Abstract
The molecular basis of Kufs disease is unknown, whereas a series of genes accounting for most of the childhood-onset forms of neuronal ceroid lipofuscinosis (NCL) have been identified. Diagnosis of Kufs disease is difficult because the characteristic lipopigment is largely confined to neurons and can require a brain biopsy or autopsy for final diagnosis. We mapped four families with Kufs disease for whom there was good evidence of autosomal-recessive inheritance and found two peaks on chromosome 15. Three of the families were affected by Kufs type A disease and presented with progressive myoclonus epilepsy, and one was affected by type B (presenting with dementia and motor system dysfunction). Sequencing of a candidate gene in one peak shared by all four families identified no mutations, but sequencing of CLN6, found in the second peak and shared by only the three families affected by Kufs type A disease, revealed pathogenic mutations in all three families. We subsequently sequenced CLN6 in eight other families, three of which were affected by recessive Kufs type A disease. Mutations in both CLN6 alleles were found in the three type A cases and in one family affected by unclassified Kufs disease. Mutations in CLN6 are the major cause of recessive Kufs type A disease. The phenotypic differences between variant late-infantile NCL, previously found to be caused by CLN6, and Kufs type A disease are striking; there is a much later age at onset and lack of visual involvement in the latter. Sequencing of CLN6 will provide a simple diagnostic strategy in this disorder, in which definitive identification usually requires invasive biopsy.
Introduction
The neuronal ceroid lipofuscinoses (NCLs) are a family of inherited, neurodegenerative disorders that are characterized by lysosomal lipopigment storage in neurons, and usually the eye, and cause progressive neurological impairment, motor and intellectual deterioration, seizures, visual failure, and early death.1,2 In the past, the NCLs have been classified according to the age at onset as infantile (INCL, Santavuori-Haltia), late-infantile (LINCL, Jansky-Bielschowsky), juvenile (JNCL, Batten disease, Spielmeyer-Vogt), and adult (Kufs disease).1–3 Mutations causing the childhood NCL forms have been reported in eight genes: PPT1 (CLN1 [MIM 256730]), TPP1 (CLN2 [MIM 204500]), CLN3 (MIM 204200), CLN5 (MIM 256731), CLN6 (MIM 601780), MFSD8 (CLN7 [MIM 610951]), CLN8 (MIM 600143), and CTSD (CLN10 [MIM 610127]).1 Although the traditional classification of age at onset is pragmatically useful, genotype-phenotype correlations have shown heterogeneity.1,3,4 For example, allelic variants of PPT1, the gene underlying most cases of the infantile form, can present in later childhood or even early-adult life with neurological deterioration and visual failure;1,5,6 a missense mutation in CLN8 causes Northern epilepsy syndrome and other mutations cause a more typical variant late-infantile NCL.3
Kufs disease is rare and differs from most other forms of NCL because the retina is not involved, vision is preserved, and onset is in adulthood.7,8 The Kufs disease locus was designated CLN4 in the 1990s but was never specifically mapped and has remained enigmatic and unsolved. The clinical presentation has been divided into two overlapping types. Type A presents with progressive myoclonus epilepsy, whereas type B presents with dementia and a variety of motor-system signs. Typically, the patients present around the age of 30, but onset has been described in patients ranging from teenagers to persons more than 50 years of age.1,8,9
In contrast to other NCL forms, which are always autosomal recessive, both recessive and dominant forms of Kufs disease have been described, and this suggests genetic heterogeneity.8,10,11 Screening genes in which mutations are known to cause other forms of NCL has generally been unrewarding.12 Determining the molecular basis of Kufs disease is complicated not only by the rarity of the disease but also by challenges in diagnosis. In the childhood forms of NCL, the characteristic lipopigment is relatively easily identified in peripheral tissues such as skin, muscle, and (sometimes) lymphocytes.13 However, in Kufs disease the distribution of the pigment is much more restricted.8,14,15 Diagnosis is most reliably made by brain biopsy, although sometimes the characteristic abnormalities can be found in neurons of the rectal mucosa or other peripheral tissues.15,16 Moreover, normal lipofuscin accumulates with age, and strict pathological criteria must be used to avoid overdiagnosis. As previously reviewed, some published cases probably do not have the disorder.8,14,16 Thus, diagnosis in life remains a challenge, and a simple molecular test is urgently required. The aim of this study was to identify the molecular basis of Kufs disease.
Subjects and Methods
Subjects
Four Kufs disease pedigrees with evidence for autosomal-recessive inheritance suggested by either consanguinity or multiple affected siblings were used for initial linkage mapping (the mapping set). After gene identification, samples from eight additional unrelated subjects or families diagnosed with Kufs disease were used for mutational testing (the validation set). All Kufs disease cases included in this study were verified pathologically by electron microscopic demonstration of fingerprint profiles or granular osmiophilic deposits in pathological material (a brain biopsy, an autopsy, or a rectal or muscle biopsy) from the subjects or from an affected first-degree relative. High-molecular-weight DNA was extracted from peripheral blood cells, skin fibroblasts, or stored autopsy material and used for mapping and sequencing analysis.
The study was approved by the Human Research Ethics Committee of Austin Health, Melbourne, Australia. Informed consent was obtained from living subjects or their relatives. Some subjects had died as much as 20 years earlier, and autopsy material was collected and stored under the appropriate regulations of other participating hospitals or universities.
Linkage Mapping
In view of the apparent absence in typical Kufs disease12 of mutations in the genes that are mutated in other NCLs, we undertook a hypothesis-free approach to map the disorder. The mapping set comprised four unrelated pedigrees of Italian ancestry; three had Kufs type A (Ku1, Ku2, and Ku3), and one had type B (Ku4, Figure 1). We genotyped six affected and eight unaffected subjects from these families by using Illumina Infinium HumanHap610W-Quad BeadChip genotyping arrays at the Australian Genome Research Facility in Melbourne, Australia.
Figure 1.
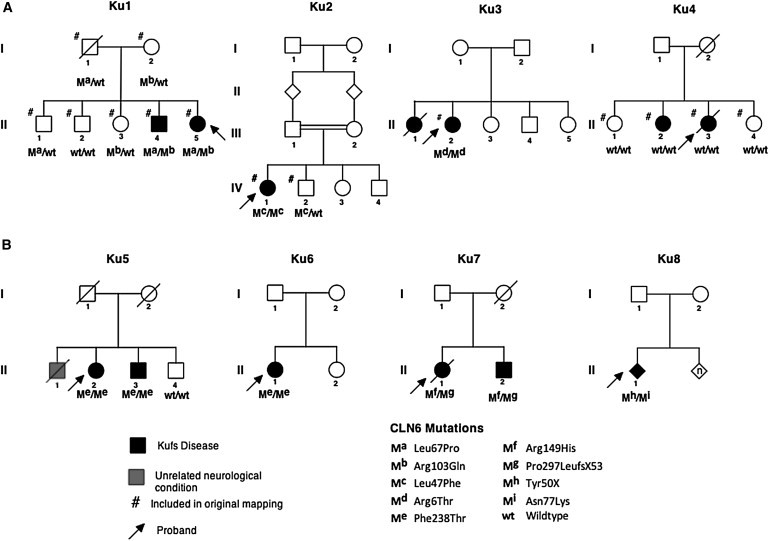
Kufs Disease Pedigrees and CLN6 Mutational Status
(A) Mapping the set of four families with the mutant allele described with the symbol “m” and with different mutations indicated by varying superscripts.
(B) Four pedigrees from the validation set with the mutations shown. The p.Ser308Thr variant in Ku8 (also found in one control) and the p.Ala34Thr variant in Ku12 are not shown.
The parents of the affected individual in Ku2 are known to be first cousins. The other pedigrees had no known consanguinity, but the parents of affected individuals came from the same small towns (in the case of Ku1 and Ku4) or from adjacent small towns (Ku3) in various parts of Italy. We estimated the inbreeding coefficient (F) of all genotyped individuals by using FEstim.17,18 We verified the relationships between genotyped individuals by using PLINK to estimate identity-by-descent (IBD) allele sharing.19
We performed multipoint parametric linkage analysis by using MERLIN20 with a fully penetrant recessive model and a disease allele frequency of 0.0001. We selected a subset of 12,323 SNP markers that have high heterozygosity and are in approximate linkage equilibrium (spaced at least 0.3 cM apart). Marker selection was performed with the Perl script linkdatagen_illumina.pl,21 which discards uninformative markers and those containing detectable Mendelian errors and generates MERLIN-style input files. Allele frequencies from the Centre d'Etude du Polymorphisme Humain (CEPH; Utah residents with ancestry from northern and western Europe) HapMap population were used. We used MERLIN to detect and remove unlikely genotypes on the basis of unlikely double recombination events and to estimate the proportion of families linked to each marker and calculate a heterogeneity LOD (hLOD) score. Finally, we used MERLIN to infer haplotypes, which were visualized with HaploPainter.22
Sequencing
All of the exons, exon-intron boundaries, and untranslated regions of ADAM10 (MIM 602192; NM_001110.2) and CLN6 (NM_017882.2) were amplified by PCR, and the PCR products were sequenced bidirectionally in triplicate with standard BigDye chemistry sequencing protocols and an ABI 3730XL sequencing platform (Life Technologies Corporation, Carlsbad, CA). ADAM10 primers were designed with Primer 3 web application. CLN6 primers were as described previously with slight modifications.23 PCR conditions and primer sequences are available on request. Sequence analysis and alignment to a reference were done with the CodonCode Aligner software (CodonCode Corporation, Dedham, MA). Variants were checked in a minimum of 360 control chromosomes.
Results
FEstim analysis validated the known first-cousin relationship of the parents in Ku2 (an estimated F = 0.067 and F = 0.093 for the two children compared to an expected value of 0.0625). The affected individual in Ku3 had an estimated F = 0.018, close to the expected value for the offspring of second cousins (0.016); we added an appropriate inbreeding loop into her family's pedigree. F for members of Ku1 and Ku4 was estimated to be 0, indicating no evidence of recent inbreeding. IBD estimation with PLINK verified all known relationships and indicated no recent relatedness between the four families.
The multipoint linkage analysis achieved a maximum hLOD score of 4.96 at 58.37 cM on chromosome 15 (Figure 2; see also Figure S1, available online). An adjacent linkage peak achieved the second highest hLOD of 3.16 at 70.67 cM. The estimated proportions of families showing linkage to these peaks were 1 and 0.74, respectively, meaning that all four families showed linkage to the highest peak, whereas only three families showed linkage to the second highest peak (the exception being Ku4, the Kufs type B family).
Figure 2.

LOD Scores Obtained by Four Individual Families in the Mapping Set and the Combined hLOD Score Across Chromosome 15
Inspection of the inferred haplotypes (not shown) revealed that the affected individuals from Ku2 and Ku3 were homozygous by descent for a haplotype but not for the same haplotype. Affected individuals from Ku1 and Ku4 each carry two different haplotypes; no haplotype appears in more than one family. The breakpoint that marks the start of the first peak is provided by Ku2, whereas Ku1 provides the breakpoint that marks the end of the second peak. Ku3 has an internal double recombination event that causes the drop in hLOD score between the two peaks. The overlap of critical regions for the four families is shown in Figure 3.
Figure 3.
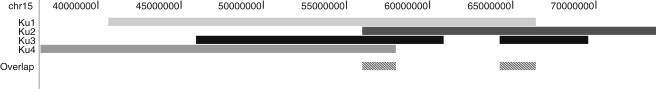
Overlap of the Critical Regions on Chromosome 15 from the Four Families in the Mapping Set
The darker the rectangle is, the greater the contribution of this family to the LOD score.
Inspection of genotypes for all markers on the SNP genotyping arrays (excluding uninformative SNPs or those containing Mendelian errors) revealed that the two affected individuals from Ku2 and Ku3 are both homozygous by state for 1143 consecutive markers in a 4.87 Mb region that overlaps the first linkage peak and for another 948 consecutive markers in a 5.29 Mb region overlapping the second linkage peak. This matches the expected homozygous linkage results from the linkage analysis and the inbreeding estimates. This information allowed the first linkage peak to be refined to 55.37–61.07 cM (rs873393–rs12907068, 2.58 Mb, 5.7 cM) and the second linkage peak to be refined to 67.27–71.27 cM (rs1477799–rs1838544, 2.37 Mb, 4 cM). We identified candidate genes in these two regions by using the NCBI MapViewer and UCSC Human Genome databases. The first peak contained nine genes, including two pseudogenes, whereas the second contained 14 genes, including one pseudogene.
On the basis of the mapping results, we initially focused on the interval from the mapping set linked to all four families (Ku1, Ku2, Ku3, and Ku4) and identified ADAM10, encoding an endopeptidase expressed in the brain, as a candidate gene. Extensive sequencing of this gene in six cases (Ku1 II-4 and II-5, Ku2 IV-1, Ku3 II-2, and Ku4 II-2 and II-3) did not reveal plausible mutations. Subsequently, we considered the second interval, which was linked only to the three Kufs type A families (Ku1, Ku2, and Ku3) but not the type B family (Ku4). An obvious candidate in this region was CLN6 (ENSG00000128973); mutations in this gene cause variant late-infantile NCL.1,24,25 Mutational analysis identified plausible mutations in CLN6 in all three Kufs type A families: Ku1 compound heterozygous c.200T>C and c.308G>A (p.Leu67Pro and p.Arg103Gln), Ku2 homozygous c.139C>T (p.Leu47Phe), and Ku3 homozygous c.17G>C (p.Arg6Thr).
No CLN6 mutations were identified in the Kufs type B family (Ku4). Linkage analysis of Ku4 alone produced 18 linkage peaks with the same maximum LOD score of 0.85. None of these peaks overlapped with genes in which mutations are known to cause NCL (data not shown).
We then sequenced CLN6 in eight unrelated subjects or families from the validation set; three were previously reported families.26–28 There were three Kufs type A families, who were presumed to have recessive inheritance. All three families had mutations in CLN6. Two of these unrelated families, Ku5 and Ku6, had the same homozygous mutation c.712T>A;713T>C (p.Phe238Thr). Family Ku7 had two mutations—c.446G>A and 890delC (p.Arg149His and Pro297LeufsX53); presumed compound heterozygosity could not be confirmed because DNA from other family members was not available. Among three additional Kufs families for whom classification into type A or type B was not possible, no CLN6 variants were detected for two families (Ku10 and Ku11), whereas three variants were identified in the third, Ku8—c.150C>G (p.Tyr50X), 231C>G (Asn77Lys), and 923G>C (Ser308Thr). No CLN6 variants were detected for the single case of dominant Kufs disease (Ku9), whereas a heterozygous change was identified in a Kufs type B case (Ku12, c.100G>A [pAla34Thr]).
Sequence traces for these variants are shown in Figure S2. Variants found in cases are listed in Table 1, whereas the familial segregation of variants is shown in Figure 1. When DNA from other family members was available, all variants identified were found to segregate with the pattern of disease. Segregation could not be tested for Ku3, Ku6, and Ku8 because only DNA from the proband was available.
Table 1.
Clinical Summary and CLN6 Variants in 12 Pedigrees with Kufs Disease
| Family | Country of Origin | Casea | Kufs Type | Age at Onset (Year) and Sex | Clinical Summary | CLN6 Mutationb | Inferred Protein Change |
|---|---|---|---|---|---|---|---|
| Mapping set | |||||||
| Ku1 | Italy | II-5 | A | 16 F | Action myoclonus initially, tonic-clonic seizures 8 years later, followed by dementia; no ataxia | c.200T>C/c.308G>A | p.Leu67Pro/p.Arg103Gln |
| II-4 | A | 36 M | Initial tonic-clonic seizures, followed by myoclonus and moderate dementia; no ataxia | c.200T>C/c.308G>A | p.Leu67Pro/p.Arg103Gln | ||
| Ku2 | Italy | IV-1 | A | 28 F | Initial tonic-clonic seizures and massive myoclonus, followed by ataxia and dementia | c.139C>T/c.139C>T | p.Leu47Phe/p.Leu47Phe |
| Ku38 | Canada (Italian ancestry) | II-2 | A | 31 F | Initial tonic-clonic seizure with photic stimulation, subsequent myoclonus and dementia 5 years later; no ataxia | c.17G>C/c.17G>C | p.Arg6Thr/p.Arg6Thr |
| Ku4 | Italy | III-3 | B | 32 F | Initial dementia followed by tonic-clonic seizures and ataxia 9 years later | WT/WT | WT/WT |
| Validation set | |||||||
| Ku5 | Australia (Maltese ancestry) | II-2 | A | 46 F | Initial myoclonus, ataxia, and cognitive decline. Episode of nonconvulsive status epilepticus | c.[712T>A;713T>C]/ c.[712T>A;713T>C] | p.Phe238Thr/p.Phe238Thr |
| II-3 | A | 51 M | Presented with ataxia and myoclonus. Later mild cognitive decline | c.[712T>A;713T>C]/ c.[712T>A;713T>C] | p.Phe238Thr/p.Phe238Thr | ||
| Ku6 | Italy | II-1 | A | 17 F | Initial action myoclonus, one tonic-clonic seizure, ataxia, and cognitive decline. Scholastic difficulties from age 12 | c.[712T>A;713T>C]/ c.[712T>A;713T>C] | p.Phe238Thr/p.Phe238Thr |
| Ku726 | Ireland | II-1 | A | 35 F | Initial tonic-clonic seizures, then tremor, dementia, and ataxia | c.446G>A/c.890delC | p.Arg149His/p.Pro297LeufsX53 |
| II-2 | ? | 43 M | Initially presented with ataxia, then dementia. No tonic-clonic seizures. No data on myoclonus | c.446G>A/c.890delC | p.Arg149His/p.Pro297LeufsX53 | ||
| Ku8 | USA | II-1 | ? | unknown | No details available | c.150C>G/c.231C>G/ | p.Tyr50X/ p.Asn77Lys/ |
| Ku9 | Australia (Maltese Ancestry) | - | A dominant | 16 M | Presented with tonic-clonic seizures. Subsequent mild cognitive changes. No ataxia present | WT/WT | WT/WT |
| Ku10 | French-Canada | - | ? | 25 F | Focal seizures followed by dementia | WT/WT | WT/WT |
| Ku1127 | Italy | - | ? | 62 F | Myoclonic jerks then dementia. No ataxia | WT/WT | WT/WT |
| Ku1228 | Italy | - | B | 37 F | Initially presented with dementia followed by focal motor seizures | c.100G>A/WT | p.Ala34Thr/WT |
The following abbreviations are used: M, male; F, female; WT, wild-type. A “?” indicates that Kufs types A and B could not be differentiated on the basis of the clinical history.
Symbols under “case” for families 1–8 refer to Figure 1; cases for families 9–12 were the probands.
The reference sequence used for CLN6 was NM_017882.2.
A minimum of 360 control chromosomes were screened for the 11 CLN6 variants identified in this study; none of these variants have been previously reported in dbSNP. The p.Ser308Thr change found in Ku8 was found in one control chromosome, whereas none of the other variants were detected at all. The p.Ser308Thr change affects a nonconserved amino acid (data not shown), suggesting that it is a rare nonpathogenic variant and that the Ku8 case is a compound heterozygote for the p.Tyr50X and p.Asn77Lys variants. The p.Ala34Thr change (in Ku12) also affects a nonconserved amino acid. It was detected in a heterozygous state, and no other CLN6 changes were found for Ku12; both factors suggest that it is also a rare nonpathogenic variant. The eight other detected changes occurred in highly conserved amino acids and were deemed to be mutations (Table S1). A ninth change (Leu67Pro in Ku1 II-4 and II-5) is conserved in mammals but substituted with isoleucine, a very similar nonpolar amino acid, in lower animals and was therefore also deemed a mutation.
We used the HumVar-trained PolyPhen2 v2.0.2329 to predict the biological impact of the seven missense mutations identified (Table S1). PolyPhen 2 predicted the p.Leu47Phe and p.Asn77Lys mutations to be probably damaging and five other missense mutations to be possibly damaging. The p.Phe238Thr mutation in Ku5 and Ku6 was predicted to be benign, but its occurrence in two independent families and its pattern of segregation strongly suggests it is causative.
Discussion
Although genes in which mutations cause most forms of early-onset NCLs have been identified, the molecular basis of adult-onset NCL and particularly Kufs disease, which has no visual failure, remains a mystery. A separate locus (CLN4) has been tentatively reserved for Kufs disease; however, there has been no report of a positive-linkage result identifying its chromosomal location.1 Some authors have suggested that “mild” mutations in the genes causing the early-onset NCLs might result in phenotypes with a later onset.1,30 Consistent with this hypothesis, there are two reports of adult-onset NCL caused by mutations in CLN1 (PPT1)5,6 and two reports of early-adult-onset NCL disease caused by mutations in CLN5.12,31 However, these all had visual failure and retinal involvement and so represent unusual cases of adult-onset NCL rather than true Kufs disease, which has no visual failure. One case diagnosed with Kufs disease was later found to carry mutations in SGSH (MIM 605270), which causes the more severe and unrelated disease lysosomal storage disorder MPSIIIA.31
We found mutations in CLN6 in six pedigrees with recessive Kufs type A disease and in one case where phenotype information was unavailable; these findings indicate that mutations in this gene are a major cause of recessive Kufs type A disease. Mutations in CLN6 are well known to cause variant late-infantile NCL.1,23–25,32,33 Variant late-infantile CLN6 disease presents between 18 months and 5 years of age. Seizures and visual loss are usually early symptoms; cases with onset after 4 years of age can present with cognitive and motor decline, ataxia, and/or myoclonus epilepsy.1,24,33 Visual loss is a characteristic, although one reported case had no visual impairment by age 17.33
CLN6 encodes a 311 amino acid transmembrane protein localized in the endoplasmic reticulum.24,25,34 It contains a cytoplasmic N terminus, a luminal C terminus, and seven transmembrane domains.35 CLN6 is conserved among vertebrates, and its function is unknown. Lysosomal pH can be elevated in cells from CLN6 disease patients,36 but the activities of at least some lysosomal enzymes appear unaffected, although lysosomal degradation of an endocytosed protein was reduced.37 CLN6 can bind to the NCL protein CLN538 and to collapsing response mediator protein-2 (CRMP-2).39 In vitro analysis of certain mutations associated with variant late-infantile NCL suggests the abnormal protein is more rapidly degraded.34 Of the nine different mutations reported in this study, eight have not been reported previously. One (p.Pro297LeufsX53) from a compound heterozygous case was previously described in variant late-infantile NCL (NCL Resource—A Gateway for Batten Disease).
The nine CLN6 mutations we identified in Kufs type A in this study (in six out of six families tested) and in Ku8 (where phenotypic assignment was unavailable) are highly likely to be causative. They are missense and nonsense mutations affecting highly evolutionarily conserved amino acid residues. This was supported by analysis with PolyPhen2. Of the seven missense mutations, six were predicted to be possibly or probably damaging. The remaining change, p.Phe238Thr, is likely to be causative because it was detected as a homozygous change in two independent families. A tenth variant (p.Ala34Thr in Ku12) was thought to probably be nonpathogenic and rare because it changed a nonconserved amino acid and was heterozygous, although it was classified as possibly damaging by PolyPhen2.
Figure 4 shows the Kufs disease mutations described here and those the literature describes in variant late-infantile NCL 23–25,32,33,40–44 (NCL Resource—A Gateway for Batten Disease). There is no obvious difference in their location within CLN6, although two of the missense variants are located in less conserved regions: p.Arg6Thr in the first 42 amino acids of the N terminus and p.Leu67Pro in the short exon encoding amino acids 67–72. The marked difference in the age at onset and the disparity in eye involvement between variant late-infantile NCL and Kufs disease is striking and not easily explained. However, in approximately 40% of variant late-infantile NCL cases with CLN6 mutations, both alleles have mutations predicting protein truncation or a severely abnormal protein,23–25,32,33 whereas such mutations were present in only two Kufs cases and in both cases were present on only one allele, suggesting that patients with Kufs disease could have significant residual CLN6 function. Directly determining whether mutations in Kufs disease are “milder” or whether the phenotypic differences are due to modifying genes must await the development of an understanding of the function of CLN6 and a functional assay.1,30 A role for modifiers is suggested by the wide range in age at onset (teens to age 51) in these Kufs cases with CLN6 mutations, even among those with the same mutation (see Ku1, Ku5, and Ku6 in Table 1). Modifiers could also explain the variation within families in the temporal appearance of symptoms (see Ku5 and Ku7 in Table 1); similar variation in the symptom sequence occurs in LINCL caused by mutations in CLN6.23,33
Figure 4.

Schematic Representation of CLN6 with Mutations
The numbered blue boxes represent each exon. The numbers below each exon represent the amino acid number within the CLN6 protein. The symbols above, colored in red, are mutations reported here for Kufs disease. The grey symbols below are previously described mutations in CLN6 in variant late-infantile NCL (NCL Resource—A Gateway for Batten Disease). The black arrow indicates the mutation described in both phenotypes.
On the basis of these results, CLN6 appears to be the major gene for recessive Kufs type A, the form that presents as a progressive myoclonus epilepsy. Indeed, the discovery of homozygous or compound heterozygous CLN6 mutations in our type A, but not type B, cases supports the validity of the clinical classification, although the distinction between type A and B cases is not always clear cut.8,16,28 Notably, the proband of family Ku7 clearly had the type A phenotype, whereas her sibling could have been type B, and unfortunately the phenotypic assignment of Ku8 as type A or B could not be made because of a lack of data (see Table 1). Furthermore, the role of the heterozygous CLN6 variant in Ku12 could not be determined, and it is possible that this variant influences the disease arising from mutations in another locus. The major gene or genes for Kufs type B, as well as for the dominant form of adult NCL, await discovery. However, linkage mapping of Ku4 suggests that Kufs type B is not caused by a mutation in the same genes mutated in other forms of NCL.
Diagnosis of Kufs disease is challenging and often requires invasive biopsy. A relatively inexpensive CLN6 mutation screening should now be considered as an initial diagnostic step in suspected Kufs disease cases, especially when progressive myoclonus epilepsy is the presenting feature.
Acknowledgments
S.F.B. was supported by an NHMRC Australia Fellowship and an NHMRC Program Grant. M.B. was funded by an NHMRC Career Development Award and an NHMRC Program Grant. We thank the Batten Disease Support and Research Association for additional financial support (to S.M.) and the families themselves.
Contributor Information
Melanie Bahlo, Email: bahlo@wehi.edu.au.
Samuel F. Berkovic, Email: samuelfb@unimelb.edu.au.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Online Mendelian Inheritance in Man, http://www.omim.org
NCBI MapViewer, http://www.ncbi.nlm.nih.gov/projects/mapview/
NCL Resource—A Gateway for Batten Disease, http://www.ucl.ac.uk/ncl/cln6.shtml
Primer 3, http://frodo.wi.mit.edu/primer3
UCSC Genome Browser, http://www.genome.ucsc.edu
References
- 1.Mole, S.E., and Williams, R.E. (2010). Neuronal Ceroid-Lipofuscinoses. Gene Reviews, http://www.ncbi.nlm.nih.gov/books/NBK1428/ [DOI] [PubMed]
- 2.Santavuori P. Neuronal ceroid-lipofuscinoses in childhood. Brain Dev. 1988;10:80–83. doi: 10.1016/s0387-7604(88)80075-5. [DOI] [PubMed] [Google Scholar]
- 3.Mole S.E., Williams R.E., Goebel H.H. Correlations between genotype, ultrastructural morphology and clinical phenotype in the neuronal ceroid lipofuscinoses. Neurogenetics. 2005;6:107–126. doi: 10.1007/s10048-005-0218-3. [DOI] [PubMed] [Google Scholar]
- 4.Wisniewski K.E., Zhong N., Philippart M. Pheno/genotypic correlations of neuronal ceroid lipofuscinoses. Neurology. 2001;57:576–581. doi: 10.1212/wnl.57.4.576. [DOI] [PubMed] [Google Scholar]
- 5.van Diggelen O.P., Thobois S., Tilikete C., Zabot M.T., Keulemans J.L., van Bunderen P.A., Taschner P.E., Losekoot M., Voznyi Y.V. Adult neuronal ceroid lipofuscinosis with palmitoyl-protein thioesterase deficiency: first adult-onset patients of a childhood disease. Ann. Neurol. 2001;50:269–272. doi: 10.1002/ana.1103. [DOI] [PubMed] [Google Scholar]
- 6.Ramadan H., Al-Din A.S., Ismail A., Balen F., Varma A., Twomey A., Watts R., Jackson M., Anderson G., Green E., Mole S.E. Adult neuronal ceroid lipofuscinosis caused by deficiency in palmitoyl protein thioesterase 1. Neurology. 2007;68:387–388. doi: 10.1212/01.wnl.0000252825.85947.2f. [DOI] [PubMed] [Google Scholar]
- 7.Kufs H. Über eine Spätform der amaurotischen Idiotie und ihre heredofamiliären Grundlagen. Z ges Neurol Psychiat. 1925;95:169–188. [Google Scholar]
- 8.Berkovic S.F., Carpenter S., Andermann F., Andermann E., Wolfe L.S. Kufs' disease: a critical reappraisal. Brain. 1988;111:27–62. doi: 10.1093/brain/111.1.27. [DOI] [PubMed] [Google Scholar]
- 9.Constantinidis J., Wisniewski K.E., Wisniewski T.M. The adult and a new late adult forms of neuronal ceroid lipofuscinosis. Acta Neuropathol. 1992;83:461–468. doi: 10.1007/BF00310021. [DOI] [PubMed] [Google Scholar]
- 10.Boehme D.H., Cottrell J.C., Leonberg S.C., Zeman W. A dominant form of neuronal ceroid-lipofuscinosis. Brain. 1971;94:745–760. doi: 10.1093/brain/94.4.745. [DOI] [PubMed] [Google Scholar]
- 11.Nijssen P.C., Ceuterick C., van Diggelen O.P., Elleder M., Martin J.J., Teepen J.L., Tyynelä J., Roos R.A. Autosomal dominant adult neuronal ceroid lipofuscinosis: a novel form of NCL with granular osmiophilic deposits without palmitoyl protein thioesterase 1 deficiency. Brain Pathol. 2003;13:574–581. doi: 10.1111/j.1750-3639.2003.tb00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xin W., Mullen T.E., Kiely R., Min J., Feng X., Cao Y., O'Malley L., Shen Y., Chu-Shore C., Mole S.E. CLN5 mutations are frequent in juvenile and late-onset non-Finnish patients with NCL. Neurology. 2010;74:565–571. doi: 10.1212/WNL.0b013e3181cff70d. [DOI] [PubMed] [Google Scholar]
- 13.Carpenter S., Karpati G., Andermann F., Jacob J.C., Andermann E. The ultrastructural characteristics of the abnormal cytosomes in Batten-Kufs' disease. Brain. 1977;100:137–156. doi: 10.1093/brain/100.1.137. [DOI] [PubMed] [Google Scholar]
- 14.Goebel H.H., Braak H. Adult neuronal ceroid-lipofuscinosis. Clin. Neuropathol. 1989;8:109–119. [PubMed] [Google Scholar]
- 15.Pasquinelli G., Cenacchi G., Piane E.L., Russo C., Aguglia U. The problematic issue of Kufs disease diagnosis as performed on rectal biopsies: a case report. Ultrastruct. Pathol. 2004;28:43–48. [PubMed] [Google Scholar]
- 16.Sadzot B., Reznik M., Arrese-Estrada J.E., Franck G. Familial Kufs' disease presenting as a progressive myoclonic epilepsy. J. Neurol. 2000;247:447–454. doi: 10.1007/s004150070174. [DOI] [PubMed] [Google Scholar]
- 17.Leutenegger A.L., Prum B., Génin E., Verny C., Lemainque A., Clerget-Darpoux F., Thompson E.A. Estimation of the inbreeding coefficient through use of genomic data. Am. J. Hum. Genet. 2003;73:516–523. doi: 10.1086/378207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leutenegger A.L., Labalme A., Genin E., Toutain A., Steichen E., Clerget-Darpoux F., Edery P. Using genomic inbreeding coefficient estimates for homozygosity mapping of rare recessive traits: Application to Taybi-Linder syndrome. Am. J. Hum. Genet. 2006;79:62–66. doi: 10.1086/504640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 21.Bahlo M., Bromhead C.J. Generating linkage mapping files from Affymetrix SNP chip data. Bioinformatics. 2009;25:1961–1962. doi: 10.1093/bioinformatics/btp313. [DOI] [PubMed] [Google Scholar]
- 22.Thiele H., Nürnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21:1730–1732. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- 23.Sharp J.D., Wheeler R.B., Parker K.A., Gardiner R.M., Williams R.E., Mole S.E. Spectrum of CLN6 mutations in variant late infantile neuronal ceroid lipofuscinosis. Hum. Mutat. 2003;22:35–42. doi: 10.1002/humu.10227. [DOI] [PubMed] [Google Scholar]
- 24.Gao H., Boustany R.M., Espinola J.A., Cotman S.L., Srinidhi L., Antonellis K.A., Gillis T., Qin X., Liu S., Donahue L.R. Mutations in a novel CLN6-encoded transmembrane protein cause variant neuronal ceroid lipofuscinosis in man and mouse. Am. J. Hum. Genet. 2002;70:324–335. doi: 10.1086/338190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wheeler R.B., Sharp J.D., Schultz R.A., Joslin J.M., Williams R.E., Mole S.E. The gene mutated in variant late-infantile neuronal ceroid lipofuscinosis (CLN6) and in nclf mutant mice encodes a novel predicted transmembrane protein. Am. J. Hum. Genet. 2002;70:537–542. doi: 10.1086/338708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Callagy C., O'Neill G., Murphy S.F., Farrell M.A. Adult neuronal ceroid lipofuscinosis (Kufs' disease) in two siblings of an Irish family. Clin. Neuropathol. 2000;19:109–118. [PubMed] [Google Scholar]
- 27.Gambardella A., Pasquinelli G., Cittadella R., Bono F., Oliveri R.L., Valentino P., Zappia M., Quattrone A., Aguglia U. Kufs' disease presenting as late-onset epilepsia partialis continua. Neurology. 1998;51:1180–1182. doi: 10.1212/wnl.51.4.1180. [DOI] [PubMed] [Google Scholar]
- 28.Zini A., Cenacchi G., Nichelli P., Zunarelli E., Todeschini A., Meletti S. Early-onset dementia with prolonged occipital seizures: an atypical case of Kufs disease. Neurology. 2008;71:1709–1712. doi: 10.1212/01.wnl.0000335164.02634.f6. [DOI] [PubMed] [Google Scholar]
- 29.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lauronen L., Munroe P.B., Järvelä I., Autti T., Mitchison H.M., O'Rawe A.M., Gardiner R.M., Mole S.E., Puranen J., Häkkinen A.M. Delayed classic and protracted phenotypes of compound heterozygous juvenile neuronal ceroid lipofuscinosis. Neurology. 1999;52:360–365. doi: 10.1212/wnl.52.2.360. [DOI] [PubMed] [Google Scholar]
- 31.Sleat D.E., Ding L., Wang S., Zhao C., Wang Y., Xin W., Zheng H., Moore D.F., Sims K.B., Lobel P. Mass spectrometry-based protein profiling to determine the cause of lysosomal storage diseases of unknown etiology. Mol. Cell. Proteomics. 2009;8:1708–1718. doi: 10.1074/mcp.M900122-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore S.J., Buckley D.J., MacMillan A., Marshall H.D., Steele L., Ray P.N., Nawaz Z., Baskin B., Frecker M., Carr S.M. The clinical and genetic epidemiology of neuronal ceroid lipofuscinosis in Newfoundland. Clin. Genet. 2008;74:213–222. doi: 10.1111/j.1399-0004.2008.01054.x. [DOI] [PubMed] [Google Scholar]
- 33.Cannelli N., Garavaglia B., Simonati A., Aiello C., Barzaghi C., Pezzini F., Cilio M.R., Biancheri R., Morbin M., Dalla Bernardina B. Variant late infantile ceroid lipofuscinoses associated with novel mutations in CLN6. Biochem. Biophys. Res. Commun. 2009;379:892–897. doi: 10.1016/j.bbrc.2008.12.159. [DOI] [PubMed] [Google Scholar]
- 34.Kurze A.K., Galliciotti G., Heine C., Mole S.E., Quitsch A., Braulke T. Pathogenic mutations cause rapid degradation of lysosomal storage disease-related membrane protein CLN6. Hum. Mutat. 2010;31:E1163–E1174. doi: 10.1002/humu.21184. [DOI] [PubMed] [Google Scholar]
- 35.Heine C., Quitsch A., Storch S., Martin Y., Lonka L., Lehesjoki A.E., Mole S.E., Braulke T. Topology and endoplasmic reticulum retention signals of the lysosomal storage disease-related membrane protein CLN6. Mol. Membr. Biol. 2007;24:74–87. doi: 10.1080/09687860600967317. [DOI] [PubMed] [Google Scholar]
- 36.Holopainen J.M., Saarikoski J., Kinnunen P.K., Järvelä I. Elevated lysosomal pH in neuronal ceroid lipofuscinoses (NCLs) Eur. J. Biochem. 2001;268:5851–5856. doi: 10.1046/j.0014-2956.2001.02530.x. [DOI] [PubMed] [Google Scholar]
- 37.Heine C., Koch B., Storch S., Kohlschütter A., Palmer D.N., Braulke T. Defective endoplasmic reticulum-resident membrane protein CLN6 affects lysosomal degradation of endocytosed arylsulfatase A. J. Biol. Chem. 2004;279:22347–22352. doi: 10.1074/jbc.M400643200. [DOI] [PubMed] [Google Scholar]
- 38.Lyly A., von Schantz C., Heine C., Schmiedt M.L., Sipilä T., Jalanko A., Kyttälä A. Novel interactions of CLN5 support molecular networking between Neuronal Ceroid Lipofuscinosis proteins. BMC Cell Biol. 2009;10:83. doi: 10.1186/1471-2121-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benedict J.W., Getty A.L., Wishart T.M., Gillingwater T.H., Pearce D.A. Protein product of CLN6 gene responsible for variant late-onset infantile neuronal ceroid lipofuscinosis interacts with CRMP-2. J. Neurosci. Res. 2009;87:2157–2166. doi: 10.1002/jnr.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Muhaizea M.A., Al-Hassnan Z.N., Chedrawi A. Variant late infantile neuronal ceroid lipofuscinosis (CLN6 gene) in Saudi Arabia. Pediatr. Neurol. 2009;41:74–76. doi: 10.1016/j.pediatrneurol.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Cismondi I.A., Kohan R., Ghio A., Ramirez A.M., Halac I.N. Gene symbol: CLN6. Disease: Neuronal ceroid lipofuscinosis, late infantile. Hum. Genet. 2008;124:323–324. [PubMed] [Google Scholar]
- 42.Kousi M., Siintola E., Dvorakova L., Vlaskova H., Turnbull J., Topcu M., Yuksel D., Gokben S., Minassian B.A., Elleder M. Mutations in CLN7/MFSD8 are a common cause of variant late-infantile neuronal ceroid lipofuscinosis. Brain. 2009;132:810–819. doi: 10.1093/brain/awn366. [DOI] [PubMed] [Google Scholar]
- 43.Siintola E., Topcu M., Kohlschütter A., Salonen T., Joensuu T., Anttonen A.K., Lehesjoki A.E. Two novel CLN6 mutations in variant late-infantile neuronal ceroid lipofuscinosis patients of Turkish origin. Clin. Genet. 2005;68:167–173. doi: 10.1111/j.1399-0004.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- 44.Teixeira C.A., Espinola J., Huo L., Kohlschütter J., Persaud Sawin D.A., Minassian B., Bessa C.J., Guimarães A., Stephan D.A., Sá Miranda M.C. Novel mutations in the CLN6 gene causing a variant late infantile neuronal ceroid lipofuscinosis. Hum. Mutat. 2003;21:502–508. doi: 10.1002/humu.10207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


