Abstract
In this paper, we present a method for the ultrasensitive detection of microRNAs (miRNAs) utilizing an antibody that specifically recognizes DNA:RNA heteroduplexes, using a silicon photonic microring resonator array transduction platform. Microring resonator arrays are covalently functionalized with DNA capture probes that are complementary to solution phase miRNA targets. Following hybridization on the sensor, the anti-DNA:RNA antibody is introduced and binds selectively to the heteroduplexes, giving a larger signal than the original miRNA hybridization due to the increased mass of the antibody, as compared to the 22 oligoribonucleotide. Furthermore, the secondary recognition step is performed in neat buffer solution and at relatively higher antibody concentrations, facilitating the detection of miRNAs of interest. The intrinsic sensitivity of the microring resonator platform coupled with the amplification provided by the anti-DNA:RNA antibodies allows for the detection of microRNAs at concentrations as low as 10 pM (350 attomoles). The simplicity and sequence generality of this amplification method position it as a promising tool for high-throughput, multiplexed miRNA analysis, as well as a range of other RNA based detection applications.
INTRODUCTION
MicroRNAs (miRNAs) comprise a class of small, noncoding RNAs that are incredibly important regulators of gene translation.1, 2 Although the exact mechanisms of miRNA action are still being elucidated, they are known to play an important regulatory role in a number of biological functions, including cell differentiation and proliferation,3–7 developmental timing,8–11 neural development,12 and apoptosis.13 Given their importance in such transformative processes, it is not surprising that aberrant miRNA levels are found to accompany many diseases, such as diabetes,14 cancer,15–17 and neurodegenerative disorders,18, 19 and thus these small RNAs have been proposed as informative targets for both diagnostic and therapeutic applications.20
Despite their critical role in cellular processes and promise as biomarkers, the short sequence lengths, low abundance, and high sequence similarity of miRNAs all conspire to complicate detection using conventional RNA analysis techniques, such as Northern blotting, reverse transcriptase polymerase chain reaction (RT-PCR), and cDNA microarrays.21. Numerous approaches have been employed to adapt these methods to the specific challenges of miRNA analysis, and while offering increased measurement performance, many suffer from significant complexity.22–29 The analysis of miRNAs is further complicated by the complex nature by which miRNAs affect translation, wherein multiple miRNA sequences can be required to regulate a single mRNA and/or a particular miRNA may regulate multiple mRNAs.30, 31 Given this complexity, robust, multiplexed methods of miRNA analysis that feature high target specificity, sensitivity and dynamic range will be essential to fully unraveling the biological mechanisms of miRNA function, and may also find utility in the development of robust in vitro diagnostic platforms.
Microring optical resonators are an emerging class of sensitive, chip-integrated biosensors that have recently been demonstrated for the detection of a wide range of biomolecular targets.32–37 These optical microcavities support resonant wavelengths that are highly sensitive to biomolecule binding-induced changes in the local refractive index. In particular, the combined high Q-factor and small footprint of microring resonators make them an attractive choice for both sensitive and multiplexed biosensing. Most relevant to this report, we recently demonstrated the direct, label-free detection of miRNAs with a limit of detection of 150 fmol.33 While this is sufficient for many miRNA applications, we were interested in developing methods to further extend the sensitivity without adding undue complexity or introducing sequence-specific bias to the assay, which would compromise the generality and multiplexing capabilities of the platform.
Monoclonal and polyclonal antibodies recognizing RNA:RNA and DNA:RNA duplexes have been previously developed and utilized in hybridization based assays for the detection of nucleic acid targets including viral nucleic acids and E.coli small RNA.38–43 Of particular relevance here is an anti-DNA:RNA antibody, named S9.6, which specifically recognizes RNA:DNA heteroduplexes and has been utilized to detect RNA in a conventional fluorescence-based microarray format.44–47
In this paper, we combine the utility of the S9.6 anti-DNA:RNA antibody with the appealing detection capabilities of silicon photonic microring resonators to demonstrate the sensitive detection of mammalian miRNAs. Importantly, the S9.6 binding response is significantly larger than that observed for the miRNA itself, allowing the limit of detection to be lowered by ~3 orders of magnitude, down to 350 attomoles. We apply this approach to the multiplexed quantitation of four miRNA targets both from standard solutions as well as the total RNA extract from mouse brain tissue. These results indicate that this strategy is appealing for the multiplexed detection of miRNAs in a simple and reasonably rapid assay format that does not require RT-PCR amplification schemes.
Importantly, during the preparation of this manuscript, Šípová and co-workers reported a similar S9.6 miRNA detection assay on a grating-coupled surface plasmon resonance platform.48 Focusing on the detection of single miRNA, the authors report a similar limit of detection, further highlighting the broad utility of the S9.6 antibody in PCR-less assay formats.
EXPERIMENTAL
Materials
The silane, 3-N-((6-(N'-Isopropylidene-hydrazino))nicotinamide)propyltriethyoxysilane (HyNic Silane), and succinimdyl 4-formyl benzoate (S-4FB) were purchased from SoluLink. PBS was reconstituted with deionized water from Dulbecco's Phosphate Buffered Saline packets purchased from Sigma-Aldrich (St. Louis, MO), and the buffer pH adjusted to pH 7.4 (PBS-7.4) or pH 6.0 (PBS-6) with sodium hydroxide or hydrochloric acid. A 20× saline-sodium phosphate-EDTA buffer (SSPE) was purchased from USB Corp. for use in a high stringency hybridization buffer. All other reagents were purchased from Fisher, unless otherwise noted, and used as received.
Fabrication of Silicon Photonic Microring Resonators and Measurement Instrumentation
The fabrication of sensor chips and operational principles of the measurement instrumentation have been previously reported.34, 49 Briefly, sensor substrates, each containing 32 uniquely-addressable microring resonators within a 6×6 mm footprint, were fabricated at a commercial-scale silicon foundry on 8” silicon-on-insulator wafers using conventional deep-UV photolithography and dry etching methods, before being diced into individual chips. After immobilization of DNA capture probes (described below), the sensor chips are loaded into a biosensor scanner (Genalyte, Inc.), and the wavelengths of optical resonance of the entire array of microring elements are monitored in near real-time using an external cavity laser, integrated control hardware, and data acquisition software.
Nucleic Acid Sequences
All synthetic nucleic acids were obtained from Integrated DNA Technologies. DNA capture probes were HPLC purified prior to use, while synthetic RNA probes were RNase Free HPLC purified. The sequences of all nucleic acid strands used in this work are listed in Table 1.
Table 1.
Sequences of synthetic nucleic acids in described experiments. Bases in bold indicate the substitution of a locked nucleic acid.
| Sequence | |
|---|---|
| hsa miR-16 | 5'-UAGCAGCACGUAAAUAUUGGCG-3' |
| hsa miR-21 | 5'-UAGCUUAUCAGACUGAUGUUGA-3' |
| hsa miR-24-1 | 5'-UGGCUCAGUUCAGCAGGAACAG-3' |
| hsa miR-26a | 5'-UUCAAGUAAUCCAGGAUAGGCU-3' |
| DNA Capture Probe for hsa miR-16 | 5'-NH2 – (CH2)12 – ATC GTC GTG CATTTATAACCGC-3' |
| DNA Capture Probe for hsa miR-21 | 5'-NH2 – (CH2)12 – ATCGAATAGTCTGACTACAACT-3' |
| DNA Capture Probe for hsa miR-24-1 | 5'-NH2 – (CH2)12 – CTGTTCCTGCTGAACTGAGCCA-3' |
| DNA Capture Probe for hsa miR-26a | 5'-NH2 – (CH2)12 – AAGTTCATTAGGTCCTATCCGA-3' |
| 10mer RNA | 5'-AAAGGUGCGU-3' |
| 20mer RNA | 5'-AAAGGUGCGUUUAUAGAUCU-3' |
| 40mer DNA Modular Capture Probe | 5'-NH2 – (CH2)12 – TAGTTGCTGCAACCTAGTCTAGATCTATAAACGCACCTTT-3' |
| LNA Capture Probe for hsa miR-24-1 | 5'-NH2 – (CH2)12 – CTGTTCCTGCTGAACTGAGCCA-3' |
Modification of ssDNA Capture Probes
DNA capture probes, synthesized with amines presented on the 5' end of the sequence, were resuspended in PBS-7.4 and then buffer exchanged three times with a new PBS-7.4 solution utilizing Vivaspin 500 Spin columns (MWCO 5000, Sartorius) to remove residual ammonium acetate from the solid phase synthesis. A solution of succinimidyl-4-formyl benzoate (S-4FB, Solulink) in N,N-dimethylformamide was added in 4-fold molar excess to the DNA capture probe, and allowed to react overnight at room temperature. The 4FB-DNA solution was subsequently buffer exchanged three additional times into PBS-6 to remove any unreacted S-4FB.
Chemical and Biochemical Modification of Silicon Photonic Microring Resonator Surfaces
Prior to functionalization, sensor chips were cleaned in a freshly-prepared Piranha solution (3:1 solution of 16 M H2SO4:30% wt H2O2) for 1 min, and subsequently rinsed with copious amounts of water. (Warning: Piranha solutions can react explosively with trace organics—use with caution!) Sensor chips were then sonicated for 7 min in isopropanol, dried with a stream of N2, and stored until further use.
To attach DNA capture probes, sensor chips were immersed in a 1 mg/mL solution of HyNic Silane in ethanol for 30 min, rinsed and sonicated for 7 min in absolute ethanol, and dried with a stream of N2. Small aliquots (15 μL) of 4FB-modified-DNA were then carefully deposited onto the chips so as to cover only specific sets of microrings, and the solution droplets incubated overnight in a humidity chamber. Prior to hybridization experiments, the substrates were sonicated in 8 M urea for 7 min to remove any non-covalently immobilized capture probe.
Addition of Target miRNA to Sensor Surface
Target miRNA solutions were suspended in a high stringency hybridization buffer, consisting of 30% formamide, 4× SSPE, 2.5× Denhardt's solution (USB Corporation), 30 mM ethylenediaminetetraacetic acid, and 0.2% sodium dodecyl sulfate. Aliquots (35 μL) of target miRNA solutions were recirculated across the sensor surface at a rate of 24 μL/min for 1 hr utilizing a P625/10K.133 miniature peristaltic pump (Instech). Solutions were directed across to the surface via a 0.007” Mylar microfluidic gasket sandwiched between a Teflon cartridge and the sensor chip. Gaskets were laser etched by RMS Laser in various configurations to allow for multiple flow patterns.
Surface Blocking and Addition of S9.6
Following hybridization of the target miRNA to the sensor array, the surface was blocked with Starting Block™ (PBS) Blocking Buffer (Thermo Scientific) for 30 min at 10 μL/min, as controlled with a 11 Plus syringe pump (Harvard Apparatus) operated in withdraw mode, followed by rinsing with PBS-7.4 with 0.05% Tween for 7 min at 30 μL/min. Following surface blocking, a 2 μg/mL solution of S9.6 in PBS-7.4 with 0.05% Tween was flowed across the sensor surface for 40 min at a rate of 30 μL/min.
Generation and Purification of the S9.6 Antibody
The S9.6 antibody was harvested from the medium from cultured HB-8730 cells, a mouse hybridoma cell line obtained from the American Type Culture Collection (ATCC). Cells were cultured according to manufacturer instructions and the S9.6 antibody was purified using protein G by the Immunological Resource Center in the Carver Biotechnology Center at the University of Illinois at Urbana-Champaign. The purified antibody was aliquoted at 0.94 mg/mL in PBS-7.4, and stored at −20°C until use.
Data Analysis
To utilize the S9.6 response for quantitative purposes, we used the net sensor response after 40 min of exposure to a 2 μg/mL solution of S9.6. Control rings functionalized with a non-complementary DNA capture probe were employed to monitor non-specific hybridization-adsorption of the target miRNA as well as the non-specific binding of the S9.6 antibody. The signal from temperature reference rings (rings buried underneath a polymer cladding layer on the chip) was subtracted from all sensor signals to account for thermal drift.
Calibration data over a concentration range from 10 pM to 40 nM, with the exception of miRNA miR-16 (in which the 40 pM and 10 pM points were not obtained), was fit to the logistic function:
where A1 is the initial value limit, A2 is the final value limit, and c and p describe the center and power of the fit, respectively.
miRNA Expression Levels in Mouse Tissue
50 μg of total mouse brain RNA (Clontech) was diluted 1:5 with hybridization buffer and recirculated overnight prior to amplification with S9.6. The net sensor response after 40 min exposure to 2 μg/mL S9.6 was calibrated to each miRNA to account for variable Tm values and any secondary structure that would influence the hybridization kinetics.
RESULTS and DISCUSSION
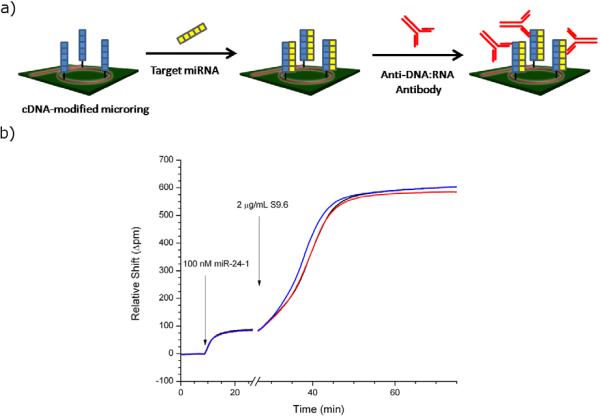
A schematic of the S9.6 assay is shown in Figure 1a. The microrings are initially functionalized with ssDNA capture probes complementary to the target miRNAs of interest. A solution containing the miRNA is flowed across the sensor surface, after which the surface is blocked to prevent non-specific protein adsorption, and subsequently exposed to the S9.6 antibody. Representative real-time shifts in the resonance wavelength for three DNA-modified microrings first to the hybridization of a complementary miRNA and then the S9.6 anti-DNA:RNA antibody are shown in Figure 1b. Notably, we do not observe any significant response when S9.6 is flowed across a surface containing only single-stranded DNA capture probes or double stranded DNA duplexes, confirming the specificity of this amplification strategy (Figure S1).
Figure 1.
(a) Schematic of the S9.6 amplification assay, in which an DNA-modified microring is sequentially exposed to complementary miRNA followed by the S9.6 antibody. (b) Signal responses from 3 separate microrings corresponding to the schematic in (a) illustrate the heightened sensitivity achieved via the S9.6 antibody.
Apparent from the data shown in Figure 1b is an unusual kinetic binding response for the S9.6 antibody binding to the DNA:RNA heteroduplex-presenting surface. Rather than display classical Langmuir-type behavior in which the rate of binding is fastest initially, the measured response appears sigmoidal, with the rate of binding actually increasing for the first 10–15 minutes, after which the curves begins to level off. Suspecting either a steric or cooperative binding explanation, we varied the concentration of DNA capture probe immobilized onto sensors, incubated complementary miRNA at saturating concentrations, and then performed S9.6 enhancement. Since the amount of underlying DNA was varied, the sensors supported different saturation levels of miRNA hybridization, but saturation was always reached by flowing a very high (40 nM) solution of the target across the surface prior to blocking and introducing S9.6. Interestingly, the shape of the S9.6 binding response varied as a function of the underlying DNA capture probe loading, as shown in Figure 2, transitioning from sigmoidal, at high relative DNA loadings, to the expected logarithmic response. We preliminarily attribute this behavior to some sort of cooperative binding effect, wherein initial S9.6 binding is sterically restricted by the high density of capture probes but subsequent binding events become increasingly favorable. As described in greater detail below, we have evidence that multiple anti-DNA:RNA antibodies can bind to a single heteroduplex and therefore one can imagine that the footprint established by the first bound S9.6 allows greater access for subsequent interactions at the same DNA:miRNA duplex. Although we do not yet fully understand the mechanism of this binding interaction, we do observe that the highest capture probe densities result in the largest observed S9.6 responses over all target miRNA concentrations measured at a fixed 40 minute time point. Therefore, for the purposes of sensitive detection, rather than kinetic analysis, we chose to functionalize our sensors with the highest achievable levels of capture probes.
Figure 2.
Capture probe density plot, showing the S9.6 response to varied ssDNA capture probe concentrations with a constant miR-24-1 target concentration (40 nM). The binding transition from cooperative binding to Langmuir binding kinetics is evident as steric limitations are relaxed as the concentration of capture probe deposition solutions is lowered below 125 nM.
After the initial verification of S9.6 binding and amplification potential, we sought to better understand the applicability and limitations of the antibody. In particular, one interesting aspect of the antibody was the large signal amplification we observed upon S9.6 binding to our sensor surfaces, especially under non-saturating conditions. As shown in Figure 1b, the net sensor shift for the hybridization-adsorption of a 100 nM solution of miR-24-1 (a concentration that will saturate binding sites) onto the sensor surface is ~80 pm. The S9.6 response for amplification is ~520 pm, limited by steric crowding of the antibody. However this secondary amplification becomes even more dramatic at lower miRNA conditions, increasing the response over 60-fold (Figure S2). Since the miRNA and antibody differ in mass by a factor of approximately 21, this suggests that ~3 S9.6 antibodies can bind to a single DNA:miRNA duplex.
To confirm our hypothesis that a single DNA:RNA heteroduplex could be bound by multiple S9.6 antibodies, we functionalized a sensor surface with a ssDNA capture probe that was complementary to 10- and 20-mer RNA test sequences. As seen in Figure S3, the primary hybridization response for the 20-mer RNA sequence is almost exactly twice as large as for the 10-mer. The observed S9.6 binding response is again larger for the 20-mer target, but the response is ~2.5 times that of the 10-mer, suggesting that two and sometimes three S9.6 antibodies can bind to the 20-mer target. Experiments performed with a 40-mer test RNA sequence confirm that multiple S9.6 antibodies can bind to single heteroduplexes, and also reveal that longer strand responses are accompanied by more complex steric binding considerations. These results, while preliminary, suggest that the S9.6 binding epitope is on the order of 6 base pairs in size, which is considerably smaller than the 15 nucleotide epitope previously proposed.44 Importantly, the small size of the epitope allows multiple S9.6 antibodies to bind to single DNA:miRNA heteroduplexes, which in turn allows for greater signal amplification.
To demonstrate the quantitative utility of this signal enhancement scheme, we performed S9.6 experiments for sensor arrays exposed to different concentrations of four different perfectly complementary target miRNAs. The resulting calibration curves for each of the target miRNAs were generated using synthetic miRNAs in buffer on separate chips, and quantitated based on the net S9.6 binding response measured after 40 minutes (Figure 3). The concentration dependent responses were obtained over 3 orders of magnitude down to a concentration of 10 pM, given the 35 μL sample volume this corresponds to a detection limit of 350 amol. One exception to this is miR-16, which only gave consistent measurements down to 160 pM. While the reason for this difference is not yet fully understood, we preliminarily attribute it to specific secondary structures of the miRNA target and capture probe. A key advantage of S9.6 for miRNA detection is the fact that it is a universal recognition element, in contrast to sequence-specific RT-PCR primers, and thus the addition of a single reagent can be used to enhance detection for all miRNA species being interrogated. This is especially valuable for multiplexed detection platforms, such as the arrays of microring resonators used herein. Sensor chips can therefore be derivatized to present multiple, sequence-specific capture probes specific to multiple miRNAs, exposed to the sample of interest, and the signal for each target can be simultaneously enhanced in a single step with the addition of S9.6.
Figure 3.
(a) Overlay of the signal responses achieved for each concentration of target miRNA. Concentrations utilized were 40 nM (black), 10 nM (red), 2.56 nM (green), 640 pM (blue), 160 pM (pink), 40 pM (cyan), 10 pM (orange), and a blank (with the exception of miR-16, which did not contain the 40 pM and 10 pM calibration points). (b) Calibration curves for the S9.6 response for miR-16, miR-21, miR-24-1, and miR-26a. Plots were constructed from the relative shifts at 40 min. The red curves represent the logistic fits to the data points. Error bars represent ±1 standard deviation for between 4 and 12 independent measurements at each concentration.
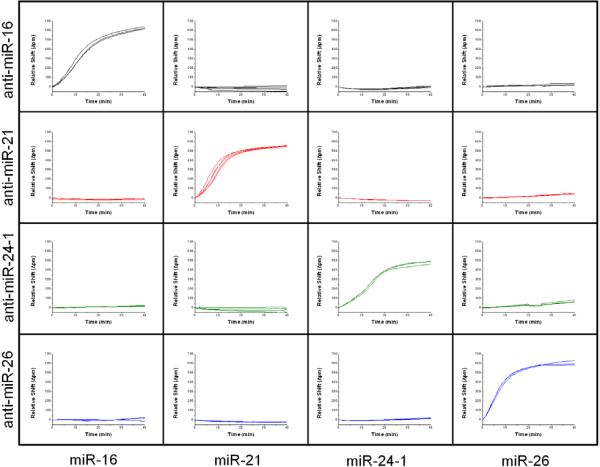
As a proof-of-concept, we functionalized discrete regions of four sensor arrays with different ssDNA capture probes that were perfectly complementary towards miR-16, miR-21, miR-24, and miR-26a. Sensor chips were then exposed to solutions containing only one of the target miRNAs at 40 nM, rinsed, blocked, and exposed to S9.6. This process was repeated for each of the four target miRNAs, each on its own chip, and the compiled responses are shown in Figure 4. Each column in the figure represents a different sensor array incubated with a specific target sequence. Taken together, it is clear that the S9.6 antibody does not introduce any cross-talk even at high target concentrations, and that the specificity of complementary probe hybridization, as reinforced through the use of a high stringency buffer, is reflected in the enhanced S9.6 assay.
Figure 4.
Simultaneous amplification of a panel of 4 miRNA targets (columns) hybridized to four complementary capture probes (rows). A panel of 4 chips was functionalized towards all four miRNA, and a single 40 nM target solution was introduced to each, followed by 2 μg/mL S9.6. Only those channels containing complementary capture probes and target miRNAs elicit an S9.6 response, allowing multiplexed miRNA analysis.
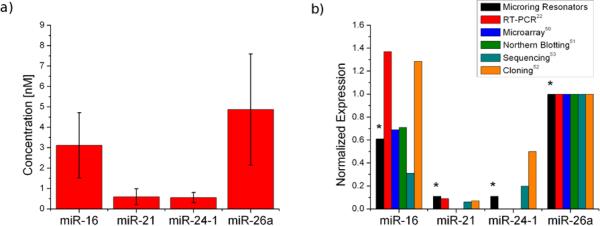
To demonstrate the applicability of the S9.6 amplification methodology to the analysis of a relevant biological system, we simultaneously examined the relative expression profiles of the four aforementioned miRNAs in mouse total brain RNA. We chose to utilize mouse brain RNA due to the previously reported relative expression levels of many miRNAs. Two of the sequences have been found to be highly overexpressed in the mouse brain relative to other tissues, while the others are expressed at lower levels.22, 50–53 We analyzed the expression of the four miRNAs in total mouse brain RNA, and after calibration and accounting for the 5-fold dilution in hybridization buffer, original expression levels were determined to be 3.12 ± 1.60 nM, 0.60 ± 0.39 nM, 0.56 ± 0.25 nM, and 4.87 ± 2.72 nM for miR-16, miR-21, miR-24-1, and miR-26a, respectively (Figure 5a and S4). The overexpression of miR-16 and miR-26a relative to miR-21 and miR-24-1 is consistent with previous literature reports across a number of different profiling techniques, and is well within the variation observed between those studies, as shown in Figure 5b.
Figure 5.
(a) Comparison of the concentrations for each of the four target miRNAs in total mouse brain RNA. Five-fold sample dilution and individual calibration plots were taken into account to calculate the final concentrations. b) Microring resonator-based relative miRNA expression profiles, normalized to miR-26a expression levels, correlate well with literature results from a variety of detection techniques. Both the technique-to-technique variability and incomplete expression profiles of currently accepted techniques highlight the need for highly multiplexed and accurate profiling methods.
While slightly beyond the scope of this manuscript, we also investigated the utility of the S9.6 signal enhancement strategy for locked-nucleic acid (LNA)-containing DNA:miRNA heteroduplexes. LNAs are non-natural nucleotide analogs of DNA that contain a 2'-O, 4'-C-methylene bridge which confer added rigidity to the resulting duplex.54 Importantly, it has been previously shown that the incorporation LNAs into DNA capture probes can increase both the selectivity and sensitivity of miRNA hybridization assays.55 We found that the S9.6 antibody can bind to LNA-containing DNA capture sequences, albeit with a slightly lower efficiency, compared to an equally miRNA-saturated ssDNA(only)-modified sensor surface (Figure S5). Although the bulk of the miRNA detection experiments described herein utilized purely DNA-based capture probes, we feel as though the demonstrated amenability of the S9.6 signal enhancement strategy to LNA-containing capture probes may be of future utility for small RNA detection.
CONCLUSION
The recently understood role of miRNAs in maintaining biological homeostasis and the plethora of disease states resulting from their disregulation has heightened the need for sensitive, multiplexed, high-throughput technologies for their analysis. In particular, the ease by which an assay can be performed affects its acceptance and utilization by other researchers. We have demonstrated a simple, highly sensitive method for the multiplexed detection of miRNAs utilizing an anti-DNA:RNA antibody with arrays of silicon photonic microring resonators. The simplicity of the assay, the ability to simultaneously read-out multiple miRNAs in a single amplification step, and the potential to utilize the antibody in complex media make the methodology extremely appealing.
Future work will focus on applying this methodology towards deciphering the role of miRNAs in myriad biological systems. We will also explore ways to further increase the amplification provided by S9.6 by conjugating the antibody with tags having advantageous dielectric properties, such as nanoparticles, which have previously been used to lower detection limits in related sensing applications.28, 56–58 The ability to utilize S9.6 with LNA capture probes also provides the potential for incorporating highly stringent and specific capture probes with this assay, possibly improving its performance characteristics in biologically complex samples.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge financial support from the National Institutes of Health (NIH) Director's New Innovator Award Program, part of the NIH Roadmap for Medical Research, through grant number 1-DP2-OD002190-01, the Center for Advanced Study at the University of Illinois at Urbana-Champaign, and the Camille and Henry Dreyfus Foundation. AJQ was supported by a fellowship from the Eastman Chemical Company. The authors also thank the Immunological Resource Center, part of the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign, for assistance in expressing and purifying the anti-DNA:RNA antibody.
References
- (1).Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- (2).Orom UA, Nielsen FC, Lund AH. Molecular Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- (3).Chen CZ, Li L, Lodish HF, Bartel DP. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- (4).Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. EMBO J. 2009;28:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Wang Y, Blelloch R. Cancer Res. 2009;69:4093–4096. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- (7).Gangaraju VK, Lin H. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Poethig RS. Curr Opin Genet Dev. 2009;19:374–378. doi: 10.1016/j.gde.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Alvarez-Garcia I, Miska EA. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- (10).Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- (11).Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- (12).Fineberg SK, Kosik KS, Davidson BL. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- (13).Chan JA, Krichevsky AM, Kosik KS. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- (14).Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- (15).Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- (16).Nicoloso MS, Jasra B, Calin GA. Cancer Treat Res. 2009;145:169–181. doi: 10.1007/978-0-387-69259-3_10. [DOI] [PubMed] [Google Scholar]
- (17).Calin GA, Croce CM. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- (18).Meola N, Gennarino VA, Banfi S. Pathogenetics. 2009;2:7. doi: 10.1186/1755-8417-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Tsai LM, Yu D. Clin Exp Pharmacol Physiol. 2010;37:102–107. doi: 10.1111/j.1440-1681.2009.05269.x. [DOI] [PubMed] [Google Scholar]
- (20).Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- (21).Qavi AJ, Kindt JT, Bailey RC. Analytical and Bioanalytical Chemistry. 2010;398:2535–2549. doi: 10.1007/s00216-010-4018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Li J, Yao B, Huang H, Wang Z, Sun C, Fan Y, Chang Q, Li S, Wang X, Xi J. Anal Chem. 2009;81:5446–5451. doi: 10.1021/ac900598d. [DOI] [PubMed] [Google Scholar]
- (25).Streit S, Michalski CW, Erkan M, Kleeff J, Friess H. Nat Protoc. 2009;4:37–43. doi: 10.1038/nprot.2008.216. [DOI] [PubMed] [Google Scholar]
- (26).Ohtsuka E, Nishikawa S, Fukumoto R, Tanaka S, Markham AF, Ikehara M, Sugiura M. European Journal of Biochemistry. 1977;81:285–291. doi: 10.1111/j.1432-1033.1977.tb11950.x. [DOI] [PubMed] [Google Scholar]
- (27).Romaniuk E, McLaughlin LW, Neilson T, Romaniuk PJ. European Journal of Biochemistry. 1982;125:639–643. doi: 10.1111/j.1432-1033.1982.tb06730.x. [DOI] [PubMed] [Google Scholar]
- (28).Fang S, Lee HJ, Wark AW, Corn RM. J Am Chem Soc. 2006;128:14044–14046. doi: 10.1021/ja065223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Nat Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- (30).Friedman RC, Farh KKH, Burge CB, Bartel DP. Genome Research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, Zhan R, He X. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- (32).Luchansky MS, Bailey RC. Analytical Chemistry. 2010;82:1975–1981. doi: 10.1021/ac902725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Qavi AJ, Bailey RC. Angewandte Chemie-International Edition. 2010;49:4608–4611. doi: 10.1002/anie.201001712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Washburn AL, Gunn LC, Bailey RC. Analytical Chemistry. 2009;81:9499–9506. doi: 10.1021/ac902006p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Washburn AL, Luchansky MS, Bowman AL, Bailey RC. Analytical Chemistry. 2010;82:69–72. doi: 10.1021/ac902451b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Luchansky MS, Washburn AL, Martin TA, Iqbal M, Gunn LC, Bailey RC. Biosensors & Bioelectronics. 2010;26:1283–1291. doi: 10.1016/j.bios.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Byeon JY, Bailey RC. Analyst. 2010 doi: 10.1039/c0an00853b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Casebolt DB, Stephensen CB. Journal of Clinical Microbiology. 1992;30:608–612. doi: 10.1128/jcm.30.3.608-612.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Fliss I, St Laurent M, Emond E, Simard RE, Lemieux R, Ettriki A, Pandian S. Appl Microbiol Biotechnol. 1995;43:717–724. doi: 10.1007/BF00164779. [DOI] [PubMed] [Google Scholar]
- (40).Lafer EM, Sousa R, Ali R, Rich A, Stollar BD. J Biol Chem. 1986;261:6438–6443. [PubMed] [Google Scholar]
- (41).Riley RL, Addis DJ, Taylor RP. J Immunol. 1980;124:1–7. [PubMed] [Google Scholar]
- (42).Stollar BD. FASEB J. 1994;8:337–342. doi: 10.1096/fasebj.8.3.7511550. [DOI] [PubMed] [Google Scholar]
- (43).Stollar BD, Rashtchian A. Anal Biochem. 1987;161:387–394. doi: 10.1016/0003-2697(87)90467-2. [DOI] [PubMed] [Google Scholar]
- (44).Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH. Nucl. Acids Res. 2006;34:e52-. doi: 10.1093/nar/gkl142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Székvölgyi L, Rákosy Z, Bálint BL, Kókai E, Imre L, Vereb G, Bacsó Z, Goda K, Varga S, Balázs M, Dombrádi V, Nagy L, Szabó G. Proceedings of the National Academy of Sciences. 2007;104:14964–14969. doi: 10.1073/pnas.0702269104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Kinney JS, Viscidi RP, Vonderfecht SL, Eiden JJ, Yolken RH. Journal of Clinical Microbiology. 1989;27:6–12. doi: 10.1128/jcm.27.1.6-12.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Journal of Immunological Methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- (48).Sipova H, Zhang S, Dudley AM, Galas D, Wang K, Homola J. Anal Chem. 2010;82:10110–10115. doi: 10.1021/ac102131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Iqbal M, Gleeson MA, Spaugh B, Tybor F, Gunn WG, Hochberg M, Baehr-Jones T, Bailey RC, Gunn LC. IEEE Journal of Selected Topics in Quantum Electronics. 2010;16:654–661. [Google Scholar]
- (50).Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Rna-a Publication of the Rna Society. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Genome Biology. 2004;5:-. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien MC, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju JY, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo SJ, Babiarz JE, Blelloch R, Schroth GP, Nusbaum C, Bartel DP. Genes & Development. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kaur H, Arora A, Wengel J, Maiti S. Biochemistry. 2006;45:7347–7355. doi: 10.1021/bi060307w. [DOI] [PubMed] [Google Scholar]
- (55).Roberts P. Nature Methods. 2006;3 [Google Scholar]
- (56).Yang WJ, Li XB, Li YY, Zhao LF, He WL, Gao YQ, Wan YJ, Xia W, Chen T, Zheng H, Li M, Xu SQ. Anal Biochem. 2008;376:183–188. doi: 10.1016/j.ab.2008.02.003. [DOI] [PubMed] [Google Scholar]
- (57).Luchansky MS, Washburn AL, McClellan MS, Bailey RC. Lab on a Chip. 2011;11:2042–2044. doi: 10.1039/c1lc20231f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Zhou WJ, Chen YL, Corn RM. Analytical Chemistry. 2011;83:3897–3902. doi: 10.1021/ac200422u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.