Abstract
We report the fabrication and characterization of gold-coated etched glass array substrates for surface plasmon resonance imaging (SPRi) analysis with significantly enhanced performances, in particular image contrast and sensitivity. The etching of the glass substrate induces a variation in the resonance condition and thus in the resonance angle between the etched wells and the surrounding area, leading to the isolation of the array spot resonance with a significant reduction of the background signal. FDTD simulations show arrays with large spots and minimal spot-to-spot spacing yield ideal differential resonance conditions, which are verified by experimental results. Simulations also indicate the etched well structure exhibits enhanced SPR electric field intensity by three-fold as compared to standard planar gold chips. Changes in the bulk sensitivity of the etched arrays have been obtained at the 10−4 RIU level based on image intensity difference. The strong image contrast allows for improved microarray imaging analysis with easily distinguished signals from background resonance. The etched array chips are demonstrated for SPRi detection of bacterial toxins through the coating of an ultrathin SiO2 film for direct vesicle fusion that establishes a supported membrane-based biosensing interface. Protein detection with cholera toxin (CT) at 5 nM is obtained, making this chip one of the most sensitive SPR imaging substrates ever reported without a post-binding amplification scheme. Furthermore, the surface can be regenerated by Triton X-100 for repeated cycles of membrane formation, protein binding, and biomolecular removal. The reusability and enhanced performance of the etched glass array chips should find a broad range of applications, opening up new avenues for high-throughput SPR imaging detection with convenience and marked surface sensitivity.
INTRODUCTION
The development of high-throughput analytical tools is a burgeoning field.1,2 While most array based technologies have focused on fluorescence detection, there has been a steady expansion in the new microarray methods with the label-free technique of surface plasmon resonance imaging (SPRi).3,4 SPRi enables real-time multiplexed monitoring of different biological interactions ranging from viruses to carbohydrates.5,6 It is a highly attractive technical platform not only for its capability to interrogate a large surface but also for its ability to produce sensorgrams for determination of kinetic/equilibrium constants, which encompasses the primary usage of SPR spectroscopy. The importance of scientific advancement with regard to SPR and SPRi has sparked a flurry of publications in the area including several recent reviews.7–9
Typical SPR imaging experiments involve data collection at a fixed angle.10 Recent work also demonstrated angle-scanning11,12 and wavelength-scanning13 formats. However, very little attention has been given to novel chip design for improving the quality of SPR detection. Using planar gold chips, the sensitivity of SPR imaging is known to be lower than conventional SPR spectroscopy due to less compact optical configuration, use of intensity-based measurements and less sensitive detectors.6,14 Even with the improved instrumentation,6 the detection sensitivity still lags considerably behind fluorescence methods. Additionally, development of imaging-specific SPR substrates is generally lacking. At present, the contrast, uniformity, and quality of SPR images from spot-to-spot on planar gold are still low15,16 and have not been the focus of recent studies. Difference images are broadly used for data reporting17 with rendered false color presentation where the background is subtracted to zero.18–20 By ignoring the background, a large amount of analytical signal and hence information becomes unavailable, severely limiting the dynamic range and sensitivity of these substrates.
The image contrast, sensitivity and large background signal often present in SPR imaging measurements are greatly influenced by the array fabrication method. Conventional arrays for SPRi have been fabricated by the use of contact printing,21 microcontact printing,22 non-contact printing,23 microfluidics,24 and continuous flow microspotting.25 Recently, photolithography has been utilized for creation of patterns and array templates for array imaging analysis.26 In addition to the creation of arbitrary patterns on a surface,26 photolithography is capable of bulk scale patterning with short processing time compared to other lithographic techniques,27 and the patterns can be made on the surface to mimic the size of the biomolecule to be deposited through the use of different photomasks.
We set out to use photolithography to generate new biochips with improved SPRi performance. One key element in this task is to minimize the background signal without constantly taking difference images. Chemical approaches to suppress non-specific adsorption are widely adopted, including the use of peptides,28 self-assembled monolayers,29 poly(ethylene glycol) (PEG),30 and PEG-polymer brushes.31 Approaches to reduce non-specific adsorption have been the focus of a recent review.32 While all these chemical methods effectively reduce the non-specific adsorption to various levels, they have limited effect on the metal surface resonance around the spots or regions of interest (ROIs). Propagating surface plasmon resonance occurs on the whole surface giving rise to significant background signals that cannot be reduced with chemical methods. To address this problem, an interesting approach by photolithography is the fabrication of gold islands or isolated gold spots on a glass substrate.24,33 Although the gold island platform provides brightly resonant ROIs with reduced background resonance, the background remains sensitive to refractive index changes due to the presence of a decaying electric field at the glass-dielectric interface.
In addition to background suppression, sensitivity improvement is another important aspect for new biochip designs. Nanohole arrays34,35 in particular are garnering significant interest given their sensitivity to small changes in refractive index. Recent work by Masson et al. indicates these arrays can enhance sensitivity of planar gold SPR films by a factor of two.34 A variety of work has appeared focusing on enhancing the evanescent field in the conventional Kretschmann configuration,36 including periodic metal nanowires,37 gold nanoposts,38 gold nanogratings,39 and new prism materials.40 It should be noted that simply making patterns and structures ultra-small is often not the most productive route for array based biological applications.32 Instead, the pattern dimensions should be carefully optimized with regard to both the biological system targeted and ease of fabrication, where large patterns with dimensions of hundreds of micrometers are often more appropriate.
In this work we report the development of new micron-scale glass etched arrays for SPR imaging analysis. Three main aspects are specifically addressed and featured: simple fabrication, suppression of background resonance, and enhancement of the electric field on the arrayed spots for increasing surface sensitivity. Figure 1 shows the scheme of the array structure, fabrication process, and optical images of the chips. The arrays differ from conventional planar gold substrates in that a glass substrate is etched down to micrometer levels and then coated with a gold film. This approach is also different from a previous report with a 15 nm grating.39 The etching process serves two main purposes: creating a differential resonance by severely reducing background resonance and enhancing the electric field amplitude inside the wells for higher surface sensitivity to biological interactions. The concept of reducing background resonance has been attempted with a polymer micropatterned array,41 where a micrometer-scale polymeric optical screen (microPOS) was used to passivate the region deposited with polymer to reduce background SPR signals. However, no appreciable sensitivity enhancement was observed. The gold coated etched glass array used in our work, on the other hand, demonstrates a significant electric field amplitude enhancement. This unique feature of the etched glass arrays is supported with both FDTD simulations and experimental results, demonstrating bulk and surface sensitivity enhancement compared to planar gold substrates and gold island arrays.
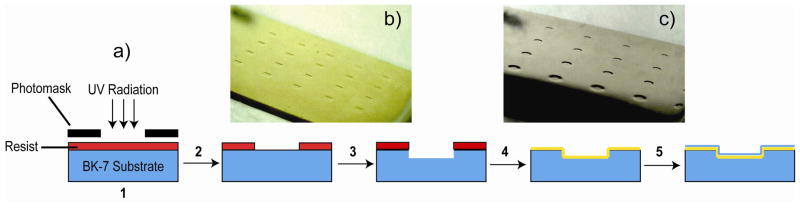
Figure 1.
a) Fabrication procedure for the etched glass array SPRi substrate. 1 BK7 glass slides were patterned using standard photolithography with a photomask containing 800 μm circles and squares with 1.6 mm center-to-center spacing; 2 The patterned substrate was developed in the exposed UV irradiated areas; 3 BOE etching to different micrometer level etch depths completed and thereafter photoresist is removed with acetone; 4 Deposition of 2 nm chromium adhesion layer, followed by a 51 ± 2 nm gold layer via e-beam evaporation; 5 For the CT/GM1 surface sensitivity study, deposition of 3–6 nm silicon dioxide (light blue) by plasma enhanced chemical vapor deposition (PECVD) at 300 ºC. b) Sample optical image of the square gold coated glass etched array (etched well: 800 μm). c) Sample optical image of the circular gold coated glass etched array (etched well: 800 μm).
EXPERIMENTAL
Materials
The metals (gold, silver, chromium) used for electron-beam evaporation were acquired as pellets of 99.99% purity from Kurt J. Lesker (USA). BOE etchant (6 parts 40% NH4F and 1 part. 49% HF) was obtained from Shipley Co. (Marlborough, MA). Ethanol (200 proof) was obtained from Gold Shield Chemical Co. (Hayward, CA). BK-7 glass substrates were from Corning (Painted Post, NY). L-α-phosphatidylcholine (PC) was purchased from Avanti Polar Lipids (Alabaster, AL). Cholera toxin (CT) from Vibrio cholera and Triton X-100 were obtained from Sigma-Aldrich (St. Louis, MO). The monosialoganglioside receptor (GM1) was obtained from Matreya (Pleasant Gap, PA). All lipids were made into stock solutions in chloroform and stored in a −80ºC freezer unless otherwise noted.
Fabrication of Etched Glass Arrays
A photolithographic method was used for pattern fabrication with high precision. A five-step process including photolithography, developing, wet chemical etching with buffer oxide etchant (BOE), e-beam evaporation of gold, and PECVD of SiO2 (optional) has been developed and utilized for this work (Figure 1a). First, positive photoresist AZ5214E was spin coated on the cleaned BK7 glass substrate and then patterned by exposing the substrate to UV light for ~10 s. Thereafter, the arrays were developed with 4:1 ratio of water to AZ400K developer solution to expose the patterned areas. The substrate was rinsed with water and dried with nitrogen for subsequent wet chemical etching.
To etch the patterned glass substrate, standard wet chemical etching using buffered hydrofluoric acid (BOE) was used (Caution!), which presents many advantages.42 First, etch depth calibration measurements were performed (Supplemental Figure S1) to a reproducible level for 1–15 μm etching in the patterned areas. This process requires control of BOE exposure time, which allows the production of desired etch depths based on simulation results. It is advantageous to employ BOE rather than HF for the etching process as BOE maintains a slower and more stable etch rate by controlling the pH and replenishing the depletion of the fluoride ions (where the buffer acts as source of extra fluorine).43 For micrometer sized etches, BOE provides good uniformity, little precipitation, and high stability of the photoresist.44 After the etching, the glass arrays were rinsed with water. Acetone was then used to remove the remaining photoresist. The etched arrays were dried under a stream of nitrogen gas and etch depth measurements were taken using a Veeco Dektek 8 profilometer (Santa Barbara, CA). The etched glass arrays were then coated with a 2-nm thick chromium adhesion layer, followed by a 51 ± 2 nm thick gold layer via electron beam evaporation. The resulting fabricated etched glass chips coated with gold are shown for square (Figure 1b) and circular (Figure 1c) 800 μm patterns. The surface roughness of the array wells were measured to be < 3 nm via AFM images obtained using a Veeco Dimension 5000 atomic force microscope (Santa Barbara, CA) with manufacturer-provided software. All images were obtained in tapping mode, and rms surface roughness values were obtained by averaging multiple 25 μm2 areas across the array well at a scan rate of 1 Hz. They were stored in a vacuum desiccator until use.
For surface sensitivity measurements the etched glass arrays were made hydrophilic by plasma enhanced chemical vapor deposition (PECVD) of 3–6 nm silicon dioxide using a Unaxis Plasmatherm 790 system (Santa Clara, CA) on top of the gold layer. All chips were stored under vacuum.
Simulation and Computational Modeling of the Arrays
Simulation was performed with 3-D Finite-Difference Time-Domain (FDTD) method and commercial software EM Explorer. This method is based on the direct solution of Maxwell’s curl equations by discretization in space and time.45 The simulation was performed using Yee cell size of 20 nm, giving an accuracy of 1–2%. Data points were taken every 0.05 deg. with 4000 iterations for high accuracy reflectivity curves. The structure (length: 94 μm, etch width: 90 μm, etch depth: 0–15 μm) was illuminated with an incident plane wave (λ= 648 nm) with Perfectly Matched Layer (PML) absorbing boundary conditions applied in all directions. The dielectric permittivities and thickness (d) of gold (ε= −14.81 + i0.77, d=51 nm), chromium (ε=8.60+ i8.19, d=2 nm) and silicon dioxide (ε=2.12, d = 6 nm) were determined by fitting the theoretical reflectivity curves obtained by FDTD calculations with the experimental curves obtained using NanoSPR6 reported in a recent work by our group.46
Instrumentation for SPRi analysis
A detailed description of the instrumentation setup was provided in previous work.47 Briefly, the etched BK7 glass array substrate coated with gold was mounted on an optical stage containing a 300 μl flow cell. An equilateral SF2 triangular prism (n=1.616) was then put in contact with the array substrate with a refractive index matching liquid (n=1.610). The optical stage was fixed to a goniometer that allows the tuning of the incident angle. A red light emitting diode (LED, 648 nm) was used for excitation and the reflected images were captured by a cooled 12-bit CCD camera (Retiga 1300 from QImaging) with a resolution of 1.3 MP (1280 × 1024 pixels) and 6.7 μm × 6.7 μm pixel size. The injection of sample solutions into the flow cell was monitored in real time by recording changes in the reflectance every 300 ms inside the etched glass array wells and on the surroundings. A sensorgram was collected using a home-built LabView program, and the images of the array were taken automatically using p-polarized light and s-polarized light alternatively and streamed to the computer. Differences images were obtained by digitally subtracting one image from another. All the experiments were carried out at room temperature (23 ºC) and used 20 mM phosphate buffered saline (PBS) buffer (pH 7.4 with 150 mM NaCl) as the running buffer.
Preparation of Lipid Vesicles
Vesicles were prepared from stock solutions in chloroform. The appropriate mole percent of each lipid was mixed together in a small vial and then dried with nitrogen to form a dry lipid film. Thereafter the lipid containing vial was placed in a vacuum desiccator for 4 h in order to completely remove all solvent. The lipid was then resuspended in 20 mM PBS buffer solution (150 mM NaCl; pH 7.4) to a lipid concentration of 1.0 mg/mL. After vigorously vortexing to remove all lipid remnants from the walls of the vial, the vesicles were probe sonicated for 20 min. The resuspended lipids were then centrifuged at 8000 rpm for 6 min to remove any titanium particles from the probe tip during sonication. Then the supernatant was extruded through a polycarbonate filter (100 nm) to produce vesicles of uniform size. Small unilamellar vesicles (SUV) prepared by this method were 125 ± 4 nm in diameter as determined by dynamic light scattering (DLS) using a particle sizing analyzer from Brookhaven Instruments Corp. (Holtsville, NY).
Test of Stripping Methods on Etched Glass Array Substrates
The NanoSPR 6: Model 321 (Chicago, Illinois) was used for all SPR measurements. This instrument uses a GaAs semiconductor laser (λ = 650 nm) and 30 μl dual-channel flow cell for high sensitivity refractive index measurements. 20 mM phosphate buffered saline solution (pH 7.4 with 150 mM NaCl) was used as both the running buffer and dilution buffer, and the flow rate was 6 mL/hr unless otherwise noted. Once a smooth baseline was established, PC or PC/GM1 vesicles (1.0 mg/mL in PBS) were injected across the SiO2-coated gold sensor chip. Instant vesicle fusion on the hydrophilic surface was observed and the membrane-laden surface was allowed to incubate until complete vesicle rupture and bilayer formation was observed (ca. 15 min.) before finally rinsing the surface with PBS to wash away any non-specifically adsorbed vesicles. For the biomolecular interaction analysis (BIA) consistent to experiments taking place on the SPR imager, cholera toxin (0.085–100 μg/mL in PBS) was injected and incubated to allow for stable lipid-protein binding. Once a stable signal was observed, the surface was rinsed with PBS again. To regenerate the sensor surface, a variety of solutions including NaOH and Triton X-100 have been tested (Supplemental Figure S2). For complete removal of the lipid-protein complex a modified protocol using 5% Triton X-100 from our previous work on calcinated nanoglassy substrates was selected.48 Briefly, after 5% Triton X-100 reached the surface, the flow rate was increased 4 times for 30s to remove bound protein/membrane from the hydrophilic surface, resulting in a return of the signal to the sensor baseline, which indicated the removal of all bound biomolecules. This process of membrane formation, CT binding, and biomolecule removal with surfactant could be easily repeated for numerous cycles and carried over to the SPR imaging format.
Calibration Assay with ethanol and SPRi
To test bulk sensitivity of the etched glass arrays various concentrations of EtOH in DI water (0.1% to 20% w/v) were made. Refractive index values for each solution were determined using an Abbe refractometer from American Optics (Ontario, Canada). All solutions were injected under flow conditions at a flow rate of 6 mL/hr in the previously described flow cell. Values for spot intensity were converted to change in percent reflectivity to correct for spot to spot variations in light intensity via the equation %ΔR = (Ip/Is) × 100 where Ip and Is represent the p-polarized and s-polarized light intensity of each spot, respectively.
CT/GM1 Biointeraction Assay with SPRi
This assay on the SPR imager was run in the same manner as the SPR spectroscopy method explained above and the surface was regenerated using 5% Triton-X 100 for subsequent biosensing cycles.
RESULTS AND DISCUSSION
Differential Resonance at the Microscale
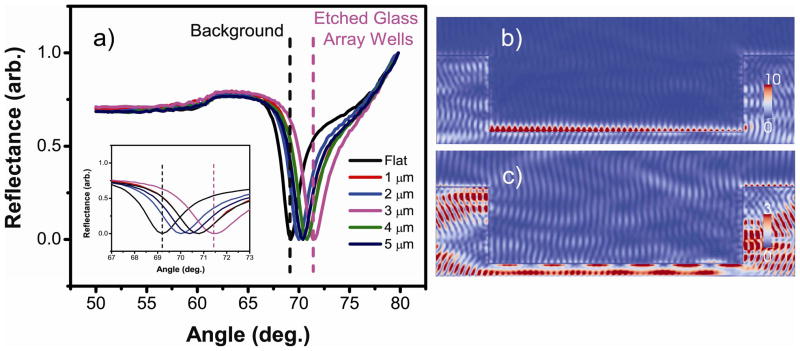
A number of resonance enhancement methods exist, which generally take advantage of localized SPR (LSPR) and surface enhanced Raman spectroscopy (SERS).49 However, research effort utilizing microstructured substrates to manipulate resonance to improve propagating SPR for SPR imaging applications has been very limited. Our approach focuses on separating the background resonance from the main etched glass array plane and therefore offers a “differential resonance” with improved performance. In these arrays the background resonance is separated from the array well resonance leading to lower background signals. To demonstrate the effectiveness of this design, FDTD simulations were carried out with parameters set for micrometer sized etches on gold substrates under aqueous conditions (Figure 2). While various etch depths have led to distinct differential resonance, the best resonance separation was obtained at the low micrometer level, specifically at ~ 3 μm, as shown in Figure 2a. The strong resonance in the wells originates from the SPR confinement and back reflection on the walls of the well. This phenomenon leads to a higher field on the left side due to SPR propagation direction and the large size of the wells.46 At higher etch levels (up to 25 μm) differential resonance was obtainable but there is no enlarged resonance separation. When a nanometric-scale depth (~ 500 nm) was used, the resonance separation was small and the chip displayed almost no differential resonance patterns. This is due to the fact that at sub-wavelength depths, surface plasmon polaritons propagate beyond the abrupt nanometric discontinuity separating the wells from the surrounding area. For wells with 3 μm etch depth, a very strong electric field is observed inside the wells at the resonance angle (Figure 2b) that is not present on the background plane. However, at a slightly lower angle, no electric field is observed inside the wells while a weak field is present on the background (Figure 2c). These results clearly demonstrate differential SPR excitation in the array wells only at a specific incident angle. It is also important to note that the scale on Figure 2b is more than three times larger than that of Figure 2c, thus the resonance in the wells is much stronger than that in the background region. The large difference in the electric field strength between the etched glass array and planar gold substrate has been quantified as well (Supplemental Table S1). In that table, it is worth noting that at the 3 μm etch depth, a 3-fold electric field enhancement is observed in the etched glass array wells compared to planar gold substrates, which presents an exciting and important feature for our design. Previous studies have indicated that higher amplitude of the electric field often leads to higher detection sensitivity for SPR imaging.38,39
Figure 2.
Simulated reflectivity curves for the etched glass array coated with gold in water. a) Reflectivity curves for both the etched glass array and gold island array substrates. Inset: Magnified region around each reflectivity curve minimum. b) Electric field distribution in a 3 μm etched glass array spot at the resonance angle. c) Electric field distribution in a 3 μm etched glass array spot off the resonance angle. The scale for b) is more than three times larger than that of c).
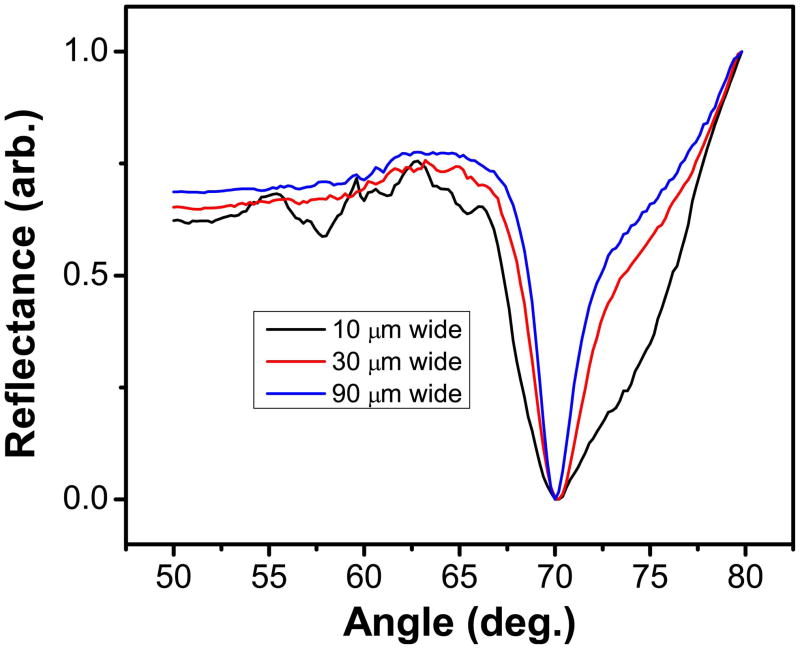
To understand this enhancement effect, simulations were carried out at a fixed micrometer level etch depth whereas the array well widths were varied. The resulting reflectivity curves are displayed in Figure 3. For wells with 3 μm etch depth, the electric field values increase with larger well widths (Supplemental Table S2), showing a nearly 2.5 fold higher field for a 90 μm well as compared to a 10 μm well. Narrow etch widths exhibit substantial interference from the side walls, resulting in larger full width half maximum (FWHM) in reflectivity curves. The large FWHM value can lead to less precise reflectivity curves, weak differential resonance effect, and possibly less detection sensitivity. In addition, when the SPR active area (well surface) increases, the contribution from the background area to the reflected light decreases. All these data suggest that large etches and small spot-to-spot distance, which leads to high signal intensity and improved signal to background ratio, would yield the strongest differential resonance pattern. The fabrication of high performance arrays is thus focused on generating large spots at micrometer etch depths with small spot-to-spot distance, which should yield two exciting features particularly useful for SPRi biosensing: differential resonance with severely reduced background resonance and marked electric field enhancement compared to existing array platforms.
Figure 3.
FDTD simulation depicting the electric field enhancement in 2 μm etched glass arrays based on different etch widths.
Characterization of Etched Glass Arrays with SPR Imaging
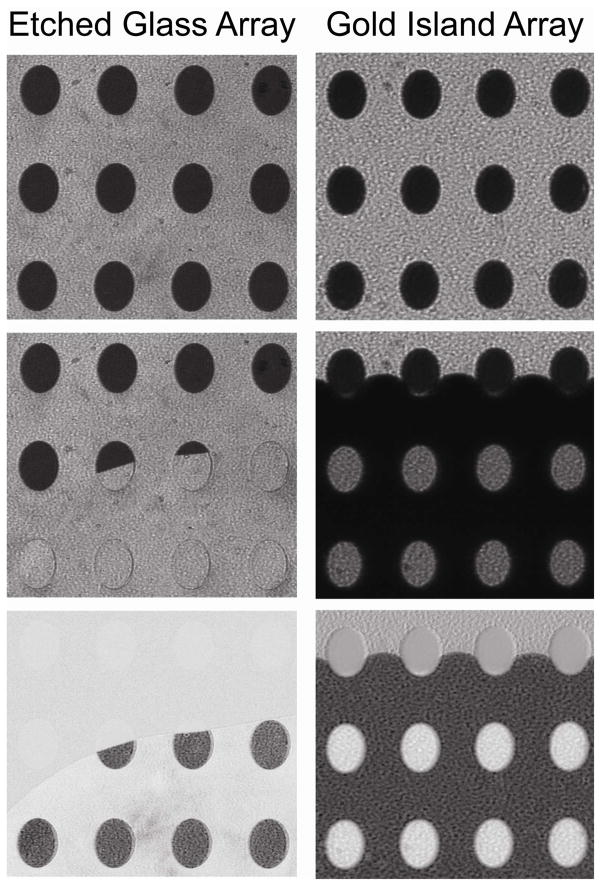
To demonstrate the utility of these chips for SPR imaging a series of experiments were conducted for imaging analysis under various conditions when the flow cell was filled with water for high-density, 800-μm spot arrays (Figure 4). For comparison purposes, chips of gold island arrays were tested under identical conditions. Gold island arrays represent an advanced form of SPR imaging substrate and have been used in a number of commercial instruments and by several research groups.4,50,51 On the left side of Figure 4 are the SPR images of the etched glass array while the gold islands arrays are depicted on the right side of the figure. Also in Figure 4 from top to bottom are SPR p-polarized image in air, p-polarized image as water fills the flow cell, and difference image for each type of the array chip. Upon injection of water the background signal is minimally affected as the etched array spots become brighter, nearly matching the background resonance. For the gold island array, however, the resonance signal changes differently as the spots go from dark to bright and vice-versa for the background. This switching background effect is due to the evanescent field at the glass-fluid interface, and change of the RI upon addition of water that drives the system out of the ATR condition. This feature of the gold island array represents a number of significant drawbacks including poor quantitation of biointeractions with SPR imaging along with a limited dynamic range.10 The image contrast flip indicates the background area supports a considerable evanescent field and thus is sensitive to changes occurring at the surface. By contrast, the etched glass arrays demonstrate a differential resonance pattern, making the signal outside the spots (background) nearly negligible and thus irrelevant even in the presence of non-specific adsorption.
Figure 4.
SPR images of an etched glass array (left column) and gold island array (right column) as water fills the flow cell. For each column, from top to bottom, initial SPR image taken in air, water covering the substrate surface, and the resulting difference image.
Not only are the etched glass arrays capable of reducing background resonance, they exhibit improved sensitivity. One approach to improve SPR imaging sensitivity is to increase the signal-to-noise (S/N) ratio.52 Upon examining the line profiles for the SPR difference images displayed in Supplemental Figure S3, the increase in S/N is easily demonstrated compared to the gold island arrays. Line profiles from SPR images of the etched glass array exhibit at least a twofold increase in S/N ratio compared to the gold island array (Table 1). Furthermore, by severely reducing the background signal (Supplemental Figure S3), the signal-to-background (S/B) ratio for the etched glass arrays is five times higher than the gold island array (Table 1). This unique feature of the etched glass arrays should enable a larger dynamic range and higher detection sensitivity by uncovering signal that remains lost among the background on the gold island arrays. Additionally, the reproducibility of the differential resonance patterning on the etched glass arrays is demonstrated with less than 1.5% RSD, more than two fold lower than that of the gold island array (Table 1). With the advent of a reproducible differential resonance pattern, this substrate is at the forefront of an exciting new class of SPR mode supported sensing materials.
Table 1.
Comparison of analytical features of etched glass array versus gold island array
| Array Characteristics | Etched Glass Array | Gold Island Array |
|---|---|---|
| Signal/Noise (S/N) | 72 | 30 |
| Signal/Background (S/B) | 10.6 | 2.1 |
| %RSD | 1.4 | 3.3 |
| Background Resonance | Weak | Strong |
| Switching Background | No | Yes |
The etched glass arrays not only eliminate the switched background effect and consequently the strong background resonance, but also show almost no appreciable reflectivity under s-polarization (Supplemental Figure S3). Under a normal SPR imaging experiment the s-polarization image is collected and then subtracted out to create a difference image devoid of inhomogeneities that may be present on the substrate. With these etched glass array chips, the need for continual retrieval of difference images is negated. The gold island arrays, however, show a signal under s-polarization due to the difference in reflection between glass and gold. Along with the presence of a weak evanescent field at the glass-dielectric interface, background signals are considerable more apparent on the gold island arrays compared to the etched glass arrays.
Bulk Sensitivity of Etched Glass Arrays
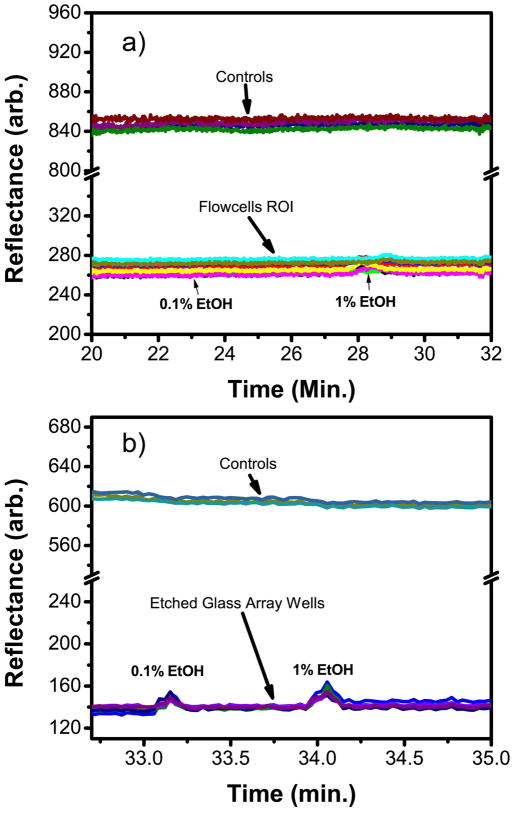
To determine if electric field enhancement in the etched wells leads to greater sensitivity, a bulk calibration test was performed with ethanol (EtOH) solutions. As shown in Figure 5, 0.1% and 1% EtOH are considerably below the detection range of the planar gold substrate while the etched glass arrays can easily observe a strong signal for 0.1% EtOH (Figure 5b). This minute shift in of refractive index of < 10−4 RIU demonstrates a high level of bulk sensitivity for the etched glass array better than existing SPR imaging substrates.
Figure 5.
Sensorgrams from SPR imaging calibration experiments for low concentrations of ethanol for both a planar gold substrate (a) and an etched glass array substrate (b).
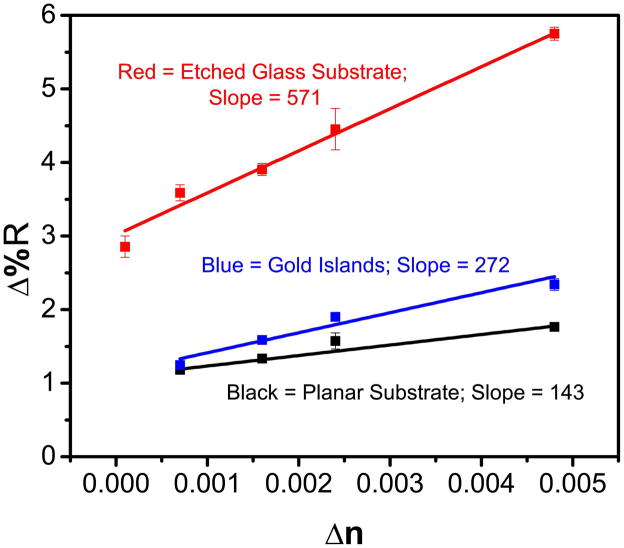
To quantify the analysis, calibration curves were completed for all three types of arrays (planar, gold island, and etched glass arrays) and the results are shown in Figure 6. The etched glass array substrate is at least four times more sensitive than the planar substrate and roughly 2 times more sensitive than the gold island array. The etched glass array also has the largest dynamic range for the tested samples. The marked improvement in bulk sensitivity can be directly attributed to the unique substrate design which creates a differential resonance pattern in the etched wells while also enhancing the electric field.
Figure 6.
SPRi calibration curves for ethanol standards on a planar gold substrate (black), gold island array (blue), and etched glass array (red) where Δn represents the change in refractive index.
Test of Surface Sensitivity with Cholera Toxin-GM1 Interactions
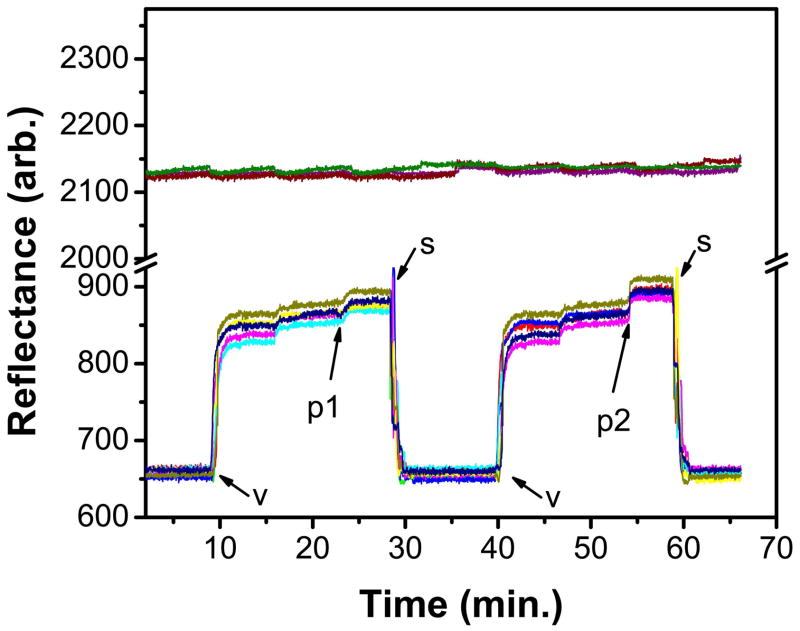
To demonstrate sensitivity for biointeraction on the etched glass array substrate, a surface sensitivity study was conducted for the detection of cholera toxin (CT) using ganglioside GM1 embedded supported lipid membranes. Cholera toxin (CT) is a protein enterotoxin secreted by the bacterium Vibrio cholerae. It is a cytotoxin with pentameric B subunits that recognize and bind to the pentasaccharide moiety of GM1 on the cell’s membrane and this interaction has been widely used as a model membrane system.53,54 To allow for direct vesicle fusion of a lipid membrane on the etched glass array a thin layer of SiO2 was deposited on the surface.55 A mixture of lipids containing 5% GM1 was then fused to the SiO2 coated gold etched glass array. The addition of 10 μg/mL CT yielded a strong binding signal on the resonant spot while the signal in the background was virtually unchanged (Figure 7). The sensor surface can be regenerated with the addition of 5% Triton X-100 using a modified procedure from previous work in our lab.48 This procedure removes the protein/membrane complex without damaging the underlying substrate. Sensorgrams showing two cycles of lipid vesicle fusion, CT binding, and protein/membrane removal are given in Figure 7.
Figure 7.
SPR sensorgram from SPR imaging analysis on SiO2-coated gold etched array demonstrating biomolecular interactions. v: vesicles, 1 mg/mL PC/GM1 (95/5); p1: 10 μg/mL cholera toxin; p2: 20 μg/mL cholera toxin; s: surfactant, 5% Triton X-100. Two control responses (located ca. 2100 arb. on the y-axis) based off background signals on the same chip are shown as well. The small signal increase after each vesicle incubation is due to surface rinsing of non-specifically adsorbed vesicles.
Additionally, a calibration curve was measured for CT binding to the GM1 incorporated membrane (Supplemental Figure S4) and the limit of detection was determined to be < 5 nM. At this concentration a discernible signal can be observed for CT on the sensorgram data from the SPR images (Supplemental Figure S5a) while no signal is observed on the control (Supplemental Figure S5b). Although a signal at 1 nM CT can be observed, the S/N is only ~2, below the standard confidence level of detection. Nevertheless, the 5 nM detection limit is multi-fold lower than the previous work using a tethered array format (260 nM)56 and roughly 60% lower than the calcinated silicate film we previously reported (12 nM).57 To the best of our knowledge, this etched glass array exhibits the most sensitive result for CT binding without post-binding amplification.
CONCLUSIONS
Through the use of both simulations and experimental data we have demonstrated the design of a unique array substrate that yields high performance for SPR imaging analysis. With differential resonance patterns, the etched glass array substrate greatly reduces the background signal, resulting in a signal-to-background ratio five times better than the gold island substrates. Furthermore, simulations indicate a 3-fold electric field enhancement compared to the best SPR imaging substrates available. This field enhancement was verified with the interaction of CT with GM1 where the surface sensitivity was an order of magnitude better than any other reported SPR imaging substrate without the use of post-binding signal amplification. This sensitivity improvement over existing SPR imaging substrates signifies a remarkable advantage for label-free biosensing. In addition to sensitivity enhancement, the fabrication process is relatively easy, and the substrate can be reused with a mild stripping buffer, minimizing both the cost and labor of array fabrication. These unique attributes of the etched glass array chip could inspire new SPR applications that demand higher throughput and better detection sensitivity. The simple chip fabrication method allows user-defined shape and depths to be used, enabling new exploration in cellular analysis and microfluidics. In addition to SPR imaging studies, other optical platforms such as total internal reflection spectroscopy (such as total internal reflection fluorescence) will benefit from this unique substrate design, changing the face of both label and label-free high throughput analysis.
Supplementary Material
Acknowledgments
This work was supported by the US National Science Foundation (CHE-0719224) and the National Institute of Health (1R21EB009551-01A2). MJL would like to acknowledge that this work was supported by the American Chemical Society, Division of Analytical Chemistry Fellowship, sponsored by Procter & Gamble.
Footnotes
Calibration curve for BOE etching, protein stripping protocol for SPR spectroscopy, SPR images comparing the etched glass array vs. gold island array, calibration curve for CT/GM1 interactions on the etched glass array, limit of detection (LOD) sensorgrams for cholera toxin binding, and two tables indicating the electric field enhancement for different etch depths and widths. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Davies MC, Alexander MR, Hook AL, Yang J, Mei Y, Taylor M, Urquhart AJ, Langer R, Anderson DG. J Drug Target. 2010;18:741–751. doi: 10.3109/1061186X.2010.521941. [DOI] [PubMed] [Google Scholar]
- 2.Markovitz-Bishitz Y, Tauber Y, Afrimzon E, Zurgil N, Sobolev M, Shafran Y, Deutsch A, Howitz S, Deutsch M. Biomaterials. 2010;31:8436–8444. doi: 10.1016/j.biomaterials.2010.07.050. [DOI] [PubMed] [Google Scholar]
- 3.Hu WH, Liu YS, Lu ZS, Li CM. Adv Funct Mater. 2010;20:3497–3503. [Google Scholar]
- 4.Ouellet E, Lausted C, Lin T, Yang CWT, Hood L, Lagally ET. Lab Chip. 2010;10:581–588. doi: 10.1039/b920589f. [DOI] [PubMed] [Google Scholar]
- 5.Wang SP, Shan XN, Patel U, Huang XP, Lu J, Li JH, Tao NJ. Proc Natl Acad Sci U S A. 2010;107:16028–16032. doi: 10.1073/pnas.1005264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, Chen Y, Yan MD. Analyst. 2008;133:1268–1273. doi: 10.1039/b804235g. [DOI] [PubMed] [Google Scholar]
- 7.Scarano S, Mascini M, Turner APF, Minunni M. Biosens Bioelectron. 2010;25:957–966. doi: 10.1016/j.bios.2009.08.039. [DOI] [PubMed] [Google Scholar]
- 8.Linman MJ, Abbas A, Cheng Q. Analyst. 2010;135:2759–2767. doi: 10.1039/c0an00466a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbas A, Linman MJ, Cheng Q. Biosens Bioelectron. 2011;26:1815–1824. doi: 10.1016/j.bios.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brockman JM, Nelson BP, Corn RM. Annu Rev Phys Chem. 2000;51:41–63. doi: 10.1146/annurev.physchem.51.1.41. [DOI] [PubMed] [Google Scholar]
- 11.Ruemmele JA, Golden MS, Gao Y, Cornelius EM, Anderson ME, Postelnicu L, Georgiadis RM. Anal Chem. 2008;80:4752–4756. doi: 10.1021/ac702544q. [DOI] [PubMed] [Google Scholar]
- 12.Beusink JB, Lokate AMC, Besselink GAJ, Pruijn GJM, Schasfoort RBM. Biosens Bioelectron. 2008;23:839–844. doi: 10.1016/j.bios.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 13.Otsuki S, Ishikawa M. Biosens Bioelectron. 2010;26:202–206. doi: 10.1016/j.bios.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 14.Jung JM, Shin YB, Kim MG, Ro HS, Jung HT, Chung BH. Anal Biochem. 2004;330:251–256. doi: 10.1016/j.ab.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Manera MG, Spadavecchia J, Taurino A, Rella R. J Opt. 2010;12:035003 (8)–035003 (8). [Google Scholar]
- 16.Liu C, Cui D, Li H. Biosens Bioelectron. 2010;26:255–261. doi: 10.1016/j.bios.2010.06.041. [DOI] [PubMed] [Google Scholar]
- 17.Steiner G. Anal Bioanal Chem. 2004;379:328–331. doi: 10.1007/s00216-004-2636-8. [DOI] [PubMed] [Google Scholar]
- 18.Ladd J, Taylor AD, Piliarik M, Homola J, Jiang S. Anal Chem. 2008;80:4231–4236. doi: 10.1021/ac800263j. [DOI] [PubMed] [Google Scholar]
- 19.Kim M, Jung SO, Park K, Jeong EJ, Joung HA, Kim TH, Seol DW, Chung BH. Biochem Biophys Res Commun. 2005;338:1834–1838. doi: 10.1016/j.bbrc.2005.10.155. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Yang B. MicroRNA Expression Detection Methods. Springer Berlin Heidelberg; 2010. pp. 199–206. [Google Scholar]
- 21.Linman MJ, Yu H, Chen X, Cheng Q. ACS Appl Mater Interfaces. 2009;1:1755–1762. doi: 10.1021/am900290g. [DOI] [PubMed] [Google Scholar]
- 22.Qin D, Xia YN, Whitesides GM. Nat Protoc. 2010;5:491–502. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 23.Ceriotti L, Buzanska L, Rauscher H, Mannelli I, Sirghi L, Gilliland D, Hasiwa M, Bretagnol F, Zychowicz M, Ruiz A, Bremer S, Coecke S, Colpo P, Rossi F. Soft Matter. 2009;5:1406–1416. [Google Scholar]
- 24.Krishnamoorthy G, Carlen ET, Bomer JG, Wijnperle D, deBoer HL, van den Berg A, Schasfoort RBM. Lab Chip. 2010;10:986–990. doi: 10.1039/c000705f. [DOI] [PubMed] [Google Scholar]
- 25.Raz SR, Liu H, Norde W, Bremer M. Anal Chem. 2010;82:8485–8491. doi: 10.1021/ac101819g. [DOI] [PubMed] [Google Scholar]
- 26.Petrou PS, Chatzichristidi M, Douvas AA, Argitis P, Misiakos K, Kakabakos SE. Biosens Bioelectron. 2007;22:1994–2002. doi: 10.1016/j.bios.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 27.Ganesan R, Kratz K, Lendlein A. J Mater Chem. 2010;20:7322–7331. [Google Scholar]
- 28.Bolduc OR, Pelletier JN, Masson JF. Anal Chem. 2010;82:3699–3706. doi: 10.1021/ac100035s. [DOI] [PubMed] [Google Scholar]
- 29.Nogues C, Leh H, Langendorf CG, Law RHP, Buckle AM, Buckle M. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charles PT, Stubbs VR, Soto CM, Martin BD, White BJ, Taitt CR. Sensors. 2009;9:645–655. doi: 10.3390/s90100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu W, Liu Y, Lu Z, Li CM. Adv Funct Mater. 2010;20:3497–3503. [Google Scholar]
- 32.Ekblad T, Liedberg B. Curr Opin Colloid Interface Sci. 2010;15:499–509. [Google Scholar]
- 33.Gifford LK, Sendroiu IE, Corn RM, Luptak A. J Am Chem Soc. 2010;132:9265–9267. doi: 10.1021/ja103043p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Live LS, Bolduc OR, Masson JF. Anal Chem. 2010;82:3780–3787. doi: 10.1021/ac100177j. [DOI] [PubMed] [Google Scholar]
- 35.Masson JF, Murray-Methot MP, Live LS. Analyst. 2010;135:1483–1489. doi: 10.1039/c0an00053a. [DOI] [PubMed] [Google Scholar]
- 36.Shalabney A, Abdulhalim I. Sens Actuators, A. 2010;A159:24–32. [Google Scholar]
- 37.Byun KM, Shuler ML, Kim SJ, Yoon SJ, Kim D. J Lightwave Technol. 2008;26:1472–1478. [Google Scholar]
- 38.Malic L, Cui B, Veres T, Tabrizian M. Opt Lett. 2007;32:3092–3094. doi: 10.1364/ol.32.003092. [DOI] [PubMed] [Google Scholar]
- 39.Hoa XD, Kirk AG, Tabrizian M. Biosens Bioelectron. 2009;24:3043–3048. doi: 10.1016/j.bios.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 40.Gupta G, Kondoh J. Sens Actuators, B. 2007;B122:381–388. [Google Scholar]
- 41.Jung J, Yuk JS, Na K, Kim O, Lee J, Yun K, Ha KS, Hyun J. Langmuir. 2007;23:10403–10406. doi: 10.1021/la7014836. [DOI] [PubMed] [Google Scholar]
- 42.Coze A, Philit G, Le Tiec Y, Rigaudiere J, Butterbaugh JW. Solid State Technology. 2000;43:146–149. [Google Scholar]
- 43.Spierings G. J Mater Sci. 1993;28:6261–6273. [Google Scholar]
- 44.Buhler J, Steiner FP, Baltes H. J Micromech Microeng. 1997;7:R1–R13. [Google Scholar]
- 45.Yee KS. Ieee Transactions on Antennas and Propagation. AP14. 1966. pp. 302–307. [Google Scholar]
- 46.Abbas A, Linman MJ, Cheng Q. Anal Chem. 2011;83:3147–3152. doi: 10.1021/ac200190b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkop T, Wang ZZ, Cheng Q. Langmuir. 2004;20:11141–11148. doi: 10.1021/la048177k. [DOI] [PubMed] [Google Scholar]
- 48.Linman MJ, Culver SP, Cheng Q. Langmuir. 2009;25:3075–3082. doi: 10.1021/la803835a. [DOI] [PubMed] [Google Scholar]
- 49.Wustholz KL, Henry AI, McMahon JM, Freeman RG, Valley N, Piotti ME, Natan MJ, Schatz GC, Van Duyne RP. J Am Chem Soc. 2010;132:10903–10910. doi: 10.1021/ja104174m. [DOI] [PubMed] [Google Scholar]
- 50.Luo YQ, Yu F, Zare RN. Lab Chip. 2008;8:694–700. doi: 10.1039/b800606g. [DOI] [PubMed] [Google Scholar]
- 51.Zordan MD, Grafton MMG, Acharya G, Reece LM, Cooper CL, Aronson AI, Park K, Leary JF. Cytometry A. 2009;75A:155–162. doi: 10.1002/cyto.a.20692. [DOI] [PubMed] [Google Scholar]
- 52.Pillet F, Thibault C, Bellon S, Maillart E, Trevisiol E, Vieu C, Francois JM, Leberre VA. Sens Actuators, B. 2010;147:87–92. [Google Scholar]
- 53.Shi J, Yang T, Cremer PS. Anal Chem. 2008;80:6078–6084. doi: 10.1021/ac800912f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi JJ, Yang TL, Kataoka S, Zhang YJ, Diaz AJ, Cremer PS. J Am Chem Soc. 2007;129:5954–5961. doi: 10.1021/ja069375w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castellana ET, Cremer PS. Surf Sci Rep. 2006;61:429–444. doi: 10.1016/j.surfrep.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor JD, Linman MJ, Wilkop T, Cheng Q. Anal Chem. 2009;81:1146–1153. doi: 10.1021/ac8023137. [DOI] [PubMed] [Google Scholar]
- 57.Phillips KS, Wilkop T, Han JH, Wu JJ, Al-Kaysi RO, Cheng Q. J Am Chem Soc. 2006;128:9590–9591. doi: 10.1021/ja0628102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.