Abstract
Plant hormones regulate many aspects of plant growth and development. Both auxin and cytokinin have been known for a long time to act either synergistically or antagonistically to control several significant developmental processes, such as the formation and maintenance of meristem. Over the past few years, exciting progress has been made to reveal the molecular mechanisms underlying the auxin–cytokinin action and interaction. In this review, we shall briefly discuss the major progress made in auxin and cytokinin biosynthesis, auxin transport, and auxin and cytokinin signaling. The frameworks for the complicated interaction of these two hormones in the control of shoot apical meristem and root apical meristem formation as well as their roles in in vitro organ regeneration are the major focus of this review.
Keywords: Auxin, cytokinin, interaction, shoot meristem, root meristem, development
INTRODUCTION
Auxin and cytokinin play fairly important roles in many aspects of plant growth and development. The interaction between auxin and cytokinin is particularly important to control a few developmental processes, such as the formation and maintenance of meristems that are essential to establish the whole plant body. For example, the shoot meristems give rise to the above-ground parts of a plant, whereas the root meristems produce the below-ground parts. Many recent studies have provided important information for the understanding of the molecular mechanisms of auxin–cytokinin interaction in the regulation of meristem development.
Maintenance of the cellular optimum auxin concentration can be controlled at multiple levels, such as biosynthesis, transport, perception, and signaling. These multiple regulation pathways contribute to the differential auxin distribution within tissues at different developmental stages. Thus far, one tryptophan (trp)-independent pathway and four trp-dependent pathways for the biosynthesis of auxin/IAA have been proposed in Arabidopsis (Zhao, 2010). These four trp-dependent pathways include the indole-3-acetamide (IAM) pathway, the indole-3-acetaldoxime (IAQx) pathway, the tryptamine (TAM) pathway, and the indole-3-pyruvic acid (IPA) pathway. Two of these pathways—the TAM pathway, considered to be rate-limited through the YUCCA family, and the IPA pathway—have also been highlighted (Vanneste and Friml, 2009). Auxin polar transport is required to direct auxin flows and to form auxin gradients in plants, which are critical for developmental pattern formation. In Arabidopsis, three protein families are required to mediate auxin transport between cells: auxin efflux PINFORMD (PIN) proteins, MULTIDRUG RESISTANCE (MDR)-p-glycoprotein (PGP) proteins, and auxin influx AUXIN RESISTANT 1 (AUX1)/LIKE AUX1 (LAX) proteins (Benjamins and Scheres, 2008; Gao et al., 2008; Zazímalová et al., 2010).
Besides the biosynthesis and transport of auxin, auxin signaling through receptors and downstream signaling components has also been suggested to be the regulating mechanism for many developmental processes. One of the important auxin receptors in Arabidopsis has been identified as the TRANSPORT INHIBITOR REPONSE 1 (TIR1) protein. TIR1 protein is an F-box protein, a component of an SCFTIR1 ubiquitination E3 complex. This E3 complex is involved in proteasome-mediated protein degradation (Ruegger et al., 1998; Gray et al., 2001; Quint and Gray, 2006). Analysis of quadruple tir1-related mutants highlights the role of TIR1 and at least three other TIR1-related AUXIN BINDING F-BOX PROTEINs (AFB1-3) in the auxin signaling for plant development (Dharmasiri et al., 2005). Thus far, two classes of transcriptional regulators represent the core of auxin signaling: the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) proteins and AUXIN RESPONSE FACTOR (ARF) proteins (Liscum and Reed, 2002; Quint and Gray, 2006). Aux/IAA proteins are known as a family of transcriptional repressors to negatively regulate auxin signaling (Ulmasov et al., 1997b). Recent studies have provided more extensive evidence that Aux/IAA proteins are the targets of SCFTIR1 complex (Paciorek and Friml, 2006; Benjamins and Scheres, 2008; Vanneste and Friml, 2009). Aux/IAAs interact with ARF proteins, a class of transcription factors that mediate auxin-dependent transcriptional regulation (Paciorek and Friml, 2006; Ulmasov et al., 1997b). The ARFs could function as either activators or repressors in the regulation of auxin-induced gene expression (Ulmasov et al., 1997a, 1999).
Like auxin, cytokinin is also a key regulator for various aspects of plant growth and development. Cytokinin homeostasis is spatially and temporally regulated by a fine balance between synthesis and catabolism. The first enzyme identified in the Arabidopsis cytokinin biosynthetic pathway is adenosine phosphate-isopentenyltransferases (IPTs). The IPTs are believed to catalyze the transfer of an isopentenyl group from dimethylallyl diphosphate to an adenine nucleotide (ATP, ADP, or AMP) (Kakimoto, 2001; Takei et al., 2001). Another landmark is the identification of two cytochrome P450 monooxygenases, CYP735A1 and CYP735A2, which catalyze the hydroxylation at the prenyl side chain of the iP-nucleotides to synthesize tZ-nucleotides (Takei et al., 2004). Furthermore, a cytokinin-activating enzyme in rice, LONELY GUY (LOG), has been recently identified to catalyze the last step of cytokinin biosynthesis. This step is involved in converting cytokinin-nucleotides produced by IPTs and CYP735As to the free-base form (Kurakawa et al., 2007). Besides biosynthesis, cytokinin homeostasis is also controlled by its catabolism process through CYTOKININ OXIDASE/DEHYDROGENASEs (CKXs) (Werner et al., 2003).
In contrast to auxin, cytokinin is perceived in plants through a multi-step phosphorelay pathway similar to the bacterial two-component signaling system (Kakimoto, 2003; To and Kieber, 2008). In Arabidopsis, three transmembrane histidine kinases have been identified as cytokinin receptors, the ARABIDOPSIS HIS KINASE 2 (AHK2), AHK3 and CYTOKININ RESPONSE1 (CRE1)/AHK4 (Hwang and Sheen, 2001; Inoue et al., 2001; Riefler et al., 2006; To and Kieber, 2008). Recent analyses on these three receptors have revealed a largely overlap expression pattern and partially redundant functions in cytokinin perception (Higuchi et al., 2004; Nishimura et al., 2004). Following the initial cytokinin perception, AHKs auto-phosphorylate themselves and transfer the phosphate group to members of the ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEINs (AHPs) family. AHPs subsequently translocate to the nucleus to phosphorylate the ARABIDOPSIS RESPONSE REGULATORs (ARRs) proteins of either type-A or type-B (Heyl and Schmülling, 2003; Kakimoto, 2003; Ferreira and Kieber, 2005). Based on the analysis of loss- and gain-of-function mutants, at least six of 11 type-B ARRs (ARR1, ARR2, ARR10–ARR12, and ARR18) have overlapping functions in the positive regulation of the cytokinin response (Mason et al., 2005; Yokoyama et al., 2007). Furthermore, the arr1, arr10, arr12 triple mutants show a strong reduction in cytokinin induction of multiple type-A ARR transcripts (Mason et al., 2005). Mutation analysis has indicated that at least eight of the 10 type-A ARRs are negative regulators of cytokinin signaling again with overlapping functions (To et al., 2004; To and Kieber, 2008).
For the past few years, genetic and molecular evidence has revealed the interaction between auxin and cytokinin during plant development (Dettmer et al., 2009; Růžičkaa et al., 2009; Wolters and Jürgens, 2009; Zhao et al., 2010). Many recent biochemical and genetic investigations have further confirmed that the intricate cross-talk and integration of hormone signaling are required for differentiation and maintenance of plant meristems. Hereafter, we shall focus our discussion on the interaction between auxin and cytokinin in the regulation of meristem development.
AUXIN AND CYTOKININ REGULATE MERISTEM FORMATION IN EARLY EMBRYOGENESIS
Auxin and cytokinin control events of major cell specification during embryogenesis (Müller and Sheen, 2008; Möller and Weijers, 2009). The first step of embryonic patterning is the establishment of the apical–basal axis, in which asymmetric distribution of auxin mediated by PIN proteins plays a major role. A zygote undergoes an asymmetric division to produce a smaller apical cell and a larger basal cell. The apical cell will generate the pro-embryo, while the basal cell will give rise to the suspensor (Mansfield and Briarty, 1991; Laux and Jürgens, 1997). At this two-cell embryo stage, PIN7 is expressed in the basal cell to transport auxin to the apical cell (Friml et al., 2003). After two more rounds of cell division, PIN7 localizes to the apical membrane of suspensor cells, resulting in the accumulation of auxin in the whole pro-embryos (Friml et al., 2003; Jenik and Barton, 2005). At the 16-cell globular stage, WUSCHEL (WUS) is switched on in the four inner cells of the pro-embryo, and it is an early molecular marker that represents initiation of the shoot apical meristem (SAM) in the embryo (Weigel and Jürgens, 2002). This WUS induction might be related to auxin accumulation. However, PIN7 polarity is reversed at the 32-cell stage, resulting in the transport of auxin towards the suspensor cells (Friml et al., 2003; Jenik et al., 2007). The transported auxin accumulates in the uppermost cell of the suspensor to form the hypophysis, the founder of the stem-cell niche of the embryonic root (Friml et al., 2003). At a later transition stage of the embryo, auxin is directed towards the center of the cotyledon primordia in the apical domain to establish the cotyledons. This early heart-stage embryo shows a cleft where the SAM will form (Weigel and Jürgens, 2002). Therefore, auxin transport is critical for the maintenance of the polar axis and the formation of two types of meristems in the embryo.
MONOPTEROS (MP)/ARF5 and BODENLOS (BDL)/IAA12 are central to auxin response during embryogenesis in Arabidopsis (Hamann et al., 2002; Weijers et al., 2006). MP has been proposed to mediate the establishment of the embryonic axial pattern by modulating TARGET OF MP 5 (TMO5) and TMO7 functions in response to auxin signals (Schlereth et al., 2010). Loss-of-function of MP or gain-of-function of BDL causes the aberrant specification of the apical cell, and prevented the formation of the embryonic root (Weijers et al., 2006). MP promotes the expression of PIN1 in pro-vascular cells of the globular embryos, leading to auxin accumulation at the basal pole of pro-embryos (Wolters and Jürgens, 2009). Embryos of the mp mutants are abnormal at their early globular stages. In addition, heart-stage mp embryos lack the central pro-vascular cylinder. As a result, no hypocotyl and primary root meristems are formed in mp mutants (Berleth and Jürgens, 1993). The gain-of-function bdl mutants show milder defects than mp mutants and have a reduced vascular system and a hypocotyl of variable length without primary root meristem (Hamann et al., 2002; Mattsson et al., 2003). Therefore, the primary auxin response mediated by MP and BDL is essential for root meristem initiation. MP and BDL are also known factors for activating quiescent centre (QC)-specific WUSCHEL RELATED HOMEOBOX5 (WOX5) and auxin-responsive PLETHORA (PLT) genes (Wolters and Jürgens, 2009). The double mutants wox8 wox9 displayed abnormal PIN1 expression pattern and abnormal auxin response in the embryo. WOX2 and WOX8 act redundantly with MP to promote PIN1 expression and to regulate localized auxin gradients (Breuninger et al., 2008). Therefore, auxin transport and response are required to trigger the specification of the root meristem founder cell.
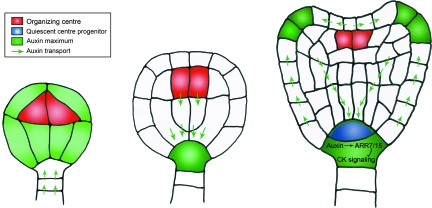
Even though auxin has been known for a long time to play a crucial role in specification of root stem-cell during embryogenesis, the function of cytokinin in early embryogenesis is recently suggested for a transient and antagonistic interaction between auxin and cytokinin (Müller and Sheen, 2008). Cytokinin signaling components are first detected in the hypophysis, the founder cell of the root meristem at the early globular stage of the embryo. After the first division, the apical daughter cell of the hypophysis remains to maintain the phophorelay activity of cytokinin signaling and is the precursor of the QC, whereas the basal daughter cell represses cytokinin signaling. Interestingly, in the basal cell of the hypophysis, auxin antagonizes cytokinin signaling by directly activating the repressors of cytokinin signaling, ARR7 and ARR15 (Figure 1) (Müller and Sheen, 2008). Thus, to sustain the activity of the embryonic root stem-cell niche, auxin mediates the suppression of cytokinin signaling in the basal cell of the hypophysis.
Figure 1.
Interaction of Auxin and Cytokinin in Embryogenesis.
At the 16-cell stage of pro-embryo, auxin is transported from suspensor cells to the apical pro-embryo and accumulates in whole pro-embryo, in which WUS is initially expressed in four inner cells of this early-globular embryo. In the late-globular embryo, directional auxin transport is reversed towards the suspensor cells, leading to the accumulation of auxin in the uppermost cell of the suspensor to form the hypophysis. By the transition stage, the hypophysis has undergone asymmetrical cell division to result in the formation of the upper small cell and the large basal daughter cell. The upper cell maintains phosphorelay activity of cytokinin signaling. In the large basal cell, auxin represses cytokinin signaling through an ARR7/15-dependent pathway. This antagonistic interaction between auxin and cytokinin in both cells controls the establishment of the embryonic root stem-cell niche.
CYTOKININ–AUXIN CROSS-TALK CONTROLS SHOOT MERISTEM DEVELOPMENT
The SAM arises during embryogenesis and generates almost all of the aerial parts of a plant. It can be subdivided into different regions, including the central zone, peripheral zone, and rib zone (Fletcher and Meyerowitz, 2000; Clark, 2001; Sablowski, 2007). The central zone is located in the center and at the summit of the meristem in which cells divide more slowly. It provides cells to both the peripheral zone and the rib zone. The surrounding peripheral zone has a higher cell division rate and gives rise to lateral organs. The rib zone is below the central zone from which the tissues of the stem are generated. Leaf primordia originate from a group of cells in the peripheral zone of the SAM, which is then replenished by cell division in both the peripheral zone itself and the central zone. Active cell division and cell differentiation occur in SAM (Clark, 2001; Williams and Fletcher, 2005; Reddy, 2008). Thus, balance between cell division and cell differentiation is necessary to control the size and structure of SAM. The size of SAM is maintained by a negative feedback loop involving WUS and CLAVATA3 (CLV3) in Arabidopsis (Brand et al., 2000; Schoof et al., 2000). WUS-expressing cells of the organizing center (OC) maintain the overlying cells as the stem cells within the central zone (Mayer et al., 1998; Schoof et al., 2000). SHOOTSTEMLESS (STM) is also required for the maintenance of stem cells in the meristem, and its expression in the whole shoot meristem prevents stem cells from switching to organ-specific cells (Clark et al., 1996; Endrizzi et al., 1996; Long et al., 1996).
It has been known for a long time that cytokinin plays a major role in regulating meristem function. Studies on the classical chemical regulation of growth and organ formation in in vitro plant tissues have indicated that excess of cytokinin over auxin promotes shoot formation from callus (Skoog and Miller, 1957). This suggests a positive action of cytokinin on SAM activity. Recent studies have shown that cytokinin deficiency reduced shoot meristem size and activity (Werner et al., 2003; Higuchi et al., 2004; Werner and Schmülling, 2009). Similarly, mutations in AtIPTs also cause reduction of the SAM size, further supporting the positive role of cytokinin in controlling shoot meristem activity (Miyawaki et al., 2006). These observations are consistent with the role of cytokinin in meristem development.
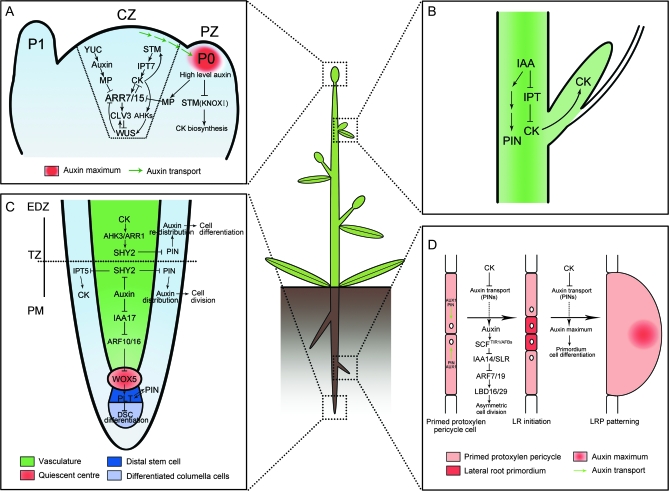
The mutual regulation of cytokinin and stem-cell-related genes can explain the regulation of cytokinin production and signaling in the SAM (Figure 2A). STM, a member of Class I KNOTTED-like homeobox (KNOXI) proteins, induces cytokinin biosynthesis through activating AtIPT7 in Arabidopsis (Jasinski et al., 2005; Yanai et al., 2005). Cytokinin triggers a rapid increase in mRNA levels of the KNOXI genes (Rupp et al., 1999), indicating a positive feedback loop between STM and cytokinin signaling (Figure 2A) (Wolters and Jürgens, 2009). KNOX transcription factors repress the biosynthesis of the growth regulator gibberellin (GA) to maintain normal meristem function. Cytokinin also stimulates the expression of genes involved in GA catabolism to reinforce the low-GA levels established by the KNOX proteins within the SAM (Jasinski et al., 2005; Wolters and Jürgens, 2009). Thus, KNOX proteins are essential for meristem development as the key growth-regulators by simultaneously activating CK and repressing GA biosynthesis. In addition, many other studies have indicated that type-A ARRs (ARR7/15), the main response genes of cytokinin signaling, are required for CLV3 expression (Zhao et al., 2010). Since CLV3 limits the expression of WUS, which, in turn, down-regulates the expression of ARR5, ARR7, and ARR15 (Leibfried et al., 2005), a negative regulation loop exists between type-A ARRs and WUS in the shoot apical meristem. Recent studies suggest that cytokinin signaling partially regulates WUS expression through this CLV-dependent pathway (Gordon et al., 2009). The induction of WUS transcripts in the clv mutants after cytokinin treatment reveals a CLV-independent mechanism of cytokinin-induced WUS expression. This investigation demonstrates that cytokinin-induced increase of WUS transcripts is mediated primarily through an AHK2/AHK4-dependent pathway (Figure 2A) (Gordon et al., 2009).
Figure 2.
Molecular Mechanisms of Auxin and Cytokinin Interaction in the Regulation of Plant Meristem Development.
(A) In CZ of shoot meristem, ARR7 and ARR15 act as integrative factors in auxin and cytokinin signaling pathways. Auxin represses the expression of ARR7 and ARR15 while cytokinin promotes their expression through a STM-dependent pathway. Both of them regulate the expression of WUS in a negative feedback loop, critical for stem-cell formation. During the formation of lateral organ primordia, a high level of auxin transported from CZ blocks the biosynthesis of cytokinin by suppressing KNOXI function in PZ. CZ, central zone; PZ, peripheral zone; P0/P1, organ primordia.
(B) Auxin is transported from the shoot apex to repress cytokinin biosynthesis, leading to the inhibition of axillary bud growth.
(C) In the root meristem, auxin promotes the expression of PINs through the degradation of SHY2 proteins, resulting in the maintenance of an auxin gradients and cell division. In contrast, cytokinin impedes the expression of PINs by stimulating the expression of SHY2, leading to auxin redistribution and cell differentiation. Auxin also plays an important role in the differentiation of root DSC by mediating the expression of WOX5 and PLT. PM, proximal meristem; EDZ, elongation differentiation zone; TZ, transition zone; DSC, distal stem cell.
(D) In certain xylempole pericycle cells, the transport and perception of auxin trigger an asymmetric cell division critical for the LR initiation and LRP patterning. By contrast, cytokinin negatively regulates the LR initiation and LRP patterning by inhibiting the expression of PINs and the auxin distribution gradients. LR, lateral roots; LRP, lateral root primordia.
Auxin also plays a critical role in the maintenance of shoot meristem. Auxin produced by YUCCA genes accumulates in the central zone of meristem at an optimal level to stimulate downstream auxin-induced genes through the Aux/IAA–ARF signaling pathway (Zhao, 2008). Interestingly, a dramatic increase in ARR7 and ARR15 expression was observed in the SAM of the yucca mutants, the pin1 mutants, the pinoid mutants, as well as plants treated with N-1-naphthylphthalamic acid (NPA), indicating that ARR7 and ARR15 activation can be directly induced by the loss of local auxin accumulation (Zhao et al., 2010). Because auxin suppresses the expression of STM that promotes cytokinin biosynthesis in the shoot meristem (Heisler et al., 2005; Jasinski et al., 2005; Yanai et al., 2005), auxin might function on the repression of ARR7 and ARR15 through the STM-mediated pathway. Another pathway has been identified through the analysis of ARR7 and ARR15 expression in the mp mutants. ARR7 and ARR15 are ectopically expressed in the central zone as well as in the peripheral zone in mp mutants, but absent in wild-type (Zhao et al., 2010). This suggests that MP limits the ARR7 and ARR15 expression both in the central zone and the peripheral zone of the meristem. Therefore, ARR7 and ARR15 act not only as suppressors of cytokinin signaling, but also as targets of MP-mediated auxin signaling in the central zone. These results also suggest that auxin and cytokinin signaling converge on ARR7 and ARR15 in the central zone of meristem during the development of shoot apical meristem (Figure 2A). In contrast to cytokinin, auxin accumulates at a relatively high level in the peripheral zone of the shoot meristem to trigger organ initiation (Benková et al., 2003; Reinhardt et al., 2003). Auxin has recently been shown to rapidly down-regulate cytokinin biosynthesis in the shoot (Nordström et al., 2004). In addition, auxin accumulation, facilitated by the efflux or influx carriers to various organ initiation sites, suppresses the expression of STM, which acts as an inhibitor of stem-cell differentiation (Furutani et al., 2004; Heisler et al., 2005). Because STM has been proved to be a positive factor of cytokinin biosynthesis (Jasinski et al., 2005; Yanai et al., 2005), a model is proposed that auxin antagonizes cytokinin for organ initiation in the peripheral zone of the meristem (Figure 2A).
Cytokinin also functions on the organ initiation (Shani et al., 2006; Perilli et al., 2010). At the site of organ initiation both in the embryonic SAM and in the inflorescence meristem, cytokinin levels are reduced by auxin (Wolters and Jürgens, 2009). Thus, cytokinin responses are also negatively regulated by auxin. High levels of cytokinin are required for the maintenance of stem cells in the meristem, but not for organ initiation. Similarly to ARR7 in Arabidopsis, ABERRANT PHYLLOTAXY1 (ABPH1), encoding a type-A ARR, is expressed in a specific pattern in the maize SAM. Mutation in ABPH1 leads to significantly larger SAM and an altered phyllotaxy (Giulini et al., 2004). These phenotypes suggest that ABPH1 restricts the size of the central zone of shoot meristem through negatively regulating cytokinin signaling. Since PIN1 is significantly down-regulated in the abph1 mutants, ABPH1 may be a convergent point of auxin signaling and cytokinin signaling to define the position of leaf primordia in maize (Lee et al., 2009). In addition, cytokinin–auxin interaction is also involved in the predominant shoot apex growth, which inhibits the outgrowth of axillary bud. In pea, it is demonstrated that auxin flows basipetally, mediated by PsPINs from the shoot apex to repress PsIPT expression, which is the gene for cytokinin biosynthesis (Figure 2B) (Shimizu-Sato et al., 2009). Consequently, the reduced levels of cytokinin increase apical dominance and inhibit axillary bud growth. MicroRNAs (miRNAs), which are post-transcriptional negative regulators in plants, are thought to play important roles in regulating shoot branching. The regulation of shoot branch production by miRNAs might correlate with the activities of auxin and cytokinin (Wang et al., 2010).
AUXIN–CYTOKININ INTERACTION REGULATES ROOT MERISTEM DEVELOPMENT
Post-embryonic root tip growth is sustained by the root meristem, which is divided into the proximal meristem (PM), the elongation differentiation zone (EDZ), and the transition zone (TZ) (Dello Ioio et al., 2007). QC is located in the region at the tip of PM, and gives rise to stem cells. Root stem cells generate daughter cells that subsequently undergo division in the PM or differentiation deviating from the PM to become the members of TZ cells. Therefore, a balance between the rate of cell division and differentiation is crucial for the maintenance of root meristem. Studies have shown that WOX5 (expressed in the QC cells) and PLTs (expressed in the stem cells surrounding the QC), major regulators of stem-cell formation and maintenance, act downstream of auxin signaling for the maintenance of distal stem-cell activity. Indeed, auxin signaling for the fate of the distal stem cells requires IAA17/AUXIN RESISTANT3 (AXR3) as well as auxin response factors (ARF10 and ARF16). Both ARF10 and ARF16 negatively regulate WOX5 transcription and restrict WOX5 transcripts to the QC center, thereby suppressing PLT gene expression and mediating the differentiation of distal stem cells in roots (Figure 2C) (Ding and Friml, 2010).
The auxin–cytokinin cross-talk also controls the root meristem development (Werner et al., 2003; Dello Ioio et al., 2007). Several studies have shown that cytokinin and auxin mutually regulate their signaling pathways or their metabolisms through certain integrators, which are the basis of interaction between these two hormones to determine a specific developmental output in root meristem (Dello Ioio et al., 2008; Moubayidin et al., 2009; Růžičkaa et al., 2009). Recently, a genetic framework has shown that antagonistic interaction between cytokinin and auxin is responsible for the control of cell division and cell differentiation in the root meristem (Dello Ioio et al., 2008; Moubayidin et al., 2009). This antagonistic interaction has been demonstrated to occur through a simple regulatory circuit via the SHORT HYPOCOTYL 2 (SHY2/IAA3), which is a member of the Aux/IAA gene family (Tian et al., 2003; Dello Ioio et al., 2008). In the wild-type roots, transcription of SHY2 in the vascular tissues of root meristem TZ is enhanced by cytokinin application. ARR1, a member of cytokinin signaling regulators, directly binds to the promoter of SHY2 (Dello Ioio et al., 2008). Moreover, no up-regulation of SHY2 expression in the roots of arr1 after cytokinin treatment suggests an ARR1-mediated cytokinin-positive regulation of SHY2 in the vascular tissues of TZ. Furthermore, the SHY2 gain-of-function or loss-of-function mutants show smaller or larger root meristems, respectively, confirming that SHY2 is necessary for the control of cytokinin over the size of root meristem (Dello Ioio et al., 2008). Activation of SHY2 results in repression of PIN genes expressed in the TZ, leading to the redistribution of auxin for cell differentiation (Figure 2C) (Dello Ioio et al., 2008; Moubayidin et al., 2009). Conversely, auxin mediates degradation of SHY2 protein, which is necessary for the transcription of PIN genes (Tian et al., 2003; Dello Ioio et al., 2008). PIN proteins collectively mediate auxin optimal distribution to regulate cell division and cell expansion in the root meristem, through a positive regulatory loop with the major regulators of stem-cell activity genes, PLTs (Blilou et al., 2005; Galinha et al., 2007; Grieneisen et al., 2007). Thus, this pathway mediated by PINs positively regulates cell division in the TZ to maintain the root meristem size.
Thus, cytokinin and auxin interact antagonistically to control the balance of cell division and differentiation mainly in the vascular tissue of TZ (Dello Ioio et al., 2008; Moubayidin et al., 2009). SHY2 seems to be an integrating factor that mediates not only the regulation of cytokinin-to-cell differentiation, but also the regulation of auxin-to-cell division (Figure 2C). Other results demonstrate that cytokinin regulates root meristem through the modulation on the PIN expression (Růžičkaa et al., 2009). It has been shown that cytokinin controls meristem size and PIN1 expression through the AHK-mediated cytokinin signaling (Růžičkaa et al., 2009). Additionally, SHY2 also represses the activation of IPT5 gene in the root meristem (Dello Ioio et al., 2008).
An antagonistic interaction between auxin and cytokinin biosynthesis has also been suggested in both developing roots and shoots. A recent study has shown that ectopic cytokinin results in a rapid increase in auxin biosynthesis in young roots and shoots (Jones et al., 2010). In contrast, the reduced cytokinin level represses auxin biosynthesis. This phenomenon indicates a cytokinin-mediated positive regulation of auxin synthesis. Together with the previous results, a model has been proposed for a homeostatic feedback regulatory loop involving in auxin and cytokinin signaling in developing roots and shoots tissue (Jones et al., 2010). According to this model, cytokinin functions as a positive regulator of auxin biosynthesis and auxin, however, represses cytokinin biosynthesis.
Studies on Arabidopsis and other plant species have revealed the roles of auxin and cytokinin in the formation of lateral roots (LR). Physiological and genetic data have demonstrated that auxin promotes LR initiation and lateral root primordium (LRP) development (Fukaki and Tasaka, 2009; Peret et al., 2009). First, auxin signals are transported by AUX1 and PINs to the protoxylem pericycle cells (Ditengou et al., 2008), and such signals are perceived by the F-box auxin receptors, TIR1 and AFBs (AFB1, AFB2, and AFB3) (Dharmasiri et al., 2005; Pérez-Torres et al., 2008). Second, the perception of auxin results in the degradation of Aux/IAA repressor proteins IAA14/ SOLITARY-ROOT (SLR) through SCFTIR1/AFBs complexes and 26S proteasomes (Fukaki et al., 2002). The de-repression of the ARF protein activity (ARF7/ARF19) activates the target genes LATERAL ORGAN BOUNDARIES-DOMAIN 16 (LBD16)/ASYMMETRIC LEAVES2-LIKE 18 (ASL18), LBD29/ASL16, and other targets required for asymmetric cell division during LR initiation (Wilmoth et al., 2005; Okushima et al., 2007; Fukaki and Tasaka, 2009; Peret et al., 2009). In contrast, cytokinin negatively regulates LR formation, probably through inhibiting auxin-induced expression of PIN genes and perturbing the establishment of an auxin gradient for LR initiation (Figure 2D) (Laplaze et al., 2007; Fukaki and Tasaka, 2009). Laplaze et al. (2007) have also shown that exogenous cytokinin inhibits the expression of several PINs in LRP, suggesting that cytokinin prevents the PIN-mediated auxin accumulation required for normal LRP patterning (Figure 2D).
AUXIN–CYTOKININ INTERACTION REGULATES IN VITRO ORGANOGENESIS
Regeneration of a patterned multi-cellular organism from the adult somatic tissue is a well-known phenomenon. Compared with animals, plants have a profound capacity to regenerate organs from their differentiated somatic tissues through the manipulation of plant hormones. The pioneering work has shown that a high auxin/cytokinin ratio induces root regeneration, whereas a low ratio promotes shoot induction (Skoog and Miller, 1957). This indicates that auxin and cytokinin might have a cross-talk during in vitro organogenesis. So far, the molecular mechanism of such interaction between auxin and cytokinin in the in vitro formation of meristem remains mostly unknown.
During in vitro establishment of the shoot meristem from the cultured root explants, cytokinin triggers the induction of ectopic WUS expression within the callus, as WUS expression is sufficient for inducing shoot regeneration (Gordon et al., 2009). Auxin pretreatment of Arabidopsis root explants leads to the up-regulation of a cytokinin receptor gene AHK4 expression during the callus formation. The increased AHK4 transcription is required for WUS activation during the shoot induction (Gordon et al., 2009). Buechel et al. (2009) have characterized the roles of ARR7 and ARR15 in shoot regeneration. They have found that overexpression of ARR7 and ARR15 suppresses shoot regeneration, whereas loss-of-function of these two genes strongly stimulates callus formation on auxin-rich callus-inducing medium (CIM) and promotes shoot induction on cytokinin-rich shoot-inducing medium (SIM) (Buechel et al., 2009). Take together, the previous studies indicate that exogenous auxin up-regulates the expression of AHK4 on CIM. Then, AHK4 enhances the response to exogenous cytokinin when callus is transferred to SIM. High cytokinin response promotes WUS expression, which is essential to induce the de novo formation of shoot meristem.
Another study has shown the role of cytokinin in the auxin-induced organ formation using hypocotyl explants (Pernisová et al., 2009). Based on this study, treatment with exogenous auxin triggers the organogenic processes, accompanied by the production of endogenous cytokinin and tissue-specific activation of cytokinin signaling. Endogenous cytokinin modulates this auxin-induced organogenesis via a negative regulation on the expression of PIN proteins, leading to an optimal auxin distribution for the induction of root-like organ (Pernisová et al., 2009). This study suggests that auxin is capable of inducing organogenesis in hypocotyl explants. Cytokinin modulates auxin distribution through its regulation on auxin efflux during this type of organogenesis.
We have reported the effect of the cytokinin/auxin ratio on WUS expression in inflorescence formation from Arabidopsis pistil explants (Cheng et al., 2010). Our results suggest that a high ratio of cytokinin/auxin promotes shoot regeneration through the induction on WUS expression. Our recent studies have also shown that the local establishment of auxin gradients mediated by the tissue-specific auxin biosynthesis and the dynamic changes of PIN proteins is important for shoot regeneration within callus (unpublished data). Moreover, the local biosynthesis of cytokinin in this regeneration system is required for the regional distribution of cytokinin and WUS expression. Thus, it is critical to identify the crucial signaling components that integrate the auxin and cytokinin signaling for callus induction and shoot regeneration. Auxin-regulated WUS expression is necessary for SAM formation during somatic embryogenesis (Su et al., 2009). The establishment of auxin gradients is correlated with induced WUS expression and subsequent SAM formation. These results might shed new light on the cross-talk between the two hormones in regulating shoot and root stem-cell formation during somatic embryogenesis.
CONCLUDING REMARKS AND PERSPECTIVES
Understanding of the auxin and cytokinin interaction in the regulation of plant development and organogenesis has advanced considerably over more than a half-century. Many early experiments have revealed the essential roles of both hormones in the cell proliferation and new organ regeneration. It is amazing that the decision of cell fate in specific tissues depends on the ratio between auxin and cytokinin, which maintains the cell proliferation or stimulates cell differentiation to form new organs, such as shoots or roots. Studies on hormones over the past two decades have been mainly focused on analyzing the mutants of genes involved in hormone synthesis and catabolism, and those encoding for receptors and signaling components. Owing to the extensive functional redundancy among gene family members, analyzing the feedback regulation of both hormone pathways is still complicated work to do. Recently, research has been geared to identifying the key factors involved in the interaction of these two hormones to control specific aspects of plant development. The cross-talk between cytokinin and auxin in the shoot and root meristem is highlighted in Figure 2. In the root meristem, auxin induces the meristematic cell division, whereas cytokinin promotes the cell to switch from the meristematic to differentiated state through inhibiting auxin signaling. In contrast, in the shoot meristem, cytokinin promotes stem-cell proliferation and inhibits stem-cell differentiation, whereas auxin triggers organ primordium initiation through repressing cytokinin biosynthesis. Thus, the antagonistic interactions are the key regulators for the cell differentiation and maintenance in the root meristem transition zone or shoot meristem organ initiation sites.
The models proposed in this review are still not sufficient to fully understand the mechanisms of hormonal interaction between auxin and cytokinin. Because most regulators involved in auxin and cytokinin signaling function in a tissue-specific manner, we may use micro-dissection or other techniques to harvest individual cells from specific tissue sections to aid such studies. By analyses of genome-wide profiling for epigenetics, transcriptomics, and proteomics in the harvested cells of different tissues, new information should be provided about tissue-specific gene regulatory networks involved in the hormonal cross-talk. Further studies on the structural analysis of the key integrators will be also critical to reveal how transcription factors in the auxin signaling pathway potentially interface with those in cytokinin signaling through protein–protein or protein–DNA interactions. For example, the crystal structure of auxin receptor TIR1 has been presented to explain how this protein is in complexes with auxin and the domain II region of an Aux/IAA protein (Tan et al., 2007). Additionally, the in vitro organogenesis may be a powerful system to study the mechanisms of hormonal cross-talk during plant organogenesis, because the regenerated organs can be induced by the known ratio of auxin and cytokinin (Skoog and Miller, 1957). Also, the single type of organs, such as shoots or roots, can be regenerated from the differentiated tissues to investigate the mechanisms involved in hormone-regulated organ regeneration.
FUNDING
The research on hormone-regulating plant organogenesis in the lab of X.S. Zhang was supported by grants from the Ministry of Science and Technology (MOST) of China (2007CB948200) and the National Natural Science Foundation (NNSF) of China (90917015, 31000652, and 30770217). No conflict of interest declared.
References
- Benjamins R, Scheres B. Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 2008;59:443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- Benková E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Berleth T, Jürgens G. The role of the monopteros gene in organizing the basal body region of the Arabidopsis embryo. Development. 1993;118:575–587. [Google Scholar]
- Blilou I, et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher J, Hobe M, Meyerowitz E, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T. Differential expression of WOX genes mediates apical–basal axis formation in the Arabidopsis embryo. Dev. Cell. 2008;14:867–876. doi: 10.1016/j.devcel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Buechel S, et al. Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur. J. Cell Biol. 2009;89:279–284. doi: 10.1016/j.ejcb.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Cheng ZJ, Zhu SS, Gao XQ, Zhang XS. Cytokinin and auxin regulates WUS induction and inflorescence regeneration in vitro in Arabidopsis. Plant Cell Rep. 2010;29:927–933. doi: 10.1007/s00299-010-0879-8. [DOI] [PubMed] [Google Scholar]
- Clark SE. Cell signalling at the shoot meristem. Nat. Rev. Mol. Cell Biol. 2001;2:276–284. doi: 10.1038/35067079. [DOI] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, et al. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 2007;17:678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- Dettmer J, Elo A, Helariutta Y. Hormone interactions during vascular development. Plant Mol. Biol. 2009;69:347–360. doi: 10.1007/s11103-008-9374-9. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Ding ZJ, Friml J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl Acad. Sci. U S A. 2010;107:12046–12051. doi: 10.1073/pnas.1000672107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditengou FA, et al. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc. Natl Acad. Sci. U S A. 2008;105:18818–18823. doi: 10.1073/pnas.0807814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996;10:967–979. doi: 10.1046/j.1365-313x.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- Ferreira FJ, Kieber JJ. Cytokinin signaling. Curr. Opin. Plant Biol. 2005;8:518–525. doi: 10.1016/j.pbi.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Meyerowitz EM. Cell signaling within the shoot meristem. Curr. Opin. Plant Biol. 2000;3:23–30. doi: 10.1016/s1369-5266(99)00033-3. [DOI] [PubMed] [Google Scholar]
- Friml J, et al. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tasaka M. Hormone interactions during lateral root formation. Plant Mol. Biol. 2009;69:437–449. doi: 10.1007/s11103-008-9417-2. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development. 2004;131:5021–5030. doi: 10.1242/dev.01388. [DOI] [PubMed] [Google Scholar]
- Galinha C, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- Gao X, Nagawa S, Wang G, Yang Z. Cell polarity signaling: focus on polar auxin transport. Mol. Plant. 2008;1:899–909. doi: 10.1093/mp/ssn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulini A, Wang J, Jackson D. Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature. 2004;430:1031–1034. doi: 10.1038/nature02778. [DOI] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl Acad. Sci. U S A. 2009;106:16529–16534. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AFM, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- Hamann T, Benková E, Bäurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Heyl A, Schmülling T. Cytokinin signal perception and transduction. Curr. Opin. Plant Biol. 2003;6:480–488. doi: 10.1016/s1369-5266(03)00087-6. [DOI] [PubMed] [Google Scholar]
- Higuchi M, et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl Acad. Sci. U S A. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- Inoue T, et al. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Jasinski S, et al. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 2005;15:1560–1565. doi: 10.1016/j.cub.2005.07.023. [DOI] [PubMed] [Google Scholar]
- Jenik PD, Barton MK. Surge and destroy: the role of auxin in plant embryogenesis. Development. 2005;132:3577–3585. doi: 10.1242/dev.01952. [DOI] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W. Embryonic patterning in Arabidopsis thaliana. Annu. Rev. Cell Dev. Biol. 2007;23:207–236. doi: 10.1146/annurev.cellbio.22.011105.102609. [DOI] [PubMed] [Google Scholar]
- Jones B, et al. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell. 2010;22:2956–2969. doi: 10.1105/tpc.110.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001;42:677–685. doi: 10.1093/pcp/pce112. [DOI] [PubMed] [Google Scholar]
- Kakimoto T. Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 2003;54:605–627. doi: 10.1146/annurev.arplant.54.031902.134802. [DOI] [PubMed] [Google Scholar]
- Kurakawa T, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Laplaze L, et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell. 2007;19:3889–3900. doi: 10.1105/tpc.107.055863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Jürgens G. Embryogenesis: a new start in life. Plant Cell. 1997;9:989–1000. doi: 10.1105/tpc.9.7.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, et al. Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol. 2009;150:205–216. doi: 10.1104/pp.109.137034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002;49:387–400. [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can. J. Bot. 1991;69:461–476. [Google Scholar]
- Mason MG, et al. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17:3007–3018. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J, Ckurshumova W, Berleth T. Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 2003;131:1327–1339. doi: 10.1104/pp.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Miyawaki K, et al. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl Acad. Sci. U S A. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller B, Weijers D. Auxin control of embryo patterning. Cold Spring Harbor Perspect. Biol. 2009;1:a001545. doi: 10.1101/cshperspect.a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sabatini S. Cytokinin–auxin crosstalk. Trends Plant Sci. 2009;14:557–562. doi: 10.1016/j.tplants.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell. 2004;16:1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström A, et al. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin–cytokinin-regulated development. Proc. Natl Acad. Sci. U S A. 2004;101:8039–8044. doi: 10.1073/pnas.0402504101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Friml J. Auxin signaling. J. Cell Sci. 2006;119:1199–1202. doi: 10.1242/jcs.02910. [DOI] [PubMed] [Google Scholar]
- Peret B, et al. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 2009;14:399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Pérez-Torres CA, et al. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell. 2008;20:3258–3272. doi: 10.1105/tpc.108.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Moubayidin L, Sabatini S. The molecular basis of cytokinin function. Curr. Opin. Plant Biol. 2010;13:21–26. doi: 10.1016/j.pbi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Pernisová M, et al. Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc. Natl Acad. Sci. U S A. 2009;106:3609–3614. doi: 10.1073/pnas.0811539106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Gray WM. Auxin signaling. Curr. Opin. Plant Biol. 2006;9:448–453. doi: 10.1016/j.pbi.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GV. Live-imaging stem-cell homeostasis in the Arabidopsis shoot apex. Curr. Opin. Plant Biol. 2008;11:88–93. doi: 10.1016/j.pbi.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, et al. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426:255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp HM, Frank M, Werner T, Strnad M, Schmülling T. Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J. 1999;18:557–563. doi: 10.1046/j.1365-313x.1999.00472.x. [DOI] [PubMed] [Google Scholar]
- Růžičkaa K, et al. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl Acad. Sci. U S A. 2009;106:4284–4289. doi: 10.1073/pnas.0900060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R. The dynamic plant stem cell niches. Curr. Opin. Plant Biol. 2007;10:639–644. doi: 10.1016/j.pbi.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Schlereth A, et al. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464:913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Shani E, Yanai O, Ori N. The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 2006;9:484–489. doi: 10.1016/j.pbi.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S, Tanaka M, Mori H. Auxin–cytokinin interactions in the control of shoot branching. Plant Mol. Biol. 2009;69:429–435. doi: 10.1007/s11103-008-9416-3. [DOI] [PubMed] [Google Scholar]
- Skoog F, Miller CO. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 1957;54:118–130. [PubMed] [Google Scholar]
- Su YH, Zhao XY, Liu YB, Zhang CL, O'Neill SD, Zhang XS. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 2009;59:448–460. doi: 10.1111/j.1365-313X.2009.03880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Sugiyama T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001;276:26405–26410. doi: 10.1074/jbc.M102130200. [DOI] [PubMed] [Google Scholar]
- Takei K, Yamaya T, Sakakibara H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J. Biol. Chem. 2004;279:41866–41872. doi: 10.1074/jbc.M406337200. [DOI] [PubMed] [Google Scholar]
- Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Tian Q, Nagpal P, Reed JW. Regulation of Arabidopsis SHY2/IAA3 protein turnover. Plant J. 2003;36:643–651. doi: 10.1046/j.1365-313x.2003.01909.x. [DOI] [PubMed] [Google Scholar]
- To JPC, Kieber JJ. Cytokinin signaling: two-components and more. Trends Plant Sci. 2008;13:85–92. doi: 10.1016/j.tplants.2007.11.005. [DOI] [PubMed] [Google Scholar]
- To JPC, et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997a;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997b;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Wang L, Mai YX, Zhang YC, Luo Q, Yang HQ. MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol. Plant. 2010;3:794–806. doi: 10.1093/mp/ssq042. [DOI] [PubMed] [Google Scholar]
- Weigel D, Jürgens G. Stem cells that make stems. Nature. 2002;415:751–754. doi: 10.1038/415751a. [DOI] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jürgens G. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell. 2006;10:265–270. doi: 10.1016/j.devcel.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Werner T, Schmülling T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009;12:527–538. doi: 10.1016/j.pbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15:2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Fletcher JC. Stem cell regulation in the Arabidopsis shoot apical meristem. Curr. Opin. Plant Biol. 2005;8:582–586. doi: 10.1016/j.pbi.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Wilmoth JC, et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43:118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- Wolters H, Jürgens G. Survival of the flexible: hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009;10:305–317. doi: 10.1038/nrg2558. [DOI] [PubMed] [Google Scholar]
- Yanai O, et al. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005;15:1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, et al. Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 2007;48:84–96. doi: 10.1093/pcp/pcl040. [DOI] [PubMed] [Google Scholar]
- Zazímalová E, Murphy AS, Yang HB, Hoyerová K, Hošek P. Auxin transporters. Why so many? Cold Spring Harbor Perspect. Biol. 2010;2:a001552. doi: 10.1101/cshperspect.a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YD. The role of local biosynthesis of auxin and cytokinin in plant development. Curr. Opin. Plant Biol. 2008;11:16–22. doi: 10.1016/j.pbi.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Zhao YD. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, et al. Hormonal control of the shoot stem-cell niche. Nature. 2010;465:1089–1092. doi: 10.1038/nature09126. [DOI] [PubMed] [Google Scholar]