Abstract
In the decade and a half since Biswal’s fortuitous discovery of spontaneous correlations in functional imaging data, the field of functional connectivity (FC) has seen exponential growth resulting in the identification of widely-replicated intrinsic networks and the innovation of novel analytic methods with the promise of diagnostic application. As such a young field undergoing rapid change, we have yet to converge upon a desired and needed set of standards. In this issue, Habeck and Moeller begin a dialogue for developing best practices by providing four criticisms with respect to FC estimation methods, interpretation of FC networks, assessment of FC network features in classifying sub-populations, and network visualization. Here, we respond to Habeck and Moeller and provide our own perspective on the concerns raised in the hope that the neuroimaging field will benefit from this discussion.
Keywords: Functional connectivity, Classification, Diagnosis, Independent component analysis, Seed-voxel analysis
Introduction
We begin by thanking Habeck and Moeller for initiating a worthwhile discussion and express our gratitude to the editors for providing a forum in which it may take place. Only with critical reflection on our own research practices can we begin to address the challenges of this rapidly growing field. We respond to their four criticisms in point/counterpoint format, and refrain from raising (too many) new questions.
A network by any other name
In their first point, Habeck and Moeller raise some important questions about network identification and the attribution of functional relevance. In particular, they question 1) the face validity of networks obtained using blind decomposition techniques like principal component analysis (PCA) and independent component analysis (ICA) and the lack of evidence in the form of behavioral or diagnostic correlates of network activity, 2) the assessment of robustness and replication in independent data, and 3) the labeling of networks based on topography without demonstration of functional relevance. In the subsequent paragraphs we put forth our own perspective on each of these concerns.
We agree that activation patterns derived purely by the virtue of statistical independence (ICA), orthogonality (PCA), or correlation with a given seed need not have any neuroscientific meaning. However, their criticism of face validity seems to ignore the last decade of research during which an overwhelming number of studies have demonstrated the behavioral relevance of components and elucidated their functional roles. We mention a handful of examples. Using ICA, Carvalho et al. (2006) investigated the effects of intoxication on simulated driving and showed dose-related deterioration in attention and driving performance associated with activity in cerebellar and motor networks. Eichele et al. (2008) demonstrated that activity in multiple ICA networks including frontal and default-mode regions predicted performance errors in a flanker task involving conflict. In an impressive meta-analysis of peak activation coordinates obtained from a large and heterogeneous collection of experiments, Smith and colleagues (2009) showed close correspondence between ICA networks obtained from resting-state decompositions and spatial activation patterns linked with task paradigms. Along with functional relevance, ICA components have been demonstrated to covary with demographic variables like age and gender (Allen et al., 2011; Biswal et al., 2010) and neuropsychiatric disorders such as Alzheimer’s disease (Greicius et al., 2004), schizophrenia, and bipolar disorder (Calhoun et al., 2008b). It is our impression that “additional evidence” of meaningful networks is presented regularly.
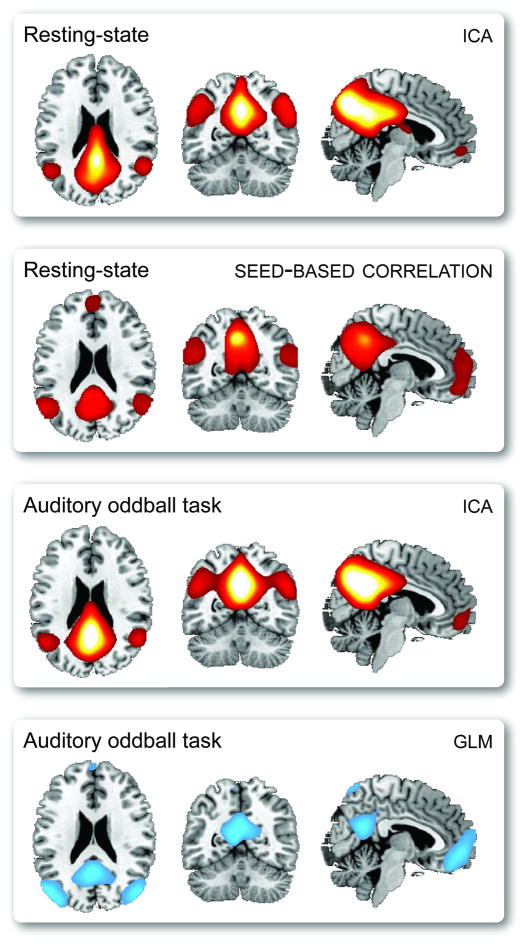
Regarding their complaint of a lack of replication and assessment of robustness, we note that components and seed-based maps have been widely replicated within and between studies. Resting-state networks are highly reproducible at the group level (Damoiseaux et al., 2006) and show reasonably high reliability at the level of the individual (Shehzad et al., 2009; Zuo et al., 2009). Furthermore, corresponding networks are found consistently in a variety of behavioral tasks (or lack thereof in resting-state paradigms), as demonstrated by Calhoun and colleagues (2008a) who show similar components in ICA decompositions of resting and task-related data. Finally, comparable networks can be obtained with seed-based and ICA approaches, and network connectivity strengths for individual subjects are correlated between the methods (Van Dijk et al., 2010). As an illustration of the robustness of FC components, we show in Figure 1 that the default mode network is easily identified regardless of experimental paradigm (auditory oddball task or resting-state) or analysis method (general linear model (GLM), ICA, or seed-based correlation).
Figure 1.
A demonstration that the default mode network can be estimated regardless of experimental paradigm (auditory oddball task or resting-state) or analysis method (GLM, ICA, or seed-based). For seed-based analysis, the seed location is in posterior cingualte cortex (MNI=[−6, −52, 43]). For both ICA decompositions, the model order is 20. For GLM results, we display only negative activations to a novel–standard contrast.
The authors raise an additional concern regarding the topographic labeling of spatial components. We agree that labels derived from topography or other conventions do not bestow relevance or inherent meaning to components. However, as discussed above, FC networks are highly reproducible across studies, and the putative importance of components can often be inferred by spatial correspondence to previously investigated networks or by localization to brain regions with well-established functions (such as visual, motor, and auditory areas). We would additionally argue that in some cases, a topographic label might be preferable to one based on a presumed function, particularly when considering a component that might subserve a variety of functions in different contexts. For example, one might denote a component as a “left lateralized fronto-parietal network”, rather than a “memory network”. Although these areas are often implicated in explicit memory processes (Iidaka et al., 2006), they undoubtedly have other functions as well, such as in language and semantic processing (Smith et al., 2009).
In summary, spatial patterns of connectivity identified with any analytic method need not have inherent significance. However, a wealth of literature demonstrates that FC networks are associated with directed behaviors and individual traits. Researchers should use this foundation of knowledge to inform their own hypotheses and interpretations regarding the roles of components.
Seeds of confusion
In their second point, Habeck and Moeller recognize that the display of seed-based correlation maps (as in Fig 1, second panel), encourages misinterpretation with respect to correlations between non-seed regions. That is, it is intuitive but incorrect to expect that regions correlated with the seed are also correlated with each other. Since such connectivity maps are often called a “network”, Habeck and Moeller call for more clarity on what generally constitutes a network.
To address the second point first, context determines the meaning and interpretation of the word “network” in brain imaging analysis. For example, both the GLM and seed-based method define a network as a subset of voxels whose timeseries are significantly correlated with a reference signal. Using graph theory, a network may be defined as a connectivity matrix between nodes, which represent voxels, areas, or components. Because there are many existing ways to characterize and estimate a “network”, we feel it is unreasonable that the community “fine-tune” a single definition. Rather, each author should explicitly state the interpretation implicit in the method.
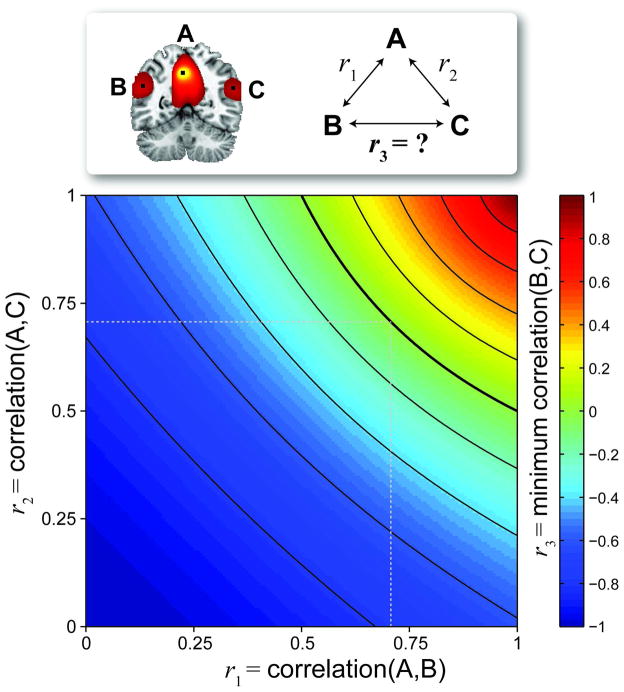
To elaborate Habeck and Moeller’s point on the misinterpretation of seed-based correlation maps, we provide a theoretical account of possible correlations between pairs of voxels, given their known association with a seed. Consider Figure 2, where a coronal slice is shown from a thresholded seed-based correlation map. Label the seed voxel “A”, and any other two voxels “B” and “C”. Given that you know r1 = corr(A,B) and r2 = corr(A,C), what is r3 = corr(B,C)? The distribution for r3 can’t be estimated without additional information or assumptions, however, we can calculate a theoretic minimum for r3, as displayed in Figure 21. From the image, it can be seen that if r1×r2 ≥ 0.5, then r3 ≥ 0. Thus, we may observe rather high correlations r1 = r2 = 0.707 with almost no information for the value of r3, 0 ≤ r3 ≤ 1. Note that we have focused on the magnitude of correlation, rather than its statistical significance, since correlation magnitude is meaningful and interpretable, indicating linear-relatedness. Correlation significance is used strictly for making a decisions regarding specific hypotheses and is largely determined by sample size (and must, in this case, be corrected for time series autocorrelation).
Figure 2.
From a seed voxel “A”, correlations with two arbitrary voxels “B” (r1 = corr(A,B)) and “C” (r2 = corr(A,C)) are known, however the correlation between “B” and “C” is not. We plot the minimum possible value for r3 = corr(B,C). Note that if r1×r2 ≥ 0.5 (above the bold arc), then r3 ≥ 0.
To summarize, it is almost instinctual to look at a seed-based map and assume that voxels that are correlated with the seed are also correlated with each other, but no such claim can be made. A fuller description of the relationships among all voxels can be determined with multivariate methods, such as ICA and graph theoretic approaches. Here too, resulting maps and networks must be clearly described. For example, if voxels A, B, and C appear in a thresholded ICA map, one can correctly infer that the timeseries of these voxels share common features. However it is not necessarily true that the timeseries of voxels A, B, and C will be highly correlated, since each may contain unique features (potentially shared with other voxels) that may comprise additional components. Thus, the precise meaning of maps generated with each method will be different, and it is the author’s responsibility to explicitly provide the correct interpretation for readers. In light of these cautionary notes, we add that seed-based maps can strongly resemble ICA components (e.g., Figure 1), suggesting that these approaches, and many others, estimate similar phenomena.
Diagnose my classification
In Point 3 of their critique, Habeck and Moeller discuss the use of FC-based biomarkers for the purpose of diagnostic classification. They recommend that innovative network measures should be habitually compared to preexisting biomarkers using out-of-sample prediction. We agree with this suggestion. Whenever possible, researchers should present comparative assessments of biomarkers in a single study. Comparisons between studies can be misleading since classification performance is affected by data quality, sampling variability, and the selected classifier and optimization procedure. By comparing the performance of several measures on the same set of data using the identical classification scheme, researchers can obtain a clearer picture of the discriminative information in each measure, and ultimately accelerate the development of better diagnostic tools. We note that in such comparisons, novel FC measures need not perform “at least as well, or better, than what is already available.” There are numerous scenarios where FC-based measures with equivalent or inferior performance would be of great interest. Traditional measures may be more costly or difficult to acquire or may expose subjects to possible harm, e.g., with radioactive tracers or carcinogenic contrast agents that must be introduced intravenously. Safe and easily acquired measures with relatively low discriminability can also be combined (potentially with those from other modalities) to form a classifier with superior performance. Thus, along with classification sensitivity and specificity, the feasibility of acquisition and possible costs to subject safety must be considered when determining diagnostic potential.
Habeck and Moeller make a second point regarding the choice of FC measures for classification purposes. Namely, that more complex or “derived” measures will have greater “statistical noise” which may hinder predictive performance. Assuming the “statistical noise” to which they refer is synonymous with estimation variance, we agree: it is generally true that estimation of higher order statistics requires more observations2. As a useful rule of thumb, J. W. Tukey suggests that the calculation of the kth moment ought to be based on at least 5k observations (as found in (Sachs, 1992), p. 172). However, estimation uncertainty is not directly related to the discriminative capabilities of a measure. Here, the appeal to simplicity may be misguided.
In our opinion, a diagnostic classifier is valuable if it (1) has features that discriminate between sub-populations of interest, (2) is sensitive and specific, and (3) can be replicated. Robustness in itself is not of value in a classification framework; biomarkers must simply be robust enough to replicate prediction across samples. The information contained in higher-order statistics should be exploited for classification, not avoided because of a fear of greater “statistical noise” or number of “processing steps.” While the authors offer Occam’s razor to shorten the reins on otherwise unbridled enthusiasm for complexity, we note Karl Menger’s Law Against Miserliness, which reminds us that “entities must not be reduced to the point of inadequacy.” Ultimately, replication is the gold-standard of science. If researchers compare and validate biomarkers with out-of-sample prediction, the most suitable classifiers will emerge naturally, regardless of their complexity.
To illustrate the disassociation between biomarker complexity and predictive power, we present a classification experiment complementing that of Habeck and Moeller. Following their example, we computed simple measures from resting-state fMRI data to predict subjects’ age (young/old) and, in a separate sample, subjects’ diagnosis (healthy/schizophrenia). We considered five minutes of resting-state data acquired on a 3T Trio Tim Siemens scanner (TR = 2 sec) from 40 young subjects (age range 14–15 years, mean ± SD age = 14.6 ± 0.50 years; 20 females) and 40 older subjects (40–55 years, 45.7 ± 3.8 years; 21 females). In addition, we considered data from 40 middle-aged subjects diagnosed with schizophrenia (range 20–39 years, 9 females) and 40 healthy subjects that were matched to the schizophrenia group for age and gender. Further details on data collection and preprocessing can be found in (Allen et al., 2011). For each dataset we derived three features: 1) a mean map, as in Habeck and Moeller, 2) a low frequency amplitude map, computed by taking the Fourier transform of each voxel timeseries and averaging amplitude in [0.01, 0.1] Hz, and 3) Fisher z-transformed seed-based correlation maps. Rather than a single seed, we used a set of four seeds to identify two so-called task-positive networks (FEF, MNI = [34, −17,54]; MT+, MNI = [−46, −69, −9]) and two task-negative networks (mPFC, MNI = [−1,53, −3]; PCC, MNI = [−6, −52,43]), as described by Fox et al. (2005). Before computing pairwise correlations, voxel timeseries were orthogonalized with respect to ventricular, white matter, and motion parameter time series and bandpass filtered within [0.01, 0.125 Hz].
For classification, we used a support vector machine (SVM) with a radial basis kernel. Within each classification problem (i.e., age or diagnosis) SVMs were trained on 60% (48 subjects) and test performance was evaluated on the remaining 40% (32 subjects); training and testing were iterated on 100 sub-population balanced samples. To reduce the high number of potential features (i.e., ≥ 68,000 voxels), we used PCA for dimension reduction. Thus, features were the subject scores for principal components of mean, amplitude, or seed correlation maps. In the training phase, PC scores were ranked by two-sample t-statistics to discriminate between groups. PC scores were then added to the SVM model in order of decreasing t-statistic magnitude and the final set of features was determined as the model with the maximum accuracy using 5-fold cross-validation of the training set. SVM model parameters were optimized with a grid search via cross-validation, and the trained model was used to predict age or diagnosis in the test set. Prediction performance was quantified with the balanced accuracy rate, that is, the mean of the true positive rate and true negative rate.
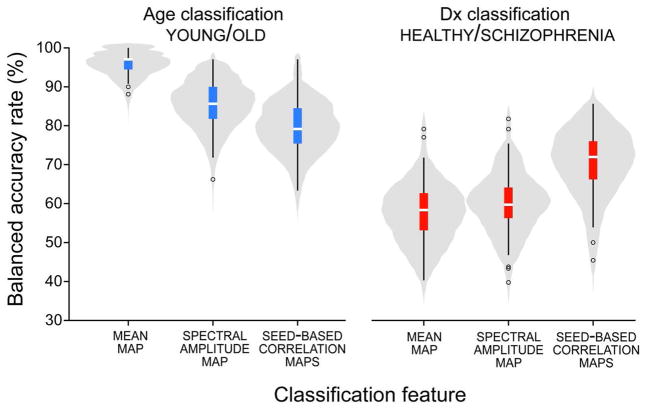
Balanced accuracy rates for prediction of age and diagnosis are shown in Figure 3. Replicating the results of Habeck and Moeller we find that for age, mean maps give the best prediction, followed by the low frequency amplitude and seed-based correlation maps. For diagnosis, the trend is reversed with seed-based correlation maps outperforming mean and amplitude maps. The opposing trends demonstrate unambiguously and empirically that predictive power can be completely disassociated from feature complexity. Classifier performance depends first and foremost on the discriminative information contained in its features relevant to the sub-populations of interest. It is hardly surprising that large differences in age can be very well predicted by the mean map since tissue relaxation constants are known to change dramatically with age, e.g., due to iron-depositions in sub-cortical nuclei (Siemonsen et al., 2008). Likewise, we expect biomarkers capturing patterns of functional connectivity to perform better in the diagnosis of schizophrenia than simpler measures since the pathophysiology of the disease is believed to involve impaired coordination between regions rather than localized deficits (Andreasen et al., 1998). In general, we should exploit prior information and carefully select features to enhance classification.
Figure 3.
Balanced accuracy rates for out-of-sample prediction of age (young/old, left) and diagnosis (healthy/schizophrenia, right). The distribution of accuracy over 100 training/testing iterations is depicted with a violin pot and overlayed boxplot.
Think before you ink
As their final point, Habeck and Moeller remind us of Tufte’s principle to maximize the “data-ink ratio” in graphical displays. Specifically, they make the observation that figures of neuroimaging analyses often have remarkably low data-ink ratios due to the prominence of visually appealing (though non-data) templates. We applaud Habeck and Moeller for initiating a discussion on existing practices of data visualization and join them in advocating for general adherence to Tufte’s ideas on maximizing data-ink, as well as his principles on maximizing data-density and minimizing chartjunk (Tufte, 2001). We take this opportunity to more deeply address the challenges associated with the effective display of neuroimaging data, and how we might, as a field, works towards a set of improved standards.
Neuroscientists face a formidable challenge in concisely displaying high-dimensional and spatially-related data. While low data-ink is regrettable, detailed structural templates serve as an important coordinate reference frame for the functional data of interest. A template provides information regarding physical boundaries and disambiguates localization in a manner that labeling schemes and wordy descriptions cannot. Consider climate scientists, who also work with highly multivariate data that span atmosphere and ocean strata. Just as the physical borders of coastlines, rivers, and mountains provide context to understand climate patterns, so too do gyri, sulci, and ventricles provide context for neuroimaging observations. While we welcome improvements to the conventional display, it is difficult to find an alternative for presenting high-dimensional information in a spatial context that others can easily interpret. A table listing maximum activation coordinates loses spatial relationships and activation extents, and other displays, such as bar charts, do not show the same information. Overall, we believe the merits of a template display outweigh its criticisms.
Generally, as much thought should go into data visualization (as a form of communication) as went into experiment design and analysis. We have a duty to prioritize the clear, undistorted presentation of the data, particularly given evidence that results presented on brain images can be perceived as more persuasive than similar information presented in less attractive formats (McCabe and Castel, 2008). For any display, researchers should ask, does the image prioritize data content? Is the figure consistent with the model or hypothesis being tested? Is the effect size accurately represented and could it be misinterpreted (such as when using a nonlinear scale when a linear scale may be assumed)? Is the figure visually appealing and accessible, encouraging inspection, or is it cluttered, disorganized, and overwhelming? For map displays, do the selected colors in the figure provide a natural interpretation and is the color map clearly defined? Furthermore, while rarely done, associated uncertainty (confidence surfaces) can be represented in maps, for example, by coding quantitative effects with hues and coding uncertainty with white or increasing alpha (transparency) (Hengl and Toomanian, 2006). Our diligence in honest visual portrayal is imperative both within the field and for the science reporters who take our neuroimaging results to the public.
Ultimately, the responsibility for good figures lies with the authors, peer-reviewers, and editors involved in the publication process. Together, we should agree upon a set of best practices for creating visual displays and hold ourselves to these standards. Journals can provide specific style recommendations beyond those affecting print quality. Researchers regularly have questions about how to present information, for example, when should one report the standard deviation versus the standard error of the mean3, and when is it appropriate to use a “fancy” 3D chart for two-dimensional or categorical data4. Formal responses to such frequently asked questions and explicit examples of different chart types exemplifying Tufte’s principles will benefit the field as a whole.
Conclusion
By and large we agree with many of the criticisms raised by Habeck and Moeller. Some of their points benefited from a more rigorous treatment, such as the interpretive challenges of seed-based correlation maps (Point 2), the determination of a classifier’s diagnostic potential (Point 3), and the call for higher standards of graphical visualization (Point 4). However other criticisms, in our opinion, are unjustified. For instance, the behavioral relevance of several FC networks has been demonstrated repeatedly (Point 1), and we feel that the evidentiary standard to which they appeal has largely been met. Regarding the use of FC in classification (Point 3), we agree that out-of-sample validation should be used regularly. Yet we find there is no need for an initial “robustness analysis”. Potential classifiers should not be prioritized or penalized by virtue of their complexity.
Based on this discussion, we feel that it may be too early for convergence to a set of best practices in the context of FC analysis and display. However, it is easy to identify better practices as those that make careful and thoughtful interpretations of written and graphical results and help to steer the reader away from misinterpretation. As with Habeck and Moeller, we hope our commentary contributes to the progress in this field and that, more importantly, the dialogue continues.
Acknowledgments
This research was supported by funding from NIH 1R01-EB006841, NIH 1R01-EB005846, NIH 2R01-EB000840, NIH 1 P20 RR021938-01, and DOE DE-FG02-08ER64581 to VDC.
Abbreviations
- FC
functional connectivity
- PCA
principal component analysis
- ICA
independent component analysis
- GLM
general linear model
- SVM
support vector machine
Footnotes
Because a covariance matrix (e.g., a correlation matrix) is positive definite, the minimum for r3 is found when the smallest eigenvalue of the 3-by-3 correlation matrix between A, B, and C is zero.
The technical details relate to relative rates of convergence of moments based on the law of large numbers and central limit theorem (Hall, 1982).
(Holopigian and Bach, 2010) suggest using the standard deviation to describe a sample distribution and the standard error of the mean for comparisons involving the mean. This applies for both graphical and numerical summaries.
Never (Jones, 2006).
Disclosure Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Allen EA, Erhardt EB, Damaraju E, Gruner W, Segall JM, Silva RF, Havlicek M, Rachakonda S, Fries J, Kalyanam R, Michael AM, Caprihan A, Turner JA, Eichele T, Adelsheim S, Bryan AD, Bustillo J, Clark VP, Feldstein Ewing SW, Filbey F, Ford CC, Hutchison K, Jung RE, Kiehl KA, Kodituwakku P, Komesu YM, Mayer AR, Pearlson GD, Phillips JP, Sadek JR, Stevens M, Teuscher U, Thoma RJ, Calhoun VD. A baseline for the multivariate comparison of resting-state networks. Front Syst Neurosci. 2011:4–5. doi: 10.3389/fnsys.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen N, Paradiso S, O’Leary D. “Cognitive dysmetria” as an integrative theory of schizophrenia: a dysfunction in cortical-subcortical-cerebellar circuitry? Schizophrenia Bulletin. 1998;24(2):203. doi: 10.1093/oxfordjournals.schbul.a033321. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, Dogonowski AM, Ernst M, Fair D, Hampson M, Hoptman MJ, Hyde JS, Kiviniemi VJ, Kotter R, Li SJ, Lin CP, Lowe MJ, Mackay C, Madden DJ, Madsen KH, Margulies DS, Mayberg HS, McMahon K, Monk CS, Mostofsky SH, Nagel BJ, Pekar JJ, Peltier SJ, Petersen SE, Riedl V, Rombouts SA, Rypma B, Schlaggar BL, Schmidt S, Seidler RD, Siegle GJ, Sorg C, Teng GJ, Veijola J, Villringer A, Walter M, Wang L, Weng XC, Whitfield-Gabrieli S, Williamson P, Windischberger C, Zang YF, Zhang HY, Castellanos FX, Milham MP. Toward discovery science of human brain function. Proc Natl Acad Sci U S A. 2010;107(10):4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Kiehl KA, Pearlson GD. Modulation of Temporally Coherent Brain Networks Estimated using ICA at Rest and During Cognitive Tasks. Hum Brain Mapp. 2008a;29(7):828–838. doi: 10.1002/hbm.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Pearlson GD, Maciejewski P, Kiehl KA. Temporal Lobe and ‘Default’ Hemodynamic Brain Modes Discriminate Between Schizophrenia and Bipolar Disorder. Hum Brain Map. 2008b;29(11):1265–1275. doi: 10.1002/hbm.20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho K, Pearlson GD, Astur RS, Calhoun VD. Simulated Driving and Brain Imaging: Combining Behavior, Brain Activity, and Virtual Reality. CNS Spectrum. 2006;11(1):52–62. doi: 10.1017/s1092852900024214. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci U S A. 2006;103(37):13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichele T, Debener S, Calhoun VD, Specht K, Engel AK, Hugdahl K, Cramon DY, Ullsperger M. Prediction of human errors by maladaptive changes in event-related brain networks. Proc Natl Acad Sci U S A. 2008;105(16):6173–6178. doi: 10.1073/pnas.0708965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102(27):9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci USA. 2004;101(13):4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall P. Bounds on the Rate of Convergence of Moments in the Central Limit Theorem. The Annals of Probability. 1982;10:4. [Google Scholar]
- Hengl T, Toomanian N. Maps are not what they seem: representing uncertainty in soil- property maps. Proceedings of the 7th International Symposium on Spatial Accuracy Assessment in Natural Resources and Environmental Sciences; Lisboa. 2006. [Google Scholar]
- Holopigian K, Bach M. A primer on common statistical errors in clinical ophthalmology. Documenta ophthalmologica. 2010;121(3):215–222. doi: 10.1007/s10633-010-9249-7. [DOI] [PubMed] [Google Scholar]
- Iidaka T, Matsumoto A, Nogawa J, Yamamoto Y, Sadato N. Frontoparietal network involved in successful retrieval from episodic memory. Spatial and temporal analyses using fMRI and ERP. Cereb Cortex. 2006;16:1349–1360. doi: 10.1093/cercor/bhl040. [DOI] [PubMed] [Google Scholar]
- Jones GE. How to lie with charts: LaPuerta. 2006. [Google Scholar]
- McCabe DP, Castel AD. Seeing is believing: the effect of brain images on judgments of scientific reasoning. cognition. 2008;107(1):343–352. doi: 10.1016/j.cognition.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Sachs L. Angewandte Statistik: Anwendung statistischer Methoden. Berlin: Springer; 1992. [Google Scholar]
- Shehzad Z, Kelly AM, Reiss PT, Gee DG, Gotimer K, Uddin LQ, Lee SH, Margulies DS, Roy AK, Biswal BB, Petkova E, Castellanos FX, Milham MP. The resting brain: unconstrained yet reliable. Cereb Cortex. 2009;19(10):2209–2229. doi: 10.1093/cercor/bhn256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemonsen S, Finsterbusch J, Matschke J, Lorenzen A, Ding XQ, Fiehler J. Age-dependent normal values of T2* and T2′ in brain parenchyma. AJNR Am J Neuroradiol. 2008;29(5):950–955. doi: 10.3174/ajnr.A0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106(31):13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. Journal of Neurophysiology. 2010;103(1):297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Kelly C, Adelstein JS, Klein DF, Castellanos FX, Milham MP. Reliable Intrinsic Connectivity Networks: Test-Retest Evaluation Using ICA and Dual Regression Approach. NeuroImage. 2009;49(3):2163–2177. doi: 10.1016/j.neuroimage.2009.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]