial of tacrolimus monotherapy versus triple therapy. Liver Transpl. 2009;15:1783–91. doi: 10.1002/lt.21907. [DOI] [PubMed] [Google Scholar]
- 11.Demetris AJ, et al. Recurrent hepatitis C in liver allografts: prospective assessment of diagnostic accuracy, identification of pitfalls, and observations about pathogenesis. Am J Surg Pathol. 2004;28:658–69. doi: 10.1097/00000478-200405000-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berenguer M, et al. A model to predict severe HCV-related disease following liver transplantation. Hepatology. 2003;38:34–41. doi: 10.1053/jhep.2003.50278. [DOI] [PubMed] [Google Scholar]
- 13.Mas VR, et al. Differentially expressed genes between early and advanced hepatocellular carcinoma (HCC) as a potential tool for selecting liver transplant recipients. Mol Med. 2006;12:97–104. doi: 10.2119/2006-00032.Mas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Archer KJ, et al. Identifying genes for establishing a multigenic test for hepatocellular carcinoma surveillance in hepatitis C virus-positive cirrhotic patients. Cancer Epidemiol Biomarkers Prev. 2009;18:2929–32. doi: 10.1158/1055-9965.EPI-09-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park MY, Hastie T. Penalized logistic regression for detecting gene interactions. Biostatistics. 2008;9:30–50. doi: 10.1093/biostatistics/kxm010. [DOI] [PubMed] [Google Scholar]
- 16.R Development Core Team. R: A Language and Environment for Statistical Computing [computer program] Vienna (Austria): R Foundation for Statistical Computing; 2005. 2011 version available at: http://www.R-project.org. [Google Scholar]
- 17.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishibashi M, Wakita T, Esumi M. 2′,5′-Oligoadenylate synthetase-like gene highly induced by hepatitis C virus infection in human liver is inhibitory to viral replication in vitro. Biochem Biophys Res Commun. 2010;392:397–402. doi: 10.1016/j.bbrc.2010.01.034. [DOI] [PubMed] [Google Scholar]
- 19.Arjona A, Sarkar DK. The circadian gene mPer2 regulates the daily rhythm of IFN-gamma. J Interferon Cytokine Res. 2006;26:645–9. doi: 10.1089/jir.2006.26.645. [DOI] [PubMed] [Google Scholar]
- 20.Wang M, Windgassen D, Papoutsakis ET. A global transcriptional view of apoptosis in human T-cell activation. BMC Med. Genomics. 2008;1:53. doi: 10.1186/1755-8794-1-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higashitsuji H, et al. A novel protein over-expressed in hepatoma accelerates export of NF-kappa B from the nucleus and inhibits p53-dependent apoptosis. Cancer Cell. 2002;2:335–46. doi: 10.1016/s1535-6108(02)00152-6. [DOI] [PubMed] [Google Scholar]
- 22.Kim MS, Shigenaga J, Moser A, Grunfeld C, Feingold KR. Suppression of DHEA sulfotransferase (Sult2A1) during the acute-phase response. Am J Physiol Endocrinol Metab. 2004;287:E731–8. doi: 10.1152/ajpendo.00130.2004. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Dai JC, Orellana SA, Greenfield EM. Endogenous protein kinase inhibitor gamma terminates immediate-early gene expression induced by cAMP-dependent protein kinase (PKA) signaling: termination depends on PKA inactivation rather than PKA export from the nucleus. J Biol Chem. 2005;280:2700–7. doi: 10.1074/jbc.M412558200. [DOI] [PubMed] [Google Scholar]
- 24.Condamine T, et al. Tmem176B and Tmem176A are associated with the immature state of dendritic cells. J Leukoc Biol. 2010;88:507–15. doi: 10.1189/jlb.1109738. [DOI] [PubMed] [Google Scholar]
- 25.Mori M. Regulation of nitric oxide synthesis and apoptosis by arginase and arginine recycling. J Nutr. 2007;137:1616S–20S. doi: 10.1093/jn/137.6.1616S. [DOI] [PubMed] [Google Scholar]
- 26.Kenyon NJ, Bratt JM, Linderholm AL, Last MS, Last JA. Arginases I and II in lungs of ovalbumin-sensitized mice exposed to ovalbumin: sources and consequences. Toxicol Appl Pharmacol. 2008;230:269–75. doi: 10.1016/j.taap.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmermann N, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest. 2003;111:1863–74. doi: 10.1172/JCI17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Keyzer D, et al. Increased PAFAH and oxidized lipids are associated with inflammation and atherosclerosis in hypercholesterolemic pigs. Arterioscler Thromb Vasc Biol. 2009;29:2041–6. doi: 10.1161/ATVBAHA.109.196592. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y, et al. Role of lipoprotein-associated phospholipase A2 in leukocyte activation and inflammatory responses. Atherosclerosis. 2007;191:54–62. doi: 10.1016/j.atherosclerosis.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Chen R, Yang L, McIntyre TM. Cytotoxic phospholipid oxidation products: cell death from mitochondrial damage and the intrinsic caspase cascade. J Biol Chem. 2007;282:24842–50. doi: 10.1074/jbc.M702865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.John K, Alla V, Meier C, Putzer BM. GRAMD4 mimics p53 and mediates the apoptotic function of p73 at mitochondria. Cell Death Differ. 2010;18:874–86. doi: 10.1038/cdd.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regev A, et al. Reliability of histopathologic assessment for the differentiation of recurrent hepatitis C from acute rejection after liver transplantation. Liver Transpl. 2004;10:1233–9. doi: 10.1002/lt.20245. [DOI] [PubMed] [Google Scholar]
- 33.D’Errico-Grigioni A, et al. Tissue hepatitis C virus RNA quantification and protein expression help identify early hepatitis C virus recurrence after liver transplantation. Liver Transpl. 2008;14:313–20. doi: 10.1002/lt.21375. [DOI] [PubMed] [Google Scholar]
- 34.Grassi A, et al. Detection of HCV antigens in liver graft: relevance to the management of recurrent post-liver transplant hepatitis C. Liver Transpl. 2006;12:1673–81. doi: 10.1002/lt.20882. [DOI] [PubMed] [Google Scholar]
- 35.Unitt E, Gelson W, Davies SE, Coleman N, Alexander GJ. Minichromosome maintenance protein-2-positive portal tract lymphocytes distinguish acute cellular rejection from hepatitis C virus recurrence after liver transplantation. Liver Transpl. 2009;15:306–12. doi: 10.1002/lt.21680. [DOI] [PubMed] [Google Scholar]
- 36.Jain A, et al. Characterization of CD4, CD8, CD56 positive lymphocytes and C4d deposits to distinguish acute cellular rejection from recurrent hepatitis C in post-liver transplant biopsies. Clin Transplant. 2006;20:624–33. doi: 10.1111/j.1399-0012.2006.00528.x. [DOI] [PubMed] [Google Scholar]
- 37.Schmeding M, et al. C4d in acute rejection after liver transplantation: a valuable tool in differential diagnosis to hepatitis C recurrence. Am J Transplant. 2006;6:523–30. doi: 10.1111/j.1600-6143.2005.01180.x. [DOI] [PubMed] [Google Scholar]
- 38.Schmeding M, et al. ELISA-based detection of C4d after liver transplantation: a helpful tool for differential diagnosis between acute rejection and HCV-recurrence. Transpl Immunol. 2010;23:156–60. doi: 10.1016/j.trim.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 39.MacQuillan GC, et al. Hepatocellular MxA protein expression supports the differentiation of recurrent hepatitis C disease from acute cellular rejection after liver transplantation. Clin Transplant. 2010;24:252–8. doi: 10.1111/j.1399-0012.2009.01068.x. [DOI] [PubMed] [Google Scholar]
- 40.Borgogna C, et al. Expression of the interferon-inducible proteins MxA and IFI16 in liver allografts. Histopathology. 2009;54:837–46. doi: 10.1111/j.1365-2559.2009.03311.x. [DOI] [PubMed] [Google Scholar]
- 41.Cabrera R, et al. Using an immune functional assay to differentiate acute cellular rejection from recurrent hepatitis C in liver transplant patients. Liver Transpl. 2009;15:216–22. doi: 10.1002/lt.21666. [DOI] [PubMed] [Google Scholar]
- 42.Rubinas T, Dellon E, Henel G. Use of an immune functional assay to differentiate recurrent hepatitis C from acute cellular rejection in liver transplant patients. Liver Transpl. 2009;15:1157. doi: 10.1002/lt.21774. [DOI] [PubMed] [Google Scholar]
- 43.Hashimoto K, et al. Measurement of CD4+ T-cell function in predicting allograft rejection and recurrent hepatitis C after liver transplantation. Clin Transplant. 2010;24:701–8. doi: 10.1111/j.1399-0012.2009.01169.x. [DOI] [PubMed] [Google Scholar]
- 44.Maluf DG, Archer KJ, Villamil F, Stravitz RT, Mas V. Hepatitis C virus recurrence after liver transplantation: biomarkers of disease and fibrosis progression. Expert Rev Gastroenterol Hepatol. 2010;4:445–58. doi: 10.1586/egh.10.39. [DOI] [PubMed] [Google Scholar]
- 45.Asaoka T, et al. Differential transcriptome patterns for acute cellular rejection in recipients with recurrent hepatitis C after liver transplantation. Liver Transpl. 2009;15:1738–49. doi: 10.1002/lt.21883. [DOI] [PubMed] [Google Scholar]
- 46.Sreekumar R, Rasmussen DL, Wiesner RH, Charlton MR. Differential allograft gene expression in acute cellular rejection and recurrence of hepatitis C after liver transplantation. Liver Transpl. 2002;8:814–21. doi: 10.1053/jlts.2002.35173. [DOI] [PubMed] [Google Scholar]
- 47.Zekry A, et al. Intrahepatic cytokine profiles associated with posttransplantation hepatitis C virus-related liver injury. Liver Transpl. 2002;8:292–301. doi: 10.1053/jlts.2002.31655. [DOI] [PubMed] [Google Scholar]
- 48.McCaughan GW, Shackel NA, Bertolino P, Bowen DG. Molecular and cellular aspects of hepatitis C virus reinfection after liver transplantation: how the early phase impacts on outcomes. Transplantation. 2009;87:1105–11. doi: 10.1097/TP.0b013e31819dfa83. [DOI] [PubMed] [Google Scholar]
- 49.Ramirez S, Perez-Del-Pulgar S, Forns X. Virology and pathogenesis of hepatitis C virus recurrence. Liver Transpl. 2008;14(Suppl 2):S27–35. doi: 10.1002/lt.21644. [DOI] [PubMed] [Google Scholar]
- 50.Krukemeyer MG, et al. Description of B lymphocytes and plasma cells, complement, and chemokines/receptors in acute liver allograft rejection. Transplantation. 2004;78:65–70. doi: 10.1097/01.tp.0000132324.14207.8b. [DOI] [PubMed] [Google Scholar]
- 51.Ohnmacht C, et al. Constitutive ablation of dendritic cells breaks self-tolerance of CD4 T cells and results in spontaneous fatal autoimmunity. J Exp Med. 2009;206:549–59. doi: 10.1084/jem.20082394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.