Abstract
Background
sabR is a pleiotropic regulatory gene which has been shown to positively regulate the nikkomycin biosynthesis and negatively affect the sporulation of Streptomyces ansochromogenes. In this study, we investigate the mechanism of SabR on modulating nikkomycin production in Streptomyces ansochromogenes.
Results
The transcription start point of sabR was determined by high-resolution S1 nuclease mapping and localized at the nucleotide T at position 37 bp upstream of the potential sabR translation start codon (GTG). Disruption of sabR enhanced its own transcription, but retarded the nikkomycin production. Over-expression of sabR enhanced nikkomycin biosynthesis in Streptomyces ansochromogenes. EMSA analysis showed that SabR bound to the upstream region of sanG, but it did not bind to the upstream region of its encoding gene (sabR), sanF and the intergenic region between sanN and sanO. DNase 1 footprinting assays showed that the SabR-binding site upstream of sanG was 5'-CTTTAAGTCACCTGGCTCATTCGCGTTCGCCCAGCT-3' which was designated as SARE. Deletion of SARE resulted in the delay of nikkomycin production that was similar to that of sabR disruption mutant.
Conclusions
These results indicated that SabR modulated nikkomycin biosynthesis as an enhancer via interaction with the promoter region of sanG, and expanded our understanding about regulatory cascade in nikkomycin biosynthesis.
Background
Two-thirds of all the known antibiotics are produced by Streptomyces which possess complex morphological differentiation [1]. Antibiotic biosynthesis is highly regulated and generally occurs in a growth-phase-dependent manner [2]. Moreover, the regulation of antibiotic biosynthesis involves complex networks that consist of pathway-specific regulatory genes, pleiotropic regulatory genes and global regulatory genes [3-5]. Over a decade of years, many transcriptional regulators have been identified and their biological functions have been revealed. Among them, the best known system under γ-butyrolactone control has been characterized in S. griseus [5]. Previous studies reported a model describing how A-Factor and its receptor-ArpA mediate pleiotropic effects on morphological differentiation and biosynthesis of secondary metabolites in Streptomyces. Binding of A-Factor to ArpA derepresses the expression of adpA that encodes a global transcriptional activator. AdpA initiates the expression of pathway-specific regulatory genes, such as strR in streptomycin biosynthesis, griR in grixazone biosynthesis and other genes (sprA, sprB, sprD, sprT [6]and sgmA [7]) related to aerial mycelium formation [8,9]. Streptomyces antibiotic regulatory proteins (SARPs) are the most common activators of antibiotic biosynthetic gene clusters. Thus, SARPs are potentially the ultimate target for some quorum-sensing signaling pathways that switch on antibiotic biosynthesis [10-16].
The peptidyl nucleoside antibiotic nikkomycin, produced by Streptomyces ansochromogenes 7100 [17] and Streptomyces tendae Tü 901 [18], is a promising antibiotic against phytopathogenic fungi and human pathogens. In recent years, considerable progress has been made in understanding nikkomycin biosynthesis [13,17-21]. The san gene cluster for the nikkomycin biosynthesis includes over 20 open reading frames (ORFs) consisting of three deduced transcriptional units (sanO-V, sanN-I and sanF-X) and a pathway-specific regulatory gene (sanG). Among them, the role of sanG has been studied in S. ansochromogenes [13,22]. The previous work proved that sanG regulated nikkomycin production by controlling the transcription of the sanO-V and sanN-I operons directly, but did not control the expression of sanF-X operon [13]. The non-coding region of sanG extends to 1 kb upstream of sanG contains five binding sites of AdpA-L which positively controls the transcription of sanG [23]. Except AdpA-L, no any other factors triggering the transcriptional changes of sanG have been reported up to now.
A regulatory gene (sabR) outside of san cluster was cloned from S. ansochromogenes previously. Disruption of sabR retarded nikkomycin production in liquid media containing glucose or glycerol as carbon source and enhanced the sporulation of S. ansochromogenes [24]. The deduced product of sabR belongs to a large family of TetR-like proteins and it is similar to γ-butyrolactone receptor which has the features with helix-turn-helix (HTH) motif located in the N-termini and butyrolactone-binding motif in the C-termini. Most proteins of this family act as repressors of secondary metabolism in Streptomyces [25,26]. Recently, several genes encoded this family proteins have been found to play a positive role during morphological development and secondary metabolism, such as tarA [27], crpA [28] and spbR [15]. In this study, the function of SabR on the regulation of sanG expression was studied. These results will expand the limited understanding of regulatory mechanism during nikkomycin biosynthesis.
Results
Disruption of sabR enhanced its own transcription
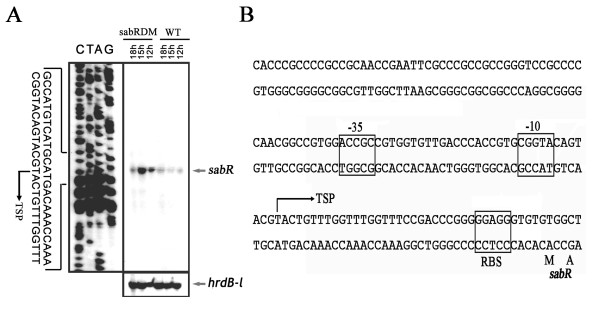
To determine the transcription start point (TSP) of sabR and to investigate whether sabR regulates its own transcription, S1 nuclease protection assay was performed. Total RNAs isolated from S. ansochromogenes and sabR disruption mutant with different time points were hybridized with 32P-labelled probe (see Methods and Table 1). The result showed that sabR has a single transcription start point (tsp), which is localized at the nucleotide T at position 37 bp upstream of the potential sabR translational start codon (GTG) (Figure 1A and 1B). Disruption of sabR quickly enhanced its own transcriptional level in the SP medium at 12, 15 and 18 h, whereas the transcriptional levels of sabR in wild-type strain tend to be weaker and constant at the same conditions (Figure 1A). After 18 h, the transcription of sabR in its disruption mutant was decreased to the same level as wild-type strain (data not shown). These results suggested that the expression of sabR could repress its own transcription at the early stage of growth.
Table 1.
Primers used in this study
| Gene and primer | Positions* | Sequence (5'-3') |
|---|---|---|
| sabR | ||
| sab1-F | +38 to +57 | CATATGGCTCAGCAGGACCGAGC |
| sab1-R | +697 to +681 | CTCGAGGCAGGCGATGCCCGACA |
| sab2-F | +38 to +57 | CATATGGCTCAGCAGGACCGAGC |
| sab2-R | +700 to +681 | GGATCCTCAGCAGGCGATGCCCGACA |
| ER-F | -132 to -115 | CCCCGTGGCACCGTTCAT |
| ER-R | +143 to +160 | GACACCGGCCCGCTTGAG |
| S1R-F | -457 to -440 | GCCAGAGCCGACACCACA |
| S1R-R | +44 to +60 | ACGGCTCGGTCCTGCTG |
| sanG | ||
| Gare1-F | -1035 to -1016 | GGAATTCCGCCCGGCAGCAGCTGGACT |
| Gare1-R | -62 to -85 | GGGGTACCAAGTGGCTCCATGTATCCGCGACC |
| Gare2-F | -34 to -12 | GCTCTAGACCAGCTCAGGAGAATGCTCGATA |
| Gare2-R | +772 to +753 | CCCAAGCTTCTCCGCGACCTCGTCATCAT |
| EG0-F | +1404 to +1423 | AGGCCACCCTGCAGACGTAC |
| EG0-R | +1741 to +1724 | GAGGAGCGTGTCGGCTTG |
| EG1-F | -415 to -396 | GCGGGAGAACGTCACCTGTT |
| EG1-R | +104 to +86 | CGGGTCGGCTGTGGTGAGT |
| EG2-F | +340 to +360 | GCTCCGAGACCGTGACGAAAG |
| EG2-R | +763 to +743 | CTCGTCATCATCAGCGTGGGT |
| EG3-F | +743 to +763 | ACCCACGCTGATGATGACGAG |
| EG3-R | +1218 to +1198 | GACGCGGTTTGTTGCTCTTGA |
| RealG-F | +1405 to +1423 | GGCCACCCTGCAGACGTAC |
| RealG-R | +1536 to +1519 | CGGGACAGGTCGAACGTG |
| sanN and sanO | ||
| ENO-F | -114 to -98 | TCTTGGTCGCCAGGTCC |
| ENO-R | +135 to +153 | CTTCGGATGCTGAATGTGC |
| sanF | ||
| EF-F | -267 to -250 | CGCGCAGGTCGGCCAGGT |
| EF-R | +234 to +213 | TACTGCTTCTCGTGCTTCGGGT |
| RealF-F | +70 to +87 | GGTGCTGACGCTCGACTC |
| RealF-R | +257 to +238 | TGAGGTCCACGAGGTTCATC |
| hrdB | ||
| S1H-F | -820 to -803 | GGGTACGCCCCGTCAGTG |
| S1H-R | +241 to +224 | AGCCTTTCCCCGCTCAAT |
| RealH-F | +290 to +308 | ACTGAGTGGCCGGAATCTG |
| RealH-R | +225 to +206 | GTCTCTGTCATGGCGCTCAT |
* The nucleotide positions for sanG, sanN, sanO, sanF, sabR and hrdB are given with respect to their transcription start points as +1. The enzyme recognition sites are underlined in the primer sequence.
Figure 1.
Transcriptional analysis of sabR. A, High resolution S1 nuclease mapping of sabR. The sabR transcripts were detected at 12, 15 and 18 h of growth from the wild-type strain (WT) and sabR disruption mutant (sabRDM). The arrowhead indicates the transcription start point (TSP) of sabR. The S. ansochromogenes hrdB-l encoding a principal sigma factor was used as control. B, The nucleotide sequence of promoter region of sabR.
Over-expression of sabR accelerated nikkomycin production
In order to over-express sabR, the pIJ8600::sabR (pIJ8600R) in which the PtipA replaced the promoter of sabR was constructed, then this recombinant plasmid was introduced into the Streptomyces ansochromogenes (wild-type strain) by conjugal transfer as described previously [23]. The resulting transformant was designated as 8600R. The nikkomycin bioassay and phenotype showed that the over-expression of sabR accelerated nikkomycin production and delayed the morphological differentiation of 8600R in the presence of thiostrepton, whereas nikkomycin production and the morphological differentiation have no obvious difference comparing with the control in the absence of thiostrepton (Figure 2A, B and 2C). The delayed morphological differentiation of 8600R grown on MMG medium was observed by scanning electron microscopy after incubation for 96 h, and the difference was gradually disappeared after incubation for 120 h. It seems that glucose used as carbon source played an associated role in this regulatory process since the phenotype was not appeared in the media using mannitol as carbon source. These results further confirmed that sabR regulates the nikkomycin biosynthesis positively and morphological differentiation negatively under certain conditions.
Figure 2.
Effects of over-expresson of sabR on nikkomycin biosynthesis and morphological differentiation of S. ansochromogenes. A, Nikkomycin bioassay of fermentation filtrates from different strains with induction of thiostrepton (the left side) or without induction of thiostrepton as control (the right side). Thiostrepton (10 μg ml-1) was added to the cultures after incubation for 12 h in SP medium. B, Phenotype of the sabR overexpressed strain (8600R) with induction of thiostrepton (the left side) or without induction of thiostrepton as control (the right side). Thiostrepton (10 μg ml-1) was added to the medium. C, Scanning electron micrographs of 8600R and 8600 which were grown at 28°C for 96 h in different media. MMM, MMG and MS media supplemented with thiostrepton (10 μg ml-1) were used. 8600, the wild-type strain carrying pIJ8600. MMM, minimal medium (MM) containing mannitol (0.5 %, w/v) as carbon source; MMG, MM containing glucose (1 %, w/v) as carbon source; MS, Mannitol soya flour medium.
Disruption of sabR decreased the transcription of sanG and sanF
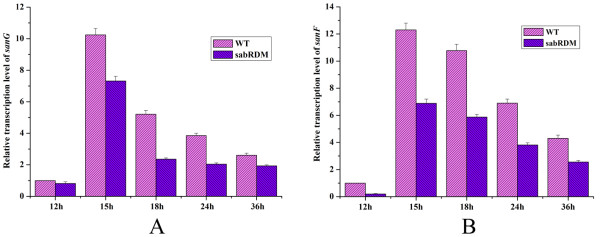
In order to know how SabR regulates nikkomycin biosynthesis in S. ansochromogenes, the effect of sabR on the transcriptions of sanG and sanF-X operon was measured by real-time quantitative PCR. The transcripts of sanG and sanF were lower in the sabR disruption mutant in comparison with that in the wild-type strain after fermentation for 12 h to 36 h (Figure 3). Especially, the transcripts of sanG and sanF were almost reduced to 50% in the sabR disruption mutant (sabRDM) in contrast to wild-type strain (WT) at 18 h. After 36 h, the transcripts of sanG and sanF in sabRDM gradually restored to the same level of WT (data not shown), suggesting that sabR could positively regulate the nikkomycin biosynthesis by modulating the transcription of sanG and sanF at the early stage of cell growth.
Figure 3.
Transcriptional analysis of sanG (A) and sanF (B) by real-time RT-PCR. The sanG and sabF transcriptional levels were detected after fermentation for 12, 15, 18, 24 and 36 h in wild-type strain (WT) and sabR disruption mutant (sabRDM). Error bars were calculated from three independent samples in each reaction.
SabR bound to the upstream region of sanG
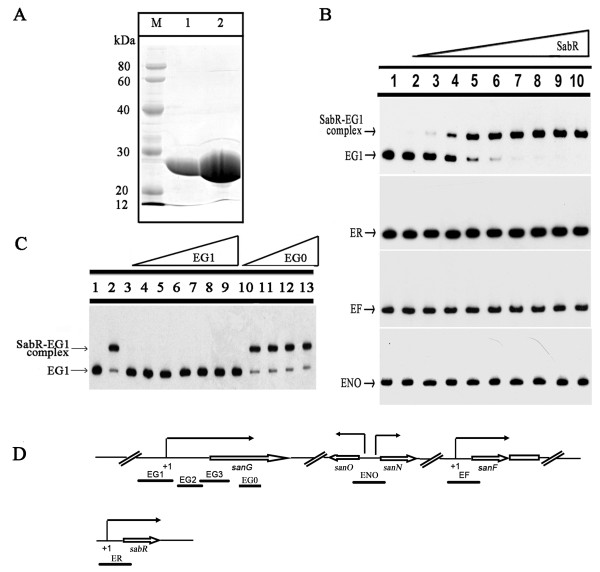
To determine the role of SabR in the regulation of nikkomycin biosynthesis, a series of EMSAs were performed. SabR was over-expressed in E. coli as His6-tagged protein and purified to near homogeneity by a single chromatography on Ni-NTA resin (Figure 4A). The sanG probes (EG1, EG2 and EG3), sabR probe ER, sanF probe EF, as well as one probe ENO covering the transcription start points of sanN and sanO were used (Figure 4D). EMSAs showed that the purified His6-tagged SabR bound to the probe EG1 of sanG to form a complex, but no complex was formed to the probe EG2 and EG3 of sanG. Meanwhile, no significant shift was found for probes sabR, sanF, sanN and sanO, suggesting that SabR regulated the transcription of sabR and sanF indirectly (Figure 4B). EMSAs with unlabelled specific and non-specific competitor DNA were used as controls (Figure 4C). These results indicated that SabR regulated nikkomycin biosynthesis directly by interaction with the upstream region of sanG. As SanG controls the transcription of sanN and sanO, SabR regulates the transcription of sanN and sanO via directly modulating the transcription of sanG.
Figure 4.
EMSA analysis of SabR binding to the upstream of sanG, sabR, sanN, sanO and sanF. A, Purification of the SabR-His6 from E. coli. M, protein marker; 1 and 2, purified SabR-His6 protein. B, The upstream region of sanG, sabR, sanN, sanO or sanF was incubated with or without increasing amounts of SabR-His6 (lanes 1-10 contain 0, 52, 104, 130, 208, 260, 390, 520, 650 and 780 nM, respectively). C, Competition assays using unlabeled specific DNA EG1 and nonspecific competitor DNA EG0. Lanes 3-9, EMSA of 208 nM SabR-His6 with labeled probe and unlabeled specific competitor EG1. Lanes 10-13, EMSA of 208 nM SabR-His6 with labeled probe and nonspecific competitor EG0. The arrows indicate the free probe and SabR -DNA complexes. D, The gene organization of sanG, sanNO, sanF and sabR.
Detection of the SabR-binding sites
To identify the specific binding sites of SabR in the upstream region of sanG, DNase 1 footprinting assay was carried out using [γ-32P]-labeled probe. One region at positions -64 to -29 nucleotides was protected by SabR from DNase 1 digestion, its sequence was 5'-CTTTAAGTCACCTGGCTCATTCGCGTTCGCCCAGCT-3' (Figure 5A and 5B). This sequence showed resemblance to the reported ARE which were bound by γ-butyrolactone receptors described previously (Figure 5C), and it was designated as SARE. These results confirmed that SabR regulated nikkomycin biosynthesis by interaction with SARE sequences upstream of sanG directly.
Figure 5.
DNase 1 footprinting analysis of SabR binding to the upstream of sanG. A, DNase 1 footprinting experiments. The amounts of SabR-His6 used in lane 1 to 7 were 0, 208, 260, 390, 520, 650 and 780 nM, respectively. The region protected against DNase 1 digestion by SabR was indicated by solid line. B, Nucleotide sequence of sanG promoter and SabR-binding sites. The transcription start point (TSP) of sanG is indicated by an arrow. The nucleotide sequence of SARE protected against DNase 1 digestion by SabR is underlined. C, Comparison of SARE with the ARE consensus sequence recognized by the Streptomyces γ-butyrolactone receptors. Identical residues are highlighted in black. Arrows indicate the position of the 22 bp inverted repeat sequence identified as a consensus sequence (ARE box) recognized by the γ-butyrolactone autoregulator receptor protein ArpA[39].
The function of SARE upstream of sanG
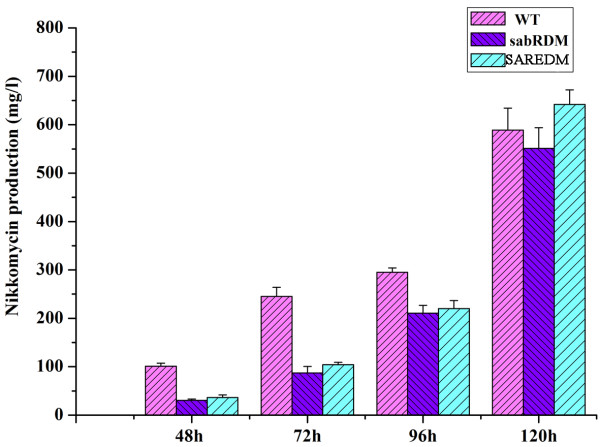
In order to know the function of SARE and its relationship with SabR in vivo, SARE deletion mutant (SAREDM) was constructed. The bioassay showed that nikkomycin production was delayed in the SAREDM as that in the SabRDM from 48 h to 96 h fermentation. After 96 h, the nikkomycin production in SAREDM gradually restored to the level of WT, even slightly higher at 120 h (Figure 6). Therefore, SARE exhibited identical effect as sabR on nikkomycin production, further confirming that SabR positively regulates nikkomycin biosynthesis by interaction with SARE region upstream of sanG positively.
Figure 6.
Analysis of nikkomycin production from 48 to 120 h fermentation of the wild-type strain (WT), sabR disruption mutant (sabRDM) and SARE deletion strain (SAREDM). Error bars were calculated from three independent samples in each experiment.
Discussion
Our results revealed that SabR played not only the positive role for nikkomycin biosynthesis but also a negative role for morphological differentiation in S. ansochromogenes. Disruption of sabR resulted in the decrease of nikkomycin production, a phenomenon identical to pristinamycin production in spbR disruption mutant of S. pristinaespiralis [15]. However, disruption of arpA led to increased streptomycin biosynthesis in S. griseus [9] and inactivation of the barA led to precocious virginiamycin biosynthesis in S. virginiae [29]. Different γ-butyrolactone receptors have different effects on the morphological differentiation. SabR and ArpA repressed the morphological differentiation of S. ansochromogenes and S. griseus [8,24], BarA did not affect the morphological differentiation of S. virginiae. These results reflected that γ-butyrolactone receptors play alternative physiological roles involved in species-specific regulatory systems. In fact, two categories of homologs of autoregulator receptors are found in Streptomyces. One group is real receptors (ArpA, BarA, FarA and ScbR) in which binding of autoregulator is confirmed either by direct binding of natural or synthetic ligands or by gel-shift assay using crude culture filtrate [30]; the second group includes regulators (CrpA, CrpB, BarB, BarZ and so on) which show similarity to the first group receptors but lack binding of any autoregulators [31,32]. The regulators belonging to the second group widely distribute in Streptomyces and are usually involved in control of secondary metabolism and/or morphological differentiation. So far, no γ-butyrolactone or its analogue has been identified in S. ansochromogenes and no any ligands of SabR were found, but SabR could bind to the SARE region without ligand (Figure 4). The lack of SabR binding to its upstream region, in spite of the clear repression on sabR expression and opposite effect on nikkomycin production, implied that SabR belongs to the second group.
The demonstration that SabR interacted with the promoter region of sanG supported that ARE existed upstream of genes involved in antibiotic biosynthesis. The results of DNase 1 footprinting showed that SabR protected a sequence similar to those protected by PapR1, TylS and CcaR and provided the experimental evidence that γ-butyrolactone receptors recognized ARE motifs [15]. However, the disability of SabR binding to the upstream region of sabR was unexpected. The lack of SabR binding to its upstream region, mild effect of sabR disruption on sanG expression and almost complete recovery of nikkomycin production in sabR disruption mutant (sabRDM) or SAREDM at 96 h or 120 h cultivation implied that there should be one or more critical regulators other than SabR to control nikkomycin biosynthesis. Further experimental analysis will hopefully elucidate the detailed regulatory relationship between SabR and nikkomycin biosynthesis.
Conclusions
In conclusion, this study presented detailed molecular and genetic analysis for sabR on the production of nikkomycin in S. ansochromogenes. The results revealed that the SabR regulated nikkomycin biosynthesis positively via interaction with the upstream region of sanG. It might be useful to expand the limited understanding of regulation exerted by SabR.
Methods
Strains, plasmids, media and growth conditions
The strains and plasmids used in this study are listed in Table 2. Escherichia coli DH5α, BL21 (DE3), ET12567 (pUZ8002), and their derivative strains were grown at 37°C in Luria-Bertani (LB) medium containing necessary antibiotics for propagating plasmids. The nikkomycin producer, Streptomyces ansochromogenes 7100 and sabR disruption mutant were incubated at 28°C. For nikkomycin production, SP medium (3 % mannitol, 1 % soluble starch, 0.75 % yeast extract, and 0.5 % soy peptone, pH 6.0) was used. Liquid medium YEME and solid medium MM were prepared according to standard procedures [33]. Alternaria longipes was used as indicator strain for nikkomycin bioassay and incubated at 28°C in PDA medium. The plasmid pUC119::kan, pET23b, pIJ8600 and their derivatives were collected in our lab. E. coli-Streptomyces shuttle vector pKC1139 used for gene disruption was kindly provided by Prof. Keith Chater (John Innes Centre, Norwich, UK).
Table 2.
Strains and plasmids used in this study
| Strains or plasmids | relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| S. ansochromogenes 7100 | Wild-type strain | [40] |
| sabRDM | The sabR disruption mutant | [24] |
| E. coli DH5α | F- recA f80 dlacZ ΔM15 | Gibco BRL |
| BL21(DE3) | F- ompT hsdS gal dcm (DE3) | Novagen |
| ET12567 (pUZ8002) | recE dam dcm hsdS Cmr Strr Tetr Kmr | [41] |
| Alternaria longipes | Indicator strain for nikkomycin bioassays | [40] |
| Plasmids | ||
| pBluescript KS+ | Routine cloning and subcloning vector | Stratagene |
| pET23b | Expression vector | Novagen |
| pET23b::sabR | sabR gene cloned in pET23b | This work |
| pIJ8600 | ori pUC, oriT RK2, int ΦC31, tipAp, tsr, aprR | [33] |
| pIJ8600::sabR | sabR gene cloned in the induced vector of pIJ8600 which containing PtipA as promoter | This work |
| pKC1139 | E.coli-Streptomyces shuttle vector | [33] |
| pGARE1 | A 974 bp DNA fragment containing the left flank of SARE was inserted into pUC119::kan | This work |
| pGARE2 | A 806 bp DNA fragment containing the right flank of SARE was inserted into GAREL1 | This work |
| pGARE3 | A 2.8 kb DNA fragment containing the left and right flanks of SARE and kanamycin resistance gene from pGARE2 was inserted into pKC1139 | This work |
| pGARE4 | The 1 kb kanamycin resistance gene was deleted from pGARE3 | This work |
| pGARE5 | A 1.8 kb DNA fragment containing the left and right flanks of SARE from pGARE4 was inserted into pKC1139 | This work |
DNA manipulation and sequencing
Plasmids and genomic DNA were isolated from Streptomyces [33] or E. coli [34] according to the standard protocols. Intergeneric conjugation from E. coli ET12567 to S. ansochromogenes was carried out as described previously [33]. DNA sequencing was performed by Invitrogen Biotechnology Company. Database searching and sequence analysis were carried out using Artemis program (Sanger, UK), FramePlot 2.3 [35] and the program PSI-BLAST[36].
Construction of SARE disruption mutant
Disruption of SARE was performed by gene replacement via homologous recombination. Firstly, a 974 bp DNA fragment was amplified from the genomic DNA of S. ansochromogenes 7100 with primers Gare1-F and Gare1-R, then it was digested with KpnI-EcoRI and inserted into the corresponding sites of pUC119::kan which contains the kanamycin resistance cassette to generate pGARE1. Secondly, an 806 bp DNA fragment was amplified from the genomic DNA of S. ansochromogenes 7100 with primers Gare2-F and Gare2-R, and it was digested with HindIII-XbaI and inserted into the corresponding sites of pGARE1 to generate pGARE2. Thirdly, pGARE2 was digested by HindIII-EcoRI and the 2.8 kb DNA fragment was inserted into the corresponding sites of pKC1139 to generate a recombinant plasmid pGARE3. The plasmid pGARE3 was passed through E. coli ET12567 (pUZ8002) and introduced into S. ansochromogenes 7100 by conjugation [33]. The kanamycin resistance (KanR) and apramycin sensitivity (AprS) colonies were selected, and the SARE disruption mutant was confirmed by PCR amplification and designated as pre-SARE. Meanwhile, the 4.9 kb DNA fragment from pGARE2 digested with XbaI-KpnI was blunted by T4 DNA polymerase and self-ligated to generate pGARE4. Subsequently pGARE4 was digested with HindIII-EcoRI and inserted into the corresponding sites of pKC1139 to give pGARE5, which was then introduced into the pre-SARE strain. The kanamycin sensitive (KanS) strains were selected and the SARE disruption mutants (SAREDM) were confirmed by PCR. The fidelity of all subcloned fragments was confirmed by DNA sequencing.
Construction of a sabR over-expressing strain
In order to analyze the effects of over-expression of sabR on nikkomycin biosynthesis and morphological differentiation, a 672 bp DNA fragment containing the complete sabR was amplified using sab2-F and sab2-R as primers, and then it was inserted into the NdeI-BamHI sites of pIJ8600 to generate pIJ8600::sabR, which was subsequently integrated into the chromosomal ΦC31 attB site of S. ansochromogenes 7100 by conjugation.
RNA isolation and S1 mapping analysis
Total RNAs were isolated from both S. ansochromogenes and sabR disruption mutant after incubation in SP medium for different times as described previously [13]. Mycelium was collected, frozen quickly in liquid nitrogen and ground into fine white powder. RNAs were then extracted using the Trizol reagent (Invitrogen, USA) according to the manufacturer's protocol. Quality and quantity of RNAs were examined by UV spectroscopy and checked by agarose gel electrophoresis. To erase the chromosomal DNA contamination, each sample was treated with DNase 1 and tested by PCR to ensure that there was no chromosomal DNA. To investigate transcription of sabR during nikkomycin biosynthesis, S1 protection assays were performed using the hrdB-like gene (hrdB-l) which encoded the principal sigma factor of S. ansochromogenes and expected to express constant during the time-course as a control. The hrdB-l probe was generated by PCR using the unlabeled primer S1H-F and the primer S1H-R, which was uniquely labeled at its 5' end with [γ-32P]-ATP by T4 polynucleotide kinase (Promega, USA). For sabR, the probe was generated by PCR using the radiolabeled primer S1R-R and the unlabeled primer S1R-F. The DNA sequencing ladders were generated using the fmol DNA cycle sequencing kit (Promega, USA) with the corresponding labeled primers. Protected DNA fragments were analyzed by electrophoresis on 6 % polyacrylamide gels containing 7 M urea.
Real-time quantitative PCR analysis
RNA samples (1 μg) were reversedly transcribed using SuperScript™ III and random pentadecamers (N15) as described by the vendor of the enzyme (Invitrogen). Samples of cDNA were then amplified and detected with the ABI-PRISM 7000 Sequence Detection System (Applied Biosystems) using optical grade 96-well plates. Each reaction (50 μl) contained 0.1-10 ng of reversed-transcribed DNA, 25 μl Power SYBR Green PCR Master Mix (Applied Biosystems), 0.4 μM of both forward and reverse primers for sanG and sanF respectively. The PCR reactive conditions were maintained at 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 60°C for 1 min, fluorescence was measured at the end of each cycle. Data analysis was made by Sequence Detection Software supplied by Applied Biosystems.
Expression and purification of SabR
The coding region of sabR was amplified by using primers sab1-F and sab1-R. The amplified fragment was digested with NdeI-XhoI and inserted into pET23b to generate the expression plasmid pET23b::sabR. After confirmed by DNA sequencing, it was introduced into E. coli BL21 (DE3) for protein expression. When E. coli BL21 (DE3) harboring pET23b::sabR was grown at 37°C in 100 ml LB supplemented with 100 μg ampicillin ml-1 to an OD600 of 0.6, IPTG was added to a final concentration of 0.1 mM and the cultures were further incubated for an additional 12 h at 30°C. The cells were harvested by centrifugation at 6000 g, 4°C for 3 min, washed twice with binding buffer [20 mM Tris base, 500 mM NaCl, 5 mM imidazole, 5 % glycerol (pH 7.9)] and then resuspended in 10 ml of the same buffer. The cell suspension was treated by sonication on ice. After centrifugation (14000 g for 20 min at 4°C), the supernatant was recovered, and SabR-His6 was separated from the whole-cell lysate using Ni-NTA agarose chromatography (Novagen). After extensive washing with buffer [20 mM Tris base, 500 mM NaCl, 60 mM imidazole, 5 % glycerol (pH 7.9)], the SabR-His6 proteins were specifically eluted from the resin with 4 ml elution buffer [20 mM Tris base, 500 mM NaCl, 250 mM imidazole, 5 % glycerol (pH 7.9)] and concentrated to about 20 μg μl-1 by ultrafiltration (Millipore membrane, 3 kDa cut-off size) according to the protocol provided by the manufacturer. Protein purity was determined by Coomassie brilliant blue staining after SDS-PAGE on a 12 % polyacrylamide gel. The purified protein was stored in 5 % glycerol at -70°C.
Electrophoretic mobility-shift assays (EMSAs)
The EMSAs were performed as described previously [37]. The primers were labeled with T4 DNA polynucleotide kinase and the DNA fragments used for [γ-32P]-labeled probes were amplified by PCR, and then purified by using PCR purification kit (Qiagen). For EMSAs with SabR-His6, the sanG probes were generated by PCR using primers EG0-F, EG1-F, EG2-F, EG3-F and EG0-R, EG1-R, EG2-R, EG3-R, which were uniquely labeled at its 5' end with [γ-32P]-ATP using T4 polynucleotide kinase respectively. The sabR, sanF and sanNO probes were generated by PCR using unlabeled primers ER-F, EF-F, ENO-F and the radiolabeled primers ER-R, EF-R and ENO-R, respectively. During the EMSA, the [γ-32P]-labeled DNA probe (1000 cpm) was incubated individually with varying quantities of SabR-His6 at 25°C for 25 min in a buffer containing 1 μg of poly-(dI-dC) (Sigma), 20 mM Tris-base (pH 7.5), 1 mM DTT, 10 mM MgCl2, 0.5 μg calf BSA μl-1 and 5 % glycerol in a total volume of 20 μl. After incubation, protein-DNA complex and free DNA were separated by electrophoresis on non-denaturing 4.5 % polyacrylamide gels with a running buffer containing 45 mM Tris-HCl (pH 8.0), 45 mM boric acid and 1 mM EDTA at 10 V cm-1 and 4°C. Gels were dried and exposed to Biomax radiographic film (Kodak). As controls, unlabeled probe (25-fold, 50-fold, 75-fold, 100-fold, 150-fold, 175-fold and 200-fold specific competitor or 25-fold, 50-fold, 100-fold and 200-fold non-specific competitor) and labeled probe were mixed with SabR-His6 and incubated for 25 min at 25°C. The resulting DNA-protein complexes were then subjected to electrophoresis and autoradiography as described above. In order to quantify all probes, the probe DNA concentration was detected by ultraviolet spectrophotometer at the wavelength of 260 nm.
DNase 1 footprinting
To characterize the SabR-binding sites upstream region of sanG, a DNA fragment was amplified by PCR with the labeled primer EG1-F. The footprinting reaction mixture contained 30,000 cpm of [γ-32P]-labeled DNA probe, 6 ng to 0.3 μg of SabR-His6, 2.5 μg of poly-(dI-dC) (Sigma) and 20 mM Tris-base (pH 7.5), 1 mM DTT, 10 mM MgCl2, 0.5 μg calf BSA μl-1 and 5 % (v/v) glycerol in a total volume of 50 μl. After incubation of the mixture at 25°C for 25 min, 5.5 μl RQ1 RNase-free DNase Buffer and 0.1 U DNase 1 were added to the above reaction and the mixture was incubated for 1 min. The reaction was stopped by adding 50 μl of stop solution (20 mM EGTA, pH 8.0), and 100 μl of phenol/CH3Cl (1:1, v/v). After precipitation in ethanol, the pellet was washed with 75 % (v/v) ethanol and re-suspended in 5 μl of H2O, and then electrophoresed on a 6 % (w/v) polyacrylamide/urea gel.
Nikkomycin bioassay
Nikkomycins produced by S. ansochromogenes 7100 were measured by a disk agar diffusion method using A. longipes as indicator strain. Nikkomycins in culture filtrates were identified by HPLC analysis. For HPLC analysis, Agilent 1100 HPLC and RP C-18 were used. The detection wavelength was 290 nm. Chemical reagent, mobile phase and gradient elution process were referenced as described by Fiedler [38].
Microscopy
The experiments of scanning electron microscopy were performed exactly as described previously [23].
Abbreviations
EMSA: electrophoretic mobility-shift assay; SARE: autoregulatory element of sanG; SARP: Streptomyces antibiotic regulatory protein; TSP: transcription start point.
Authors' contributions
HRT and GL conceived the project. YYP performed the experiments, LQW, XHH and YQT conducted partial data analysis. YYP, GL and HRT wrote the paper. All authors read and approved the final manuscript. The authors declare no conflict of interest.
Contributor Information
Yuanyuan Pan, Email: panyy@im.ac.cn.
Linqi Wang, Email: wanglq98@hotmail.com.
Xihong He, Email: hexihong05@mails.gucas.ac.cn.
Yuqing Tian, Email: tianyq@im.ac.cn.
Gang Liu, Email: liug@sun.im.ac.cn.
Huarong Tan, Email: tanhr@im.ac.cn.
Acknowledgements
We are grateful to Prof. Keith Chater (John Innes Centre, Norwich, UK) for providing E. coli ET12567 (pUZ8002) and plasmids (pKC1139 and pSET152). We would like to thank Dr. Brenda Leskiw (University of Alberta, Canada) for the gift of apramycin. We thank Dr. Wenbo Ma (Assistant Professor in University of California at Riverside, CA) for critical reading and revising of the manuscript. This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 31030003 and 30970072) and the Ministry of Science and Technology of China (2009CB118905).
References
- Hopwood DA. Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology. 1999;145:2183–2202. doi: 10.1099/00221287-145-9-2183. [DOI] [PubMed] [Google Scholar]
- Chater KF. Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos Trans R Soc Lond B Biol Sci. 2006;361:761–768. doi: 10.1098/rstb.2005.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias P, Fernandez-Moreno MA, Malpartida F. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol. 1999;181:6958–6968. doi: 10.1128/jb.181.22.6958-6968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Hwang Y, Kim S, Kim E, Choi C. Effect of a global regulatory gene, afsR2, from Streptomyces lividans on avermectin production in Streptomyces avermitilis. J Biosci Bioeng. 2000;89:606–608. doi: 10.1016/S1389-1723(00)80065-1. [DOI] [PubMed] [Google Scholar]
- Horinouchi S. Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci Biotechnol Biochem. 2007;71:283–299. doi: 10.1271/bbb.60627. [DOI] [PubMed] [Google Scholar]
- Kato J, Chi WJ, Ohnishi Y, Hong SK, Horinouchi S. Transcriptional control by A-factor of two trypsin genes in Streptomyces griseus. J Bacteriol. 2005;187:286–295. doi: 10.1128/JB.187.1.286-295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, Suzuki A, Yamazaki H, Ohnishi Y, Horinouchi S. Control by A-factor of a metalloendopeptidase gene involved in aerial mycelium formation in Streptomyces griseus. J Bacteriol. 2002;184:6016–6025. doi: 10.1128/JB.184.21.6016-6025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Kameyama S, Onaka H, Horinouchi S. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol Microbiol. 1999;34:102–111. doi: 10.1046/j.1365-2958.1999.01579.x. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Yamazaki H, Kato JY, Tomono A, Horinouchi S. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci Biotechnol Biochem. 2005;69:431–439. doi: 10.1271/bbb.69.431. [DOI] [PubMed] [Google Scholar]
- Wietzorrek A, and Bibb M. A novel family of proteins that regulates antibiotic production in Streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol. 1997;25:1181–1184. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- Sheldon PJ, Busarow SB, Hutchinson CR. Mapping the DNA-binding domain and target sequences of the Streptomyces peucetius daunorubicin biosynthesis regulatory protein, DnrI. Mol Microbiol. 2002;44:449–460. doi: 10.1046/j.1365-2958.2002.02886.x. [DOI] [PubMed] [Google Scholar]
- Horinouchi S. AfsR as an integrator of signals that are sensed by multiple serine/threonine kinases in Streptomyces coelicolor A3(2) J Ind Microbiol Biotechnol. 2003;30:462–467. doi: 10.1007/s10295-003-0063-z. [DOI] [PubMed] [Google Scholar]
- Liu G, Tian YQ, Yang HH, Tan HR. A pathwayspecific transcriptional regulatory gene for nikkomycin biosynthesis in Streptomyces ansochromogenes that also influences colony development. Mol Microbiol. 2005;55:1855–1866. doi: 10.1111/j.1365-2958.2005.04512.x. [DOI] [PubMed] [Google Scholar]
- Li R, Liu G, Xie ZJ, He XH, Chen WQ, Deng ZX, Tan HR. PolY, a transcriptional regulator with ATPase activity, directly activates transcription of polR in polyoxin biosynthesis in Streptomyces cacaoi. Mol Microbiol. 2010;75:349–364. doi: 10.1111/j.1365-2958.2009.06968.x. [DOI] [PubMed] [Google Scholar]
- Folcher M, Gaillard H, Nguyen LT, Nguyen KT, Lacroix P, Bamas-Jacques N, Rinkel M, Thompson CJ. Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic Biosynthesis, and expression of a superoxide dismutase. J Biol Chem. 2001;276:44297–44306. doi: 10.1074/jbc.M101109200. [DOI] [PubMed] [Google Scholar]
- Wang LQ, Tian XY, Wang J, Yang HH, Fan KQ, Xu GM, Yang KQ, Tan HR. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc Natl Acad Sci. 2009;106:8617–8622. doi: 10.1073/pnas.0900592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HB, Wang GJ, Tian YQ, Liu G, Tan HR. SanM catalyzes the formation of 4-pyridyl-2-oxo-4-hydroxyisovalerate in nikkomycin biosynthesis by interacting with SanN. Biochem Biophys Res Commun. 2007;361:196–201. doi: 10.1016/j.bbrc.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Bruntner C, Lauer B, Schwarz W, Möhrle V, Bormann C. Molecular characterization of co-transcribed genes from Streptomyces tendae Tü901 involved in the biosynthesis of the peptidyl moiety of the peptidyl nucleoside antibiotic nikkomycin. Mol Gen Genet. 1999;262:102–114. doi: 10.1007/pl00008637. [DOI] [PubMed] [Google Scholar]
- Lauer B, Russwurm R, Schwarz W, Kálmánczhelyi A, Bruntner C, Rosemeier A, Bormann C. Molecular characterization of co-transcribed genes from Streptomyces tendae Tü901 involved in the biosynthesis of the peptidyl moiety and assembly of the peptidyl nucleoside antibiotic nikkomycin. Mol Gen Genet. 2001;264:662–673. doi: 10.1007/s004380000352. [DOI] [PubMed] [Google Scholar]
- Chen H, Hubbard BK, O'Connor SE, Walsh CT. Formation of beta-hydroxy histidine in the biosynthesis of nikkomycin antibiotics. Chem Biol. 2002;9:103–112. doi: 10.1016/S1074-5521(02)00090-X. [DOI] [PubMed] [Google Scholar]
- Niu GQ, Liu G, Tian YQ, Tan HR. SanJ, an ATP-dependent picolinate-CoA ligase, catalyzes the conversion of picolinate to picolinate-CoA during nikkomycin biosynthesis in Streptomyces ansochromogenes. Metab Eng. 2006;8:183–195. doi: 10.1016/j.ymben.2005.12.002. [DOI] [PubMed] [Google Scholar]
- He XH, Li R, Pan YY, Liu G, Tan HR. SanG, a transcriptional activator, controls nikkomycin biosynthesis through binding to the sanN-sanO intergenic region in Streptomyces ansochromogenes. Microbiology. 2010;156:828–837. doi: 10.1099/mic.0.033605-0. [DOI] [PubMed] [Google Scholar]
- Pan YY, Liu G, Yang HH, Tian YQ, Tan HR. The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol Microbiol. 2009;72:710–723. doi: 10.1111/j.1365-2958.2009.06681.x. [DOI] [PubMed] [Google Scholar]
- Li WL, Liu G, Tan HR. Disruption of sabR affects nikkomycin biosynthesis and morphogenesis in Streptomyces ansochromogenes. Biotechnol Lett. 2003;25:1491–1497. doi: 10.1023/A:1025402902098. [DOI] [PubMed] [Google Scholar]
- Novakova R, Kutas P, Feckova L, Kormanec J. The role of the TetR-family transcriptional regulator Aur1R in negative regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology. 2010;156:2374–2383. doi: 10.1099/mic.0.037895-0. [DOI] [PubMed] [Google Scholar]
- Hillerich B, Westpheling J. A new TetR family transcriptional regulator required for morphogenesis in Streptomyces coelicolor. J Bacteriol. 2008;190(1):61–67. doi: 10.1128/JB.01316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P, Scharfenstein LL, Dyer JM, Cary JW. Disruption of a gene encoding a putative γ-butyrolactone-binding protein in Streptomyces tendae affects nikkomycin production. Appl Microbiol Biotechnol. 2001;56:414–419. doi: 10.1007/s002530100621. [DOI] [PubMed] [Google Scholar]
- Onaka H, Nakagawa T, Horinouchi S. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol Microbiol. 1998;28:743–753. doi: 10.1046/j.1365-2958.1998.00832.x. [DOI] [PubMed] [Google Scholar]
- Nakano H, Takehara E, Nihira T, Yamada Y. Gene replacement analysis of the Streptomyces virginiae barA Gene encoding the butyrolactone autoregulator receptor reveals that BarA acts as a repressor in virginiamycin biosynthesis. J Bacteriol. 1998;180:3317–3322. doi: 10.1128/jb.180.13.3317-3322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano E. g-Butyrolactones Streptomyces signaling molecules regulating antibiotic production and differentiation. Curr Opin Microbiol. 2006;9:1–8. doi: 10.1016/j.mib.2005.12.015. [DOI] [PubMed] [Google Scholar]
- Nishida H, Ohnishi Y, Beppu T, Horinouchi S. Evolution of gamma-butyrolactone synthases and receptors in Streptomyces. Environ Microbiol. 2007;9(8):1986–1994. doi: 10.1111/j.1462-2920.2007.01314.x. [DOI] [PubMed] [Google Scholar]
- Xu GM, Wang J, Wang LQ, Tian XY, Yang HH, Fan KQ, Yang KQ, Tan HR. "Pseudo" gamma-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J Biol Chem. 2010;285(35):27440–27448. doi: 10.1074/jbc.M110.143081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: The John lnnes Foundation. 2000.
- Sambrook J, Fritsch T, Maniatis EF. Molecular Cloning: A laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. 1989.
- Ishikawa J, Hotta K. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol Lett. 1999;174:251–253. doi: 10.1111/j.1574-6968.1999.tb13576.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;125:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HH, Wang LQ, Xie ZJ, Tian YQ, Liu G, Tan HR. The tyrosine degradation gene hppD is transcriptionally activated by HpdA and repressed by HpdR in Streptomyces coelicolor, while hpdA is negatively autoregulated and repressed by HpdR. Mol Microbiol. 2007;65:1064–1077. doi: 10.1111/j.1365-2958.2007.05848.x. [DOI] [PubMed] [Google Scholar]
- Fiedler HP. Screening for new microbial products by high performance liquid chromatography using a photodiode array detector. J Chromatogr. 1984;316:487–494. doi: 10.1016/s0021-9673(00)96176-4. [DOI] [PubMed] [Google Scholar]
- Onaka H, Horinouchi S. DNA-binding activity of the A-factor receptor protein and its recognition DNA sequences. Mol Microbiol. 1997;24:991–1000. doi: 10.1046/j.1365-2958.1997.4081772.x. [DOI] [PubMed] [Google Scholar]
- Zeng HM, Tan HR, Li JL. Cloning and function of sanQ: a gene involved in nikkomycin biosynthesis of Streptomyces ansochromogenes. Curr Microbiol. 2002;45:175–179. doi: 10.1007/s00284-001-0115-4. [DOI] [PubMed] [Google Scholar]
- Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2) J Bacteriol. 1999;181:204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]