Abstract
Background
The luxS gene in Shewanella oneidensis was shown to encode an autoinducer-2 (AI-2)-like molecule, the postulated universal bacterial signal, but the impaired biofilm growth of a luxS deficient mutant could not be restored by AI-2, indicating it might not have a signalling role in this organism.
Findings
Here, we provide further evidence regarding the metabolic role of a luxS mutation in S. oneidensis. We constructed a luxS mutant and compared its phenotype to a wild type control with respect to its ability to remove AI-2 from the medium, expression of secreted proteins and biofilm formation. We show that S. oneidensis has a cell-dependent mechanism by which AI-2 is depleted from the medium by uptake or degradation at the end of the exponential growth phase. As AI-2 depletion is equally active in the luxS mutant and thus does not require AI-2 as an inducer, it appears to be an unspecific mechanism suggesting that AI-2 for S. oneidensis is a metabolite which is imported under nutrient limitation. Secreted proteins were studied by iTraq labelling and liquid chromatography mass spectrometry (LC-MS) detection. Differences between wild type and mutant were small. Proteins related to flagellar and twitching motility were slightly up-regulated in the luxS mutant, in accordance with its loose biofilm structure. An enzyme related to cysteine metabolism was also up-regulated, probably compensating for the lack of the LuxS enzyme. The luxS mutant developed an undifferentiated, loosely-connected biofilm which covered the glass surface more homogenously than the wild type control, which formed compact aggregates with large voids in between.
Conclusions
The data confirm the role of the LuxS enzyme for biofilm growth in S. oneidensis and make it unlikely that AI-2 has a signalling role in this organism.
Keywords: quorum sensing, autoinducer-2, Shewanella oneidensis, luxS, biofilm, secretome
Background
Shewanella oneidensis is a Gram-negative Gammaproteobacterium isolated from the sediment of fresh water habitats [1], and occasionally from water columns and clinical specimens [2]. The most investigated characteristic of S. oneidensis is its ability to use a broad spectrum of electron acceptors [3]. In our previous study, Shewanella species were shown to produce autoinducer-2, proposed to be a universal signal molecule in bacteria [4], and to contain its synthesis gene, luxS [5]. As S. oneidensis readily forms biofilms and lives in bacterial communities and since AI-2 is the product of the widely distributed luxS enzyme, AI-2 signalling would represent a possibility for this species to react to the bacterial density in its environment.
However it is under debate if the signalling role of AI-2 can be generalized. First, LuxS is not a designated enzyme producing AI-2 but rather an enzyme in the central S-adenosyl-methionine cycle [6]. Second, other bacterial species have no genes homologous to the AI-2 receptor of Vibrio species and seem just to transport AI-2 into the cells under nutrient limitation. There is an ABC-transporter in Escherichia coli and in Salmonella typhimurium which is specific for AI-2, but AI-2 taken up into the cell regulates only the expression of this transporter [7]. Recently, a luxS mutant of S. oneidensis strain MR-1 was shown to form a slightly reduced biofilm during the first 16 h of growth [8]. Since the wild type biofilm could not be restored by complementation with AI-2, the authors concluded that disruption of the activated methyl cycle rather than signalling by AI-2 caused the observed change in biofilm growth. This was supported by the observation of impaired growth of the luxS mutant on medium containing methionine, a component of the activated methyl cycle, as the sole sulfur source [8].
Here, we provide independent observations regarding the phenotype of a luxS mutant of S. oneidensis in comparison to a wild type control.
Results and discussion
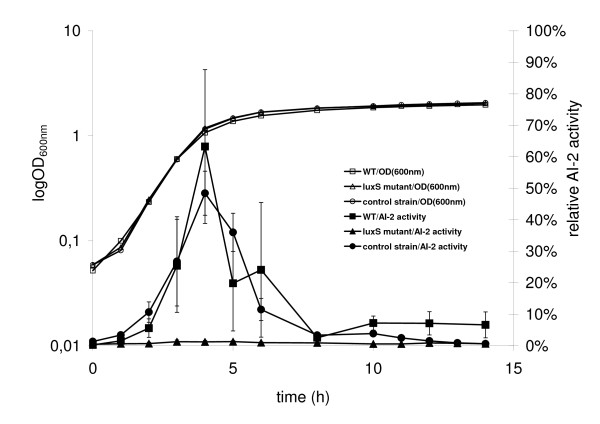
By targeted single-crossingover homologous recombination, the luxS mutant and the WT control carrying the same kanamycin cassette were constructed. The luxS mutant and the WT control grew at almost identical growth-rates in LB medium under aerobic conditions. The luxS mutant did not produce AI-2 (approx. 2% of positive control), while the WT control produced the same amount of AI-2 as the wild type. Furthermore, as observed previously with the wild type strain [5], the WT control accumulated AI-2 in the exponential phase in the medium and depleted it in the early stationary phase (Figure 1.).
Figure 1.
AI-2 production of S. oneidensis wild type, luxS mutant and WT control strain. All three strains were grown in LB at 30°C with shaking. Periodically, the growth was measured (OD600 nm) and sterile culture supernatants were collected and used to determine the produced AI-2. The AI-2 amount in the supernatant was detected by the V. harveyi bioassay and expressed in relative AI-2 amount. All strains grew at identical growth rates. The AI-2 production of the MR-1 wild type strain (WT) and the WT control strain (control strain) are indistinguishable, while that of the luxS mutant was at background level.
AI-2 removal from the culture supernatant
As AI-2 depletion from the medium is AI-2 dependent in S. typhimurium and in E. coli [7], we tested this possibility in S. oneidensis.
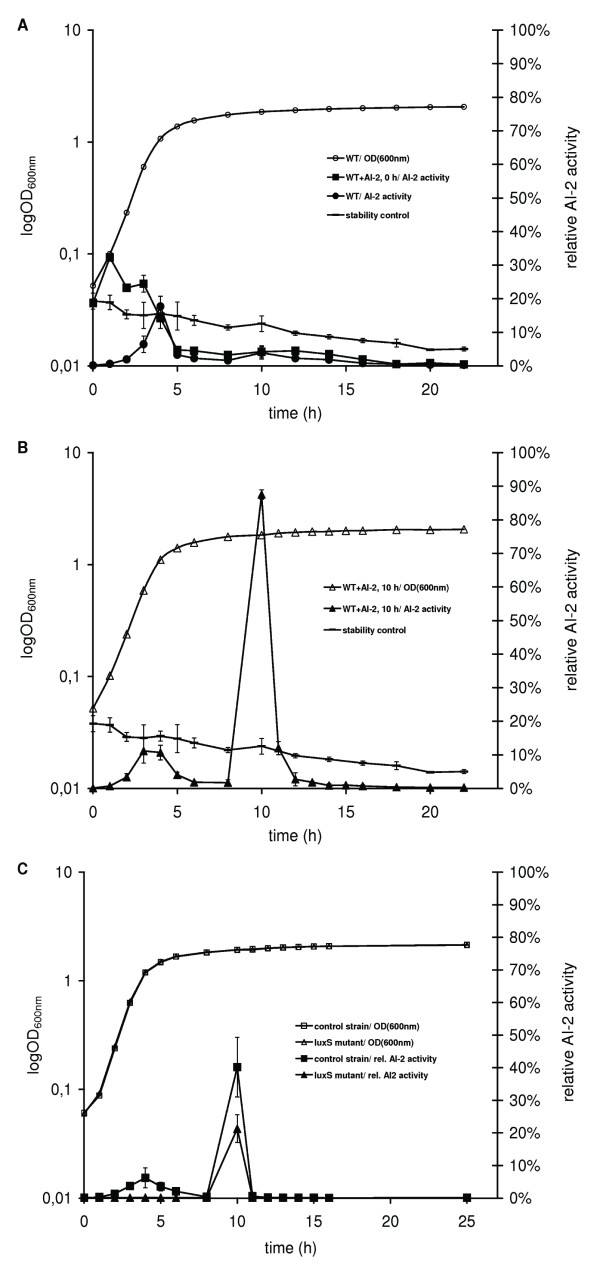
As a first test, S. oneidensis wild-type was supplemented with AI-2 at two different time points: at the beginning of growth, to see if AI-2 depletion can be pre-induced by AI-2 before its natural onset, and after 10 h of growth, when AI-2 cannot be detected anymore in the medium, to determine if it is still depleted. As a result, AI-2 was depleted at the same time point in the native and supplemented cultures, indicating that AI-2 did not induce its depletion. Thus, its depletion appears to be growth-phase dependent (Figure 2A). When added after 10 h of growth, AI-2 disappeared within one hour, indicating that AI-2 depletion was still very active (Figure 2B).
Figure 2.
AI-2 depletion by S. oneidensis wild type, WT control and luxS mutant. (A) S. oneidensis depleted AI-2 added at the beginning of growth (WT+AI-2, 0 h) at the end of the logarithmic phase like the control culture (WT). (B) When the wild type strain was supplemented with AI-2 after 10 h of growth (WT+AI-2, 10 h), as indicated by the dashed line, AI-2 activity increased strongly. Still, the wild type depleted AI-2 within one hour. AI-2 had a low decomposition rate under the tested conditions (stability control). (C) When the luxS mutant and the WT control were supplemented with AI-2 after 10 h of growth, both strains depleted AI-2 within one hour.
To test if the luxS mutant lost this capability, synthetic AI-2 was added to both, the luxS mutant and WT control, after 10 h of growth and AI-2 levels were measured over time. Just like the wild type strain, both strains depleted AI-2 within 1 h after its addition, thus clearly indicating that AI-2 decrease is independent of the presence of AI-2 in this bacterium (Figure 2C).
By contrast, in S. typhimurium and E. coli, the expression of the ABC-transporter is AI-2 dependent and this transporter is specific for AI-2 [7]. Therefore, AI-2 uptake in S. oneidensis might be caused by a transporter with a broad substrate spectrum or a degradation mechanism which is expressed independently of AI-2 at the end of the exponential phase. This finding supports the hypothesis, that AI-2 for S. oneidensis is a metabolite which is imported under nutrient limitation.
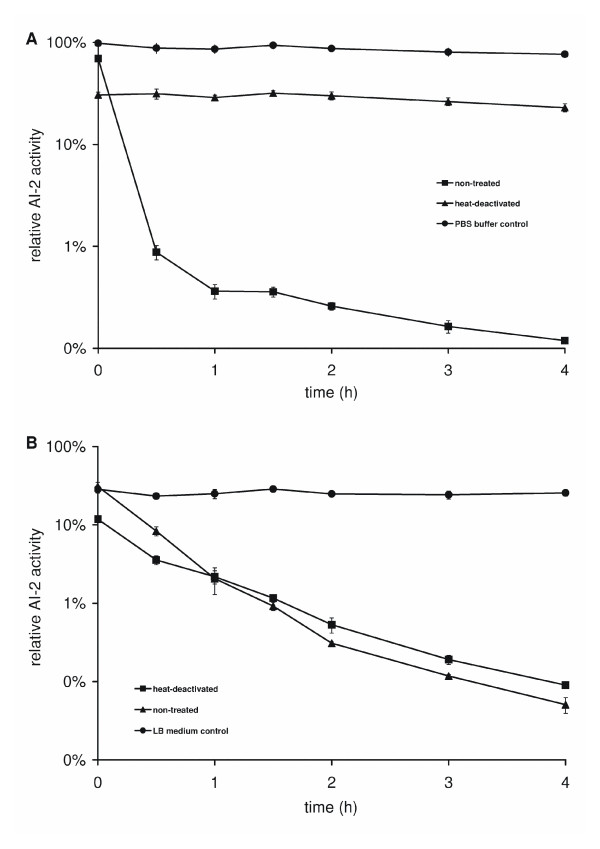
To investigate if AI-2 is depleted by the cells or degraded extracellularly, the cells were pelleted and resuspended in PBS buffer after 10 h of growth, and then this cell suspension, its cell-free supernatant, and heat-inactivated control solutions prepared from these solutions were tested for AI-2 decrease. The living cells removed AI-2 from the medium within one hour, while the heat-killed cells left the AI-2 concentration unaffected, and thus AI-2 depletion is clearly cell-driven in S. oneidensis. Surprisingly, AI-2 decreased substantially also in the cell-free supernatants, although at a lower rate. There was no difference between heat-inactivated and untreated sterile supernatants. This decrease must therefore be caused by the reaction of AI-2 with heat-stable components of the cell-free supernatant, rather than by an enzyme (Figure 3A and 3B).
Figure 3.
Uptake and extracellular depletion of AI-2 by S. oneidensis wild type. (A) Native cells of S. oneidensis washed in PBS depleted externally added AI-2 rapidly, while heat-killed cells were unable to do so. AI-2 was stable in PBS buffer. (B) AI-2 levels decreased in the non-treated as well as in the heat-inactivated supernatants. AI-2 was stable in LB medium.
Secretome analyses
The secretome of S. oneidensis luxS mutant and the WT control strain was compared in LB medium at the beginning of the stationary phase (OD600 nm = 1.5), shortly after the peak of AI-2 activity, when AI-2 would be expected to have induced extracellular proteins.
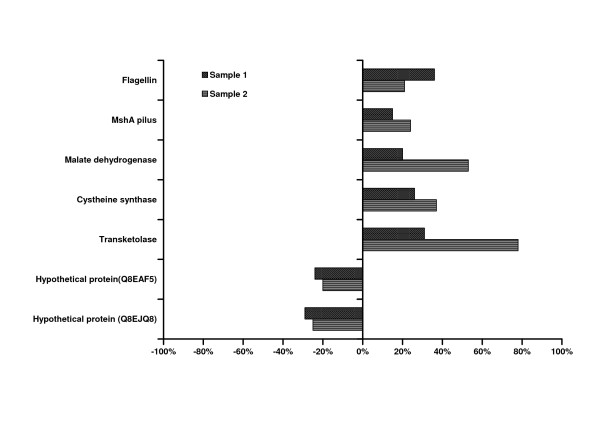
In two independent experiments, 172 proteins and 169 proteins, from which 103 were common in both experiments, were identified (see Additional file 1). Only eight proteins could be identified which showed differential regulation at moderately low cut-off limits, i.e. between 1.2 and 0.8 fold regulation (Figure 4). These cut-off limits are sufficient to detect regulation in view of the high sensitivity of the applied LC-MS method.
Figure 4.
Differentially regulated proteins in the luxS mutant. Seven proteins were slightly but consistently differentially regulated in the luxS mutant compared to the WT control in the two tested samples. Two proteins related to motility - MshA pilin and flagellin - and three metabolic enzymes (malate dehydrogenase, cysteine synthase and transketolase) were induced, while two hypothetical proteins were repressed.
Two protein components related to motility, flagellin and MshA (mannose-sensitive hemaglutinin) pilin protein were slightly upregulated. Flagellin is a major component of the flagella (Interpro of EMBL-EBI). The MshA pilin constitutes the type IV pilus related to twitching motility [9].
In other luxS mutants, for example in Campylobacter jejuni [10] and in Helicobacter pylori [11], reduced motility has been observed. Both flagellar and twitching motility are involved in biofilm development in S. oneidensis and in other species. The mshA mutant of S. oneidensis could not adhere to the glass surface appropriately, while mutants without flagella or with paralysed flagella formed only non-differentiated, flat biofilms [9]. Similarly, flagellar motility in E. coli and twitching motility in P. aeruginosa was involved in spreading on a glass surface during biofilm formation [12,13]. Therefore upregulation of these proteins in the S. oneidensis luxS mutant would indicate a better glass coverage and eventually a different structure of the mature biofilm, which was actually observed in our biofilm experiment.
Three metabolic enzymes, namely cysteine synthase, malate dehydrogenase and transketolase, were slightly upregulated. These proteins might have been upregulated in response to the disrupted methyl cycle in the luxS mutant. They could either compensate for the lack of homocysteine or channel the accumulating S-ribosyl-homocysteine (SRH) into the carbon metabolism. Cysteine synthase conceivably compensates for the lack of homocysteine, since the product of the catalysed reaction, cysteine, is easily convertible to homocysteine. The connection of this enzyme to methylation processes is also supported by the fact that the gene for cysteine synthase forms one operon with the luxS gene in Halobacillus halophilus [14] and is located at a different position but followed by an O-methyl-transferase in S. oneidensis. The other two enzymes are involved in carbon metabolism. Malate dehydrogenase is an enzyme of the citric acid cycle. Transketolase provides a link between glycolysis and the pentose-phosphate pathway.
Biofilm growth
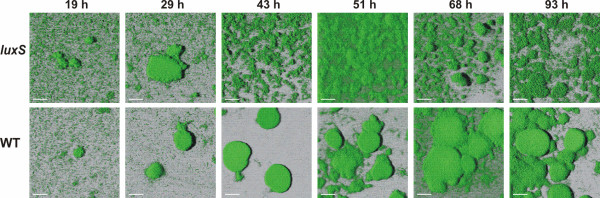
The S. oneidensis luxS mutant and the WT control strain were cultivated statically on glass slides in defined minimal medium. The biofilm of the luxS mutant showed a less-differentiated layer of loosely-bound cells which covered the surface evenly, while the WT control tended to form tight, flat and round-shaped clusters. In addition, the luxS mutant biofilm developed faster and after four days it was already in the detaching phase, while the WT biofilm was still developing (Figure 5. and Table 1.). At the early developmental stages, up to 9 h, the mutant and the control biofilms were indistinguishable: both strains displayed a homogenous single layer that gradually grew denser. On the second day, after 19 h and 29 h of growth, the mutant grew denser everywhere, while the wild type aggregated into clusters. This difference became most apparent on the second day, after 43 h, when the luxS mutant had a thicker biofilm forming a homogenous layer in contrast to the wild type clusters, as also reflected by the biofilm parameters. On the third day, after 51 h and 68 h of incubation, the mutant biofilm remained unchanged and appeared to have reached its maximum in volume and depth, while the control biofilm was still growing. On the fourth day, after 93 h, the mutant biofilm seemed to have arrived at the detachment phase due to its porous appearance and biofilm parameters, while the control biofilm was still developing.
Figure 5.
Biofilm development of the S. oneidensis luxS mutant and the WT control strain. Biofilms of both strains were grown on glass surfaces, which were placed in Petri dishes filled with SDM. At each time point, a glass slide was removed from the Petri dish and used for microscopic investigation. The pictures are selected from 2-5 records. Up to 9 h of growth, 2 D images from the fluorescence microscope were recorded with 40 × magnification, and these showed no difference between the two strains. After 19 h of growth, 3 D images could be recorded by Confocal Laser Scanning Microscopy. From 19 h until the end of the experiment, the luxS mutant formed a less-differentiated, loosely-bound biofilm, and the WT control tended to gather into tight clusters.
Table 1.
Biofilm parameters calculated by the PHLIP Matlab tool.
| Time [h] |
Total biovolume [μm3] | Substratum coverage [%] | Mean thickness [μm] | |||
|---|---|---|---|---|---|---|
| luxS mutant | WT control | luxS mutant | WT control | luxS mutant | WT control | |
| 19 | 9,3*105 | 4,5*105 | 15 | 6 | 15 | 6 |
| 29 | 2,9*106 2,8*106 |
2,3*106 1,4*106 |
15 20 |
9 7 |
16 17 |
16 15 |
| 43 | 7,2*106 8,3*106 |
2,6*106 2,4*106 |
32 65 |
21 18 |
15 14 |
9 8 |
| 51 | 4,2*106 3,2*106 |
4,6*106 | 51 30 |
35 | 25 19 |
11 |
| 68 | 7,9*106 | 9,4*106 5,2*106 |
57 | 50 43 |
22 | 12 8 |
| 93 | 3,9*106 4,0*106 |
1,1*107 8,1*106 |
39 43 |
54 81 |
14 11 |
13 17 |
Biofilm formation depends strongly on the medium used. In our hands, S. oneidensis was unable to produce substantial biofilms in complex media like LB, and also the frequently applied, complex, nutrient-limited biofilm medium LML [11,15] did not work well, but biofilm growth was observed in the defined Shewanella minimal medium (SDM). In the work of Learman et al. [8] a minimal medium different from SDM was applied for biofilm investigations. Under these conditions, the biofilm of the luxS mutant had 10% less biomass than the wild type after one day when grown in a microtitre plate under static conditions. After three days, the wild type biofilm had increased its biomass by 40%, while the mutant biofilm remained constant [8]. Under flow-through conditions, the luxS mutant initially showed a lower density of microcolonies (after 16 h), while no difference to the wild-type could be observed at a later stage of biofilm growth (48 h). These data cannot be compared to our results directly, because we used a different medium and a glass surface. However, the principle observations are similar: The dynamics of biofilm growth were different between wild-type and mutant, with the maximum of difference seen after 16 h in the flow through system of Learman et al. [8], and after 43 h in our experiment. Moreover, the mutant biofilm stopped growth earlier than the wild type in both studies, possibly indicating a problem with toxic intermediates. In contrast to Learman et al. [8] we observed a marked difference in the architecture of the mutant biofilm, which failed to develop the big clusters of the wild type.
The luxS mutation had different effects on biofilm formation in different species, but this may be the consequence of the cultivation conditions: flow-chamber vs. static conditions and type of medium used. In the flow chamber, the luxS mutant of Klebsiella pneumoniae formed a flat, more homogenous biofilm after 16-24 h of incubation in contrast to the mature, mushroom-like structures of the control [16]. The luxS mutant of Aggregatibacter actinomycetemcomitans developed a biofilm with reduced biomass and more sparse coverage after 60 h of incubation on saliva-coated coverglass in a flow-chamber [17]. Under static conditions, however, the contrary was observed. The luxS mutant of Eikenella corrodens formed a biofilm with higher biomass compared to the control on polystyrene surfaces of microtitre plates [18]. The luxS mutant biofilms of Helicobacter pylori exhibited the same structure, but contained more cells than the control [19]. And finally, the luxS mutant of Lactobacillus reuteri formed thicker biofilms than the control both in vitro and in vivo [20]. The only exception is the luxS mutant of Salmonella typhimurium that lost the ability to form biofilms on gallstones almost completely under static conditions and exhibited impaired biofilms on polystyrene pegs [21]. Remarkably, most luxS mutants investigated, except S. typhimurium, formed flatter biofilms in flow-chambers, while thicker biofilms developed in static systems compared to the wild type, and this latter observation is confirmed by the biofilm of our luxS mutant of S. oneidensis. Furthermore, all these luxS mutants tended to cover the surface more homogenously. Conceivably, the luxS mutants tends to form loosely-bound biofilms, but depending on the cultivation method they appear differently: in static systems, the cells are accumulating and form a thicker biofilm, while in a flow-chamber, the biofilms are flat, because the loosely-connected cells are washed away by the continuous flow.
Conclusion
This study shows that the luxS mutation in S. oneidensis results in the development of loosely-bound biofilms. The concomitant slight upregulation of motility related proteins, which was detected in the secretome, may be one reason for this changed biofilm architecture, because it can lead to a weaker connection between the cells. Other changes in the secretome, e.g. upregulation of a homocysteine synthase, appear to be compensating for the metabolic defect caused by the lack of the LuxS enzyme. There is a very active cell-driven mechanism for AI-2 depletion in this bacterium, which is growth-phase dependent and not induced by AI-2, thereby indicating a role for AI-2 as a metabolite imported under conditions of nutrient limitation in S. oneidensis. Taking together the results of our study with those of Learman et al. [8], it seems unlikely that AI-2 has a signalling role in S. oneidensis.
Methods
Bacterial strains, plasmids and primers
Bacterial strains, plasmids and primers are listed in Tables 2 and 3.
Table 2.
Strains and plasmids used in this study.
| Strain | Description and genotype | Sources/References |
|---|---|---|
| Shewanella oneidensis | ||
| MR-1 | Wild type strain | BCC/[2] |
| luxS-ins | pKnock-Km plasmid inserted into the luxS gene | this study |
| WTKm | pKnock-Km plasmid inserted before the luxS gene | this study |
| Escherichia coli | ||
| S17-1λpir | Biparental mating TpR SmR recA thi pro hsdR RP4-2-Tc::Mu-Km::Tn7 λpir |
Biomedal (Spain)/[29] |
| pir-116 | Maintaining of R6K ori plasmids F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ- rpsL nupG pir-116(DHFR) |
Epicentre/[30] |
| DH5α | Subcloning routine F- Φ80lacZΔM15 Δ(lacZYA-argF)U169 recAl endAI hsdR17(rk-, mk+) phoA supE44 thi-1 gyrA96 relAI λ- |
Invitrogen/[31] |
| HB101 | Carrier of the pRK2013 plasmid for triparental mating. F- supE44 lacY1 ara-14 galK2 xyl-5 mtl-1 leuB6 Δ(mcrC-mrr) recA13 rpsL20 thi-1 Δ(gpt-proA)62 hsdSB20 λ- |
[32] |
| Vibrio harveyi | ||
| BB152, BAA-1119 | luxM::Tn5; produces only AI-2 | ATCC/[33] |
| MM77 | luxM::Tn5, luxS::CmR; produces neither AI-1 nor AI-2 | ATTC/[34] |
| BB170, BAA-1117 | luxN::Tn5; senses only AI-2 | ATCC/[33] |
| Plasmids | ||
| pKnock-Km | KmR; oriT; source plasmid for the suicide vectors into Sh. oneidensis | NCCB 3407/[24] |
| pEX18Ap | AmpR; source plasmid for a replicating plasmid into Sh. oneidensis | Max Schobert, TU Braunschweig/[35] |
| pPS858 | AmpR, GmR; source of the Gentamicin-GFP (Gm-GFP) cassette | Max Schobert, TU Braunschweig/[35] |
| pRK2013 | KmR; helper plasmid for conjugation | DSMZ 5599/[36] |
| pKnock-luxS | Construction of luxS-ins KmR; pKnock-Km containing an internal fragment of luxS of Sh. oneidensis |
This study |
| pBA2106 | Construction of WTKm KmR, pKnock-Km containing an upstream fragment of luxS of Sh. oneidensis |
This study |
| pEX18ApGm | For tagging Sh. oneidensis with gfp KmR, GmR, the Gm-GFP cassette ligated into pEX18Ap |
This study |
Table 3.
Primers used in this study.
| Primer name | Primer sequencea | Anneal. temp. |
|---|---|---|
| CluxS_NotI_for | TGG CAG AGA ACT GTT TAG gcggccgc AAC AGG CTC GCT TGA CG | 66°C |
| BluxS_BamHI_rev | ATG GCA TAG AGA TCT CCA ggatcc CAG GGC GAT ACA ACG CCA C | 66°C |
| LuxS_EcoRI_beg | CAT TAC TTG gaattc TTA CCG TTG ACC ATA CTC | 60°C |
| LuxS_KpnI_mid | CAT TGG A ggtacc TC AAT AAT TTC CAC ATC | 60°C |
aNucleotides in bold indicate the built-in restriction sites.
Culture media and growth conditions
Bacteria were cultivated aerobically in Erlenmeyer flasks by shaking at 160 rpm on a rotary shaker using standard microbiological techniques. Growth was monitored by measuring optical density at 600 nm (OD600 nm) with a Pharmacia Biotech spectrophotometer. E. coli strains were grown at 37°C in Luria-Bertani (LB) medium. V. harveyi strains were grown at 30°C in autoinducer bioassay (AB) medium [22,23]. S. oneidensis was grown at 30°C in LB. For biofilm growth, S. oneidensis strains were cultivated in Shewanella defined minimal (SDM) medium (31) with 20 mM lactic acid as carbon source. For counter selection of E. coli after conjugation, solid SM medium was used. E. coli strains were selected with ampicillin (Amp) at 150 μg/ml, kanamycin (Km) at 50 μg/ml and gentamicin (Gm) at 20 μg/ml. S. oneidensis was selected with Gm at 20 μg/ml, Km at 100 μg/ml after conjugation on plates and Km at 200 μg/ml in aerobic cultures, respectively.
Experiments with addition of AI-2
In the first experiment with the wild type strain, four flasks were filled with the same volume of medium. Three of them were simultaneously inoculated with the overnight culture of the wild type. Two cultures were supplemented with AI-2 at a concentration of 5.3 μM at 0 h and 10 h of growth, respectively, while the third culture was used as the control for AI-2 production. The fourth flask contained sterile medium and was supplemented with AI-2 at 0 h of incubation to test AI-2 stability under the experimental conditions. From all cultures and the medium control, samples were taken periodically and tested for AI-2 levels. Experiments with the luxS mutant and the WT control were conducted in the same way except that AI-2 was added only after 10 h of growth.
To study AI-2 depletion in cells and culture supernatants separately, S. oneidensis wild type cells were harvested after 10 h of growth and suspended in PBS buffer. One half of the cell suspension was heated at 95°C for 20 minutes to kill the cells, while the other half remained native. The culture supernatant was filter-sterilized, and similarly to the cell suspension, one half of the supernatant was heat-inactivated, while the other half remained native. Control solutions were sterile PBS buffer and LB medium. After adjusting the temperature to 30°C, all solutions were supplemented with AI-2 at a concentration of 5.3 μM. Then samples were taken periodically and tested for AI-2 levels.
Construction of the luxS mutant and WT control in S. oneidensis
The luxS mutant and the WT control were constructed via single homologous recombination between a suicide plasmid and the chromosome of the wild type strain. The suicide plasmids were derived from the conditionally replicating plasmid pKnock-Km [24] by directional PCR cloning. For the luxS mutant, a coding fragment of the luxS gene, approximately 0.2 kbp long, was amplified with the primers LuxS_EcoRI_beg and the LuxS_KpnI_mid and then ligated into the EcoRI and KpnI restriction sites of the pKnock-Km plasmid. The resulting plasmid was first maintained in E. coli pir-116, then transformed in E. coli S17-1λpir and finally introduced into S. oneidensis wild type strain by biparental mating. The mating mixture was plated on SDM medium, as this medium lacks essential amino acids for E. coli to grow. To ensure that pure S. oneidensis colonies were obtained, the clones were streaked on LB agar with ampicillin at 150 μg/ml and kanamycin, and single colonies were selected and tested for mutation by PCR and sequencing (data not shown). For the WT control, an approximately 0.6 kbp long fragment upstream of the luxS gene with 17 bp spacing was amplified with the CluxS_NotI_for and BluxS_BamHI_rev primers and then ligated into the NotI and BamHI restriction sites of the pKnock-Km plasmid. The resulting plasmid was introduced into S. oneidensis wild type as described previously. The WT control was verified by PCR.
Conjugation
Conjugation proved to be the only method to introduce DNA into S. oneidensis. Both biparental and triparental mating were applied, but the latter method proved to be more efficient. For biparental mating, E. coli S17-1 λpir with the appropriate plasmid (donor strain), and S. oneidensis wild type (recipient strain) were grown to OD600 nm = 1, which corresponds to about 108 cells/ml. 1.5 ml of each culture was harvested by centrifugation at 4.000 × g for 3 min at room temperature (RT) and washed twice in 1 ml LB. After one more centrifugation, the pellets were dissolved in 30 μl LB. Controls of both cultures separately and conjugation mixes were prepared and spotted onto mating disks, which are sterile mixed cellulose ester membranes (Millipore, MF type without triton) placed on LB agar plates. After incubation for one day at RT, the cells were dissolved in 1 ml PBS buffer at RT, and 200-300 μl of the suspension was plated onto SM agar with Km (100 μg/ml). Appropriate orange clones appeared after 1-2 weeks at RT. For triparental mating, the donor strain was an E. coli strain, which optimally maintained the mobilizable plasmid, the conjugating strain was E. coli HB101 with the helper plasmid pRK2013, and the recipient strains was S. oneidensis. These strains were prepared for conjugation and the clones were selected as described for biparental mating.
GFP-tagging of the luxS mutant and the WT control
To enable microscopic documentation of living bacteria, the luxS mutant and the WT control were labelled with the green fluorescent protein (GFP). For this purpose, the Gentamycin-GFP (Gm-GFP) cassette of the pPS858 plasmid was inserted into the BamHI site of the pEX18Ap plasmid containing an origin of transfer, which enabled conjugation into S. oneidensis, and ColE1 ori, which enabled plasmid replication in S. oneidensis. The resulting pEX18ApGm plasmid was conjugated by triparental mating into the luxS mutant and the WT control, respectively, and selected as described above.
Detection of AI-2
AI-2 was determined in the culture supernatant using the V. harveyi bioassay with the sensor strain V. harveyi BB170 as published previously [23]. The sensor strain V. harveyi BB170 was cultivated to bright light intensity with a final OD600 of 1.0-1.1. This culture was diluted 1:5000 in AB medium, resulting in the working solution of the sensor strain. 180 μl of this working solution were added to 20 μl of the test samples, reference media and controls pipetted on white microtitre plates (NUNC, Roskilde, Denmark). Four replicates were measured per test sample and six replicates for the control. The microtitre plates were incubated at 30°C with agitation at 650 rpm. Luminescence was measured hourly in a Victor Wallac Luminescence Reader (Perkin Elmer) for 6 h. Sterile culture supernatants from V. harveyi BB152 and MM77 served as positive and negative controls, respectively. Chemically synthesized AI-2 [25-27] diluted in AB medium was used as an additional positive control at a concentration of 5.3 μM. The AI-2 amount present in the sample was expressed in relative AI-2 activity. First, the fold change of induction of luminescence was calculated by dividing the luminescence of the samples by the luminescence of the sterile reference medium. The maximum of fold induction determined during 6 h was referred to as AI-2 activity. The relative AI-2 activity was calculated by dividing the AI-2 activity of the sample by that of the positive control.
Biofilm cultivation
Biofilms of the GFP-tagged S. oneidensis luxS mutant and WT control were grown in a static system in defined Shewanella minimal medium (SDM) with 20 mM lactic acid as carbon source [1]. First, the strains were pre-cultivated at 30°C in LB medium for 4 h to reach an OD600 nm of 0.7-1. Then, 10 biofilm dishes were prepared. For each of them, 12 ml of the LB culture were pipetted on a microscopic glass slide placed in a Petri dish, and after 45 min of incubation at 30°C, each microscopic slide was transferred to another Petri dish with 12 ml of fresh SDM medium. These biofilm dishes were also incubated at 30°C. At each time point, one biofilm dish was sacrificed for microscopy: the microscopic glass slide was washed in sterile PBS buffer and covered with a glass slip. The biofilm structures on the microscopic glass slides were examined with a fluorescence and a confocal laser scanning microscope. The fluorescence microscope was an Olympus BX60 microscope equipped with Plan objectives (Olympus, 40 x, 0.65; 10 x, 0.25) and with a fluorescence lamp (Olympus, model U-ULS100Hg), for which the light source was provided by a 100 W high pressure mercury burner (Olympus, model BH2-RFL-T3). A Colorview digital camera and cellA imaging software were used for documentation. The confocal laser scanning microscope (CLSM) was a Fluoview 1000 (Olympus) microscope equipped with UPlanSApo objectives and simultaneously detected bright field and EGFP images (20 mW 561 nm solid state laser and 30 mW 488 nm multiline argon laser with 2% power respectively). The monochrome series of images of the CLSM was measured along the optical axis with 1 μm increments. The Imaris X64 5.7.2 software was used for creating the 3 D images. The biofilm parameters like biomass, substrate coverage and mean thickness, were calculated by the Phlip Matlab toolbox [28].
Secretome analysis
The S. oneidensis luxS mutant and the WT control strain were cultivated at 30°C in LB for 5-5.5 h until an OD600 nm of 1.5 was reached. Then, the whole 100 ml culture was centrifuged at 4.000 × g for 6 minutes at 4°C and the culture supernatant was sterilized by filtration (0,22 μm). For secretome analysis, proteins were precipitated by sodium deoxylate (DOC). Peptides were generated by trypsine digestion. The peptides were labelled by different iTRAQ reagents (Applied Biosystems) according to the manufacturer's instructions.
iTraq labelled peptides were first separated using strong cation exchange on a Mono S PC 1.5/5 column with an gradient over 35 minutes from 100 buffer A to 35% buffer B (A = 25% acetonitrile, 0.1% formic acid; B = 25% acetonitrile, 0.1% formic acid, 500 mM KCl) at a flow rate of 150 μl/min. Fractions of 150 μl were collected, dried in a SpeedVac centrifuge and purified using ZipTipμ RP 18 material. The separation of the peptide samples was performed using a bioinert Ultimate nano-HPLC system (Dionex). 10 μl of each sample (containing up to 1 μg peptides) were injected, and peptides were purified and concentrated on a C18-PepMap pre-column (0.3 mm i.d. × 5 mm, 100 Å pore size, 3 μm particle size) at a flow rate of 30 μl/min in 0.1% TFA. Subsequently, peptides were separated on an analytical 75 μm i.d. × 150 mm C18-PepMap column (Dionex, 100 Å pore size, 3 μm particle size) at a flow rate of 200 nl/min. The gradient (Solution A: 0.1% formic acid, 5% acetonitrile; solution B: 0.1% formic acid, 80% acetonitrile) started at 5% and ended at 60% B after 120 minutes. MS and MS/MS data were acquired using a Q-TOFmicro mass spectrometer (Waters, Milford Massachusetts; USA). Doubly and triply charged peptide ions were automatically selected by the MassLynx software (MassLynxx 4.1 1 b) and fragmented for a maximum of 18 seconds per peptide. MS data were automatically processed and peak lists for subsequent protein identification by database searches were generated by the MassLynx software. Database searches were carried out with an in house MASCOT server using the EMBL Shewanella oneidensis database. Proteins were only accepted as identified when at least one unique peptide showed an individual score above 20, which indicated identity or extensive homology (p < 0.05) using the given settings (Enzyme: Trypsin; Max. missed cleavages: 1; Fixed modification: iTRAQ (K); iTRAQ (N-term); Oxidation (M); Peptide tolerance: 0.4 Da; MS/MS tolerance 0.3 Da).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AGB and IWD planned the investigations and wrote the manuscript. AGB performed the experiments. LOJ and JOW measured the secretome samples. All authors read and approved the final manuscript.
Supplementary Material
Table S1. All proteins identified in the first and the second experiment are listed (prot_extracted). The two lists were compared and common proteins were selected (common_prot). Then the proteins were sorted according to their regulation and regulated proteins were selected (regulated_prot).
Contributor Information
Agnes M Bodor, Email: agnes.m.bodor@gmail.com.
Lothar Jänsch, Email: lothar.jaensch@helmholtz-hzi.de.
Josef Wissing, Email: josef.wissing@helmholtz-hzi.de.
Irene Wagner-Döbler, Email: irene.wagner-doebler@helmholtz-hzi.de.
Acknowledgements
We thank Andreas Dötsch and Mathias Müsken for their help in preparing and evaluating the images of the confocal laser scanning microscope and Kirsten Minkhart for conducting the LC-MSMS measurements.
References
- Myers CR, Nealson KH. Bacterial Manganese Reduction and Growth with Manganese Oxide as the Sole Electron Acceptor. Science. 1988;240:1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- Venkateswaran K, Moser DP, Dollhopf ME, Lies DP, Saffarini DA, MacGregor BJ, Ringelberg DB, White DC, Nishijima M, Sano H. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int J Syst Bacteriol. 1999;49(Pt 2):705–724. doi: 10.1099/00207713-49-2-705. [DOI] [PubMed] [Google Scholar]
- Hau HH, Gralnick JA. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol. 2007;61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- Bassler BL. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/S1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- Bodor A, Elxnat B, Thiel V, Schulz S, Wagner-Dobler I. Potential for luxS related signalling in marine bacteria and production of autoinducer-2 in the genus Shewanella. BMC Microbiol. 2008;8:13. doi: 10.1186/1471-2180-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Williams P. Bacterial cell-to-cell communication: sorry, can't talk now - gone to lunch! Curr Opin Microbiol. 2002;5:216–222. doi: 10.1016/S1369-5274(02)00304-1. [DOI] [PubMed] [Google Scholar]
- Taga ME, Semmelhack JL, Bassler BL. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol Microbiol. 2001;42:777–793. doi: 10.1046/j.1365-2958.2001.02669.x. [DOI] [PubMed] [Google Scholar]
- Learman DR, Yi H, Brown SD, Martin SL, Geesey GG, Stevens AM, Hochella MF Jr. Involvement of Shewanella oneidensis MR-1 LuxS in biofilm development and sulfur metabolism. Appl Environ Microbiol. 2009;75:1301–1307. doi: 10.1128/AEM.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann KM, Saville RM, Shukla S, Pelletier DA, Spormann AM. Initial Phases of biofilm formation in Shewanella oneidensis MR-1. J Bacteriol. 2004;186:8096–8104. doi: 10.1128/JB.186.23.8096-8104.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvers KT, Park SF. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology. 2002;148:1475–1481. doi: 10.1099/00221287-148-5-1475. [DOI] [PubMed] [Google Scholar]
- Osaki T, Hanawa T, Manzoku T, Fukuda M, Kawakami H, Suzuki H, Yamaguchi H, Yan X, Taguchi H, Kurata S. Mutation of luxS affects motility and infectivity of Helicobacter pylori in gastric mucosa of a Mongolian gerbil model. J Med Microbiol. 2006;55:1477–1485. doi: 10.1099/jmm.0.46660-0. [DOI] [PubMed] [Google Scholar]
- Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Sewald X, Saum SH, Palm P, Pfeiffer F, Oesterhelt D, Muller V. Autoinducer-2-producing protein LuxS, a novel salt- and chloride-induced protein in the moderately halophilic bacterium Halobacillus halophilus. Appl Environ Microbiol. 2007;73:371–379. doi: 10.1128/AEM.01625-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teal TK, Lies DP, Wold BJ, Newman DK. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl Environ Microbiol. 2006;72:7324–7330. doi: 10.1128/AEM.01163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrino D, Haagensen JA, Rich C, Forestier C. Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J Bacteriol. 2005;187:2870–2880. doi: 10.1128/JB.187.8.2870-2880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Lamont RJ, Demuth DR. Autoinducer 2 is required for biofilm growth of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect Immun. 2007;75:4211–4218. doi: 10.1128/IAI.00402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azakami H, Teramura I, Matsunaga T, Akimichi H, Noiri Y, Ebisu S, Kato A. Characterization of autoinducer 2 signal in Eikenella corrodens and its role in biofilm formation. J Biosci Bioeng. 2006;102:110–117. doi: 10.1263/jbb.102.110. [DOI] [PubMed] [Google Scholar]
- Cole SP, Harwood J, Lee R, She R, Guiney DG. Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol. 2004;186:3124–3132. doi: 10.1128/JB.186.10.3124-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW, Ghazally S, Walter J, Loach D, Brooks H, Cook G, Surette M, Simmers C, Bremer P, Dal Bello F. Ecological behavior of Lactobacillus reuteri 100-23 is affected by mutation of the luxS gene. Appl Environ Microbiol. 2005;71:8419–8425. doi: 10.1128/AEM.71.12.8419-8425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty AM, Schwesinger WH, Gunn JS. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect Immun. 2002;70:2640–2649. doi: 10.1128/IAI.70.5.2640-2649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg EP, Hastings JW, Ulitzur S. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Archives of Microbiology. 1979;120:87–91. doi: 10.1007/BF00409093. [DOI] [Google Scholar]
- Vilchez R, Lemme A, Thiel V, Schulz S, Sztajer H, Wagner-Dobler I. Analysing traces of autoinducer-2 requires standardization of the Vibrio harveyi bioassay. Anal Bioanal Chem. 2007;387:489–496. doi: 10.1007/s00216-006-0824-4. [DOI] [PubMed] [Google Scholar]
- Alexeyev MF. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques. 1999;26:824–6. doi: 10.2144/99265bm05. 828. [DOI] [PubMed] [Google Scholar]
- De Keersmaecker SC, Varszegi C, van Boxel N, Habel LW, Metzger K, Daniels R, Marchal K, De Vos D, Vanderleyden J. Chemical synthesis of (S)-4,5-dihydroxy-2,3-pentanedione, a bacterial signal molecule precursor, and validation of its activity in Salmonella typhimurium. J Biol Chem. 2005;280:19563–19568. doi: 10.1074/jbc.M412660200. [DOI] [PubMed] [Google Scholar]
- DeKeersmaecker SC, Vanderleyden J. Constraints on detection of autoinducer-2 (AI-2) signalling molecules using Vibrio harveyi as a reporter. Microbiology. 2003;149:1953–1956. doi: 10.1099/mic.0.C0117-0. [DOI] [PubMed] [Google Scholar]
- Semmelhack MF, Campagna SR, Federle MJ, Bassler BL. An expeditious synthesis of DPD and boron binding studies. Org Lett. 2005;7:569–572. doi: 10.1021/ol047695j. [DOI] [PubMed] [Google Scholar]
- Mueller LN, de Brouwer JF, Almeida JS, Stal LJ, Xavier JB. Analysis of a marine phototrophic biofilm by confocal laser scanning microscopy using the new image quantification software PHLIP. BMC Ecol. 2006;6:1. doi: 10.1186/1472-6785-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Bio/Technology. 1983. pp. 784–791.
- Metcalf WW, Jiang W, Wanner BL. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene. 1994;138:1–7. doi: 10.1016/0378-1119(94)90776-5. [DOI] [PubMed] [Google Scholar]
- Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer HW, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Mok KC, Wingreen NS, Bassler BL. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 2003;22:870–881. doi: 10.1093/emboj/cdg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Figurski DH, Meyer RJ, Helinski DR. Suppression of Co1E1 replication properties by the Inc P-1 plasmid RK2 in hybrid plasmids constructed in vitro. J Mol Biol. 1979;133:295–318. doi: 10.1016/0022-2836(79)90395-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. All proteins identified in the first and the second experiment are listed (prot_extracted). The two lists were compared and common proteins were selected (common_prot). Then the proteins were sorted according to their regulation and regulated proteins were selected (regulated_prot).