Abstract
Regulation of cell proliferation and motility is essential for normal development. The Rho family of GTPases plays a critical role in the control of cell polarity and migration by effecting the cytoskeleton, membrane trafficking, and cell adhesion. We investigated a recognized developmental disorder, Adams-Oliver syndrome (AOS), characterized by the combination of aplasia cutis congenita (ACC) and terminal transverse limb defects (TTLD). Through a genome-wide linkage analysis, we detected a locus for autosomal-dominant ACC-TTLD on 3q generating a maximum LOD score of 4.93 at marker rs1464311. Candidate-gene- and exome-based sequencing led to the identification of independent premature truncating mutations in the terminal exon of the Rho GTPase-activating protein 31 gene, ARHGAP31, which encodes a Cdc42/Rac1 regulatory protein. Mutant transcripts are stable and increase ARHGAP31 activity in vitro through a gain-of-function mechanism. Constitutively active ARHGAP31 mutations result in a loss of available active Cdc42 and consequently disrupt actin cytoskeletal structures. Arhgap31 expression in the mouse is substantially restricted to the terminal limb buds and craniofacial processes during early development; these locations closely mirror the sites of impaired organogenesis that characterize this syndrome. These data identify the requirement for regulated Cdc42 and/or Rac1 signaling processes during early human development.
Introduction
Members of the large family of GTPases act as molecular switches that control many aspects of cell activity through a remarkably simple biochemical mechanism of cycling between two conformational forms. The active state requires bound guanosine triphosphate (GTP) to allow interaction with one of many effector proteins, whereas the GTPase-mediated hydrolysis of GTP to guanosine diphosphate (GDP) engenders an inactive state.1 Although the Rho switch appears straightforward, the process is closely controlled by at least three classes of regulators, namely guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs) and GDP dissociation inhibitors (GDIs). The Rho GTPases, which include Cdc42 and Rac1, hold central functions in cell division, survival, and migration; alterations in expression have been widely studied in cancer and indicate a role in tumor invasion and metastasis.2 However, regulation of cell proliferation and migration are also fundamental aspects of organ formation, especially during early developmental stages. We have studied an inherited disorder characterized by abnormalities of limb development, a recognized paradigm of human organogenesis, and report a GAP regulatory defect as the primary cause.
Adams-Oliver syndrome (AOS; MIM 100300) describes the congenital absence of skin, aplasia cutis congenita (ACC), in combination with terminal transverse limb defects (TTLD) (Figure 1A). Limb abnormalities typically affect the distal phalanges or entire digits or, rarely, more proximal limb structures. Important associated anomalies include vascular cutis marmorata and cardiac and vascular abnormalities, for example pulmonary hypertension.3 Although the combination of ACC and TTLD most often occurs in sporadic cases, segregation within families is consistent with autosomal-dominant inheritance in some kindred and autosomal-recessive inheritance in others. Variability of the disease phenotype is also widely recognized and includes an absence of either of the major features in obligate carriers, indicating reduced penetrance of the disease allele. Clinically, in cases with a known family history, the presence of either ACC or TTLD has been considered sufficient to warrant the diagnosis of AOS.3
Figure 1.
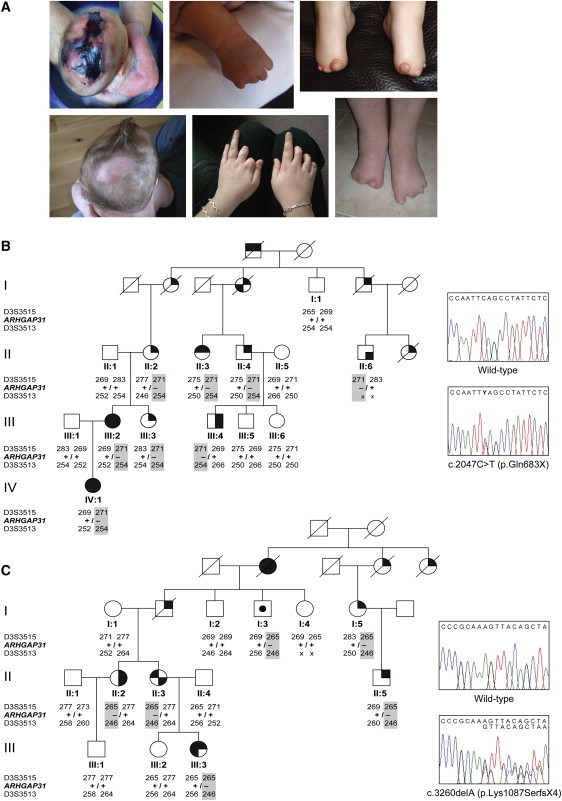
Features of ACC-TTLD and Segregation of ARHGAP31 Mutations
(A) Characteristic phenotype of ACC-TTLD showing severe ACC (left panels) and a range of TTLD defects of the hands (middle panels) and feet (right panels), including partial absence of the fingers and toes and short distal phalanxes of fingers and toes.
(B) Pedigree structure of family AOS-12 showing segregation of the c.2047C>T nonsense mutation represented in the adjacent sequence chromatogram.
(C) Segregation and sequence chromatogram of the c.3260delA frameshift mutation in family AOS-5. Mutation carriers are denoted by +/–.
Key to symbols: square, male; circle, female; upper left shading, aplasia cutis congenita; lower left shading, bony defect/abnormal fontanelle; upper right shading, terminal transverse limb defects; lower right shading, syndactyly; center shading, unsymptomatic mutation carrier; blank, unaffected.
We have now used genome-wide linkage analysis to study two kindreds with autosomal-dominant ACC-TTLD and subsequently identified heterozygous mutations in a RhoGAP family member, Rho GTPase-activating protein 31 (ARHGAP31; MIM 610911), also known as Cdc42 GTPase-activating protein (CdGAP),4 by candidate gene and exome sequencing. We determined the distribution of expression of Arhgap31 during development and verified the pathogenic effect of these mutations in primary human dermal fibroblasts from patients with ACC-TTLD. This genetic finding identifies the importance of Cdc42/Rac1 pathways in the developmental processes of scalp and limb formation.
Subjects and Methods
Clinical Ascertainment
Index subjects were recruited via the Adams-Oliver Syndrome Support Group, Deeside, UK, and through specialist clinical genetics centers from within the UK and continental Europe. Additional family members, including unaffected individuals and spouses, were then invited to participate in the study. All participants underwent a detailed physical examination by experienced clinical geneticists. A diagnosis of ACC-TTLD was based on clinical guidelines3 and supported by radiological investigations in selected patients. Kindreds AOS-5 and AOS-12 were previously reported in the medical literature5,6 and were re-examined in 2009.3 All subjects gave written informed consent in accordance with the protocol approved by the Guy's and St Thomas' NHS Foundation Trust local research ethics committee.
Genotyping, Linkage Analysis, and Mutation Detection
We extracted genomic DNA from peripheral venous blood by standard techniques or from saliva by using the Oragene DNA Self-Collection Kit (DNA Genotek). A genome-wide screen was performed for 22 individuals from two multigenerational families via the GoldenGate HumanLinkage V Panel on an iScan System (Illumina) according to the manufacturer's guidelines. Linkage analysis was performed with Merlin v1.1.2 software under an autosomal-dominant disease model with a disease allele frequency of 0.0001 and a penetrance value of 85%. Additional polymorphic markers for refinement mapping were selected with an average heterozygosity of 74%. Fluorescently tagged PCR fragments were analyzed on an ABI3730xl DNA analyzer, and genotypes were assigned via GeneMapper v3.7 software (Applied Biosystems).
All coding exons and intron-exon boundaries of the candidate genes ARHGAP31, GSK3B (MIM 605004), LSAMP (MIM 603241), and POPDC2 (MIM 605823) were screened by direct DNA sequencing. Primers were designed with Primer3 software.7 PCR products were purified with ExoSAP-IT (GE Healthcare) and sequenced with BigDye Terminator v3.1 chemistry (Applied Biosystems). Sequence traces were aligned to reference with Sequencher v4.9 software (Gene Codes Corporation).
Exome capture of subject III:2 (Figure 1B) was undertaken with the SureSelect Target Enrichment System (Agilent) and sequenced on a Genome Analyzer IIx (Illumina). Paired-end sequence reads were aligned to the reference genome (hg18) with Novoalign software (Novocraft Technologies). Duplicate reads, resulting from PCR clonality or optical duplicates and reads mapping to multiple locations were excluded from downstream analysis. Single nucleotide substitutions and small insertion deletions were identified and quality filtered with the SamTools software package8 and in-house software tools.9 Variants were annotated with respect to genes and transcripts with the SNPClassifier tool.10 Filtering of variants for novelty was performed by comparison to dbSNP131 and 1000 Genomes pilot SNP calls (March 2010). The accession numbers of the reference sequences used for mutation nomenclature are NM_020754.2 (mRNA) and NP_065805.2 (protein).
Gene-Expression Analysis
Fetal expression of ARHGAP31 was assessed with a human fetal multiple tissue cDNA (MTC) panel (Clontech). We performed PCR with standard protocols and used primers ARHGAP31_3Fw (5′ AGCTCATGTGACCTCACCAA 3′) and ARHGAP31_3Rv (5′ AGACTGGAGCAGGGAAGGAG 3′) to generate a 976 bp fragment. GAPDH primers (Clontech) were used as an internal control.
For RT-PCR, cDNA was generated from 1 μg of RNA extracted from patient and wild-type (WT) EBV-transformed lymphoblasts via the Verso cDNA Kit (ABgene). Real-time quantitative PCR was performed with ARHGAP31 Taqman gene-expression probes according to the standard protocol on a real-time PCR 7900HT (Applied Biosystems). GAPDH (Applied Biosystems) was used as an endogenous control. We calculated relative levels of gene expression by SDS v2.2 software (Applied Biosystems) by using the comparative CT method of data analysis (relative quantity = 2 − ΔΔCt).
Whole-Mount In Situ Hybridization
The genomic sequence of Arhgap31 was obtained from Ensembl. We designed primers by using Primer3 software7 to produce a PCR product of 543 bp from the 3′ UTR of Arhgap31 (genomic location: chromosome 16:38,599,795–38,600,337). T3 and T7 RNA polymerase sites were added to the 5′ end of the forward and reverse primers respectively (forward: 5′ AATTAACCCTCACTAAAGGCTGCTGGAGGAAGGTTTCTG 3′; reverse: 5′ TAATACGACTCACTATAGGCGCCTCTCCACACCATATTT 3′). Digoxigenin (DIG)-labeled (Roche Applied Science) antisense riboprobes were generated by in vitro transcription of purified PCR-amplified DNA template with T7 RNA polymerase.
CD1 mouse embryos at developmental stages 9.5, 10.5, 11.5, and 12.5 days postcoitum (dpc) were obtained from the Mary Lyon Centre, MRC Harwell, Oxfordshire, UK. Embryos were fixed overnight in 4% paraformaldehyde at 4°C, stored in methanol, and rehydrated in a series of graded methanol washes in PBS and 0.1% Tween 20 (PBST). Proteinase K (10 μg/ml) (Roche Applied Science) permeabilization was performed for 15–35 min depending on the stage of development. Embryos were washed twice in 0.1 M triethanolamine, and acetic anhydride was added to the second wash. Samples were then washed in PBST and refixed in 4% PFA/0.2% gluteraldehyde for 20 min. After washing in PBST, embryos were prehybridized for 2 hr at 60°C and hybridized for 48 hr at 60°C in hybridization buffer containing the DIG-labeled probe. Samples were washed three times for 20 min each time in 2× SSC and Tween 20 and then three times for 30 min each time in 0.2× SSC and 0.1% Tween 20 at 60°C. Samples were then washed twice for 15 min each time in maleic acid buffer (MAB) at room temperature. A final wash for 2 hr in MAB, 2% Boehringer-Mannheim blocking reagent (BMB), and 20% heat-treated lamb serum solution at room temperature was performed before overnight incubation in the same solution containing a 1/2000 dilution of DIG antibody coupled to alkaline phosphatase (Roche Applied Science). Embryos were then washed three times for 1 hr each time in MAB and color detected with 2 ml of BM purple precipitating solution (Roche Applied Science).
Optical Projection Tomography
Whole-mount in situ hybridization (WISH) was performed as described above. Embryos were mounted in 1% agarose, dehydrated in methanol, and then cleared overnight in BABB (1 part benzyl alcohol: 2 parts benzyl benzoate). We then imaged samples with a Bioptonics Optical Projection Tomography (OPT) Scanner 3001 by using brightfield to detect the staining and tissue autofluorescence (excitation 425 nm, emission 475 nm) to capture the anatomy.11 The resulting images were reconstructed with Bioptonics proprietary software and automatically thresholded and merged to a single 3D image output via Bioptonics Viewer software. The downstream digital dissection and sectioning were performed with Amira software (Visage Imaging).
Cloning and Mutagenesis
Full-length Myc-tagged ARHGAP31 was generated as previously described.4 We engineered mutant constructs by performing site-directed mutagenesis with the QuickChange kit (Stratagene) on the WT template. Primers are available on request.
Cell Culture
Cells were maintained at 37°C in a humidified incubator with 5% CO2. Human endometrioid cancer (HeLa, ATCC) and human embryonic kidney (HEK293) cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 4.5 g/ml GlutaMax and 10% heat-inactivated fetal bovine serum (FBS). EBV-transformed lymphoblasts (ECACC) from a WT control and ACC-TTLD patients with and without ARHGAP31 mutations were cultured in RPMI-1640 supplemented with 10% heat-inactivated FBS. Human primary dermal fibroblast cells were established from 4 mm tissue biopsies from a WT control individual and an ACC-TTLD patient carrying the p.Gln683X mutation. Tissues were enzymatically dissociated with accutase, and fibroblasts were grown in basal medium 106 supplemented with 2% (v/v) FBS, 1 μg/ml hydrocortisone, 10 ng/ml human epidermal growth factor, 3 ng/ml basic fibroblast growth factor, and 10 μg/ml heparin. Normal human neonatal dermal fibroblasts (HDF; Lonza), used as an additional control for proliferation assays, were cultured under the same conditions as described above. All cell culture reagents were obtained from Invitrogen. Transient transfections of HeLa cells were performed with FuGENE (Roche) in accordance with manufacturer's instructions and with a 3:1 ratio of transfection reagent to DNA.
Immunofluorescence
We plated HeLa cells and fibroblasts in 6-well plates (Corning) on acid-treated glass coverslips (Laboratory Sales Limited) and allowed them to grow until 80% confluent. Cells were fixed in ice-cold methanol, rehydrated with 1× PBS, and blocked with a 0.5% bovine serum albumin (BSA; Sigma-Aldrich) in 1× PBS solution. After blocking, we incubated cells with a polyclonal rabbit antibody raised against a peptide corresponding to amino acids 541–562 of mouse Arhgap31 (PRD1) and purified them on a protein A-sepharose column (1:500 dilution). After washing with blocking solution, a secondary rabbit-specific fluorophore-tagged antibody (Abcam) was added for 1 hr. Coverslips were rinsed and mounted on slides with hard-set mounting medium containing a DAPI nuclear stain (Vector Labs). Golgi and cytoskeleton visualizations were performed with 58K Golgi protein and monoclonal tubulin antibodies (Abcam), respectively. Antibodies were diluted according to manufacturer's instructions. PRD1 antibody specificity tests were performed with a preimmune serum from the antibody host rabbit in place of the Arhgap31 antibody. A second specificity test utilized a blocking peptide specific to the PRD1 antibody epitope used at a 10:1 concentration (peptide to antibody). We further used a blocking peptide specific to a random region of Arhgap31 as an additional negative control. Both blocking peptides were synthesized by Sigma-Aldrich. All images were acquired on a Zeiss LSM 510 confocal microscope and processed with Adobe Photoshop. Statistical comparisons for cell rounding experiments were conducted via a Fisher's exact test.
G-LISA Cdc42 Activation Assay
HEK293 cells were grown to 70% confluency on 100 mm dishes and transiently transfected with polyethyleneimine (Polysciences) and 100 ng of empty vector DNA or vector-encoding Myc-tagged WT ARHGAP31, p.Lys1087SerfsX4, or p.Gln683X. Cells were lysed 16 hr after transfection according to manufacturer's instructions (Cytoskeleton) and snap-frozen in liquid nitrogen. Levels of tagged proteins and Cdc42 were determined by immunoblotting with polyclonal Myc-specific (Cell Signaling Technology) and Cdc42-specific (Santa Cruz Biotechnology) antibodies, respectively. The relative amounts of GTP-bound Cdc42 in each condition were determined in duplicate. For each Cdc42-GTP measurement, 100 μg of protein lysate was used. To compare WT and mutant ARHGAP31 activity, we used a Student's t test with a two-tail distribution.
Immunoprecipitation
HEK293 cells were cotransfected with mouse pRK5Myc-Arhgap31 (1083–1425) and pEGFP-Arhgap31 (1–221 or 1–820). After 16 hr, cells were lysed on ice in 25 mM HEPES (pH 7.4), 100 mM NaCl, 10 mM MgCl2, 5% glycerol, 1% NP-40, 1 mM Na3VO4, 10 mM NaF, 1 mM PMSF, and protease cocktail inhibitors (Roche Applied Science). Protein lysates were centrifuged for 10 min at 14,000 g and precleared for 1 hr at 4°C with protein G-sepharose (GE Healthcare). Supernatants were incubated for 3 hr at 4°C with 2 μg of monoclonal Myc-specific antibody (9E10) and protein G-sepharose. Immune complexes were washed three times in lysis buffer and resuspended in SDS sample buffer. Proteins were resolved by SDS-PAGE and detected by immunoblotting with antibodies to GFP (A6455, Molecular Probes) and Myc.
Proliferation Assay
The CyQUANT fluorescence-based microplate assay was used for quantitation of cell number. To generate a standard calibration curve, we measured binding to cellular nucleic acids by using 485 nm (±10 nm) excitation and 530 nm (±12.5 nm) emission filters with a CytoFluor 2350 fluorescence microplate reader. The fluorescence emission of the dye-nucleic acid complexes was then correlated linearly with cell numbers from a dilution series ranging from 100 to 50 × 104 cells, measured with a hemocytometer.
Sample cells were lysed at room temperature with 1 ml of CyQUANT GR dye with lysis buffer and incubated in darkness for 2–5 min at room temperature. Six-well culture plates of cells were harvested on days 1–9 and lysed with 200 μl of CyQUANT GR dye with lysis buffer. Sample fluorescence was measured, and growth curves were plotted as cell number over time in culture. For each independent control (WT and HDF), we used an unpaired t test to compare cultured cell numbers with the patient (p.Gln683X) sample at each time point.
Wound-Healing Assay
Wound-healing assays were performed with WT and p.Gln683X primary dermal fibroblast cells plated on fibronectin-coated 35 mm tissue culture dishes with an IBIDI chamber at a density of 8500 cells/well. Cells were serum- and growth-supplement-starved for 12 hr before removing the cells from half of each well with a sterile rubber policeman. Wounding was performed after a 12 hr incubation, cultures were washed twice with 1× PBS, and wound margins were photographed (t = 0 hr). The same wound margin fields were photographed at different time points, pictures were superimposed, and areas were measured with ImageJ software.
Results
Genome-wide Mapping and Identification of ARHGAP31 Mutations
Linkage analysis identified a locus for ACC-TTLD at 3q13.31-q13.33 on chromosome 3 (Figure S1). Subsequent refinement mapping defined a 5.53 Mb critical interval flanked by markers rs714697 and D3S4523 and containing a total of 21 protein-coding genes and three open-reading frames (Figure S2). We sequenced four genes in affected members of the linked AOS-5 and AOS-12 families. In each kindred we identified within the terminal coding exon of ARHGAP31 a distinct sequence variant (c.2047C>T and c.3260delA) that segregates with the syndrome phenotype and predicts the formation of premature truncating mutations (p.Gln683X and p.Lys1087SerfsX4) (Figures 1B and 1C). No likely disease-causing sequence variants were detected in the other genes analyzed. To comprehensively exclude the existence of a pathogenic mutation in the linked interval, we performed whole-exome sequencing in one individual from family AOS-12 and verified the c.2047C>T ARHGAP31 nonsense mutation as the only novel variant within the extended 13.2 Mb linkage region (Figure S2).
We screened ARHGAP31 by DNA sequencing in affected members from three other multiplex kindreds that are unlinked to the chromosome 3 locus and from a cohort of 43 sporadic individuals with features of ACC and TTLD, either alone or in combination. A nonsynonymous polymorphism (c.2180C>T, p.Thr727Ile) was detected in two sporadic cases, but no pathogenic sequence variants were identified in this extended cohort. To exclude the possibility that the truncating variants were also polymorphisms or that ARHGAP31 harbors frequent but functionally insignificant variation, we resequenced all 12 exons in 72 unrelated control individuals. None of these individuals carried either of the likely disease-causing ARHGAP31 mutations or any other missense or splice-site variants. We additionally sequenced exon 12, the site of the putative disease-causing mutations, in a further 1138 unrelated control subjects of European origin. Although the c.2180C>T polymorphism was detected in two control subjects, neither of the ACC-TTLD truncating mutations was detected in the combined total of over 2000 chromosomes assayed.
ARHGAP31 Expression during Early Development
Analysis of ARHGAP31 transcript expression showed abundant and ubiquitous levels in all human fetal tissues examined (Figure S3). To determine the regional expression of Arhgap31 mRNA, we studied mouse embryos during early development (Figure 2). At 9.5 dpc, the strongest expression is in the developing heart, with regional localization to the ventral walls of primitive ventricle and primitive atrium (Figures 2A and 2B). By 10.5 dpc, Arhgap31 expression becomes largely restricted to the lateral walls of the developing ventricle, and expression in the primitive atrium becomes localized to its outer wall (Figures 2C and 2D). At 11.5 dpc, Arhgap31 expression is largely restricted to the surface ectoderm, and strong expression overlies the entire heart field, symmetrical regions of the head and flank, and the apical regions of the hand and foot plates (Figures 2E and 2F). By 12.5 dpc, the expression in the surface ectoderm is not detectable by WISH (data not shown).
Figure 2.
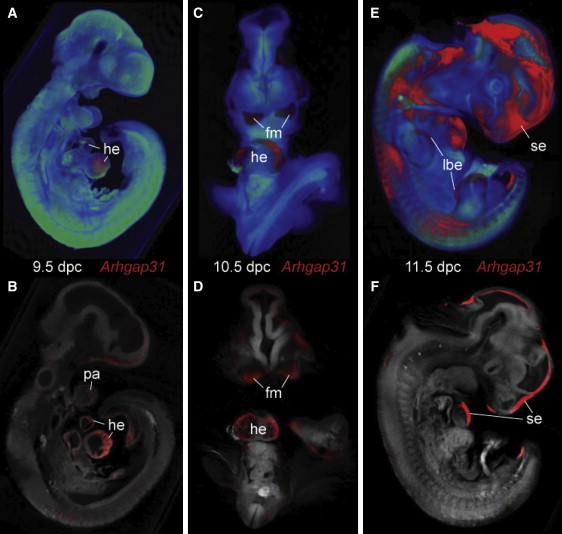
Expression of Arhgap31 during Mouse Embryogenesis
(A) Right lateral view of volume rendered OPT 3D representation of a 9.5 dpc mouse embryo showing Arhgap31 expression (in red) in developing heart (he).
(B) Digital section of same embryo as in (A) showing expression in ventral wall of the early ventricle and atrium of the heart and the first pharyngeal arch (pa).
(C) Frontal view of rendered and (D) digital coronal section of 10.5 dpc mouse embryo with expression in the lateral walls of the early ventricles of the heart and the first-pharyngeal-arch-derived facial mesenchyme (fm).
(E and F) By 11.5 dpc the expression of Arhgap31 is restricted to distinct regions of the surface ectoderm (se), including the upper and lower limb bud (lbe).
Effect of ARHGAP31 Mutations
To determine the impact of exon 12 premature termination mutations (Figure 3A), we assessed transcript stability by quantitative RT-PCR of RNA extracted from lymphoblasts. A comparison between WT control and two related subjects, both heterozygous for the c.2047C>T mutation, showed no reduction in the abundance of ARHGAP31 transcript, in support of the hypothesis that the mutant transcript is not removed by the process of nonsense-mediated decay (Figure 3B).
Figure 3.
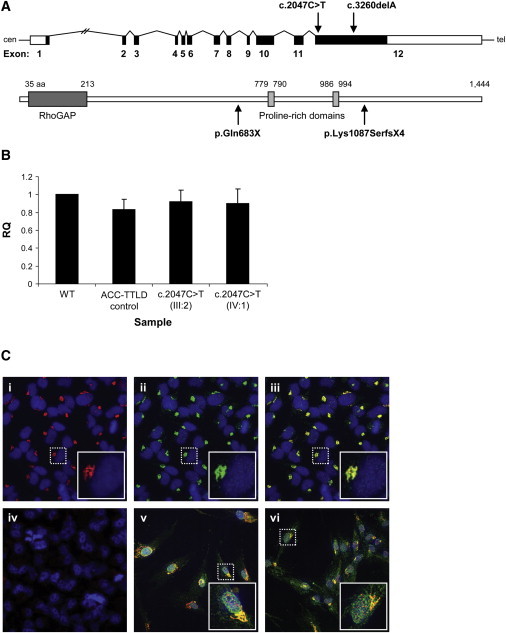
Transcript and Protein Expression in WT and Mutant Cells
(A) Schematic of the ARHGAP31 structure showing the position of the mutations identified in exon 12. The ARHGAP31 structure beneath depicts the known RhoGAP and proline-rich domains, a site of phosphorylation by GSK-3.
(B) Real-time quantitative RT-PCR is used for the examination of ARHGAP31 transcript levels in lymphoblasts from two related subjects heterozygous for the c.2047C>T nonsense mutation as compared to a genotypically normal control (WT). Patient and control samples show no appreciable difference in transcript expression. Sample identifiers refer to the pedigree structure in Figure 1B. The ACC-TTLD control is a patient with no ARHGAP31 mutation (molecular genetic basis unknown). Data represent mean ± standard deviation (SD) from three independent experiments. RQ is used as an abbreviation for relative quantification.
(C) Immunostaining of (i) endogenous ARHGAP31 (red) and (ii) Golgi (green); marked levels of colocalization to the Golgi apparatus in HeLa cells are visible (iii). The nucleus is stained in blue. (iv) The high specificity of the ARHGAP31 antibody is indicated by the absence of staining in the presence of blocking peptide to the binding epitope. In both (v) WT and (vi) mutant (p.Gln683X) fibroblasts, ARHGAP31 (green) localizes to the Golgi (red) and appears identical and of equivalent intensity. Images in the inset boxes show a 3× magnification of the single cells marked by the dashed lines.
Because the antibody to ARHGAP31 was unsuitable for protein detection by immunoblot analysis, we used immunofluorescence and found ARHGAP31 predominantly localized to the Golgi. We found no indication of protein degradation, such as loss of staining intensity or aggregate formation, and ARHGAP31 localization appeared normal in ACC-TTLD primary fibroblasts that harbor the p.Gln683X mutant protein (Figure 3C). However, subtle differences in localization or indeed organelle morphology, although not qualitatively evident, cannot be excluded.
ARHGAP31 is a member of the RhoGAP family of proteins known to inactivate the Rho GTPases Cdc42 and Rac1.4 Thus, we next investigated the impact of truncation of ARHGAP31 on GAP activity. We performed in vitro experiments in HEK293 cells and found that, relative to full-length ARHGAP31, both truncated proteins displayed a marked augmentation of GAP activity upon Cdc42, resulting in a significant downregulation of the active GTPase (Figure 4A). We conclude that both disease mutations in ARHGAP31 behave as dominant gain-of-function alleles.
Figure 4.
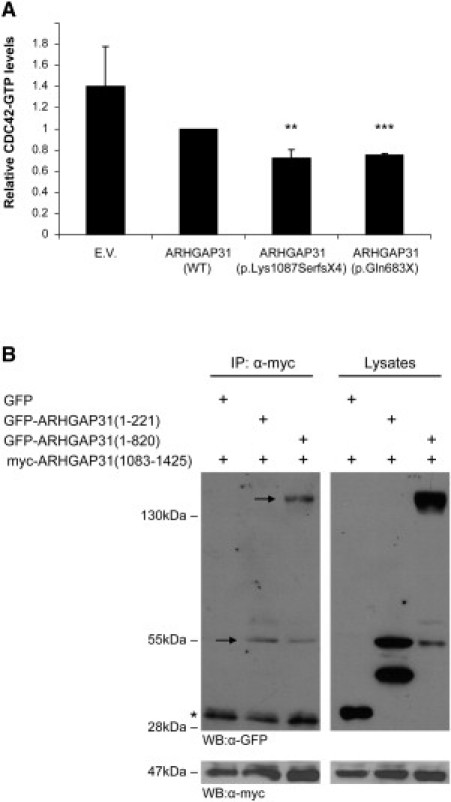
Analysis of GAP Activity in ARHGAP31 Truncations
(A) G-LISA assays measuring the relative amounts of Cdc42-GTP levels in HEK293 cells expressing Myc-tagged WT ARHGAP31 (full-length), p.Lys1087SerfsX4 or p.Gln683X. Relative Cdc42-GTP values are expressed as a ratio of Cdc42-GTP levels found in full-length ARHGAP31. Data are presented as mean ± SEM from four independent experiments. E.V. = empty vector; ∗∗p < 0.0002, ∗∗∗p < 0.00001.
(B) Immunoprecipitation of mouse ARHGAP31 deletion constructs were used to map the intramolecular interaction between C-terminal amino acids 1083-1425 and the proximal 221 residues harboring the RhoGAP domain. Full-length protein products are marked by the arrows (smaller bands represent degradation products; the asterisk [∗] indicates the IgG light chain). Levels of transfected proteins, assessed by immunoblotting of the lysates with Myc antibody, are displayed in the lower panel.
The ARHGAP31 mutations associated with the ACC-TTLD phenotype result in the removal of the C-terminal tail. To test the possibility that the C terminus of ARHGAP31 affects the GAP activity through intramolecular interactions, we generated green fluorescent protein (GFP)-tagged ARHGAP31 deletion constructs to perform protein immunoprecipitation studies in HEK293 cells with a Myc-tagged construct encoding the C terminus. Indeed, we found that the C terminus of ARHGAP31 is able to interact with the N-terminal region (amino acids 1–820), and we further refined the interaction to a region comprising the RhoGAP domain (amino acids 1–221). Although our data do not exclude the possibility that the C terminus might also bind a second motif downstream of the RhoGAP domain, these results indicate the potential for an autoregulation mechanism (Figure 4B).
In Vitro Phenotypic Analysis of Mutant ARHGAP31
Perturbation of Cdc42 and/or Rac1 signaling pathways impacts directly upon cell proliferation and migration in a cell-specific manner.12,13 Therefore, we hypothesized that the ACC-TTLD defects would have an impact upon cell proliferation and ordered cell migration. Following p.Gln683X fibroblasts over a 9 day period revealed a significantly reduced rate of cell proliferation (Figure 5A). In addition, we performed wound-healing assays with the mutant fibroblasts, which showed significant differences in cell migration, suggestive of altered cell motility (Figures 5B and 5C). The rounding of cells is a characteristic of impairment of the ordered process of actin polymerization and associated to a specific mode of cell movement during tumor cell migration.13 Transient transfection of disease-causing ARHGAP31 constructs induced a rounded phenotype in a significant proportion of HeLa cells, in keeping with recent observations of suppressed Cdc42 activity (Figures 5D and 5E).14 However, cytoskeletal organization, as assessed by F-actin staining of human fibroblasts heterozygous for the p.Gln683X mutation, was not qualitatively distinct from controls, presumably because of dosage compensation by the WT allele (data not shown).
Figure 5.
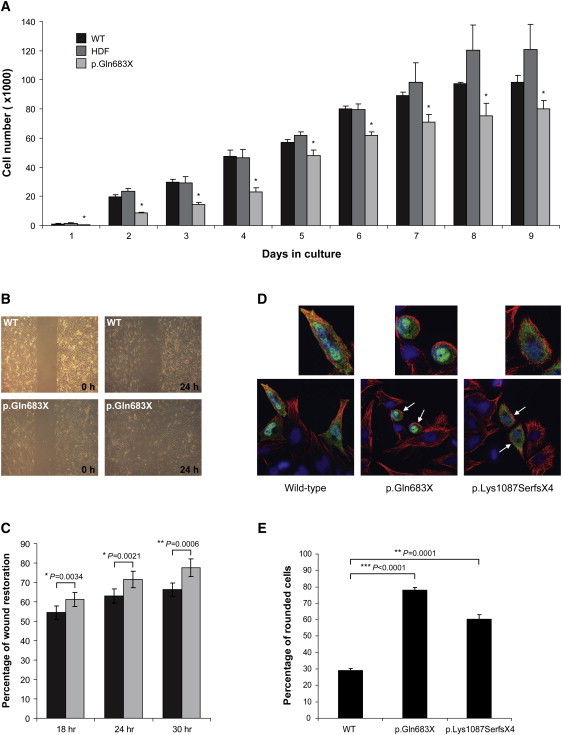
Functional Characterization of ARHGAP31 Mutations
(A) Bar chart comparing the proliferative activity of p.Gln683X primary dermal fibroblasts (light gray bars) with two distinct control fibroblast cell lines (black and dark gray bars). Data represent mean ± SEM for three independent experiments. Statistical analysis of each time point (days 1–9) revealed a significant decrease in the proliferative ability of cells carrying the p.Gln683X mutation when compared independently to each of the two unaffected controls (∗p < 0.01). The abbreviation WT indicates primary dermal fibroblasts from a tissue biopsy; HDF is used as an abbreviation for human dermal neonatal fibroblasts.
(B) Wound-healing migration assay showing coverage of a cell-free gap by primary dermal fibroblasts heterozygous for the p.Gln683X mutation and WT control fibroblasts at 24 hr after wounding.
(C) Plot showing percentage of wound restoration at 18, 24, and 30 hr after wounding. Fibroblasts heterozygous for the p.Gln683X mutation (light gray bars) migrate at a significantly faster rate than similar WT control fibroblasts (black bars). Data show mean ± SEM from three independent experiments.
(D) HeLa cells transiently transfected with Myc-tagged WT ARHGAP31, p.Gln683X, and p.Lys1087SerfsX4 constructs. Cell shape was visualized by confocal microscopy for tubulin (red) and transfected cells identified by costaining with fluorescent conjugated Myc antibody (green). DAPI was used to stain the cell nuclei (blue). Rounded cells are indicated by the white arrows and 2× higher magnifications of individual cells are shown above.
(E) Bar chart showing the mean percentage of rounded HeLa cells observed for each ARHGAP31 construct from three independent transfection experiments (error bars denote SD).
Discussion
Fundamental insight into the signaling pathways that are necessary for normal limb patterning and outgrowth has been gained primarily from the study of model organisms and inherited disorders of limb formation.15,16 The fibroblast growth factor (FGF), bone morphogenetic protein (BMP), hedgehog, and Wnt protein families have all been implicated in this important paradigm of organogenesis. Spatial and temporal expression of these signaling molecules is critical. For example, in the apical ectodermal ridge (AER), a specialized epithelium located along the distal tip of the limb bud, molecular signals generated by several members of the Fgf family control limb outgrowth and proximal-distal patterning.17 Wnt signals interact with FGFs in the AER to maintain mesenchymal progenitors in an undifferentiated, proliferative state.18 By contrast, the expression of BMP ligands regulates dorsal-ventral patterning and interdigital cell death19 and inhibits sonic hedgehog transcription through disruption of FGF and Wnt signaling.20 Further delineating the crosstalk and interaction between such genes and pathways is required for an integrative model of limb organogenesis; however, additional critical steps remain to be elucidated.15
In this study, we have used a classical positional cloning approach in conjunction with the contemporary technology of exome sequencing21 to identify distinct truncating mutations within the terminal exon of ARHGAP31. Combining candidate gene analysis with large-scale exome sequencing now provides an opportunity for rapid detection of genes in Mendelian disease. Importantly, we have shown that this strategy can be successful with exome data from a single affected individual and have been able to verify truncating mutations of ARHGAP31 as the only novel variation within our extended linkage interval.
The clinical phenotypes in the two kindreds with ARHGAP31 mutations share a number of features. Both mutations are associated with scalp aplasia cutis congenita and upper and/or lower limb defects of significant variability and reduced penetrance, including short distal phalanges, partial absence of the fingers and toes, and cutaneous syndactyly of toes 2 and 3 (Figures 1B and 1C). Using whole-mount in situ hybridization and optical projection tomography in mouse embryos, we have detected Arhgap31 transcript expression in distinct regions of the surface ectoderm, including the head and upper and lower limb buds, at 11.5 dpc. Expression in the distal tip of the limb buds during late stages of embryonic development would be consistent with a role in limb outgrowth and proximal-distal patterning. Interestingly, and despite evident expression of Arhgap31 in the developing mouse heart, no affected subjects displayed evidence of congenital cardiac abnormalities, which is a widely recognized feature of the ACC-TTLD spectrum. To address this further, we screened ARHGAP31 in an extended panel of ACC-TTLD patients with and without cardiac abnormalities. No additional mutations were identified, indicating that defects in ARHGAP31 account for only a small proportion of subjects with the ACC-TTLD spectrum of clinical features. Future studies will probably identify additional gene defects causative of AOS.
Our data suggest the c.2047C>T nonsense mutation does not activate the nonsense-mediated decay pathway, in keeping with premature termination codons downstream of the final splice junction.22 Immunofluorescence studies identified the expression and localization of mutant protein to be confined to the Golgi apparatus, a site of activity of Cdc42, at levels comparable to those observed for WT ARHGAP31.23 The Rho family members Cdc42 and Rac1 are active when GTP bound. The hydrolysis of GTP, for example by ARHGAP31, leads to inactivation of Cdc42 and Rac1 and, as such, intracellular Cdc42- and Rac1-GTP levels are inversely proportional to the activity of ARHGAP31. Measurements of GAP activity, as determined by G-LISA assays, demonstrated that both truncating mutations result in a significant downregulation of active Cdc42, compatible with a dominant gain-of-function mechanism of these disease-causing alleles.
Both the ARHGAP31 mutations associated with the ACC-TTLD phenotype are predicted to truncate the C-terminal tail. We postulated that the C terminus of ARHGAP31 is capable of interacting with the amino terminus so as to shield the RhoGAP domain, consistent with comparable autoregulatory mechanisms reported for other members of GTPase pathways, for example p50RhoGAP, N-chimaerin, and the downstream signaling effectors WASP and PAK1.24–27 In this study, we demonstrate an interaction between the C terminus of ARHGAP31 and the N-terminal RhoGAP domain, suggesting a model in which truncation of the ARHGAP31 C-terminal domain in mutant proteins would result in the exposure of a constitutively active RhoGAP catalytic site (Figure 6A).
Figure 6.
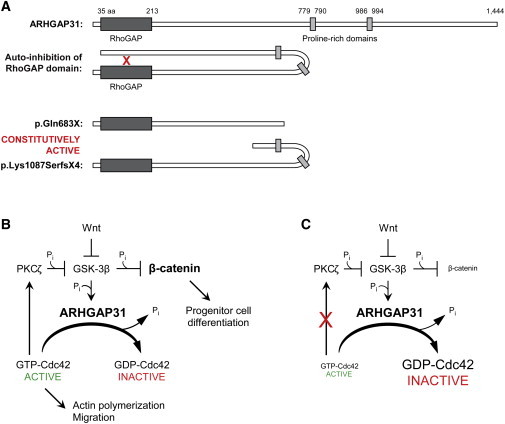
Schematic of Disrupted ARHGAP31 Signaling
(A) Schematic representing the putative mechanism of disease. The C terminus of ARHGAP31 inhibits the activity of the RhoGAP domain by specific interaction with amino acids 1–221 (red “X”). Truncated mutant proteins lacking the C terminus would therefore be incapable of autoinhibition, and this would result in a constitutively active RhoGAP domain.
(B) Schematic of the normal ARHGAP31 signaling system. ARHGAP31 cycles Cdc42 from an active to an inactive form by hydrolysis of GTP to GDP. GSK-3β upregulates ARHGAP31 levels, probably through phosphorylation at a consensus ERK1 site. Activated Cdc42 promotes actin polymerization and cellular processes, including migration, and acts to inhibit GSK-3β activity by stimulating PKCζ phosphorylation of GSK-3β. Wnt signaling is an additional negative regulator of GSK-3β activity. Downregulation of GSK-3β leads to decreased proteasomal degradation of cytosolic β-catenin. Active β-catenin translocates to the nucleus, whereupon the engagement of transcriptional cofactors controls the differentiation of progenitor cells in the skin.
(C) In ACC-TTLD mutant cells, constitutive activation of ARHGAP31 leads to an imbalance between active and inactive Cdc42. A decrease in the levels of active GTP-bound Cdc42 results in reduced activation of PKCζ with a concomitant increase in β-catenin degradation and disruption of cellular processes.
The following abbreviations are used: PKCζ, protein kinase C; GSK-3β, glycogen synthase kinase 3 beta; Pi, inorganic phosphate.
Perturbation of Cdc42 and/or Rac1 signaling impacts upon directed migration, proliferation, and differentiation in a cell-specific manner.12 Constitutive inactivation of Cdc42 by GTPase inhibitors, for example VopS, leads to cell rounding because of disruption of the actin cytoskeleton.14 Furthermore, low Rac1 activity is associated with a rounded mode of cell movement.28 In this report, we demonstrate a very similar outcome for cell morphology upon transient overexpression of ARHGAP31 mutant proteins and a significant disruption of cell migration in fibroblast mutant cells, pointing toward the unregulated suppression of Cdc42/Rac1 function. Importantly, ARHGAP31 has recently emerged as a regulator of Cdc42 and Rac1 signaling to the cytoskeleton and thereby plays a key role in controlling the temporal and spatial cytoskeletal remodeling necessary for the precise control of cell morphology and migration.29 In addition, its GAP activity is regulated in an adhesion-dependent manner and appears to be required for normal cell spreading, polarized lamellipodia formation, and cell migration.29 Although the wound-healing assays performed in this study do not measure the direction of cell movement, it is feasible that the increased GAP activity in ARHGAP31 mutant fibroblasts leads to a loss of adhesion and/or structural defects in cell protrusion formation, regulated by Cdc42 and Rac1,30 thus resulting in disorganized cell migration. A more comprehensive disease-cell-based study aimed in particular at examining cytoskeletal dysfunction is now required to build on these early observations and further define the mechanisms driving pathogenesis in this disorder.
Further insight into the wider ARHGAP31 signaling pathway is provided by a keratinocyte-specific Cdc42 knockout mouse model, which offers an integrated model for the molecular basis of the AOS phenotype.31 Mutant mice display cellular abnormalities of skin morphogenesis, phenotypically illustrated by progressive loss of hair follicles. Central to this process is the stabilization of β-catenin by Wnt and Cdc42 signaling, which together inhibit the activity of glycogen synthase kinase 3 beta (GSK-3β), a critical driver of β-catenin degradation.32 By contrast, ARHGAP31 levels are augmented by GSK-3β activity.33 We would suggest that in ACC-TTLD cells the inactivation of Cdc42 by constitutively active mutant ARHGAP31 would compromise this critical negative feedback loop and result in upregulation of mutant ARHGAP31 by GSK-3β and concomitant destabilization of β-catenin with consequent impairment of cellular processes, in particular progenitor cell differentiation, requisite for skin layer and hair follicle production (Figures 6B and 6C). In addition to this, Rac1 activity has recently been implicated in nuclear localization of β-catenin during canonical Wnt signaling,34 and keratinocyte-restricted deletion has been identified as having a critical role in hair follicle integrity.35 More importantly, genetic removal of Rac1 in the mouse embryonic limb bud ectoderm disrupts Wnt signaling and results in severe truncations of the limb. Furthermore, conditional deletion of Rac1 in the mouse limb bud mesenchyme also leads to skeletal deformities, including abnormal fusion of the skull, developmental limb defects, and syndactyly.36 These reports, combined with our ARHGAP31 study, demonstrate the crucial roles of Cdc42 and Rac1 in skin morphogenesis and limb development. Clearly, additional work, informed by our genetic findings, will now be required to further elucidate these early mechanistic insights and confirm the molecular processes proposed within this model.
Taken together, our findings demonstrate that heterozygous gain-of-function mutations in ARHGAP31 cause an autosomal-dominant form of ACC-TTLD through introduction of premature termination codons in the terminal exon of the gene. Expression of Arhgap31 during development appears confined to the limb buds, cranium, and early cardiac structures and provide a remarkable correlation with the developmental defects that define ACC-TTLD. This report generates insight into the critical pathways regulating the processes of cell proliferation and movement in vivo, and the consequences for human health with dysregulation of skin and limb formation.
Acknowledgments
The authors thank the participating families; the clinicians who recruited patients to the European AOS Consortium (L. Al-Gazali, D. Amor, F. Brancati, E. Craft, B. Dallapiccola, S. Davies, C. Deshpande, J. Dixon, S. Holden, J. Hurst, P. Itin, E. Jacquemin, D. Johnson, E. Kinning, Y. Lacassie, W. Lam, A. Lampe, P. Lapunzina, M. Maniscalco, V. McConnell, L. McGregor, V. Meiner, J. Nelson, K. Orstavik, J. Paprocka, M. Patel, S. Price, J. Prothero, E. Seemanova, M. Tekin, B. Tuysuz, A. Vandersteen, and M. Whiteford.); and A. Ridley, who critically read the manuscript. This work was supported by grants from the British Heart Foundation (BHF) to R.C.T. (RG/08/006/25302), Wellcome Trust to E.R.M. and R.C.T. (078751/Z/05/Z), European Union Framework 6 award for PULMOTENSION (LSHM-CT-2005-018725), Canadian Institute of Health Research to N.L-V. (MOP-84449), and German Research Foundation to M.Z. (ZE 524/2-2). The authors acknowledge use of BRC Core Facilities provided by the financial support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London and King's College Hospital NHS Foundation Trust. R.C.T. is a senior investigator at the NIHR. R.D.M. is a BHF Intermediate Research Fellow (FS/07/036). P.B. and M.F. are MRC career development fellows funded by the National Institute of Dental and Craniofacial Research (National Institutes of Health) Craniofacial Center Grant (P50 DE-16215-05). D.R.F. is a MRC senior clinician scientist. N.L-V. is a Fonds de la Recherche en Santé du Quebec chercheur-boursier senior.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://www.1000genomes.org
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Novoalign, http://www.novocraft.com
ImageJ, http://rsbweb.nih.gov/ij
References
- 1.Tcherkezian J., Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol. Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 2.Vega F.M., Ridley A.J. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 3.Snape K.M., Ruddy D., Zenker M., Wuyts W., Whiteford M., Johnson D., Lam W., Trembath R.C. The spectra of clinical phenotypes in aplasia cutis congenita and terminal transverse limb defects. Am. J. Med. Genet. A. 2009;149A:1860–1881. doi: 10.1002/ajmg.a.32708. [DOI] [PubMed] [Google Scholar]
- 4.Tcherkezian J., Triki I., Stenne R., Danek E.I., Lamarche-Vane N. The human orthologue of CdGAP is a phosphoprotein and a GTPase-activating protein for Cdc42 and Rac1 but not RhoA. Biol. Cell. 2006;98:445–456. doi: 10.1042/BC20050101. [DOI] [PubMed] [Google Scholar]
- 5.Verdyck P., Blaumeiser B., Holder-Espinasse M., Van Hul W., Wuyts W. Adams-Oliver syndrome: Clinical description of a four-generation family and exclusion of five candidate genes. Clin. Genet. 2006;69:86–92. doi: 10.1111/j.1399-0004.2006.00552.x. [DOI] [PubMed] [Google Scholar]
- 6.Bonafede R.P., Beighton P. Autosomal dominant inheritance of scalp defects with ectrodactyly. Am. J. Med. Genet. 1979;3:35–41. doi: 10.1002/ajmg.1320030109. [DOI] [PubMed] [Google Scholar]
- 7.Rozen S., Skaletsky H.J. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S., Misener S., editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 8.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson M.A., Irving M.D., Asilmaz E., Gray M.J., Dafou D., Elmslie F.V., Mansour S., Holder S.E., Brain C.E., Burton B.K. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat. Genet. 2011;43:303–305. doi: 10.1038/ng.779. [DOI] [PubMed] [Google Scholar]
- 10.Li K., Stockwell T.B. VariantClassifier: A hierarchical variant classifier for annotated genomes. BMC Res Notes. 2010;3:191. doi: 10.1186/1756-0500-3-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharpe J., Ahlgren U., Perry P., Hill B., Ross A., Hecksher-Sørensen J., Baldock R., Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- 12.Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 13.Sanz-Moreno V., Marshall C.J. The plasticity of cytoskeletal dynamics underlying neoplastic cell migration. Curr. Opin. Cell Biol. 2010;22:690–696. doi: 10.1016/j.ceb.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Yarbrough M.L., Li Y., Kinch L.N., Grishin N.V., Ball H.L., Orth K. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science. 2009;323:269–272. doi: 10.1126/science.1166382. [DOI] [PubMed] [Google Scholar]
- 15.Zeller R., López-Ríos J., Zuniga A. Vertebrate limb bud development: Moving towards integrative analysis of organogenesis. Nat. Rev. Genet. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- 16.Stricker S., Mundlos S. Mechanisms of digit formation: Human malformation syndromes tell the story. Dev. Dyn. 2011;240:990–1004. doi: 10.1002/dvdy.22565. [DOI] [PubMed] [Google Scholar]
- 17.Mariani F.V., Ahn C.P., Martin G.R. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature. 2008;453:401–405. doi: 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ten Berge D., Brugmann S.A., Helms J.A., Nusse R. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development. 2008;135:3247–3257. doi: 10.1242/dev.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maatouk D.M., Choi K.S., Bouldin C.M., Harfe B.D. In the limb AER Bmp2 and Bmp4 are required for dorsal-ventral patterning and interdigital cell death but not limb outgrowth. Dev. Biol. 2009;327:516–523. doi: 10.1016/j.ydbio.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Bastida M.F., Sheth R., Ros M.A. A BMP-Shh negative-feedback loop restricts Shh expression during limb development. Development. 2009;136:3779–3789. doi: 10.1242/dev.036418. [DOI] [PubMed] [Google Scholar]
- 21.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hentze M.W., Kulozik A.E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 23.Michaelson D., Silletti J., Murphy G., D'Eustachio P., Rush M., Philips M.R. Differential localization of Rho GTPases in live cells: Regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 2001;152:111–126. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moskwa P., Paclet M.H., Dagher M.C., Ligeti E. Autoinhibition of p50 Rho GTPase-activating protein (GAP) is released by prenylated small GTPases. J. Biol. Chem. 2005;280:6716–6720. doi: 10.1074/jbc.M412563200. [DOI] [PubMed] [Google Scholar]
- 25.Colón-González F., Leskow F.C., Kazanietz M.G. Identification of an autoinhibitory mechanism that restricts C1 domain-mediated activation of the Rac-GAP alpha2-chimaerin. J. Biol. Chem. 2008;283:35247–35257. doi: 10.1074/jbc.M806264200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei M., Lu W., Meng W., Parrini M.C., Eck M.J., Mayer B.J., Harrison S.C. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- 27.Devriendt K., Kim A.S., Mathijs G., Frints S.G., Schwartz M., Van Den Oord J.J., Verhoef G.E., Boogaerts M.A., Fryns J.P., You D. Constitutively activating mutation in WASP causes X-linked severe congenital neutropenia. Nat. Genet. 2001;27:313–317. doi: 10.1038/85886. [DOI] [PubMed] [Google Scholar]
- 28.Sanz-Moreno V., Gadea G., Ahn J., Paterson H., Marra P., Pinner S., Sahai E., Marshall C.J. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 29.LaLonde D.P., Grubinger M., Lamarche-Vane N., Turner C.E. CdGAP associates with actopaxin to regulate integrin-dependent changes in cell morphology and motility. Curr. Biol. 2006;16:1375–1385. doi: 10.1016/j.cub.2006.05.057. [DOI] [PubMed] [Google Scholar]
- 30.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 31.Wu X., Quondamatteo F., Lefever T., Czuchra A., Meyer H., Chrostek A., Paus R., Langbein L., Brakebusch C. Cdc42 controls progenitor cell differentiation and beta-catenin turnover in skin. Genes Dev. 2006;20:571–585. doi: 10.1101/gad.361406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willert K., Shibamoto S., Nusse R. Wnt-induced dephosphorylation of axin releases beta-catenin from the axin complex. Genes Dev. 1999;13:1768–1773. doi: 10.1101/gad.13.14.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danek E.I., Tcherkezian J., Triki I., Meriane M., Lamarche-Vane N. Glycogen synthase kinase-3 phosphorylates CdGAP at a consensus ERK 1 regulatory site. J. Biol. Chem. 2007;282:3624–3631. doi: 10.1074/jbc.M610073200. [DOI] [PubMed] [Google Scholar]
- 34.Wu X., Tu X., Joeng K.S., Hilton M.J., Williams D.A., Long F. Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell. 2008;133:340–353. doi: 10.1016/j.cell.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chrostek A., Wu X., Quondamatteo F., Hu R., Sanecka A., Niemann C., Langbein L., Haase I., Brakebusch C. Rac1 is crucial for hair follicle integrity but is not essential for maintenance of the epidermis. Mol. Cell. Biol. 2006;26:6957–6970. doi: 10.1128/MCB.00075-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki D., Yamada A., Amano T., Yasuhara R., Kimura A., Sakahara M., Tsumaki N., Takeda S., Tamura M., Nakamura M. Essential mesenchymal role of small GTPase Rac1 in interdigital programmed cell death during limb development. Dev. Biol. 2009;335:396–406. doi: 10.1016/j.ydbio.2009.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


